Abstract
Dissecting the molecular basis of quantitative traits is a significant challenge and is essential for understanding complex diseases. Even in model organisms, precisely determining causative genes and their interactions has remained elusive, due in part to difficulty in narrowing intervals to single genes and in detecting epistasis or linked quantitative trait loci. These difficulties are exacerbated by limitations in experimental design, such as low numbers of analyzed individuals or of polymorphisms between parental genomes. We address these challenges by applying three independent high-throughput approaches for QTL mapping to map the genetic variants underlying 11 phenotypes in two genetically distant Saccharomyces cerevisiae strains, namely (1) individual analysis of >700 meiotic segregants, (2) bulk segregant analysis, and (3) reciprocal hemizygosity scanning, a new genome-wide method that we developed. We reveal differences in the performance of each approach and, by combining them, identify eight polymorphic genes that affect eight different phenotypes: colony shape, flocculation, growth on two nonfermentable carbon sources, and resistance to two drugs, salt, and high temperature. Our results demonstrate the power of individual segregant analysis to dissect QTL and address the underestimated contribution of interactions between variants. We also reveal confounding factors like mutations and aneuploidy in pooled approaches, providing valuable lessons for future designs of complex trait mapping studies.
Keywords: QTL mapping, bulk segregant analysis, individual segregant analysis, next generation sequencing, yeast, reciprocal hemizygosity scanning
MOST medical and agricultural traits are complex, influenced by multiple alleles with varying effect sizes that interact to produce inherited phenotypic variation. Previous studies in model organisms (Steinmetz and Davis 2004; Ehrenreich et al. 2009; Flint and Mackay 2009; Flint 2011) have yielded insights into genetic principles that shape complex traits. These studies have shown that in addition to major QTL with large effects, many loci with smaller effects contribute to phenotypic variation. Indeed, although many alleles have been associated with complex traits in humans, their individual and cumulative effects are usually small (<10%) (Lango Allen et al. 2010). Further studies have revealed extensive context-dependent effects such as epistasis or genotype-by-sex interactions, as well as pleiotropic effects, most instances of which have likely not been detected. Hence, understanding the genetic basis of complex traits remains an open challenge (Stranger et al. 2011).
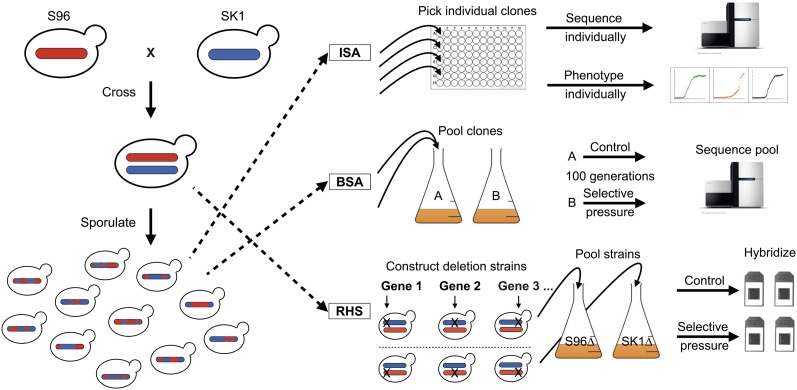
In this study, we applied three high-throughput methods for the first time to comprehensively identify causative variants underlying 11 phenotypes in two genetically distant yeast strains, S96 and SK1 (Liti et al. 2009; Schacherer et al. 2009). Each method begins with a hybrid generated by crossing these two strains. The first method is the commonly used bulk segregant analysis (BSA) (Segre et al. 2006; Birkeland et al. 2010; Ehrenreich et al. 2010; Wenger et al. 2010; Parts et al. 2011; Swinnen et al. 2012), in which millions of segregants from a hybrid undergo selection under an environmental pressure. QTL mapping is then performed by identifying regions of allelic enrichment via sequencing of the pool (Figure 1).
Figure 1.
Schematic overview of the three QTL mapping approaches used in this study. The parental strain backgrounds S96 and SK1 were used for all approaches. ISA: genotyping and phenotyping are performed on individual segregants. BSA: a pool of segregants is grown in control and selective media. By sequencing the pooled genomic DNA, the allelic enrichments in the pool are determined. RHS: in a hybrid strain, alleles are alternatively deleted, resulting in reciprocal hemizygous, isogenic hybrid strains that differ only by a single allele. DNA barcodes specific to each gene enable the pooling and parallel analysis of strain fitness on a genome-wide level. After selective growth, the barcodes are amplified and hybridized to a microarray (or sequenced), providing a proxy of fitness that can be used to measure the effects of allelic variation in each gene on the phenotype of interest.
The second method utilized here is individual segregant analysis (ISA) of 720 segregants from the hybrid. These segregants were genotyped by next-generation sequencing (Wilkening et al. 2013) and individually phenotyped to detect genomic regions linked to the phenotypes of interest (Figure 1). Most previous QTL mapping studies in yeast have been performed with sample sizes on the order of 100 segregants and up to 3000 markers (average SNP distance: 4 kb) (Steinmetz et al. 2002; Brem et al. 2005; Gatbonton et al. 2006; Foss et al. 2007; Hu et al. 2007; Marullo et al. 2007; Nogami et al. 2007; Perlstein et al. 2007; Ehrenreich et al. 2009; Li et al. 2013). Our experiment thus increases the sample size by 7-fold and the number of markers by 20-fold (average SNP distance: 150 bp). To date, only one study with a similar number of segregants (1008) has been published; however, it did not validate causative genes (Bloom et al. 2013).
To attain QTL mapping at single-gene resolution, we developed and applied a third method termed “reciprocal hemizygosity scanning” (RHS). For this method, we constructed a hemizygous deletion collection in the hybrid by deleting either the SK1 or the S96 allele and replacing it with a kanamycin resistance gene (KanMX) and a molecular barcode (Figure 1) (Winzeler 1999). This collection includes ∼75% of the essential and non-essential open reading frames (ORFs) in the yeast genome, allowing for the direct comparison of allelic variants within a single pooled experiment on a genome-wide scale (Steinmetz et al. 2002; Steinmetz and Davis 2004). This is the first report of this genome-wide approach including >19,000 hemizygous strains (∼4861 genes deleted in duplicate per background).
Overlaying QTL detected by these three methods yielded extremely high resolution, allowing us to identify putative allelic variants underlying eight phenotypes. We also discovered strong interactions between QTL and differences between the three approaches, which can partially be explained by different experimental parameters (e.g., duration of growth), but also by confounding factors such as accumulation of mutations influencing the pooled RHS and BSA approaches.
Materials and Methods
Yeast strain generation
Haploid strains from S288c (BY4742 prototrophic MATα, referred to as “S96”) and SK1 (SK1 MATa ura3∆ his3∆ flo8∆ can1∆::STE2pr-HIS3) were crossed, and an individual hybrid strain was sporulated by transferring the cells grown in YPD (yeast extract, 10 g/liter; Bacto peptone, 20 g/liter; dextrose, 20 g/liter) to 200 ml sporulation medium [0.5% (w/v) potassium acetate] and incubating them at 22° with agitation. After spreading the cells on YPD plates, 768 clones were randomly picked in eight 96-well plates, grown overnight, and stored as glycerol stocks. This set of segregants can be copied and sent to other labs upon request. For BSA, we crossed our haploid SK1 strain, in which the PMS1 ORF was exchanged with its S288c version to make it more genetically stable (Heck et al. 2006; Demogines et al. 2008b) [SK1 MATa ura3∆ his3∆ flo8∆ can1-∆::STE2pr-HIS3PMS1(S288c)] with an S96 strain (BY4742 MATα ura3∆ his3∆ can1∆::STE2pr-HIS3). Two independent crosses were grown in 100 ml YPD until OD600nm = 1 and sporulated as described above. We used the synthetic genetic array (SGA) marker system to select for MATa strains on SD plates lacking histidine and supplemented with L-canavanine (60 mg/ml) (Tong et al. 2001; Pan et al. 2004). The resulting colonies were scraped off the plates with an estimated number of 4 × 108 independent segregants per pool. Aliquots of the pool were frozen at −80° in 15% glycerol for later use. ENA6 was amplified from genomic DNA of SK1 and cloned into the p416 expression vector (Mumberg et al. 1995) using SpeI and XhoI restriction sites (Supporting Information, Table S1). The empty plasmid as well as the ENA6-containing plasmid were transformed into SK1 cells and tested in normal and high-salt conditions.
Genotyping
Both ISA and BSA analyses were performed with the same 65,234 single nucleotide polymorphisms (SNP) positions, as described before (Wilkening et al. 2013). In brief, sequences were aligned to the S288c reference genome (Saccharomyces Genome Database), using Novoalign and allowing only unique alignments. Realignment of the subsequent BAM files, SNP calling, and genotyping were performed using GATK (McKenna et al. 2010). For ISA segregants, missing genotypes were imputed with BEAGLE (Browning and Browning 2007). In total, 768 segregants were sequenced, but from the coverage aneuploidies were detected in 26 individual segregants (Wilkening et al. 2013). After exclusion of these aneuploid strains and strains with low coverage or contamination, 720 segregants were used for subsequent analyses.
QTL mapping
For ISA, we estimated the genetic map for our data set and calculated the LOD score at each position using R/qtl (Broman et al. 2003). The threshold at 5% significance level was estimated using the permutation test implemented in R/qtl.
To identify smaller-effect QTL and interactions between QTL for the high-salt and high-temperature phenotypes, we stratified the ISA samples according to the major QTL allele prior to repeating QTL analysis. In principle, this is similar to using the genotypes at major QTL as an additive covariate, as implemented in R/qtl (discussed in Broman et al. 2003). For BSA, the allele frequency was calculated at each SNP position for all conditions. The allele frequency was fitted using local polynomial regression assuming binomial distribution, and confidence intervals were called using a bootstrapping method. To determine whether the allele frequency at the peak for a given condition was significant compared to the control (YPD at 30° for 100 generations), a permutation test was performed for each peak and P-values were corrected using Benjamini–Hochberg (details in Supporting Information).
We also performed an in silico comparison of the ISA method with a simulated BSA using only the best performing strains (pool of 50 segregants with extreme phenotypes) for eight of the phenotypes analyzed in this study (Figure S1). Detailed description of the analysis can be found in File S1. Phenotype and genotype information used for QTL mapping can be found in File S2.
Estimating heritability of traits
A genomic selection method was used to estimate, for each trait, the proportion of phenotypic variance that could be explained by using all the 65,234 markers used for QTL mapping. Ridge regression best linear unbiased prediction (rrBLUP) was applied using the rrBLUP package (Endelman 2011). The model has two components of error, genetic variance (Vg) and error variance (Ve). The heritability of the trait, which is the proportion of phenotypic variance that can be explained by all genetic markers, can be estimated by calculating Vg/(Vg + Ve). For estimating narrow sense heritability, the additive kinship matrix described in the rrBLUP package was used as the relationship matrix, and, for estimating broad sense heritability, the non-additive Gaussian kernel was used.
Phenotyping
For ISA, individual strains were phenotyped in 96-well plates by growth curve analysis (Proctor et al. 2011). Cells were grown overnight in YPD to saturation to obtain similar densities for all strains. These colonies were replicated in the medium of interest in transparent 96-well plates and grown until saturation (usually 1–2 days). Doubling times were calculated from OD measurement of liquid cultures at a wavelength of 595 nm in a plate reader (Genios, Tecan) as previously described (St Onge et al. 2007). The relative fitness was calculated as (1/doubling time at stress condition)/(1/doubling time in YPD at 30°). The phenotype “fitness YPD” was calculated as 1/doubling time in YPD at 30°. “OD saturation” refers to the OD595nm at the saturation phase.
In addition, a colony-size assay was performed for the high-salt phenotype by replicating YPD overnight cultures on agar plates and growing them for 2–4 days until an average colony diameter of ∼5 mm was reached. To determine colony sizes, photos were taken of the agar plates and processed with the CellProfiler software (Carpenter et al. 2006). The relative fitness in a specific condition was calculated as follows: log (colony size treatment) − log (colony size control). To account for variability between plates, the colony sizes were normalized using the median colony size per plate. Colony shapes were determined by visual observation of the control plates (30° YPD) used for the colony-size assay.
BSA
BSA was done similarly to the approach in Parts et al. (2011). In brief, two independent pools of segregants from an S96 × SK1 cross (see Yeast strain generation) were grown in 100 ml YPD for 4 h. From this preculture, 400 ml of each specific condition medium (treatment) and of YPD (30° control) were inoculated with a starting OD600nm of 0.08 and grown until OD600nm = 2. This dilution step was repeated to keep cells in continuous exponential growth for ∼100 divisions. The cells were then collected and kept at −80° for later DNA isolation and library preparation.
Sequencing library preparation
Genomic DNA from individual (ISA) or pooled strains (BSA) was isolated from fresh and frozen cell pellets with the PrepEase kit (USB). Adapters were ligated to sonicated DNA as previously described (Wilkening et al. 2013). After size selection on an E-Gel (Invitrogen), libraries were amplified with Illumina paired-end primers, cleaned, and sequenced (105 bp paired-end) on a HiSeq 2000 (Illumina).
RHS
Both alleles of each gene in the genome were individually deleted in the SK1 × S96 hybrid and replaced with a molecular barcode and a kanMX4 cassette. The resulting RHS pools were grown for 20 generations under the following conditions: YPD at 30°, YPD at 38°, YPD + 350 mM NaCl, and YPD + 350 µM cantharidin. Genomic DNA from the pool was extracted, and the uptags and downtags containing the barcodes were amplified by PCR and hybridized to Tag4 Microarrays (Affymetrix) (Pierce et al. 2007). Fitness of each deletion strain was deduced from the signal intensity of the barcodes on the microarray. For each gene, the selection coefficient s (or relative growth rate of the strain in the pool) was estimated using the log2 fold change of normalized signal intensity between the initial and final time points (details in Supporting Information). The allelic effect at each locus was calculated as the difference between the selection coefficients (Δs = sSK1 – sS96).
Confirmation of QTL
To test the effect of a gene variant on a specific trait, the ORF ± 300 bp was deleted by homologous recombination. For S96, a CORE cassette (Storici et al. 2001) (kindly provided by Michael Knop) with KlURA3 (counterselectable) and kanMX4 (reporter) markers were inserted by standard DNA-targeting procedures (Gietz and Schiestl 2007) at the respective ORF locus. For SK1, transformation was done by electroporation (as described in http://www.koko.gov.my/CocoaBioTech/DNA%20Cells36.html). Cells were then spread on synthetic dextrose plates supplemented with geneticin (G418, 400 µg/ml) and lacking uracil for 3–4 days at 30°. The correct integration site was confirmed by colony PCR with internal and external primers (Table S1). For allele replacement experiments, cells were transformed with the ORF region ± 600 bp amplified from the strain carrying the desired allele. Counterselection for the CORE cassette excision was performed by selection on plates containing 5-fluoroorotic acid (5-FOA, 1 g/liter).
Computational detection of genetic interactions
Apart from the stratification of the samples according to the major QTL, to identify QTL acting in a specific background, the interaction distance method (ID) (Ignac et al. 2012) was applied. ID is based on merging interaction information, a generalization of mutual information to three variables, and the normalized information distance, a metric of the amount of information shared between two variables. ID was applied to measure dependence between two genetic markers and a phenotype, allowing us to detect the presence of interactions between the QTL. Positive ID values indicate redundant information between markers with strong effects on the phenotype (due to linkage or genetic redundancy); negative values indicate synergy between the markers in predicting the quantitative phenotype. To estimate the statistical significance of an ID value, we computed IDs between one million randomly generated markers given the same phenotype. A candidate interaction was considered significant if its P-value was <0.005. In applying ID, we discretized the variables: for example, the phenotype of high-temperature growth was discretized into four bins of equal size. To reduce computational requirements, we initially reduced the number of markers to 1226 by identifying blocks of highly correlated markers. Figure S2A shows the significant interaction candidates among the reduced set of markers. For the high-resolution TAO3-MKT1 interaction analysis, we then selected all markers with the appropriate coordinates from the full marker set.
Results
We performed QTL mapping on 11 distinct traits using three high-throughput approaches (BSA, ISA, and RHS). In the following, we present the QTL mapping results of these independent approaches, ranging from a simple Mendelian trait to nonselectable traits with two to three QTL with similar effect sizes to complex traits driven by many QTL with different effect sizes. Five of these traits were analyzed with all three methods, which allowed us to evaluate their performance in QTL detection. Finally, our large set of individual segregants allowed us to identify interactions between QTL within specific phenotypes.
BSA, ISA, and RHS effectively detect the causal QTL for cantharidin resistance
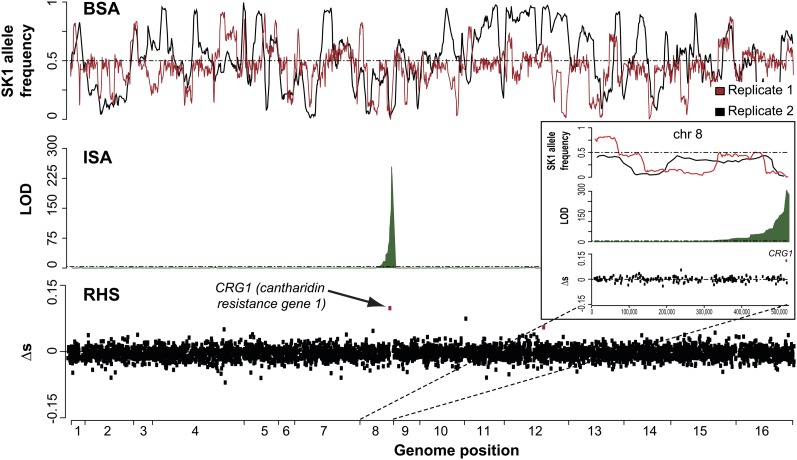
We first evaluated the three methods (ISA, BSA, and RHS) to map QTL for a Mendelian trait (cantharidin resistance), where S96 is resistant and SK1 is sensitive to the drug. With ISA, we mapped a single interval of ∼1 kb (LOD >200) on chromosome (chr) 8 (Figure 2). Within 3 kb of this interval, a BSA QTL was called, with the S96 allele highly enriched (∼95%); this QTL was found in both biological replicates, while several additional BSA peaks with similar amplitudes were not. The gene CRG1 was located in the strongest QTL peak in ISA and was also the top RHS hit (Figure 2). We confirmed the causative role of CRG1 by individually phenotyping the RHS hemizygous strains, demonstrating that deletion of the S96 CRG1 allele abolished cantharidin resistance (Figure S3). Individual and combinatorial replacement of the two nonsynonymous CRG1 SNPs (D82E and Y119C) indicated that both are necessary for cantharidin resistance in S96: 60 colonies of SK1 cells carrying both SNPs grew on cantharidin plates but none grew from the single-SNP replacements. Our results are consistent with previous reports that CRG1 confers cantharidin resistance (Niewmierzycka and Clarke 1999; Hoon et al. 2008; Lissina et al. 2011). Crg1 has been shown to mediate resistance to cantharidin by direct methylation of the compound, rendering it nontoxic for yeast (Lissina et al. 2011). Our results suggest that the enzymatic activity of Crg1 or its interaction with cantharidin is impaired by this change of either of the two amino acids. Thus, collectively, our results demonstrate that all three approaches successfully detect the true QTL for this Mendelian trait.
Figure 2.
Cantharidin resistance QTL mapped by BSA, ISA, and RHS. The top LOD score identified by ISA is located directly at the causal CRG1 gene, which was also the top hit in RHS (bottom plot). For BSA, the SK1 allele frequency (1 corresponding to 100% SK1, 0 to 100% S96) is plotted for two biological replicates. These replicates were not reproducible overall (likely due to spontaneous beneficial mutations in individual cells of the pool, as seen for 5-FU treatment), except for very few regions (including the CRG1 locus). The results on chr 8, which contains CRG1, are magnified (inset). For RHS, ∆s represents the difference between the selection coefficients of S96 and SK1.
ISA detects QTL for two nonselectable traits
We next analyzed two nonselectable traits that clearly differed among the parental strains and segregants, namely colony shape and flocculation. For these traits, BSA and RHS approaches could not be performed since they use pooled phenotyping and require a selective pressure. SK1 cells form a wrinkled colony shape on agar plates, whereas S96 cells form smooth colonies (Figure S4 and Figure S5). A total of 8.5% of the progeny formed wrinkled colonies, which suggests that the trait is conditioned by at least three or four independent genes (probability of 0.53–0.54 when assuming the same effect size). Consistent with this estimate, we identified three QTL, which, using gene deletion and allele replacement, we narrowed down to three genes (AMN1, MUC1, and SFL1) (Figure S5) required for the wrinkled SK1-like colonies.
While neither of the parental strains flocculated, one-quarter of the segregants did (23% in lactose-rich medium), suggesting that at least two independent genes condition the phenotype. Indeed, we detected two QTL (Figure S4), each of which contains a gene known to modify flocculation: the FLO1 allele (Hodgson et al. 1985) from the S96 background (disrupted in SK1 according to our sequencing data) and the SFL1 allele (Fujita et al. 1989) from the SK1 background (which harbors a premature stop codon at amino acid 477 in SK1). For SFL1, the calculated maximum LOD score in the QTL was within the gene and even close to its premature (likely causative) stop codon in SK1. By narrowing the QTL of these physical phenotypes down to the presumably causative genes, we demonstrate the ability of ISA to map QTL at high resolution in these nonselective binary traits.
From simple to complex traits
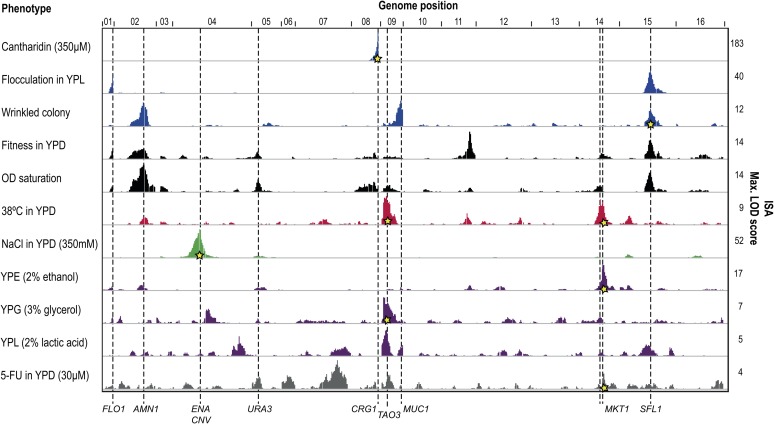
In our study, the average confidence interval size for QTL detected in ISA was 6 cM (∼18 kb). This resolution was much higher compared to previous studies, where the interval size has been around 16 cM (Steinmetz et al. 2002; Sinha et al. 2008). Given the successful identification of causative genes for less complex traits, we applied ISA to eight quantitative traits showing a continuous distribution among the segregants (Figure 3 and Table S2). We used these results to estimate the extent to which increasing the number of segregants improves the resolution of QTL detection (Figure S6 and Table S3). Our results suggest that for complex, multifactorial traits, increasing the number of segregants from 200 to at least 600 improves the resolution by more than twofold (Figure S6), but has no effect for Mendelian traits. Four of these complex growth traits [ethanol, 5-fluorouracil (5-FU), high-salt concentration, and high temperature] were also analyzed by BSA and RHS. In contrast to the physical traits, these traits are more suitable for BSA and RHS since the fittest strains can be selected via pooled growth. However, our RHS approach displayed a high false-positive rate (discussed later), and RHS results are therefore not shown.
Figure 3.
Detection of ISA QTL for 11 phenotypes. LOD scores are plotted for all phenotypes tested in this study using the IGV browser (Robinson et al. 2011) (Mendelian traits, blue; fitness traits in rich media, black; high temperature, red; high-salt, green; nonfermentable carbon sources, purple; 5-FU, gray). QTL containing putative causative variants are marked with a gray dashed line and labeled with the gene. Gene variants that were confirmed by allele exchange or by individual growth of RHS strains (for CRG1) in this study are marked with a star at the respective phenotype. Two gene variants were found to modify more than one phenotype: MKT1 (high temperature, ethanol, 5-FU) and TAO3 (high temperature, glycerol).
ISA uncovers the architecture of the complex high-salt tolerance trait
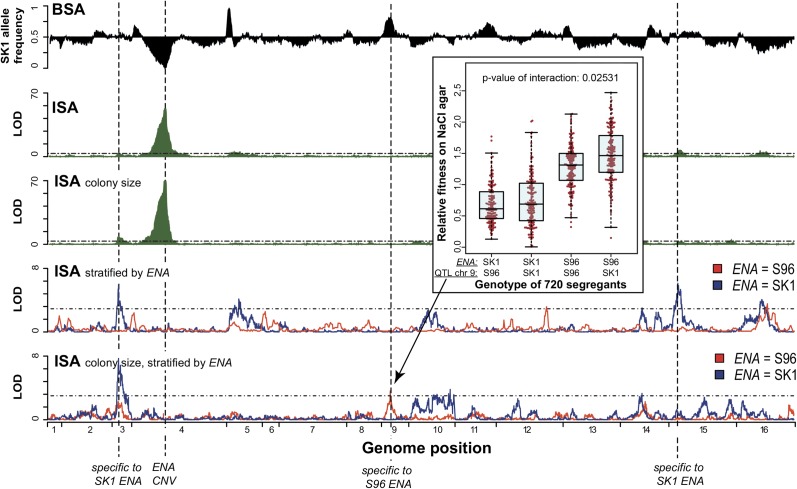
We combined ISA and BSA to dissect the high-salt tolerance trait as thoroughly as possible. At a high-salt concentration (350 mM NaCl), S96 grew faster than SK1, and their progeny showed a continuous distribution of growth rates. Six QTL were identified with ISA and eight with BSA, two of which overlapped (chr 4 and chr 16). The chr 4 QTL were the strongest identified by each approach (Figure 4, BSA; Table S2, ISA; LOD >50, cutoff LOD = 3.5). Within the 95% confidence interval of these QTL lies a cluster of ENA genes encoding sodium pumps, which are known to confer salt resistance (Haro et al. 1991). In contrast to a cluster of five highly similar ENA genes present in S96 (ENA1-5), SK1 carries only one copy of ENA6, a phylogenetically distant ENA gene (Daran-Lapujade et al. 2009). ENA copy number variation has been associated with high-salt tolerance across different yeast strains (Warringer et al. 2011). Accordingly, we found that overexpressing ENA6 increased salt resistance in SK1 cells (Figure S7). Moreover, we determined that ENA copy number accounts for 20% of the phenotypic variance. Our results thus demonstrate the ability of both ISA and BSA to successfully identify a large-effect locus.
Figure 4.
Identification of high-salt QTL. For BSA, SK1 allele frequency is plotted in black (replicates were highly reproducible and therefore only one is shown). For ISA results, the LOD scores are plotted in green. In addition to the standard method (1–2 days liquid culture), ISA phenotyping was also performed by a colony-size assay on agar (2–4 days) to account for the effects of growth duration. The major QTL identified by all approaches was the ENA locus on chr 4 (ENA CNV), which contains a cluster of genes encoding sodium pumps. By stratifying the ISA samples according to their ENA genotype (S96 ENA, red; SK1 ENA, blue), QTL specific to SK1 ENA (chr 3 and chr 15 for liquid culture) and S96 ENA (chr 9 for colony-size assay) were detected. The synergistic effect of the QTL on chr 9 in combination with ENA is also illustrated in the boxplot using the individual fitness (according to colony size) of 720 segregants. To test for interactions, we used an ANOVA test. A linear model was fitted to the data: phenotype ~ QTL1 + QTL2 + QTL1:QTL2 . The P-value for the interaction is the significance of including the interaction term (QTL1:QTL2).
However, identification of the major QTL is often not sufficient for understanding the genetic basis of a trait. If the sample size is too small, the phenotypic variance caused by a large-effect QTL like the ENA locus can be overestimated, and QTL with smaller effects will be obscured. To overcome this risk and identify additional QTL for salt tolerance, we stratified the ISA segregants according to their ENA genotype and repeated the QTL analysis. This highlighted the contribution of QTL with significant LOD scores (chr 3, 5, 14, 15, 16) even in the detrimental SK1 ENA background (Figure 4). We thus identified six QTL that explain >80% of both narrow (additive genetic factors) and broad sense heritability (all genetic factors including genetic interactions) (Visscher et al. 2008) (Table S4), suggesting that we have captured most of the causative alleles. These results demonstrate that allelic stratification can reveal additional QTL and thus enable a more comprehensive dissection of complex traits.
Six of eight BSA QTL were specific to BSA, in which cells were cultured for 5–9 days (vs. 1–2 days in ISA). To test whether the difference in QTL detection could be attributed to long-term effects, we performed a colony-size assay on agar with the ISA segregants (2–4 days in culture). In segregants with the S96 ENA background, we observed a beneficial effect on chr 9, where one of the BSA-specific QTL was also detected (Figure 4). This observation suggests that variations in experimental procedures, such as assay duration, can lead to the detection of different QTL. The combination of several methods could thus be a strategy to more thoroughly resolve the alleles responsible for a complex trait.
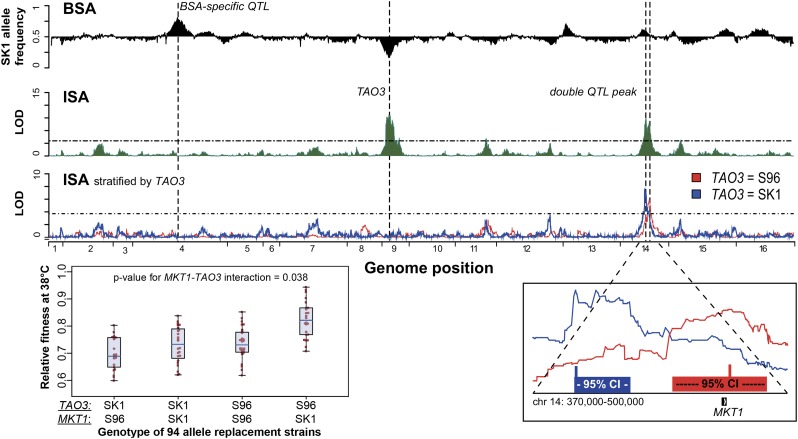
Mapping of high-temperature QTL reveals major differences between ISA and BSA
We then applied both BSA and ISA to dissect another selective phenotype, high-temperature growth. At high temperature (38°), S96 grew faster than SK1, and five QTL were identified with ISA and three with BSA. Only the QTL on chr 9 (S96 allele beneficial) was common between these two approaches (Figure 5). Within this QTL, we identified TAO3 as the gene responsible for high-temperature resistance, which we confirmed by allele replacement. By applying the stratification method described in the preceding section, we identified additional smaller-effect QTL that act specifically in the SK1-TAO3 or the S96-TAO3 background. We were able to separate a double LOD peak on chr 14 (Figure 5, bottom right): the left peak was mapped in the SK1 TAO3 background, and the right peak (including MKT1) in the S96 TAO3 background. By generating strains with all four combinations of TAO3 and MKT1 alleles in the S96 background, we confirmed that the effect of the MKT1 variant was indeed larger in combination with the S96 TAO3 allele (Figure 5, bottom left). These results show that the resolution of our ISA approach is sufficient to identify two QTL within a distance of <100 kb (Figure 5) and demonstrate the power of ISA for detecting epistatic genetic interactions. The identified QTL explain ∼59 and 47% of the narrow sense and broad sense heritability, respectively (Table S4), suggesting that several additional causative alleles remain undiscovered.
Figure 5.
Identification of high-temperature QTL. BSA and ISA results are plotted as in Figure 4. By stratifying the ISA samples by the major QTL (S96 TAO3, red; SK1 TAO3, blue), the double QTL peak on chr 14 could be separated into two individual QTL—chr 14: 390,000–410,000 for the SK1 TAO3 subset and chr 14: 480,000–500,000 for the S96 TAO3 subset. A magnification of this region is shown in the bottom right, with 95% confidence intervals and the calculated maximum shown as boxes below the separated peaks. The synergistic interaction between the S96 TAO3 and the SK1 MKT1 was confirmed in 94 allele replacement strains (bottom left boxplot). The P-value for interaction was calculated as described in the legend of Figure 4.
Mapping genetic loci associated with growth on ethanol, glycerol, and 5-FU
For growth with ethanol as the carbon source (YPE: Yeast extract, 10 g/liter; Bactopeptone, 20 g/liter; 2% v/v Ethanol), both ISA and BSA detected a QTL on chr 14, and the SK1 allele of MKT1 was confirmed as causative for improving growth by allele replacement (Figure S7 and Figure S8). In addition to its impact on high-temperature growth, we also confirmed that the SK1 allele of TAO3 (lying within a major ISA QTL on chr 9) significantly improves growth in media containing glycerol as the carbon source (YPG) (Figure S7). Finally, for growth in 5-FU, one QTL was identified by both ISA and BSA on chr 5 (likely due to a URA3 deletion in the SK1 background). None of the other BSA QTL were reproducible between the two biological replicates (Figure S8). Using ISA, however, we successfully identified and confirmed by allele replacement that MKT1 (within the QTL of chr 14) is causative for improved growth on 5-FU.
ISA allows the detection and characterization of genetic interactions
The complete dissection of complex traits can be hindered by non-additive genetic interactions, but also by the presence of closely linked alleles, which often remain undetected despite their contribution to phenotype. We previously reported one such linked region surrounding MKT1 on chr 14 (Steinmetz et al. 2002), which could explain the missing heritability (both narrow and broad) observed for high-temperature growth. In fact, the experimentally validated MKT1–TAO3 interaction responsible for high-temperature growth lies in regions of high linkage to other genes on chr 9 and 14. We observed that the LOD profile for high-temperature growth has many local peaks on chr 9 and chr 14, which carry TAO3 and MKT1, respectively (Figure S2A). If we include all markers on chr 9 and chr 14 for estimating the heritability, >65% of both narrow and broad sense heritability can be explained, suggesting the presence of closely linked, interacting QTL. To dissect these interactions at a finer resolution, we applied an interaction distance method (Ignac et al. 2012). A subset of 1226 markers was used to identify regions with the strongest interactions on chr 9 and chr 14. These regions of interaction were further analyzed with a denser marker set, and our results suggest that both regions contain more than one causative locus and that these loci interact with each other (Figure S2A).
The interaction distance method (Ignac et al. 2012) allowed us to detect both redundancy and synergistic effects between ISA QTL for growth in YPE and YPD (Table S5 and Figure S2B). Our results demonstrate that ISA is a powerful method to detect and characterize genetic interactions, which must be accounted for to explain phenotypic variance in complex traits.
Several factors confound QTL detection in pooled approaches
We next assessed the impact of experimental factors that confound QTL identification for each approach, which may partly explain their differing results (Figure S8, Figure S9, and Figure S10). While for most traits BSA QTL of both biological replicates were nearly identical, they varied widely between replicates for resistance to cantharidin and 5-FU (Figure 2 and Figure S8). Sequencing the 5-FU BSA pools revealed nonsense mutations in genes conferring resistance to the drug (FUR4, URA2), suggesting that individual cells acquired beneficial mutations and overtook the population, causing the enrichment of false-positive loci. This effect is specific to bulk selection approaches and is likely to occur for all phenotypes for which single mutations can confer a significant growth advantage. The impact of such confounding mutations would be expected to increase with the strength and length of the selection procedure.
In addition to mutations that cause resistance in BSA, we determined diploidization to be another confounding factor during long-term selection in BSA. We sequenced BSA pools at different time points during selection at high temperature. A decrease of an initially strong SK1 allele enrichment on chr 3 (from ∼100 to 50% allele frequency) was observed between generations 16 and 24 (Figure S10). This loss was detected in all of our BSA experiments and can also be observed in other studies (Ehrenreich et al. 2010). This enrichment of SK1 alleles on chr 3 corresponds to the MATa locus used for the initial selection of haploid progeny for BSA. Mating-type PCR (Huxley et al. 1990) performed on 32 individual clones after 100 generations confirmed that all cells had become diploid at this stage. This implies that a small number of MATα cells in the initial BSA pool mated with MATa cells and that these diploid cells then overtook the population.
As mentioned earlier, RHS results displayed a high rate of false-positive hits for our complex traits. Resequencing 50 of the RHS deletion strains revealed numerous chromosomal aberrations, which mostly consisted of triploidies. Twelve of 38 false-positive strains and 4 of 12 randomly selected strains were aneuploid (see Table S6 and Figure S9 for details). Since these aberrations affect many genes, their consequences are likely to obscure allelic differences at a single locus, especially when these are more subtle as for complex traits, and can thus lead to false positives.
These observations suggest that both pooled methods are vulnerable to genetic alterations that can render the detection of truly causative QTL difficult. These confounding factors should therefore be taken into account by adapting the experimental approaches, for example by decreasing the duration of selective pressure or comparing additional biological replicates.
Discussion
Despite intensive efforts, dissecting the genetic basis of complex traits is a persistent challenge. Several methods have been developed, including pooled approaches such as BSA, allowing for millions of individuals to be tested in a single experiment. Using next-generation sequencing techniques, large numbers of segregants can now be individually genotyped at a reasonable cost (Wilkening et al. 2013), enabling higher-resolution QTL mapping. To further increase resolution to the level of individual genes, we also developed and applied the RHS method. Here, we discuss the biological significance of quantitative trait gene (QTG) identification for two dissected traits (wrinkled colony shape and flocculation), confounding factors of pooled approaches such as BSA and RHS, and the importance of interactions between QTL.
QTG for wrinkled colony shape and flocculation are closely related
For wrinkled colony shape, we identified three QTG (Figure S4), which are also known to modify flocculation. Among these, AMN1 encodes a protein required for daughter-cell separation (Wang et al. 2003) and cell clumpiness (Yvert et al. 2003). It has not previously been associated with colony shape, but was recently implicated in flocculation (Li et al. 2013). Moreover, a loss-of-function mutation in the S96 AMN1 allele (D368V) has been reported to cause widespread gene expression changes (Yvert et al. 2003; Ronald et al. 2005). Another gene we linked to flocculation is MUC1 (also known as FLO11), which encodes a key cell-surface protein required for flocculation, as well as invasive and pseudohyphal growth (Lo and Dranginis 1998). Furthermore, the number of serine/threonine-rich tandem repeats in MUC1 has been linked to flocculation strength (Verstrepen et al. 2005; Liu et al. 2007), and this region is 1.1 kb shorter in SK1, corresponding to ∼12 repeats vs. 40 repeats in S96 (for primer sequences, see Table S1). MUC1 expression level has also been connected to colony shape (Barrales et al. 2008; White et al. 2011; Voordeckers et al. 2012). The third gene that we identified is SFL1, which is a known flocculation inhibitor (Fujita et al. 1989). Its deletion causes wrinkled colony shape in the Σ1278b background (Halme et al. 2004), which is consistent with our observation that the SK1 allele of SFL1 with its premature stop codon is required for wrinkled colony shape. Despite AMN1, MUC1, and FLO5 having previously been implicated in flocculation (Govender et al. 2008; Li et al. 2013) and the first two genes showing an effect on colony morphology in our study, we did not see any effect of these polymorphisms on flocculation in our background, as no QTL for flocculation were detected at these genes. However, SFL1 was detected as a QTL for both traits.
Two pleiotropic genes were identified across the traits tested
We confirmed by allele replacement that two QTG modify multiple phenotypes (TAO3, MKT1) (Figure 3). We found that the SK1 allele of the well-known pleiotropic gene MKT1 was beneficial for three growth phenotypes (high temperature, YPE, and 5-FU) (Figure 3 and Figure S7). Previous studies have found MKT1 to modify high temperature (Steinmetz et al. 2002), sporulation (Deutschbauer and Davis 2005), petite frequency (Dimitrov et al. 2009), DNA repair (Demogines et al. 2008a), and drug sensitivity (Kim and Fay 2009; Ehrenreich et al. 2010). The most likely causative polymorphism is a D30G mutation, with G being conserved across all other sequenced strains (Swinnen et al. 2011). Moreover, as reported by Zhu et al. (2008), MKT1 is a global regulator of gene expression and can therefore influence many traits. Similarly, TAO3 was identified as a QTG in two phenotypes (high temperature and YPG) (Figure 3 and Figure S7) in our study. This gene has previously been identified as a causative QTG for sporulation (Deutschbauer and Davis 2005) in the same strain background (SK1 × S96), but has not been connected to high-temperature resistance before our study. Finally, the implication of SFL1 in colony shape in this study along with its previous implication in flocculation (Fujita et al. 1989) suggests that this gene is also pleiotropic. On the other hand, QTL identified for growth in three different nonfermentable carbon sources (lactose : YPL, ethanol : YPE, glycerol : YPG) did not overlap (Figure 3), suggesting the absence of gene variants that globally influence the metabolism of nonfermentable carbon sources (e.g., enzymes of the Krebs cycle or mitochondrial respiration) in our strain background.
Analyses of pooled approaches suggests potential confounding factors
Unlike morphological traits, phenotypes that confer a growth advantage under a specific condition are especially suited for BSA and RHS, as phenotypic selection can be performed in bulk. The BSA approach is relatively fast and easy and confers a significant advantage over the other two approaches in terms of time and cost. However, we observed that spontaneous mutations conferring resistance to the selective pressure can lead to biased results. Assuming a low number of cells with an advantageous mutation in the original BSA pool, a shorter selection time (e.g., 10–30 generations) might alleviate this effect. Moreover, the analysis of multiple time points and biological replicates should help to identify true QTL for traits with a strong selective pressure.
Additionally, the use of a BSA pool with haploid segregants harbors the risk of diploidization, as seen in our time-dependent high-temperature QTL maps (Figure S10). This observation is in accordance with a recent study showing the invasion of diploids in a population of haploids despite no apparent growth advantage (Gerstein and Otto 2011). The presence of heterozygous strains diminishes the enrichment of beneficial alleles, since for dominant alleles the allele frequency would rarely reach 100% because of recessive alleles remaining in the heterozygous strains. To prevent the diploidization of haploid segregants in BSA, the strains could be diploidized in advance. The haploid segregants could also be independently phenotyped, followed by genotyping the pool of strains with extreme phenotypes. Nevertheless, even this method would still be limited by large-effect QTL masking smaller-effect QTL. Moreover, as explained earlier, genetic interactions are not detectable with BSA, nor does BSA allow the separation of linked QTL. Thus, combining the modified BSA strategies above with ISA should compensate for these limitations and lead to a more comprehensive understanding of the genetic architecture of complex traits.
The genome-wide RHS approach developed in our laboratory is based on individual gene deletions and successfully identified the causative gene for a Mendelian trait (cantharidin resistance). It should theoretically have performed best at identifying individual causative genes; nevertheless, it displayed a high false-positive rate for complex traits, most likely caused by chromosomal aberrations (Figure S9). A related recent study also reported incidences of aneuploidies and mutations, leading to high false-positive rates (Kim et al. 2012). This issue could be circumvented either by sequencing all strains and eliminating aberrant genotypes or by constructing additional replicate strains. With the emergence of more efficient gene-editing techniques, e.g., CRISPR/Cas (Cong et al. 2013), an RHS-type approach could also be feasible in the near future for human cells, which should enhance the detection of functional alleles for phenotypes with medical implications (e.g., drug resistance and cancer development and progression).
Culture duration may contribute to BSA-ISA differences
For high salt resistance, different QTL were detected between BSA and ISA. One factor likely contributing to these differences is the longer culture time in BSA. Our results using a longer-term colony assay in ISA (Figure 4) suggest that, in the BSA pool, an early selection for the advantageous S96 ENA allele occurred, followed by enrichment of beneficial alleles in this background. This effect has also been observed in two studies, which identified different QTL depending on sporulation time (Deutschbauer and Davis 2005; Ben-Ari et al. 2006). This selection for large-effect QTL and the subsequent enrichment of additional alleles in this background is a caveat to BSA studies, as QTL acting specifically in the presence of the detrimental major QTL allele would be overlooked. This problem can be avoided by using ISA, where genotypic stratification of segregants can be performed.
Gene–gene interactions and linked QTL hinder the identification of QTG
In addition to large-effect QTL masking the genetic effects of other causative genes, the difficulty of dissecting quantitative traits is increased by two factors even in an ISA approach. First, synergistic interactions can occur between functionally related genes (Perez-Perez et al. 2009). With the interaction distance method we detected synergistic gene–gene interactions, similarly to Bloom et al. (2013), as well as redundancy effects (Table S5). We found very little overlap between these interaction pairs and those found using synthetic lethality screens (Tong et al. 2001, 2004), suggesting that natural and synthetic variant interactions may shape phenotypic robustness differently. Second, a group of tightly linked genes can be responsible for large-effect QTL (Noor et al. 2001). With the high-temperature growth phenotype, we confirmed the novel TAO3–MKT1 interaction, and many more are expected from our interaction distance method (Figure S2). These results suggest that effects of linked causative genes and synergy are prevalent and should be accounted for in future efforts to map quantitative traits. Parts et al. (2011) have shown that multiple rounds of crossing from generations F1 to F12 can reduce the linkage between two loci, an approach that could thus increase QTL resolution and dissect linked QTG.
In conclusion, our study addresses the fundamental issue of how to improve quantitative trait dissection. Applying three high-throughput approaches allowed us to resolve eight potential causative genes for eight phenotypes. Nevertheless, for the complex traits, the causal alleles explained only part of the heritability (on average ∼60% of broad sense and ∼66% of narrow sense) (Table S4), suggesting the contribution of additional factors such as linked QTL and epistatic effects. To thoroughly assess the impact of these factors on phenotype, future studies should improve resolution by increasing the sample size. Future studies would also benefit from accounting for experimental differences that can influence the loci detected, for example by combining multiple approaches as we have done here. Our findings indicate that we are currently looking at the tip of the iceberg: the focus should now be placed on the development of innovative experimental and computational strategies to deepen our understanding of the complex architecture of quantitative traits.
Supplementary Material
Acknowledgments
We thank Vicent Pelechano for fruitful discussions; Michael Knop for providing materials; and Wu Wei and Leopold Parts for valuable feedback on the manuscript. This study was technically supported by the European Molecular Biology Laboratory Genomics Core Facility, where the libraries were sequenced. The research leading to these results has received funding from the Deutsche Forschungsgemeinschaft (DFG-GZ: WI 3311/2-1) (to S.W.) and from the National Institutes of Health, the Deutsche Forschungsgemeinschaft (1422/2-2), and the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. AdG-294542 (to L.M.S.).
Footnotes
Communicating editor: M. Johnston
Literature Cited
- Barrales R. R., Jimenez J., Ibeas J. I., 2008. Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae. Genetics 178: 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari G., Zenvirth D., Sherman A., David L., Klutstein M., et al. , 2006. Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet. 2: e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland S. R., Jin N., Ozdemir A. C., Lyons R. H., Jr, Weisman L. S., et al. , 2010. Discovery of mutations in Saccharomyces cerevisiae by pooled linkage analysis and whole-genome sequencing. Genetics 186: 1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. S., Ehrenreich I. M., Loo W. T., Lite T. L., Kruglyak L., 2013. Finding the sources of missing heritability in a yeast cross. Nature 494: 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R. B., Storey J. D., Whittle J., Kruglyak L., 2005. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature 436: 701–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Browning S. R., Browning B. L., 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81: 1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., et al. , 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daran-Lapujade P., Daran J. M., Luttik M. A., Almering M. J., Pronk J. T., et al. , 2009. An atypical PMR2 locus is responsible for hypersensitivity to sodium and lithium cations in the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D. FEMS Yeast Res. 9: 789–792. [DOI] [PubMed] [Google Scholar]
- Demogines A., Smith E., Kruglyak L., Alani E., 2008a Identification and dissection of a complex DNA repair sensitivity phenotype in Baker’s yeast. PLoS Genet. 4: e1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demogines A., Wong A., Aquadro C., Alani E., 2008b Incompatibilities involving yeast mismatch repair genes: a role for genetic modifiers and implications for disease penetrance and variation in genomic mutation rates. PLoS Genet. 4: e1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer A. M., Davis R. W., 2005. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 37: 1333–1340. [DOI] [PubMed] [Google Scholar]
- Dimitrov L. N., Brem R. B., Kruglyak L., Gottschling D. E., 2009. Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics 183: 365–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I. M., Gerke J. P., Kruglyak L., 2009. Genetic dissection of complex traits in yeast: insights from studies of gene expression and other phenotypes in the BYxRM cross. Cold Spring Harb. Symp. Quant. Biol. 74: 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I. M., Torabi N., Jia Y., Kent J., Martis S., et al. , 2010. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464: 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman J. B., 2011. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Gen. 4: 250–255. [Google Scholar]
- Flint J., 2011. Mapping quantitative traits and strategies to find quantitative trait genes. Methods 53: 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Mackay T. F., 2009. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 19: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss E. J., Radulovic D., Shaffer S. A., Ruderfer D. M., Bedalov A., et al. , 2007. Genetic basis of proteome variation in yeast. Nat. Genet. 39: 1369–1375. [DOI] [PubMed] [Google Scholar]
- Fujita A., Kikuchi Y., Kuhara S., Misumi Y., Matsumoto S., et al. , 1989. Domains of the SFL1 protein of yeasts are homologous to Myc oncoproteins or yeast heat-shock transcription factor. Gene 85: 321–328. [DOI] [PubMed] [Google Scholar]
- Gatbonton T., Imbesi M., Nelson M., Akey J. M., Ruderfer D. M., et al. , 2006. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein A. C., Otto S. P., 2011. Cryptic fitness advantage: diploids invade haploid populations despite lacking any apparent advantage as measured by standard fitness assays. PLoS ONE 6: e26599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 31–34. [DOI] [PubMed] [Google Scholar]
- Govender P., Domingo J. L., Bester M. C., Pretorius I. S., Bauer F. F., 2008. Controlled expression of the dominant flocculation genes FLO1, FLO5, and FLO11 in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 74: 6041–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A., Bumgarner S., Styles C., Fink G. R., 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415. [DOI] [PubMed] [Google Scholar]
- Haro R., Garciadeblas B., Rodriguez-Navarro A., 1991. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 291: 189–191. [DOI] [PubMed] [Google Scholar]
- Heck J. A., Argueso J. L., Gemici Z., Reeves R. G., Bernard A., et al. , 2006. Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. Proc. Natl. Acad. Sci. USA 103: 3256–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson J. A., Berry D. R., Johnston J. R., 1985. Discrimination by heat and proteinase treatments between flocculent phenotypes conferred on Saccharomyces cerevisiae by the genes FLO1 and FLO5. J. Gen. Microbiol. 131: 3219–3227. [DOI] [PubMed] [Google Scholar]
- Hoon S., Smith A. M., Wallace I. M., Suresh S., Miranda M., et al. , 2008. An integrated platform of genomic assays reveals small-molecule bioactivities. Nat. Chem. Biol. 4: 498–506. [DOI] [PubMed] [Google Scholar]
- Hu X. H., Wang M. H., Tan T., Li J. R., Yang H., et al. , 2007. Genetic dissection of ethanol tolerance in the budding yeast Saccharomyces cerevisiae. Genetics 175: 1479–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley C., Green E. D., Dunham I., 1990. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 6: 236. [DOI] [PubMed] [Google Scholar]
- Ignac, T. M., N. A. Sakhanenko, A. Skupin, and D. J. Galas, 2012 New methods for finding associations in large data sets: generalizing the maximal information coefficient (MIC). Tampere International Center for Signal Processing. TICSP series # 6. Proceedings of the Ninth International Workshop on Computational Systems Biology. June, 2012, Germany, pp. 39–42. [Google Scholar]
- Kim H. S., Fay J. C., 2009. A combined-cross analysis reveals genes with drug-specific and background-dependent effects on drug sensitivity in Saccharomyces cerevisiae. Genetics 183: 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. S., J. Huh, L. Riles, A. Reyes, and J. C. Fay, 2012 A noncomplementation screen for quantitative trait alleles in Saccharomyces cerevisiae. G3 2: 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H., Estrada K., Lettre G., Berndt S. I., Weedon M. N., et al. , 2010. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467: 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang L., Wu X., Fang O., Wang L., et al. , 2013. Polygenic molecular architecture underlying non-sexual cell aggregation in budding yeast. DNA Res. 20: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissina E., Young B., Urbanus M. L., Guan X. L., Lowenson J., et al. , 2011. A systems biology approach reveals the role of a novel methyltransferase in response to chemical stress and lipid homeostasis. PLoS Genet. 7: e1002332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Wang D., Wang Z. Y., He X. P., Zhang B., 2007. Genetic basis of flocculation phenotype conversion in Saccharomyces cerevisiae. FEMS Yeast Res. 7: 1362–1370. [DOI] [PubMed] [Google Scholar]
- Lo W. S., Dranginis A. M., 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9: 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marullo P., Aigle M., Bely M., Masneuf-Pomarede I., Durrens P., et al. , 2007. Single QTL mapping and nucleotide-level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res. 7: 941–952. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M., 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122. [DOI] [PubMed] [Google Scholar]
- Niewmierzycka A., Clarke S., 1999. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem. 274: 814–824. [DOI] [PubMed] [Google Scholar]
- Nogami S., Ohya Y., Yvert G., 2007. Genetic complexity and quantitative trait loci mapping of yeast morphological traits. PLoS Genet. 3: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor M. A., Cunningham A. L., Larkin J. C., 2001. Consequences of recombination rate variation on quantitative trait locus mapping studies: simulations based on the Drosophila melanogaster genome. Genetics 159: 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Yuan D. S., Xiang D., Wang X., Sookhai-Mahadeo S., et al. , 2004. A robust toolkit for functional profiling of the yeast genome. Mol. Cell 16: 487–496. [DOI] [PubMed] [Google Scholar]
- Parts L., Cubillos F. A., Warringer J., Jain K., Salinas F., et al. , 2011. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 21: 1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez J. M., Candela H., Micol J. L., 2009. Understanding synergy in genetic interactions. Trends Genet. 25: 368–376. [DOI] [PubMed] [Google Scholar]
- Perlstein E. O., Ruderfer D. M., Roberts D. C., Schreiber S. L., Kruglyak L., 2007. Genetic basis of individual differences in the response to small-molecule drugs in yeast. Nat. Genet. 39: 496–502. [DOI] [PubMed] [Google Scholar]
- Pierce S. E., Davis R. W., Nislow C., Giaever G., 2007. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat. Protoc. 2: 2958–2974. [DOI] [PubMed] [Google Scholar]
- Proctor M., Urbanus M. L., Fung E. L., Jaramillo D. F., Davis R. W., et al. , 2011. The automated cell: compound and environment screening system (ACCESS) for chemogenomic screening. Methods Mol. Biol. 759: 239–269. [DOI] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald J., Brem R. B., Whittle J., Kruglyak L., 2005. Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet. 1: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J., Shapiro J. A., Ruderfer D. M., Kruglyak L., 2009. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458: 342–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre A. V., Murray A. W., Leu J. Y., 2006. High-resolution mutation mapping reveals parallel experimental evolution in yeast. PLoS Biol. 4: e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha H., David L., Pascon R. C., Clauder-Münster S., Krishnakumar S., et al. , 2008. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics 180: 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Onge R. P., Mani R., Oh J., Proctor M., Fung E., et al. , 2007. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 39: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz L. M., Davis R. W., 2004. Maximizing the potential of functional genomics. Nat. Rev. Genet. 5: 190–201. [DOI] [PubMed] [Google Scholar]
- Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., et al. , 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330. [DOI] [PubMed] [Google Scholar]
- Storici F., Lewis L. K., Resnick M. A., 2001. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19: 773–776. [DOI] [PubMed] [Google Scholar]
- Stranger B. E., Stahl E. A., Raj T., 2011. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics 187: 367–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen S., Thevelein J. M., Nevoigt E., 2011. Genetic mapping of quantitative phenotypic traits in Saccharomyces cerevisiae. FEMS Yeast Res. [DOI] [PubMed] [Google Scholar]
- Swinnen S., Schaerlaekens K., Pais T., Claesen J., Hubmann G., et al. , 2012. Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res. 22: 975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., et al. , 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., et al. , 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813. [DOI] [PubMed] [Google Scholar]
- Verstrepen K. J., Jansen A., Lewitter F., Fink G. R., 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37: 986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., Hill W. G., Wray N. R., 2008. Heritability in the genomics era: concepts and misconceptions. Nat. Rev. Genet. 9: 255–266. [DOI] [PubMed] [Google Scholar]
- Voordeckers K., De Maeyer D., van der Zande E., Vinces M. D., Meert W., et al. , 2012. Identification of a complex genetic network underlying Saccharomyces cerevisiae colony morphology. Mol. Microbiol. 86: 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shirogane T., Liu D., Harper J. W., Elledge S. J., 2003. Exit from exit: resetting the cell cycle through Amn1 inhibition of G protein signaling. Cell 112: 697–709. [DOI] [PubMed] [Google Scholar]
- Warringer J., Zorgo E., Cubillos F. A., Zia A., Gjuvsland A., et al. , 2011. Trait variation in yeast is defined by population history. PLoS Genet. 7: e1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger J. W., Schwartz K., Sherlock G., 2010. Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet. 6: e1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. G., Piccirillo S., Dusevich V., Law D. J., Kapros T., et al. , 2011. Flo11p adhesin required for meiotic differentiation in Saccharomyces cerevisiae minicolonies grown on plastic surfaces. FEMS Yeast Res. 11: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkening S., Tekkedil M. M., Lin G., Fritsch E. S., Wei W., et al. , 2013. Genotyping 1000 yeast strains by next-generation sequencing. BMC Genomics 14: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Yvert G., Brem R. B., Whittle J., Akey J. M., Foss E., et al. , 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 35: 57–64. [DOI] [PubMed] [Google Scholar]
- Zhu J., Zhang B., Smith E. N., Drees B., Brem R. B., et al. , 2008. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat. Genet. 40: 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.