Abstract
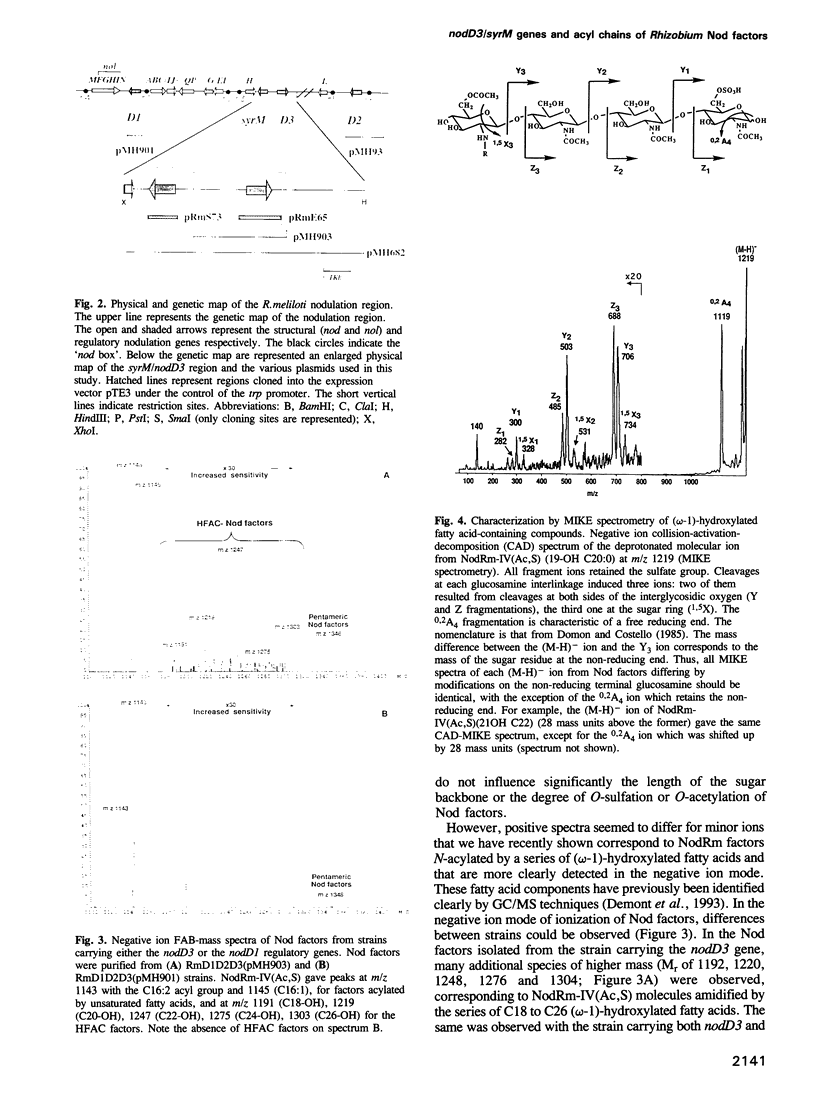
Rhizobia elicit the formation of nitrogen-fixing nodules on specific legume hosts. Rhizobium meliloti nodulation (nod) genes control a signal exchange between the two symbiotic partners during infection and the early steps of nodulation. The regulatory nodD1, nodD2 and nodD3 genes are involved in the specific perception of different plant and environmental signals and activate the transcription of the nod operons. Once activated, the structural nod genes specify the synthesis of diffusible lipo-oligosaccharides, the Nod factors, which signal back to the plant. R. meliloti Nod factors are sulfated chito-oligosaccharides which are mono-N-acylated by unsaturated C16 fatty acids or by a series of C18 to C26 (omega-1)-hydroxylated fatty acids. In this paper we show that the regulatory nodD3 gene and another symbiotic regulatory gene, syrM, which mediate bacterial responses to plant signals that differ from those involving nodD1 and nodD2, determine the synthesis of Nod factors with different acyl moieties. nodD3 and syrM are required for the synthesis of Nod factors N-acylated by the (omega-1)-hydroxylated fatty acids. This regulatory mechanism makes possible the qualitative adaptation of bacterial Nod signal production to plant signals in the course of the symbiotic process.
Full text
PDF


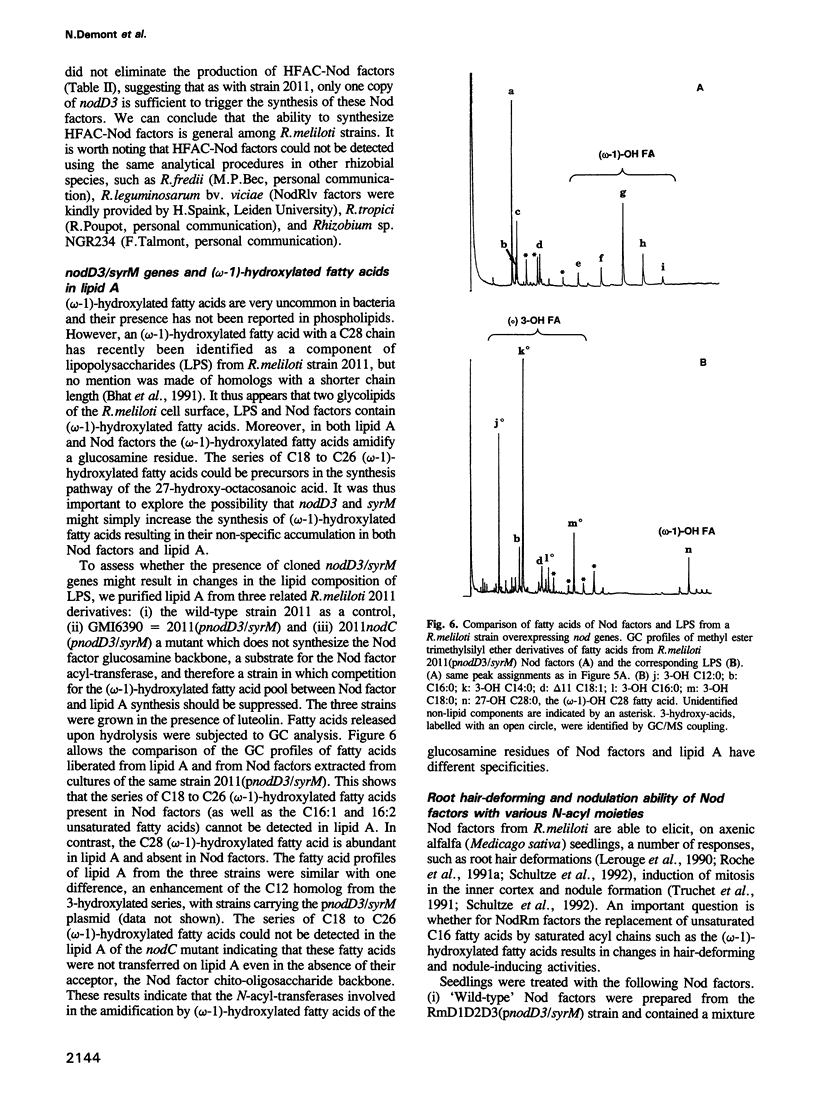
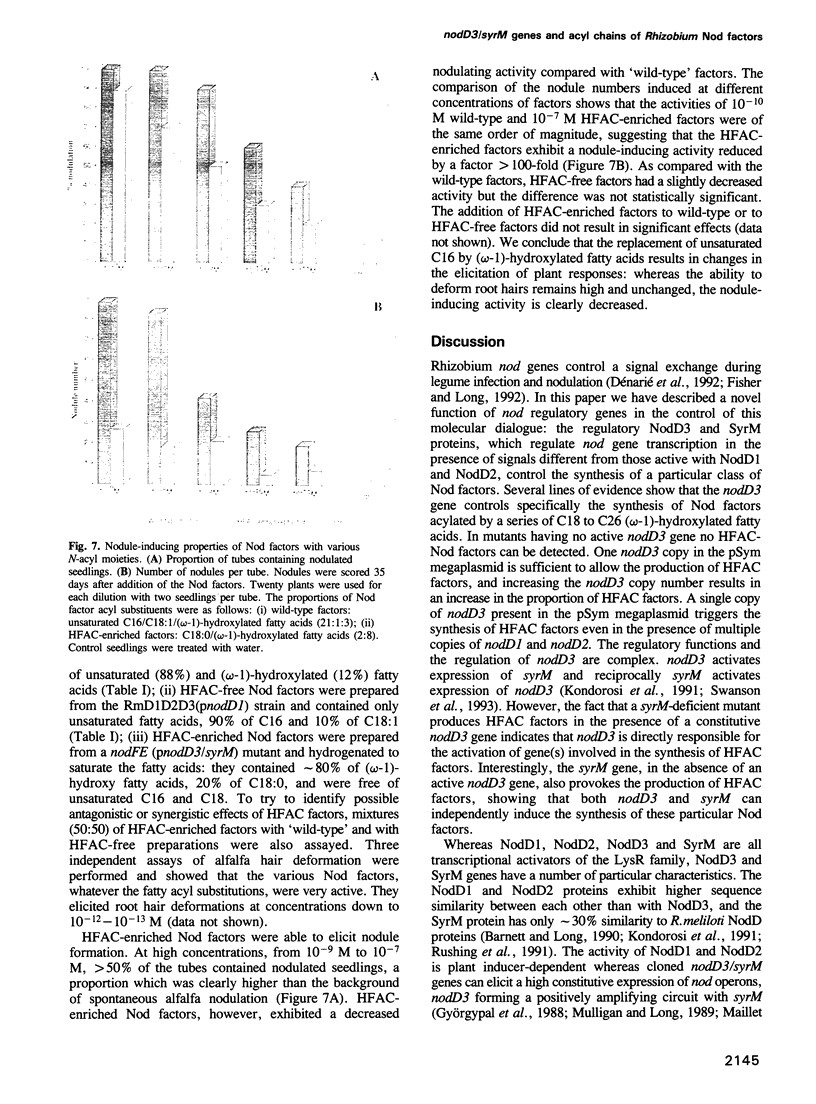




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkinson E. M., Long S. R. Homology of Rhizobium meliloti NodC to polysaccharide polymerizing enzymes. Mol Plant Microbe Interact. 1992 Sep-Oct;5(5):439–442. doi: 10.1094/mpmi-5-439. [DOI] [PubMed] [Google Scholar]
- Barnett M. J., Long S. R. DNA sequence and translational product of a new nodulation-regulatory locus: syrM has sequence similarity to NodD proteins. J Bacteriol. 1990 Jul;172(7):3695–3700. doi: 10.1128/jb.172.7.3695-3700.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat U. R., Mayer H., Yokota A., Hollingsworth R. I., Carlson R. W. Occurrence of lipid A variants with 27-hydroxyoctacosanoic acid in lipopolysaccharides from members of the family Rhizobiaceae. J Bacteriol. 1991 Apr;173(7):2155–2159. doi: 10.1128/jb.173.7.2155-2159.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debellé F., Rosenberg C., Dénarié J. The Rhizobium, Bradyrhizobium, and Azorhizobium NodC proteins are homologous to yeast chitin synthases. Mol Plant Microbe Interact. 1992 Sep-Oct;5(5):443–446. doi: 10.1094/mpmi-5-443. [DOI] [PubMed] [Google Scholar]
- Debellé F., Rosenberg C., Vasse J., Maillet F., Martinez E., Dénarié J., Truchet G. Assignment of symbiotic developmental phenotypes to common and specific nodulation (nod) genetic loci of Rhizobium meliloti. J Bacteriol. 1986 Dec;168(3):1075–1086. doi: 10.1128/jb.168.3.1075-1086.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
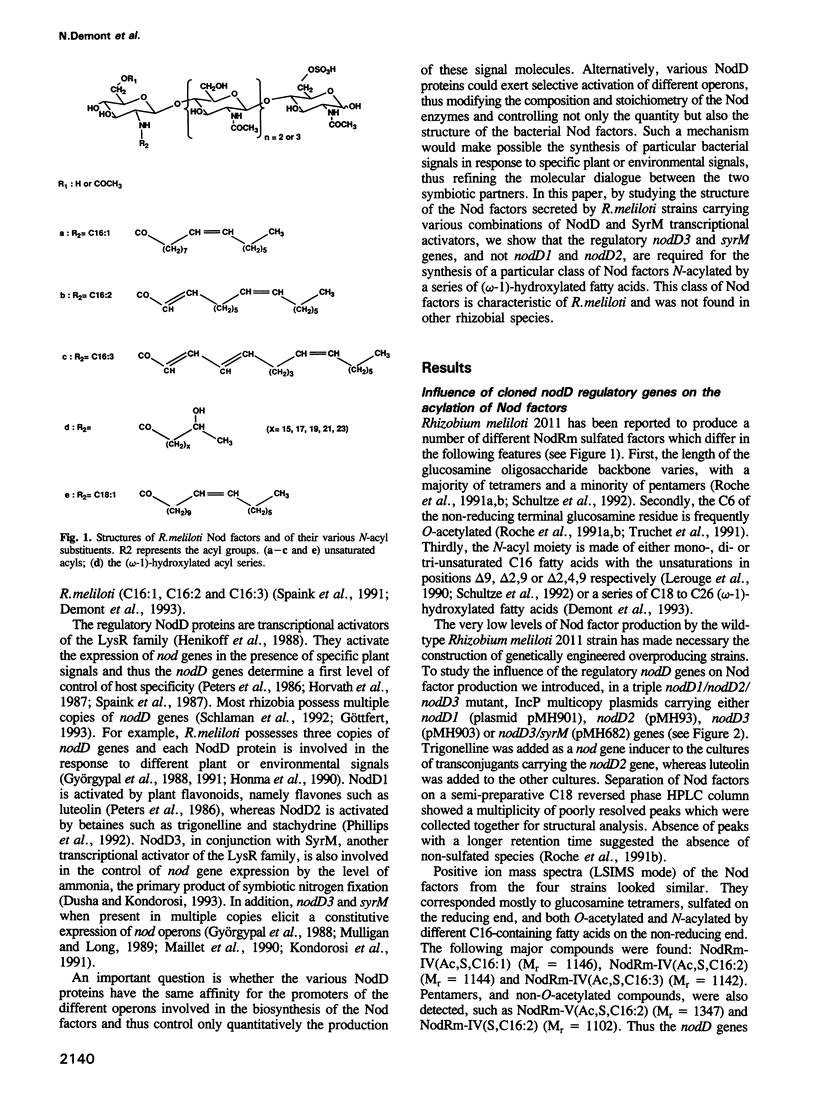
- Demont N., Debellé F., Aurelle H., Dénarié J., Promé J. C. Role of the Rhizobium meliloti nodF and nodE genes in the biosynthesis of lipo-oligosaccharidic nodulation factors. J Biol Chem. 1993 Sep 25;268(27):20134–20142. [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusha I., Kondorosi A. Genes at different regulatory levels are required for the ammonia control of nodulation in Rhizobium meliloti. Mol Gen Genet. 1993 Sep;240(3):435–444. doi: 10.1007/BF00280398. [DOI] [PubMed] [Google Scholar]
- Dénarié J., Cullimore J. Lipo-oligosaccharide nodulation factors: a minireview new class of signaling molecules mediating recognition and morphogenesis. Cell. 1993 Sep 24;74(6):951–954. doi: 10.1016/0092-8674(93)90717-5. [DOI] [PubMed] [Google Scholar]
- Dénarié J., Debellé F., Rosenberg C. Signaling and host range variation in nodulation. Annu Rev Microbiol. 1992;46:497–531. doi: 10.1146/annurev.mi.46.100192.002433. [DOI] [PubMed] [Google Scholar]
- Egelhoff T. T., Long S. R. Rhizobium meliloti nodulation genes: identification of nodDABC gene products, purification of nodA protein, and expression of nodA in Rhizobium meliloti. J Bacteriol. 1985 Nov;164(2):591–599. doi: 10.1128/jb.164.2.591-599.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. F., Egelhoff T. T., Mulligan J. T., Long S. R. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 1988 Mar;2(3):282–293. doi: 10.1101/gad.2.3.282. [DOI] [PubMed] [Google Scholar]
- Fisher R. F., Long S. R. DNA footprint analysis of the transcriptional activator proteins NodD1 and NodD3 on inducible nod gene promoters. J Bacteriol. 1989 Oct;171(10):5492–5502. doi: 10.1128/jb.171.10.5492-5502.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. F., Long S. R. Interactions of NodD at the nod Box: NodD binds to two distinct sites on the same face of the helix and induces a bend in the DNA. J Mol Biol. 1993 Oct 5;233(3):336–348. doi: 10.1006/jmbi.1993.1515. [DOI] [PubMed] [Google Scholar]
- Fisher R. F., Long S. R. Rhizobium--plant signal exchange. Nature. 1992 Jun 25;357(6380):655–660. doi: 10.1038/357655a0. [DOI] [PubMed] [Google Scholar]
- Göttfert M. Regulation and function of rhizobial nodulation genes. FEMS Microbiol Rev. 1993 Jan;10(1-2):39–63. doi: 10.1111/j.1574-6968.1993.tb05863.x. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma M. A., Asomaning M., Ausubel F. M. Rhizobium meliloti nodD genes mediate host-specific activation of nodABC. J Bacteriol. 1990 Feb;172(2):901–911. doi: 10.1128/jb.172.2.901-911.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma M. A., Ausubel F. M. Rhizobium meliloti has three functional copies of the nodD symbiotic regulatory gene. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8558–8562. doi: 10.1073/pnas.84.23.8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath B., Bachem C. W., Schell J., Kondorosi A. Host-specific regulation of nodulation genes in Rhizobium is mediated by a plant-signal, interacting with the nodD gene product. EMBO J. 1987 Apr;6(4):841–848. doi: 10.1002/j.1460-2075.1987.tb04829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath B., Heidstra R., Lados M., Moerman M., Spaink H. P., Promé J. C., van Kammen A., Bisseling T. Lipo-oligosaccharides of Rhizobium induce infection-related early nodulin gene expression in pea root hairs. Plant J. 1993 Oct;4(4):727–733. doi: 10.1046/j.1365-313x.1993.04040727.x. [DOI] [PubMed] [Google Scholar]
- John M., Röhrig H., Schmidt J., Wieneke U., Schell J. Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):625–629. doi: 10.1073/pnas.90.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondorosi E., Buiré M., Cren M., Iyer N., Hoffmann B., Kondorosi A. Involvement of the syrM and nodD3 genes of Rhizobium meliloti in nod gene activation and in optimal nodulation of the plant host. Mol Microbiol. 1991 Dec;5(12):3035–3048. doi: 10.1111/j.1365-2958.1991.tb01863.x. [DOI] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J. C., Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990 Apr 19;344(6268):781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- Leyh T. S., Vogt T. F., Suo Y. The DNA sequence of the sulfate activation locus from Escherichia coli K-12. J Biol Chem. 1992 May 25;267(15):10405–10410. [PubMed] [Google Scholar]
- Maillet F., Debellé F., Dénarié J. Role of the nodD and syrM genes in the activation of the regulatory gene nodD3, and of the common and host-specific nod genes of Rhizobium meliloti. Mol Microbiol. 1990 Nov;4(11):1975–1984. doi: 10.1111/j.1365-2958.1990.tb02047.x. [DOI] [PubMed] [Google Scholar]
- Mergaert P., Van Montagu M., Promé J. C., Holsters M. Three unusual modifications, a D-arabinosyl, an N-methyl, and a carbamoyl group, are present on the Nod factors of Azorhizobium caulinodans strain ORS571. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1551–1555. doi: 10.1073/pnas.90.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan J. T., Long S. R. A family of activator genes regulates expression of Rhizobium meliloti nodulation genes. Genetics. 1989 May;122(1):7–18. doi: 10.1093/genetics/122.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D. A., Joseph C. M., Maxwell C. A. Trigonelline and Stachydrine Released from Alfalfa Seeds Activate NodD2 Protein in Rhizobium meliloti. Plant Physiol. 1992 Aug;99(4):1526–1531. doi: 10.1104/pp.99.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupot R., Martinez-Romero E., Promé J. C. Nodulation factors from Rhizobium tropici are sulfated or nonsulfated chitopentasaccharides containing an N-methyl-N-acylglucosaminyl terminus. Biochemistry. 1993 Oct 5;32(39):10430–10435. doi: 10.1021/bi00090a019. [DOI] [PubMed] [Google Scholar]
- Price N. P., Relić B., Talmont F., Lewin A., Promé D., Pueppke S. G., Maillet F., Dénarié J., Promé J. C., Broughton W. J. Broad-host-range Rhizobium species strain NGR234 secretes a family of carbamoylated, and fucosylated, nodulation signals that are O-acetylated or sulphated. Mol Microbiol. 1992 Dec;6(23):3575–3584. doi: 10.1111/j.1365-2958.1992.tb01793.x. [DOI] [PubMed] [Google Scholar]
- Roche P., Debellé F., Maillet F., Lerouge P., Faucher C., Truchet G., Dénarié J., Promé J. C. Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo-oligosaccharide signals. Cell. 1991 Dec 20;67(6):1131–1143. doi: 10.1016/0092-8674(91)90290-f. [DOI] [PubMed] [Google Scholar]
- Roche P., Lerouge P., Ponthus C., Promé J. C. Structural determination of bacterial nodulation factors involved in the Rhizobium meliloti-alfalfa symbiosis. J Biol Chem. 1991 Jun 15;266(17):10933–10940. [PubMed] [Google Scholar]
- Rosenberg C., Boistard P., Dénarié J., Casse-Delbart F. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol Gen Genet. 1981;184(2):326–333. doi: 10.1007/BF00272926. [DOI] [PubMed] [Google Scholar]
- Rushing B. G., Yelton M. M., Long S. R. Genetic and physical analysis of the nodD3 region of Rhizobium meliloti. Nucleic Acids Res. 1991 Feb 25;19(4):921–927. doi: 10.1093/nar/19.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan J., Carlson R. W., Spaink H. P., Bhat U. R., Barbour W. M., Glushka J., Stacey G. A 2-O-methylfucose moiety is present in the lipo-oligosaccharide nodulation signal of Bradyrhizobium japonicum. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8789–8793. doi: 10.1073/pnas.89.18.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaman H. R., Okker R. J., Lugtenberg B. J. Regulation of nodulation gene expression by NodD in rhizobia. J Bacteriol. 1992 Aug;174(16):5177–5182. doi: 10.1128/jb.174.16.5177-5182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze M., Quiclet-Sire B., Kondorosi E., Virelizer H., Glushka J. N., Endre G., Géro S. D., Kondorosi A. Rhizobium meliloti produces a family of sulfated lipooligosaccharides exhibiting different degrees of plant host specificity. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):192–196. doi: 10.1073/pnas.89.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwedock J. S., Long S. R. Rhizobium meliloti genes involved in sulfate activation: the two copies of nodPQ and a new locus, saa. Genetics. 1992 Dec;132(4):899–909. doi: 10.1093/genetics/132.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwedock J., Long S. R. ATP sulphurylase activity of the nodP and nodQ gene products of Rhizobium meliloti. Nature. 1990 Dec 13;348(6302):644–647. doi: 10.1038/348644a0. [DOI] [PubMed] [Google Scholar]
- Spaink H. P. Rhizobial lipo-oligosaccharides: answers and questions. Plant Mol Biol. 1992 Dec;20(5):977–986. doi: 10.1007/BF00027167. [DOI] [PubMed] [Google Scholar]
- Spaink H. P., Sheeley D. M., van Brussel A. A., Glushka J., York W. S., Tak T., Geiger O., Kennedy E. P., Reinhold V. N., Lugtenberg B. J. A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature. 1991 Nov 14;354(6349):125–130. doi: 10.1038/354125a0. [DOI] [PubMed] [Google Scholar]
- Swanson J. A., Mulligan J. T., Long S. R. Regulation of syrM and nodD3 in Rhizobium meliloti. Genetics. 1993 Jun;134(2):435–444. doi: 10.1093/genetics/134.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J. A., Tu J. K., Ogawa J., Sanga R., Fisher R. F., Long S. R. Extended Region of Nodulation Genes in Rhizobium meliloti 1021. I. Phenotypes of Tn5 Insertion Mutants. Genetics. 1987 Oct;117(2):181–189. doi: 10.1093/genetics/117.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchet G., Debellé F., Vasse J., Terzaghi B., Garnerone A. M., Rosenberg C., Batut J., Maillet F., Dénarié J. Identification of a Rhizobium meliloti pSym2011 region controlling the host specificity of root hair curling and nodulation. J Bacteriol. 1985 Dec;164(3):1200–1210. doi: 10.1128/jb.164.3.1200-1210.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brussel A. A., Bakhuizen R., van Spronsen P. C., Spaink H. P., Tak T., Lugtenberg B. J., Kijne J. W. Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science. 1992 Jul 3;257(5066):70–72. doi: 10.1126/science.257.5066.70. [DOI] [PubMed] [Google Scholar]