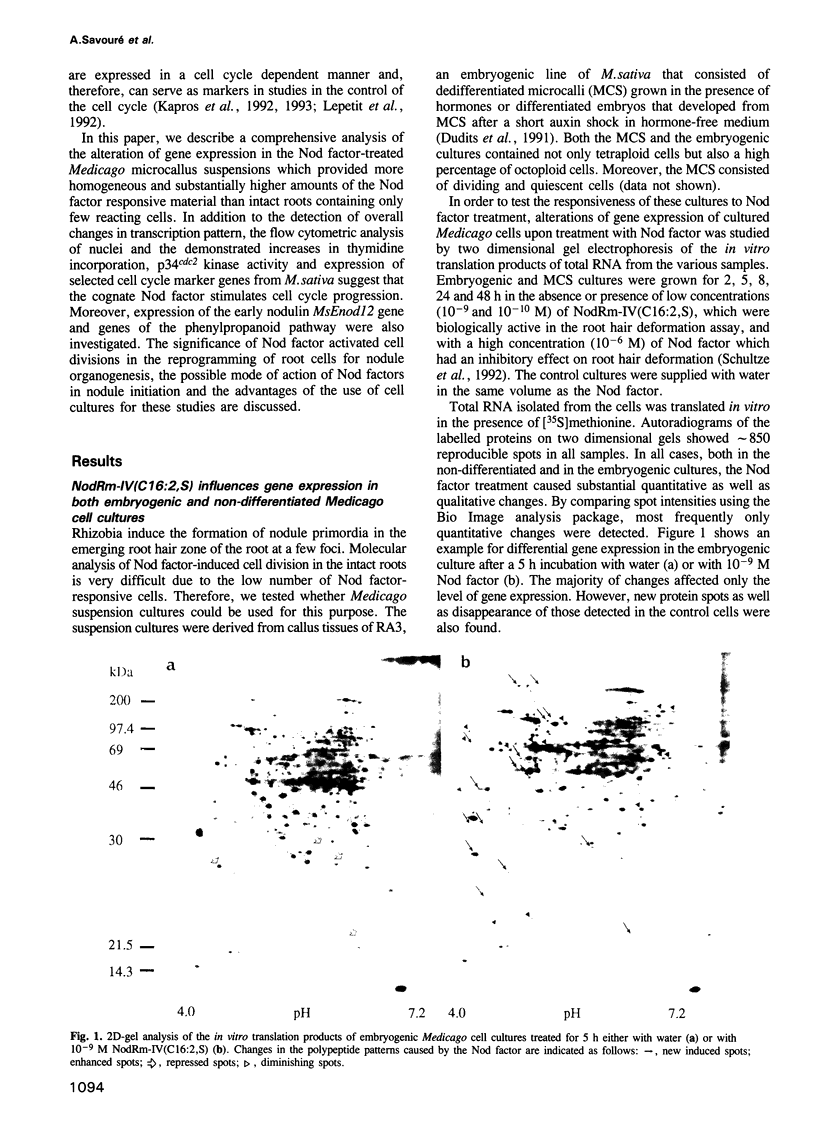
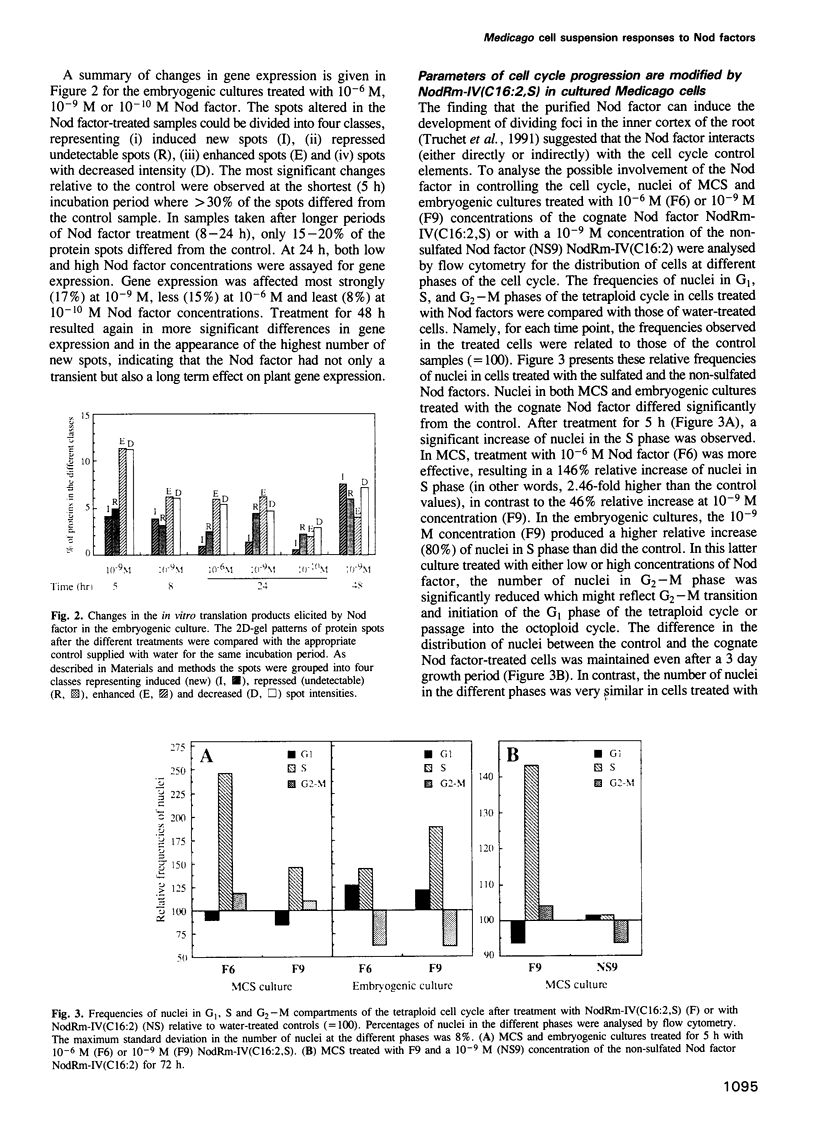
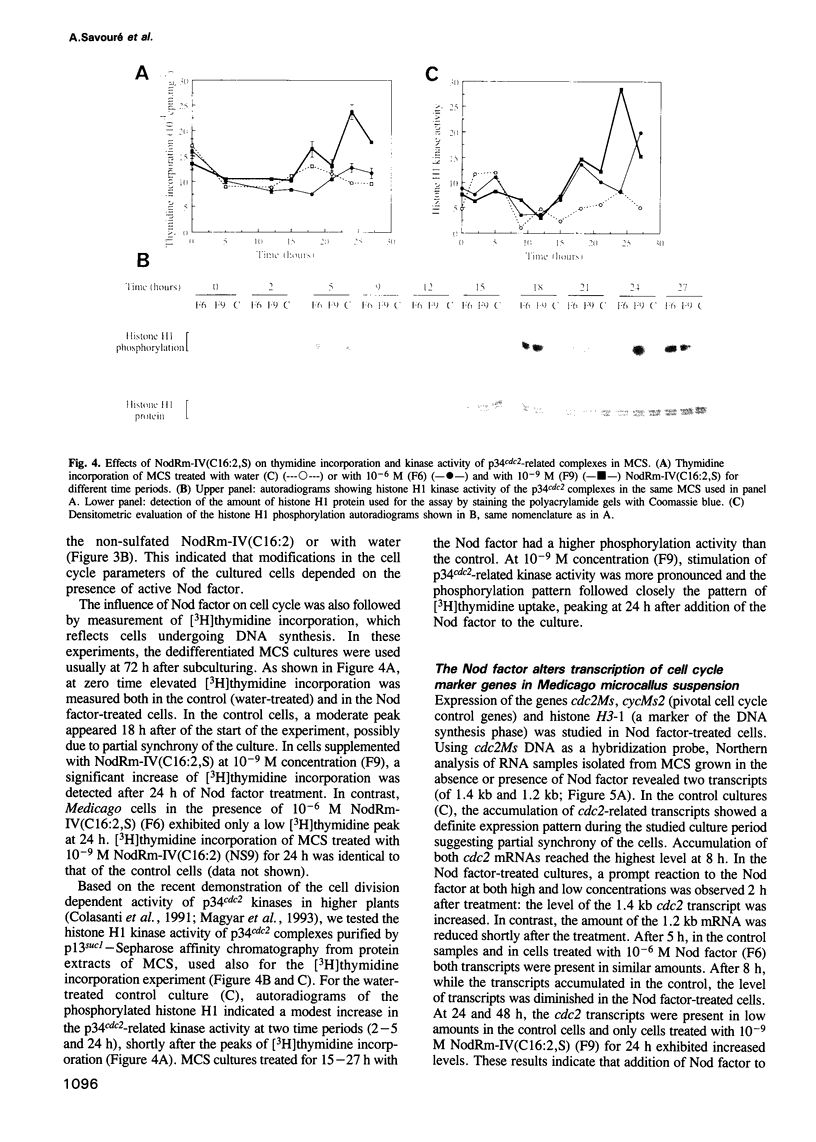
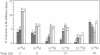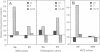Abstract
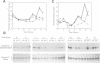
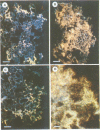
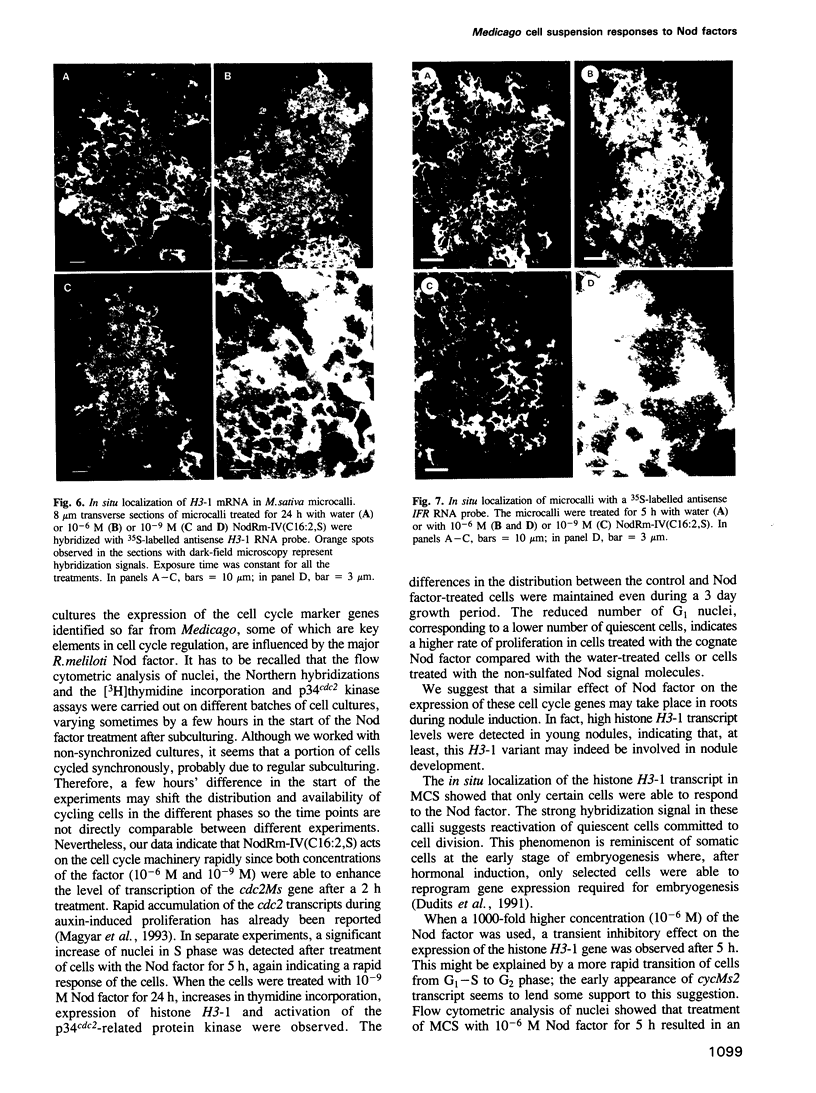
We have shown that treatment of Medicago microcallus suspensions with the cognate Rhizobium meliloti Nod signal molecule NodRm-IV(C16:2,S) can modify gene expression both qualitatively and quantitatively. At concentrations of 10(-6) - 10(-9) M, this host specific plant morphogen but not the inactive non-sulfated molecule stimulated cell cycle progression as indicated by the significantly enhanced thymidine incorporation, elevated number of S phase cells, increase in kinase activity of the p34cdc2-related complexes and enhancement of the level of expression of several cell cycle marker genes, the histone H3-1, the cdc2Ms and the cyclin cycMs2. The presented data suggest that at least part of the physiological role of the Nod factor may be linked to molecular events involved in the control of the plant cell division cycle. In situ hybridization experiments with antisense H3-1 RNA probe indicated that only certain cells of the calli were able to respond to the Nod factor. High (10(-6) M) but not low (10(-9) M) concentrations of the active Nod factors induced the expression of the isoflavone reductase gene (IFR), a marker gene of the isoflavonoid biosynthesis pathway in most callus cells. Our results indicate that Medicago cell responses to the Nod signal molecules can be investigated in suspension cultures.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Kiss G. B., Bauer P., Poiret M., Pierre M., Savouré A., Kondorosi E., Kondorosi A. Identification of two alfalfa early nodulin genes with homology to members of the pea Enod12 gene family. Plant Mol Biol. 1993 Jan;21(2):375–380. doi: 10.1007/BF00019952. [DOI] [PubMed] [Google Scholar]
- Baev N., Schultze M., Barlier I., Ha D. C., Virelizier H., Kondorosi E., Kondorosi A. Rhizobium nodM and nodN genes are common nod genes: nodM encodes functions for efficiency of nod signal production and bacteroid maturation. J Bacteriol. 1992 Dec;174(23):7555–7565. doi: 10.1128/jb.174.23.7555-7565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Colasanti J., Tyers M., Sundaresan V. Isolation and characterization of cDNA clones encoding a functional p34cdc2 homologue from Zea mays. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3377–3381. doi: 10.1073/pnas.88.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. F., Long S. R. Rhizobium--plant signal exchange. Nature. 1992 Jun 25;357(6380):655–660. doi: 10.1038/357655a0. [DOI] [PubMed] [Google Scholar]
- Hirsch A. M., Bhuvaneswari T. V., Torrey J. G., Bisseling T. Early nodulin genes are induced in alfalfa root outgrowths elicited by auxin transport inhibitors. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1244–1248. doi: 10.1073/pnas.86.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt H., Mink M., Pfosser M., Bögre L., Györgyey J., Jonak C., Gartner A., Dudits D., Heberle-Bors E. Alfalfa cyclins: differential expression during the cell cycle and in plant organs. Plant Cell. 1992 Dec;4(12):1531–1538. doi: 10.1105/tpc.4.12.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt H., Páy A., Györgyey J., Bakó L., Németh K., Bögre L., Schweyen R. J., Heberle-Bors E., Dudits D. Complementation of a yeast cell cycle mutant by an alfalfa cDNA encoding a protein kinase homologous to p34cdc2. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1636–1640. doi: 10.1073/pnas.88.5.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M., Rubery P. H. Naturally occurring auxin transport regulators. Science. 1988 Jul 15;241(4863):346–349. doi: 10.1126/science.241.4863.346. [DOI] [PubMed] [Google Scholar]
- Jacobs T. Control of the cell cycle. Dev Biol. 1992 Sep;153(1):1–15. doi: 10.1016/0012-1606(92)90087-w. [DOI] [PubMed] [Google Scholar]
- Kapros T., Bögre L., Németh K., Bakó L., Györgyey J., Wu S. C., Dudits D. Differential Expression of Histone H3 Gene Variants during Cell Cycle and Somatic Embryogenesis in Alfalfa. Plant Physiol. 1992 Feb;98(2):621–625. doi: 10.1104/pp.98.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepetit M., Ehling M., Chaubet N., Gigot C. A plant histone gene promoter can direct both replication-dependent and -independent gene expression in transgenic plants. Mol Gen Genet. 1992 Jan;231(2):276–285. doi: 10.1007/BF00279801. [DOI] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J. C., Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990 Apr 19;344(6268):781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- Norbury C. J., Nurse P. Control of the higher eukaryote cell cycle by p34cdc2 homologues. Biochim Biophys Acta. 1989 Jul 28;989(1):85–95. doi: 10.1016/0304-419x(89)90036-x. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Paiva N. L., Edwards R., Sun Y. J., Hrazdina G., Dixon R. A. Stress responses in alfalfa (Medicago sativa L.) 11. Molecular cloning and expression of alfalfa isoflavone reductase, a key enzyme of isoflavonoid phytoalexin biosynthesis. Plant Mol Biol. 1991 Oct;17(4):653–667. doi: 10.1007/BF00037051. [DOI] [PubMed] [Google Scholar]
- Pichon M., Journet E. P., Dedieu A., de Billy F., Truchet G., Barker D. G. Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell. 1992 Oct;4(10):1199–1211. doi: 10.1105/tpc.4.10.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N. P., Relić B., Talmont F., Lewin A., Promé D., Pueppke S. G., Maillet F., Dénarié J., Promé J. C., Broughton W. J. Broad-host-range Rhizobium species strain NGR234 secretes a family of carbamoylated, and fucosylated, nodulation signals that are O-acetylated or sulphated. Mol Microbiol. 1992 Dec;6(23):3575–3584. doi: 10.1111/j.1365-2958.1992.tb01793.x. [DOI] [PubMed] [Google Scholar]
- Ren Y. Y., West C. A. Elicitation of Diterpene Biosynthesis in Rice (Oryza sativa L.) by Chitin. Plant Physiol. 1992 Jul;99(3):1169–1178. doi: 10.1104/pp.99.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche P., Debellé F., Maillet F., Lerouge P., Faucher C., Truchet G., Dénarié J., Promé J. C. Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo-oligosaccharide signals. Cell. 1991 Dec 20;67(6):1131–1143. doi: 10.1016/0092-8674(91)90290-f. [DOI] [PubMed] [Google Scholar]
- Roche P., Lerouge P., Ponthus C., Promé J. C. Structural determination of bacterial nodulation factors involved in the Rhizobium meliloti-alfalfa symbiosis. J Biol Chem. 1991 Jun 15;266(17):10933–10940. [PubMed] [Google Scholar]
- Sanjuan J., Carlson R. W., Spaink H. P., Bhat U. R., Barbour W. M., Glushka J., Stacey G. A 2-O-methylfucose moiety is present in the lipo-oligosaccharide nodulation signal of Bradyrhizobium japonicum. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8789–8793. doi: 10.1073/pnas.89.18.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B., Van De Wiel C., Zalensky A., Horvath B., Spaink H., Van Eck H., Zwartkruis F., Wolters A. M., Gloudemans T., Van Kammen A. The ENOD12 gene product is involved in the infection process during the pea-Rhizobium interaction. Cell. 1990 Jan 26;60(2):281–294. doi: 10.1016/0092-8674(90)90743-x. [DOI] [PubMed] [Google Scholar]
- Schmidt J., Wingender R., John M., Wieneke U., Schell J. Rhizobium meliloti nodA and nodB genes are involved in generating compounds that stimulate mitosis of plant cells. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8578–8582. doi: 10.1073/pnas.85.22.8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze M., Quiclet-Sire B., Kondorosi E., Virelizer H., Glushka J. N., Endre G., Géro S. D., Kondorosi A. Rhizobium meliloti produces a family of sulfated lipooligosaccharides exhibiting different degrees of plant host specificity. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):192–196. doi: 10.1073/pnas.89.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink H. P., Aarts A., Stacey G., Bloemberg G. V., Lugtenberg B. J., Kennedy E. P. Detection and separation of Rhizobium and Bradyrhizobium Nod metabolites using thin-layer chromatography. Mol Plant Microbe Interact. 1992 Jan-Feb;5(1):72–80. doi: 10.1094/mpmi-5-072. [DOI] [PubMed] [Google Scholar]
- Spaink H. P., Sheeley D. M., van Brussel A. A., Glushka J., York W. S., Tak T., Geiger O., Kennedy E. P., Reinhold V. N., Lugtenberg B. J. A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature. 1991 Nov 14;354(6349):125–130. doi: 10.1038/354125a0. [DOI] [PubMed] [Google Scholar]
- Verma DPS. Signals in Root Nodule Organogenesis and Endocytosis of Rhizobium. Plant Cell. 1992 Apr;4(4):373–382. doi: 10.1105/tpc.4.4.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brussel A. A., Bakhuizen R., van Spronsen P. C., Spaink H. P., Tak T., Lugtenberg B. J., Kijne J. W. Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science. 1992 Jul 3;257(5066):70–72. doi: 10.1126/science.257.5066.70. [DOI] [PubMed] [Google Scholar]