Abstract
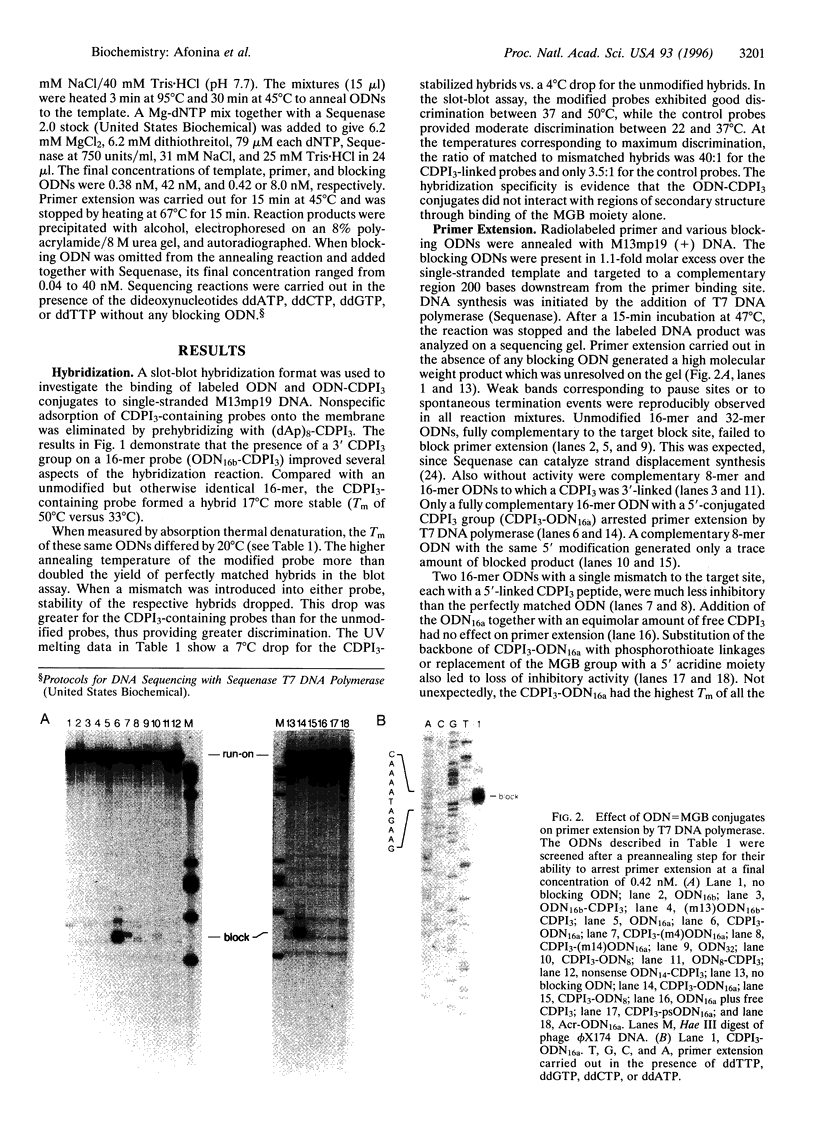
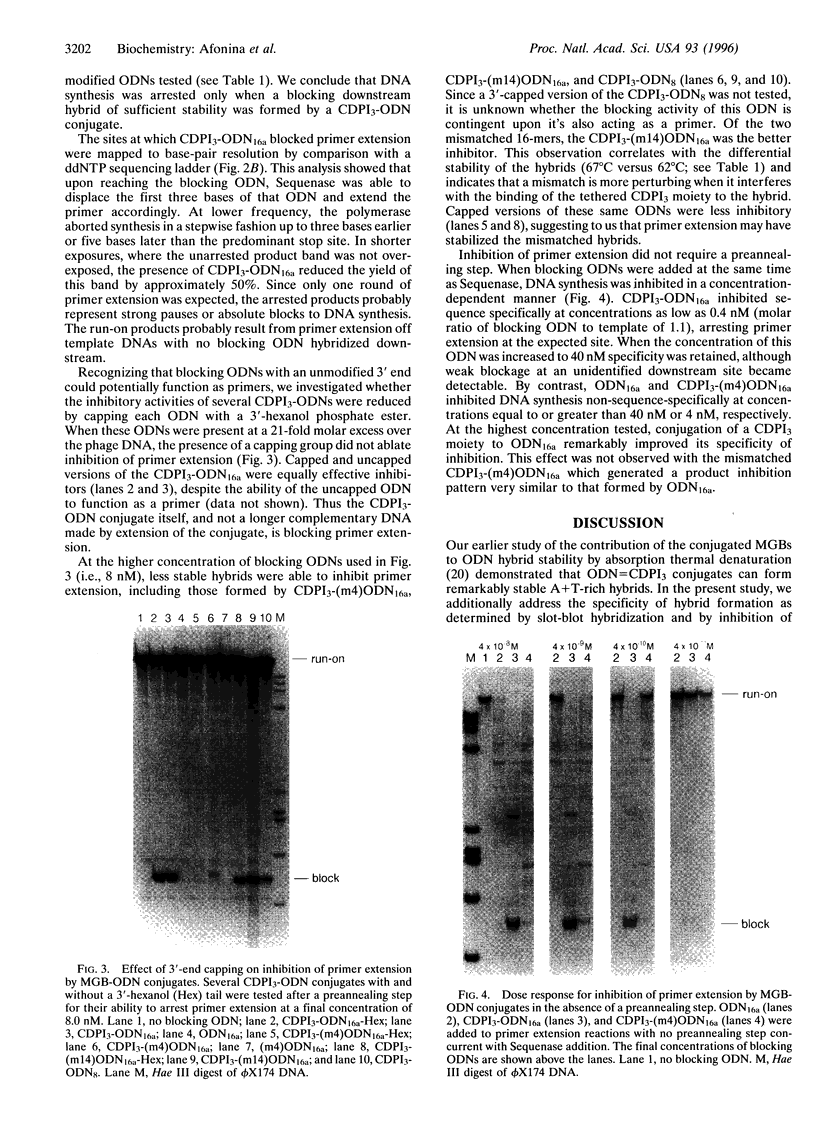
A minor groove binder (MGB) derivative (N-3-carbamoyl-1,2-dihydro-3H-pyrrolo[3,2-e]indole-7-carboxylate tripeptide; CDPI3) was covalently linked to the 5' or 3' end of several oligodeoxyribonucleotides (ODNs) totally complementary or possessing a single mismatch to M13mp19 single-stranded DNA. Absorption thermal denaturation and slot-blot hybridization studies showed that conjugation of CDPI3 to these ODNs increased both the specificity and the strength with which they hybridized. Primer extension of the same phage DNA by a modified form of phage T7 DNA polymerase (Sequenase) was physically blocked when a complementary 16-mer with a conjugated 5'-CDPI3 moiety was hybridized to a downstream site. Approximately 50% of the replicating complexes were arrested when the blocking ODN was equimolar to the phage DNA. Inhibition was unaffected by 3'-capping of the ODN with a hexanol group or by elimination of a preannealing step. Blockage was abolished when a single mismatch was introduced into the ODN or when the MGB was either removed or replaced by a 5'-acridine group. A 16-mer with a 3'-CDPI3 moiety failed to arrest primer extension, as did an unmodified 32-mer. We attribute the exceptional stability of hybrids formed by ODNs conjugated to a CDPI3 to the tethered tripeptide binding in the minor groove of the hybrid. When that group is linked to the 5' end of a hybridized ODN, it probably blocks DNA synthesis by inhibiting strand displacement. These ODNs conjugated to CDPI3 offer attractive features as diagnostic probes and antigene agents.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asseline U., Delarue M., Lancelot G., Toulmé F., Thuong N. T., Montenay-Garestier T., Hélène C. Nucleic acid-binding molecules with high affinity and base sequence specificity: intercalating agents covalently linked to oligodeoxynucleotides. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3297–3301. doi: 10.1073/pnas.81.11.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boger D. L., Johnson D. S. CC-1065 and the duocarmycins: unraveling the keys to a new class of naturally derived DNA alkylating agents. Proc Natl Acad Sci U S A. 1995 Apr 25;92(9):3642–3649. doi: 10.1073/pnas.92.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker D. J., Vickers T. A., Bruice T. W., Freier S. M., Jenison R. D., Manoharan M., Zounes M. Pseudo--half-knot formation with RNA. Science. 1992 Aug 14;257(5072):958–961. doi: 10.1126/science.1502560. [DOI] [PubMed] [Google Scholar]
- François J. C., Thuong N. T., Hélène C. Recognition and cleavage of hairpin structures in nucleic acids by oligodeoxynucleotides. Nucleic Acids Res. 1994 Sep 25;22(19):3943–3950. doi: 10.1093/nar/22.19.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper H. B., Cimino G. D., Hearst J. E. Solution hybridization of crosslinkable DNA oligonucleotides to bacteriophage M13 DNA. Effect of secondary structure on hybridization kinetics and equilibria. J Mol Biol. 1987 Sep 20;197(2):349–362. doi: 10.1016/0022-2836(87)90128-8. [DOI] [PubMed] [Google Scholar]
- Gamper H. B., Reed M. W., Cox T., Virosco J. S., Adams A. D., Gall A. A., Scholler J. K., Meyer R. B., Jr Facile preparation of nuclease resistant 3' modified oligodeoxynucleotides. Nucleic Acids Res. 1993 Jan 11;21(1):145–150. doi: 10.1093/nar/21.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannangeli C., Thuong N. T., Hélène C. Oligonucleotide clamps arrest DNA synthesis on a single-stranded DNA target. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):10013–10017. doi: 10.1073/pnas.90.21.10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacia J. G., Dervan P. B., Wold B. J. Inhibition of Klenow fragment DNA polymerase on double-helical templates by oligonucleotide-directed triple-helix formation. Biochemistry. 1994 May 24;33(20):6192–6200. doi: 10.1021/bi00186a019. [DOI] [PubMed] [Google Scholar]
- Hansen J. B., Koch T., Buchardt O., Nielsen P. E., Wirth M., Nordén B. Acridine-psoralen amines and their interaction with deoxyribonucleic acid. Biochemistry. 1983 Oct 11;22(21):4878–4886. doi: 10.1021/bi00290a003. [DOI] [PubMed] [Google Scholar]
- Latham J. A., Cech T. R. Defining the inside and outside of a catalytic RNA molecule. Science. 1989 Jul 21;245(4915):276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- Li L. H., Swenson D. H., Schpok S. L., Kuentzel S. L., Dayton B. D., Krueger W. C. CC-1065 (NSC 298223), a novel antitumor agent that interacts strongly with double-stranded DNA. Cancer Res. 1982 Mar;42(3):999–1004. [PubMed] [Google Scholar]
- Lukhtanov E. A., Kutyavin I. V., Gamper H. B., Meyer R. B., Jr Oligodeoxyribonucleotides with conjugated dihydropyrroloindole oligopeptides: preparation and hybridization properties. Bioconjug Chem. 1995 Jul-Aug;6(4):418–426. doi: 10.1021/bc00034a012. [DOI] [PubMed] [Google Scholar]
- Maine I. P., Kodadek T. Efficient unwinding of triplex DNA by a DNA helicase. Biochem Biophys Res Commun. 1994 Nov 15;204(3):1119–1124. doi: 10.1006/bbrc.1994.2578. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Origins of netropsin binding affinity and specificity: correlations of thermodynamic and structural data. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4359–4363. doi: 10.1073/pnas.84.13.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monia B. P., Johnston J. F., Ecker D. J., Zounes M. A., Lima W. F., Freier S. M. Selective inhibition of mutant Ha-ras mRNA expression by antisense oligonucleotides. J Biol Chem. 1992 Oct 5;267(28):19954–19962. [PubMed] [Google Scholar]
- Nielsen P. E., Egholm M., Buchardt O. Peptide nucleic acid (PNA). A DNA mimic with a peptide backbone. Bioconjug Chem. 1994 Jan-Feb;5(1):3–7. doi: 10.1021/bc00025a001. [DOI] [PubMed] [Google Scholar]
- Orum H., Nielsen P. E., Egholm M., Berg R. H., Buchardt O., Stanley C. Single base pair mutation analysis by PNA directed PCR clamping. Nucleic Acids Res. 1993 Nov 25;21(23):5332–5336. doi: 10.1093/nar/21.23.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds V. L., McGovren J. P., Hurley L. H. The chemistry, mechanism of action and biological properties of CC-1065, a potent antitumor antibiotic. J Antibiot (Tokyo) 1986 Mar;39(3):319–334. doi: 10.7164/antibiotics.39.319. [DOI] [PubMed] [Google Scholar]
- Samadashwily G. M., Dayn A., Mirkin S. M. Suicidal nucleotide sequences for DNA polymerization. EMBO J. 1993 Dec 15;12(13):4975–4983. doi: 10.1002/j.1460-2075.1993.tb06191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadashwily G. M., Mirkin S. M. Trapping DNA polymerases using triplex-forming oligodeoxyribonucleotides. Gene. 1994 Nov 4;149(1):127–136. doi: 10.1016/0378-1119(94)90421-9. [DOI] [PubMed] [Google Scholar]
- Scahill T. A., Jensen R. M., Swenson D. H., Hatzenbuhler N. T., Petzold G., Wierenga W., Brahme N. D. An NMR study of the covalent and noncovalent interactions of CC-1065 and DNA. Biochemistry. 1990 Mar 20;29(11):2852–2860. doi: 10.1021/bi00463a031. [DOI] [PubMed] [Google Scholar]
- Sun J. S., François J. C., Montenay-Garestier T., Saison-Behmoaras T., Roig V., Thuong N. T., Hélène C. Sequence-specific intercalating agents: intercalation at specific sequences on duplex DNA via major groove recognition by oligonucleotide-intercalator conjugates. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9198–9202. doi: 10.1073/pnas.86.23.9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J Biol Chem. 1989 Apr 15;264(11):6447–6458. [PubMed] [Google Scholar]
- Wagner R. W., Matteucci M. D., Lewis J. G., Gutierrez A. J., Moulds C., Froehler B. C. Antisense gene inhibition by oligonucleotides containing C-5 propyne pyrimidines. Science. 1993 Jun 4;260(5113):1510–1513. doi: 10.1126/science.7684856. [DOI] [PubMed] [Google Scholar]
- Wyatt J. R., Puglisi J. D., Tinoco I., Jr RNA folding: pseudoknots, loops and bulges. Bioessays. 1989 Oct;11(4):100–106. doi: 10.1002/bies.950110406. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]





