Abstract
The type IC DNA methyltransferase M.EcoR124I is a complex multisubunit enzyme that recognizes the non-palindromic DNA sequence GAAN6RTCG. Small angle X-ray scattering has been used to investigate the solution structure of the methyltransferase and of complexes of the enzyme with unmethylated and hemimethylated 30 bp DNA duplexes containing the specific recognition sequence. A major change in the quaternary structure of the enzyme is observed following DNA binding, based on a decrease in the radius of gyration from 56 to 40 A and a reduction in the maximum dimension of the enzyme from 180 to 112 A. The structural transition observed is independent of the methylation state of the DNA. CD shows that there is no change in the secondary structure of the protein subunits when DNA is bound. In contrast, there is a large increase in the CD signal arising from the DNA, suggesting considerable structural distortion which may allow access to the bases targeted for methylation. We propose that DNA binding induces a large rotation of the two HsdM subunits towards the DNA, mediated by hinge bending domains in the specificity subunit HsdS.
Full text
PDF
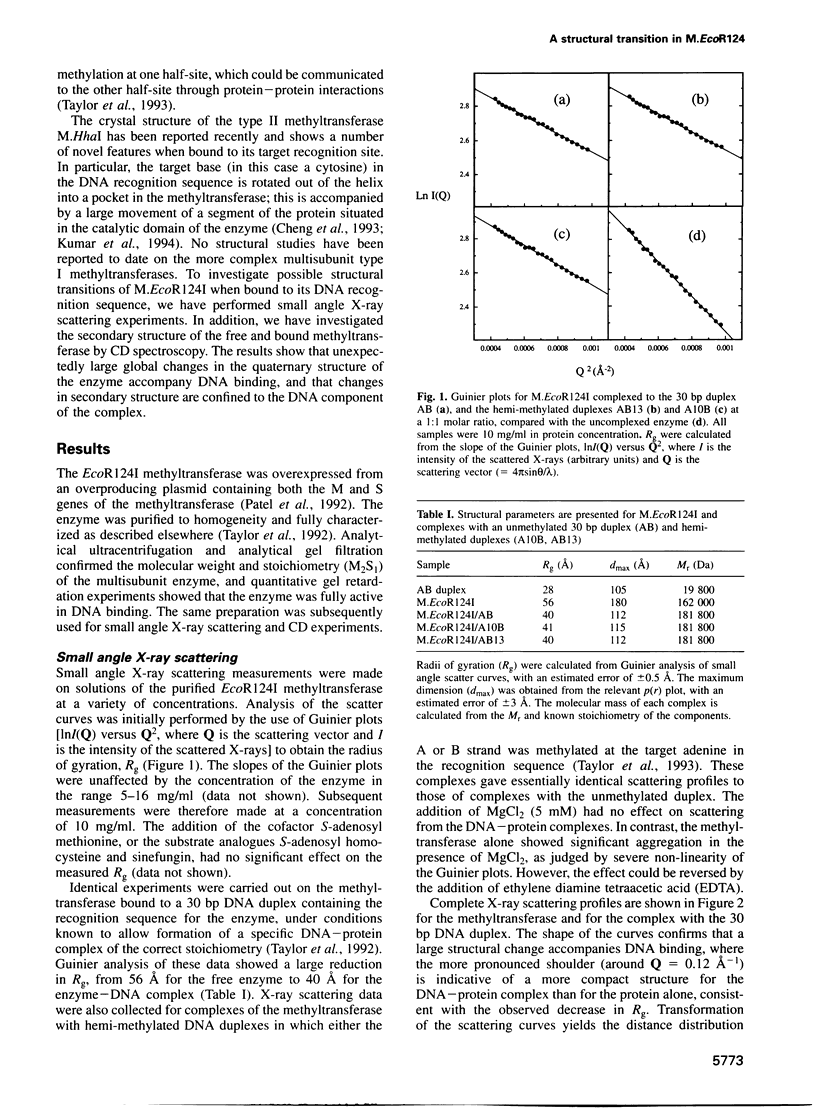
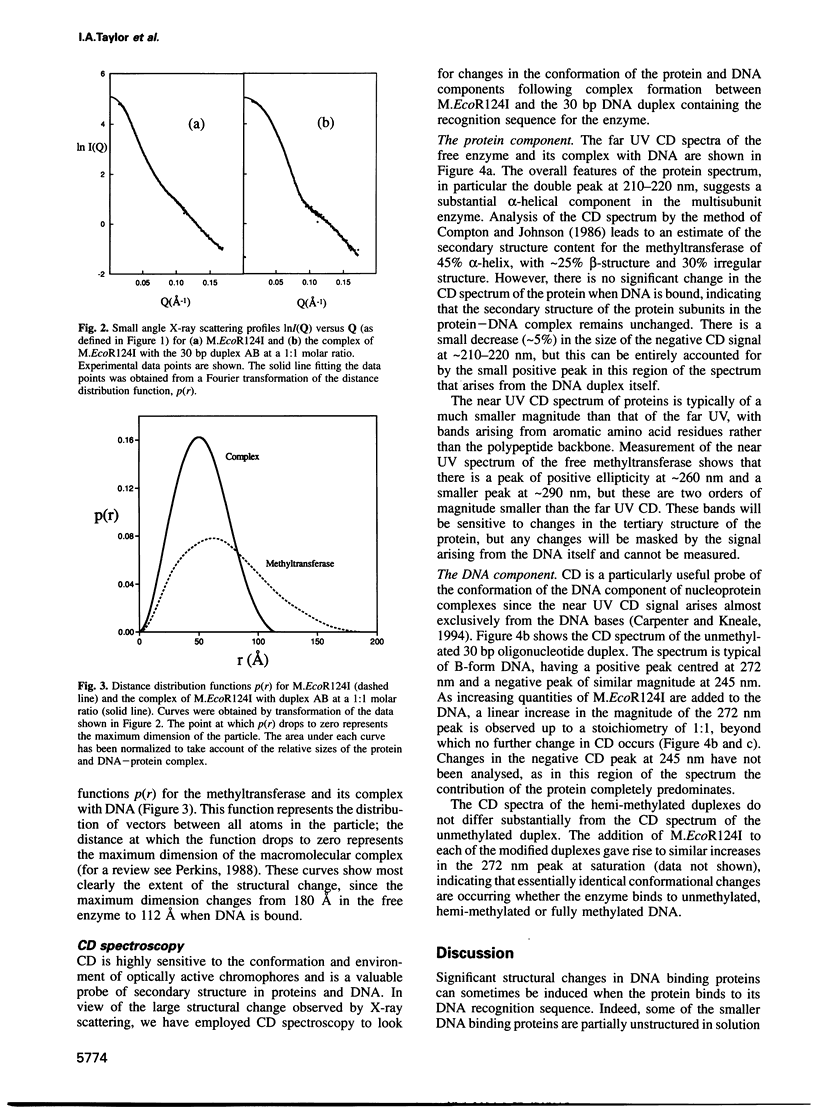
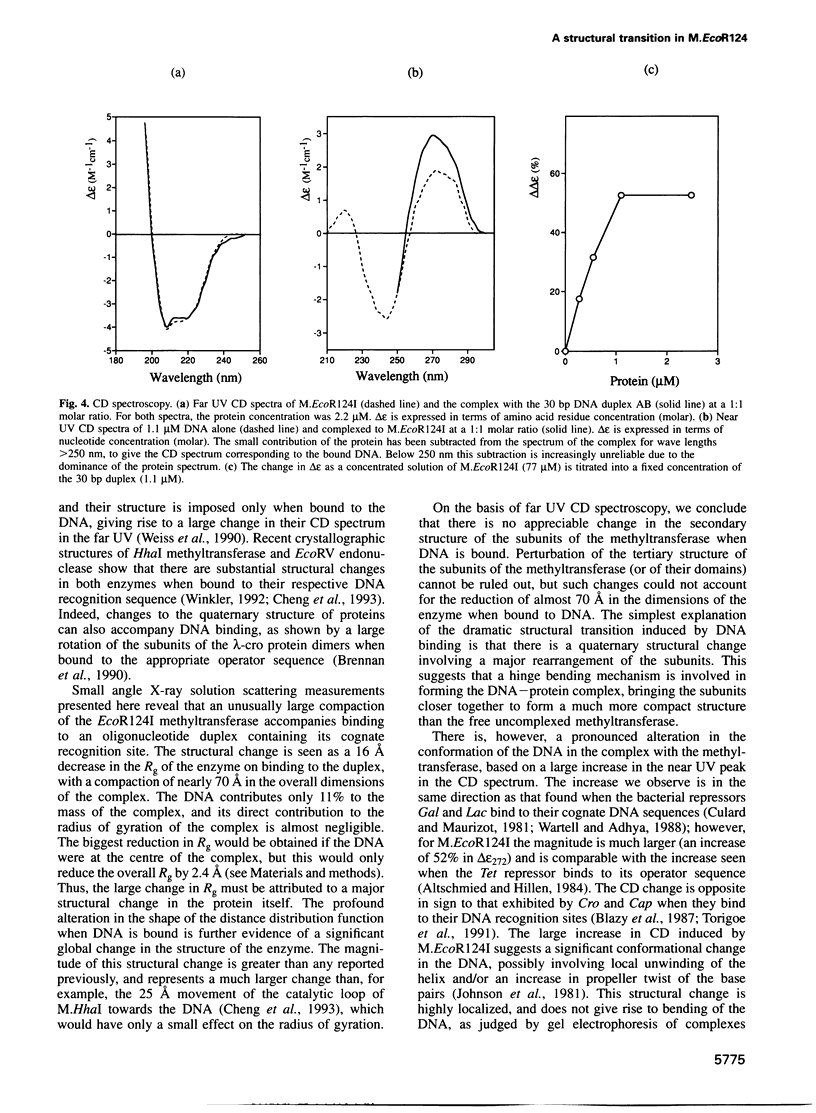
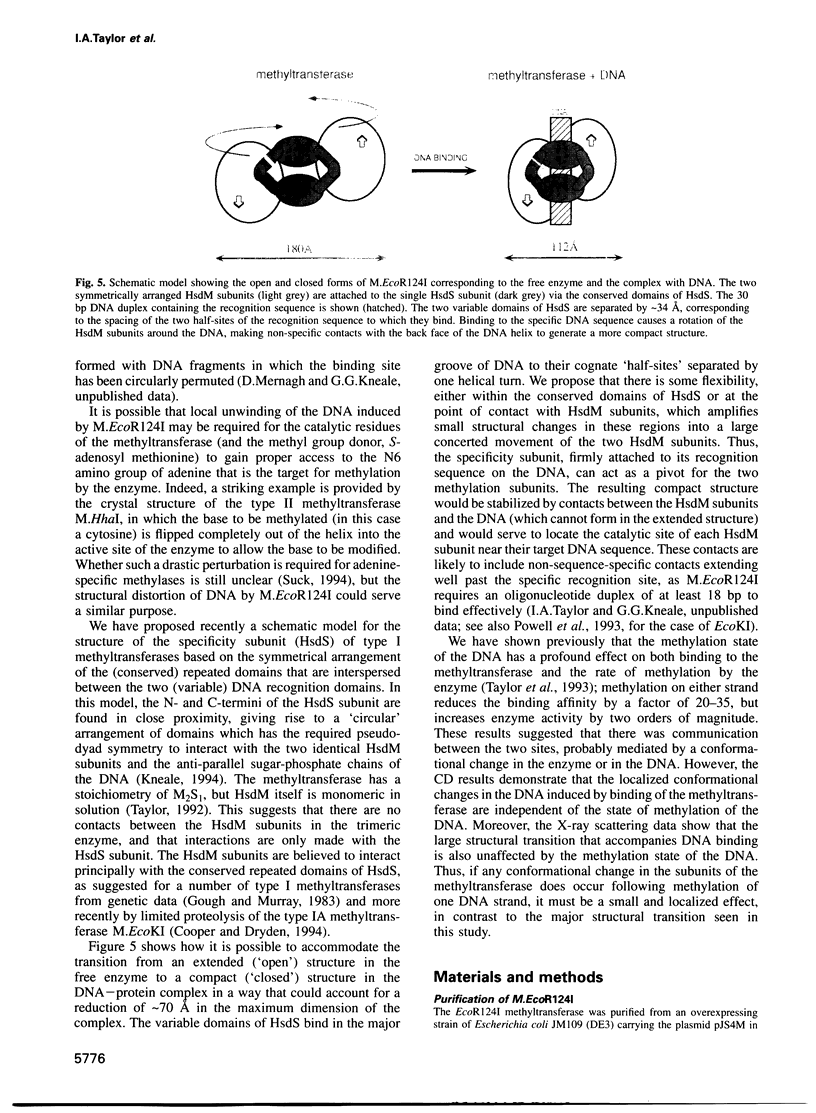


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abadjieva A., Patel J., Webb M., Zinkevich V., Firman K. A deletion mutant of the type IC restriction endonuclease EcoR1241 expressing a novel DNA specificity. Nucleic Acids Res. 1993 Sep 25;21(19):4435–4443. doi: 10.1093/nar/21.19.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschmied L., Hillen W. TET repressor.tet operator complex formation induces conformational changes in the tet operator DNA. Nucleic Acids Res. 1984 Feb 24;12(4):2171–2180. doi: 10.1093/nar/12.4.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickle T. A., Krüger D. H. Biology of DNA restriction. Microbiol Rev. 1993 Jun;57(2):434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazy B., Culard F., Maurizot J. C. Interaction between the cyclic AMP receptor protein and DNA. Conformational studies. J Mol Biol. 1987 May 5;195(1):175–183. doi: 10.1016/0022-2836(87)90334-2. [DOI] [PubMed] [Google Scholar]
- Brennan R. G., Roderick S. L., Takeda Y., Matthews B. W. Protein-DNA conformational changes in the crystal structure of a lambda Cro-operator complex. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8165–8169. doi: 10.1073/pnas.87.20.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter M. L., Kneale G. G. Analysis of DNA-protein interactions by intrinsic fluorescence. Methods Mol Biol. 1994;30:313–325. doi: 10.1385/0-89603-256-6:313. [DOI] [PubMed] [Google Scholar]
- Cheng X., Kumar S., Posfai J., Pflugrath J. W., Roberts R. J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell. 1993 Jul 30;74(2):299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- Compton L. A., Johnson W. C., Jr Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal Biochem. 1986 May 15;155(1):155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- Cooper L. P., Dryden D. T. The domains of a type I DNA methyltransferase. Interactions and role in recognition of DNA methylation. J Mol Biol. 1994 Mar 4;236(4):1011–1021. doi: 10.1016/0022-2836(94)90008-6. [DOI] [PubMed] [Google Scholar]
- Cowan G. M., Gann A. A., Murray N. E. Conservation of complex DNA recognition domains between families of restriction enzymes. Cell. 1989 Jan 13;56(1):103–109. doi: 10.1016/0092-8674(89)90988-4. [DOI] [PubMed] [Google Scholar]
- Culard F., Maurizot J. C. Lac repressor - lac operator interaction. Circular dichroism study. Nucleic Acids Res. 1981 Oct 10;9(19):5175–5184. doi: 10.1093/nar/9.19.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden D. T., Cooper L. P., Murray N. E. Purification and characterization of the methyltransferase from the type 1 restriction and modification system of Escherichia coli K12. J Biol Chem. 1993 Jun 25;268(18):13228–13236. [PubMed] [Google Scholar]
- Fuller-Pace F. V., Cowan G. M., Murray N. E. EcoA and EcoE: alternatives to the EcoK family of type I restriction and modification systems of Escherichia coli. J Mol Biol. 1985 Nov 5;186(1):65–75. doi: 10.1016/0022-2836(85)90257-8. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Murray N. E. Two DNA recognition domains of the specificity polypeptides of a family of type I restriction enzymes. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9368–9372. doi: 10.1073/pnas.83.24.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Gubler M., Braguglia D., Meyer J., Piekarowicz A., Bickle T. A. Recombination of constant and variable modules alters DNA sequence recognition by type IC restriction-modification enzymes. EMBO J. 1992 Jan;11(1):233–240. doi: 10.1002/j.1460-2075.1992.tb05046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. B., Dahl K. S., Tinoco I., Jr, Ivanov V. I., Zhurkin V. B. Correlations between deoxyribonucleic acid structural parameters and calculated circular dichroism spectra. Biochemistry. 1981 Jan 6;20(1):73–78. doi: 10.1021/bi00504a013. [DOI] [PubMed] [Google Scholar]
- Kannan P., Cowan G. M., Daniel A. S., Gann A. A., Murray N. E. Conservation of organization in the specificity polypeptides of two families of type I restriction enzymes. J Mol Biol. 1989 Oct 5;209(3):335–344. doi: 10.1016/0022-2836(89)90001-6. [DOI] [PubMed] [Google Scholar]
- Kelleher J. E., Daniel A. S., Murray N. E. Mutations that confer de novo activity upon a maintenance methyltransferase. J Mol Biol. 1991 Sep 20;221(2):431–440. doi: 10.1016/0022-2836(91)80064-2. [DOI] [PubMed] [Google Scholar]
- Kneale G. G. A symmetrical model for the domain structure of type I DNA methyltransferases. J Mol Biol. 1994 Oct 14;243(1):1–5. doi: 10.1006/jmbi.1994.1624. [DOI] [PubMed] [Google Scholar]
- Kumar S., Cheng X., Klimasauskas S., Mi S., Posfai J., Roberts R. J., Wilson G. G. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994 Jan 11;22(1):1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer H., Tovar K., Baer G., May R. P., Hillen W., Heumann H. The quaternary structure of Tet repressors bound to the Tn10-encoded tet gene control region determined by neutron solution scattering. EMBO J. 1989 Apr;8(4):1257–1263. doi: 10.1002/j.1460-2075.1989.tb03499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister J., MacWilliams M., Hübner P., Jütte H., Skrzypek E., Piekarowicz A., Bickle T. A. Macroevolution by transposition: drastic modification of DNA recognition by a type I restriction enzyme following Tn5 transposition. EMBO J. 1993 Dec;12(12):4585–4591. doi: 10.1002/j.1460-2075.1993.tb06147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Gough J. A., Suri B., Bickle T. A. Structural homologies among type I restriction-modification systems. EMBO J. 1982;1(5):535–539. doi: 10.1002/j.1460-2075.1982.tb01205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J., Taylor I., Dutta C. F., Kneale G., Firman K. High-level expression of the cloned genes encoding the subunits of and intact DNA methyltransferase, M.EcoR124. Gene. 1992 Mar 1;112(1):21–27. doi: 10.1016/0378-1119(92)90298-4. [DOI] [PubMed] [Google Scholar]
- Perkins S. J. Structural studies of proteins by high-flux X-ray and neutron solution scattering. Biochem J. 1988 Sep 1;254(2):313–327. doi: 10.1042/bj2540313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L. M., Dryden D. T., Willcock D. F., Pain R. H., Murray N. E. DNA recognition by the EcoK methyltransferase. The influence of DNA methylation and the cofactor S-adenosyl-L-methionine. J Mol Biol. 1993 Nov 5;234(1):60–71. doi: 10.1006/jmbi.1993.1563. [DOI] [PubMed] [Google Scholar]
- Price C., Pripfl T., Bickle T. A. EcoR124 and EcoR124/3: the first members of a new family of type I restriction and modification systems. Eur J Biochem. 1987 Aug 17;167(1):111–115. doi: 10.1111/j.1432-1033.1987.tb13310.x. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Kelleher J. E., Daniel A. S., Cowan G. M., Murray N. E. Roles of selection and recombination in the evolution of type I restriction-modification systems in enterobacteria. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9836–9840. doi: 10.1073/pnas.89.20.9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suck D. DNA-protein interactions. Flip out and modify. Curr Biol. 1994 Mar 1;4(3):252–255. doi: 10.1016/s0960-9822(00)00057-9. [DOI] [PubMed] [Google Scholar]
- Suri B., Bickle T. A. EcoA: the first member of a new family of type I restriction modification systems. Gene organization and enzymatic activities. J Mol Biol. 1985 Nov 5;186(1):77–85. doi: 10.1016/0022-2836(85)90258-x. [DOI] [PubMed] [Google Scholar]
- Taylor I., Patel J., Firman K., Kneale G. Purification and biochemical characterisation of the EcoR124 type I modification methylase. Nucleic Acids Res. 1992 Jan 25;20(2):179–186. doi: 10.1093/nar/20.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I., Watts D., Kneale G. Substrate recognition and selectivity in the type IC DNA modification methylase M.EcoR124I. Nucleic Acids Res. 1993 Oct 25;21(21):4929–4935. doi: 10.1093/nar/21.21.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torigoe C., Kidokoro S., Takimoto M., Kyogoku Y., Wada A. Spectroscopic studies on lambda cro protein-DNA interactions. J Mol Biol. 1991 Jun 20;219(4):733–746. doi: 10.1016/0022-2836(91)90668-v. [DOI] [PubMed] [Google Scholar]
- Tyndall C., Meister J., Bickle T. A. The Escherichia coli prr region encodes a functional type IC DNA restriction system closely integrated with an anticodon nuclease gene. J Mol Biol. 1994 Apr 1;237(3):266–274. doi: 10.1006/jmbi.1994.1230. [DOI] [PubMed] [Google Scholar]
- Wartell R. M., Adhya S. DNA conformational change in Gal repressor-operator complex: involvement of central G-C base pair(s) of dyad symmetry. Nucleic Acids Res. 1988 Dec 23;16(24):11531–11541. doi: 10.1093/nar/16.24.11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M. A., Ellenberger T., Wobbe C. R., Lee J. P., Harrison S. C., Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990 Oct 11;347(6293):575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]



