Abstract
The high level of amino acid conservation and structural similarity of the substrate-binding sites of the oxygenase domains of the nitric oxide synthase (NOS) isoforms (eNOSoxy, iNOSoxy, nNOSoxy) make the interpretation of the structural basis of inhibitor isoform specificity a challenge, and provide few clues for the design of new selective compounds. Crystal structures of iNOSoxy and nNOSoxy complexed with the neuronal NOS-specific inhibitor AR-R17447 suggest that specificity is provided by the interaction of the chlorophenyl group with an isoform-unique substrate access channel residue (L337 in rat neuronal NOS, N115 in mouse inducible NOS). This is confirmed by biochemical analysis of site-directed mutants. Inhibitors combining guanidinium-like structural motifs with long chains specifically targeting this residue are good candidates for rational isoform-specific drug design. Based on this finding, modifications of AR-R17447 to improve the specificity for the human isoforms are suggested.
Nitric oxide (NO), a ubiquitous signaling molecule, is currently one of the most intensely studied small molecules in biology because of its involvement in numerous biological events such as vasodilation, neurotransmission, and the immune response. The isozymes of NO synthase (NOS) that produce NO are dimeric multidomain polypeptides consisting of three main components: a heme-containing catalytic oxygenase domain (NOSoxy), a calmodulin binding linker, and a NADPH reductase domain. NOS transforms l-arginine to citrulline and NO in two sequential steps consuming oxygen and electrons (1). The cofactor tetrahydrobiopterin bound at the interface of the two oxygenase domains in the NOS dimer is required for NO synthesis (2, 3). In mammals, three NOS isoforms have been identified sharing 50–60% sequence identity, which differ in cellular distribution, regulation, and activity (1). Endothelial NOS (eNOS) regulates vascular tone and smooth muscle tension (4). Neuronal NOS (nNOS) produced NO functions as a diffusible neurotransmitter (5), whereas NO generated by inducible NOS (iNOS) generates cytotoxins with both protective and pathologic effects (1, 6). In line with NO's central biological role, there are a number of pathological processes associated with its over- or underproduction. For example, nNOS is implicated in stroke and migraine, and iNOS is implicated in septic shock, arthritis, and multiple sclerosis. The possibility of treating these and other conditions by inhibiting NOS has elicited intense efforts to identify or design NOS inhibitors. Because the three isoforms of NOS have unique roles in separate tissues, selective inhibition of one isozyme over the others is essential. In particular, it is important not to inhibit eNOS because of its critical role in maintaining vascular tone. Numerous inhibitors of NOS have been developed (7). The majority of the inhibitors contain amidino or ureido functional groups that mimic the guanidino group of the substrate l-arginine. The high level of amino acid conservation and striking structural similarity in the immediate vicinity of the substrate binding sites of the three NOS oxygenase domains (2, 8–11) explained the difficulty in finding selective NOS inhibitors. Nevertheless, selective inhibitors exist, such as N-(3-aminomethyl)benzyl)acetamidine (12) and N-ω-propyl-l-Arg (13), that are specific for iNOS and nNOS, respectively. Crystal structures of complexes of these compounds with the rat nNOS and mouse iNOS oxygenase domains (14) show that the structural basis for specificity is rooted in sequence differences outside the immediate active site, as well as a conformational flexibility in the active site that allows the adoption of distinct conformations in response to interactions with the inhibitors. This flexibility is also determined by isoform-specific residues outside the active site. This induced-fit-like behavior renders the rational design and development of isoform-specific inhibitors extremely difficult. To this end, sources of NOS isozyme specificity need to be identified that can be used for the inhibitor construction and optimization in a predictable manner. One of the targets that could provide such a possibility may be the substrate access channel (15) that connects the bulk solvent to heme- and pterin-binding sites and that has noticeable sequence differences in NOS isozymes.
Here, we describe crystal structures coupled with complimentary site-specific mutagenesis and biochemical analysis of the oxygenase domains of murine iNOS and rat nNOS complexed with the highly nNOS-specific inhibitor AR-R17477 [N-(4-(2-((3-chlorophenylmethyl) amino) ethyl) phenyl)-2-thiophecarboxamidine dihydrochloride] (16). AR-R17477 is known to penetrate rapidly the rat brain and to ensure effective and long-lasting inhibition of nNOS in vivo (16). The effect of this compound has also been studied in animal models of global and focal cerebral ischaemia (17, 18). The structural and biochemical data presented here suggest a promising source of isoform selectivity provided by the isoform-unique residues in the substrate access channel. Inhibitors combining guanidinium-like structural motifs with long chains specifically targeting these residues are good candidates for rational isoform-specifc drug design. Based on this finding, we suggest modifications of AR-R17447 to improve the specificity for the human isoforms.
Materials and Methods
Cloning, Mutagenesis, Protein Purification, and Crystallization. The heme oxygenase domains of murine iNOS (residues 65–498) and rat nNOS (residues 291–722) (14) were cloned, mutagenized, expressed, and purified as described (14, 19). All materials were of the highest purity available. Tetrahydrobiopterin containing iNOSoxy and nNOSoxy crystals were grown in the presence of 1 mM AR-R17477 as described (14, 20). The presence of AR-R17477 stabilized the nNOSoxy crystals significantly, which was reflected in the improved mechanical and diffraction properties; e.g., splitting observed frequently for native and other ligand complexed crystals was rare.
UV–Visible Spectroscopy. Measurements were done with a Hitachi U2010 spectrometer equipped with computer-assisted data collection software (UV Solutions, Wellesley Hills, MA). Oxygenase domain activity was measured by following H2O2-supported oxidation of Nω-hydroxy-l-Arg at 37°C for 10 min (19, 21). Binding of l-Arg, tetrahydrobiopterin, and imidazole to NOS [in 40 mM N-(2-hydroxyethyl)piperazine-N′-(3-propane sulfonic acid) pH 7.6 containing 5% glycerol and 2 mM 2-mercaptoethanol] was monitored by spectral perturbation analysis (19, 21, 22). The ferrous iNOS–CO adduct absorbing at 444 nm was used to quantitate heme protein content using an extinction coefficient of 74 mM–1·cm–1 (21, 23, 24). Kd values of imidazole were determined for full-length forms of all NOS isozymes, the oxygenase domains, and their urea-generated monomers (19, 21). For nNOS and eNOS, the Kd for imidazole is 80–100 μM, whereas for iNOS (both human and mouse), it is 40–50 μM. The effect of AR-R17477 on imidazole-bound (0.4–2 mM) NOS (low spin ferriheme, Amax 430 nm) was determined by increasing the concentration of AR-R17477 to make high spin ferri-heme (Amax 395 nm). The absorbance changes (Δ395–430) obtained after each addition of ligand were plotted against the concentration of inhibitor to get the apparent Kd (21, 22), the true Kd value was calculated from the relationship Kd(app) = Kd(true) (1+ [imidazole]/Kd (imidazole)).
Diffraction Data Collection and Refinement. Diffraction data were collected at the European Synchrotron Radiation Facility by using an ADSC-Q4 charge-coupled device detector (see supporting information, which is published on the PNAS web site) and reduced with the xds program package (25). The data are 94.5/96.5% complete to a resolution of 2.0/2.9 Å for nNOS/iNOS and have Rsym values of 9.1/5.8%, respectively. Structure determination was initiated by a round of rigid body refinement using the protein part of the iNOSoxy. l-Arg coordinates (PDB ID 1NOD) or nNOSoxy (PDB ID 1QW6) (14), respectively, as an initial model. Refinement was continued with cns (26) and shelx (27) and included simulated annealing and individual B factor refinement. During cyclic rounds of refinement and manual rebuilding, zinc ions, solvent molecules, and ligands were included in the models. The final models display good stereochemistry (see supporting information) with R/Rfree values of 19.3/22.9% and 20.7/26.2% for nNOSoxy/iNOSoxy, respectively. Structures were overlaid with the program sap (28) and adjusted manually by superpositioning the hemes.
Models of AR-R17477 Derivatives and Their Interactions with Human Isozymes. Models of chloropyridine and hydroxy-pyridine derivatives of AR-R17477 were built by modifying the inhibitor in the structure of rat nNOSoxy-AR-R17477. The compounds were analyzed in the framework of the structures of human iNOSoxy (PDB ID 4NOS), eNOSoxy (PDB ID 3NOS), and of a homology model of human nNOSoxy based on the rat nNOSoxy structure. The geometries of the chloropyridine and hydroxy-pyridine derivatives of AR-R17477 were subjected to quantum mechanics/molecular mechanics (QM/MM) semiempirical quantum chemical geometry optimization by using the PM3 method (29). Model construction and QM/MM PM3 calculations were performed with the program hyperchem (evaluation version 6.03, Hypercube, Gainesville, FL). During the optimization process, parts of AR-R17477 derivatives including the chloropyridine or hydroxy-pyridine group and the adjacent amino group were treated quantum mechanically, whereas the rest of the inhibitor and protein molecule was considered as producing a fixed potential field.
Results and Discussion
AR-R17477 is a competitive, highly selective and potent inhibitor of nNOS with IC50 inhibition values for rat nNOS, mouse iNOS, and human eNOS of 0.035, 5.0, and 3.5 μM, respectively (16). Because IC50 values depend strongly on assay conditions, we determined the binding constants of the inhibitor to full-length NOS and NOSoxy isoforms by a spectroscopic competition assay. As can be seen from Table 1, AR-R17477 binds most tightly to nNOS, and about one and two orders of magnitude more weakly to iNOS and eNOS, respectively. The compound does not bind to the monomeric form of any NOS isozyme (data not shown).
Table 1. Binding constants of AR-R17477 for wild-type and mutant NOS isoforms.
| Kd, μM
|
Critical residue
|
|||
|---|---|---|---|---|
| Protein | -H4B | +H4B | IC50, μM (16) | |
| Holoenzyme | ||||
| Rat nNOS | ND | 0.035 | 0.035 | L337 |
| Mouse iNOS | 5.0 | N115 | ||
| Human iNOS | ND | 0.15 | T121 | |
| Human eNOS | 3.5 | F105 | ||
| Oxygenase domains | ||||
| Mouse iNOSoxy | 2.5 | 0.50 | N115 | |
| Mouse iNOSoxy N115L | 1.2-1.4 | 0.09 | L115 | |
| Human iNOSoxy | 1.75 | 0.25 | T121 | |
| Human iNOSoxy T121L | 1.3 | 0.08 | L121 | |
| Rat nNOSoxy | 0.06 | 0.02 | L337 | |
| Rat nNOSoxy L337N | 1.2 | 1.5-2.0* | N337 | |
| Rat nNOSoxy L337F | ND | 2-3 | F337 | |
ND, not determined.
Spectral pertubation analysis was performed in the presence of 5 mM imidazole, because Kd (imidazole) is higher (400 μM) compared to wild-type nNOSoxy (80 μM).
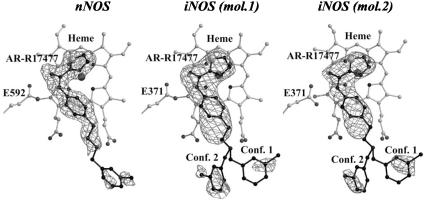
Structure of AR-R17477 Complex. The complexes of AR-R17477 with the oxygenase domains of murine iNOS and rat nNOS were obtained by cocrystallization (see supporting information for data statistics). As shown in Fig. 1, there is good electron density for the inhibitor bound to nNOSoxy and iNOSoxy. In particular, the thiophecarboxamidine and phenyl groups and the aliphatic link between the phenyl and the amino groups are well resolved. In contrast to the nNOS complex the electron density for the amino group is not well defined for iNOS and there are two conformations for the remaining part of the inhibitor. In both complexes, the chlorophenyl group is partially disordered; however, strong peaks corresponding to the chlorine atom and some density for the phenyl ring along with careful steric and chemical analysis allow unambiguous positioning of this group, which is further confirmed by x-ray crystallographic refinement (see supporting information for details). Thus, the only parts of the inhibitor that are not well defined are a carbon between the amino and the chlorophenyl groups in case of both nNOS and iNOS, and the nitrogen of the amino group in iNOS. Nevertheless, steric and conformational restraints allow defining the positions of these atoms corresponding to the equilibrium states of the inhibitor.
Fig. 1.
Fobs – Fcalc electron density omit maps contoured at 3 σ superimposed with the final model of the inhibitor AR-R17477 bound to nNOSoxy and the two crystallographically independent molecules in the asymmetric unit of iNOSoxy crystals (mol.1 and mol.2).
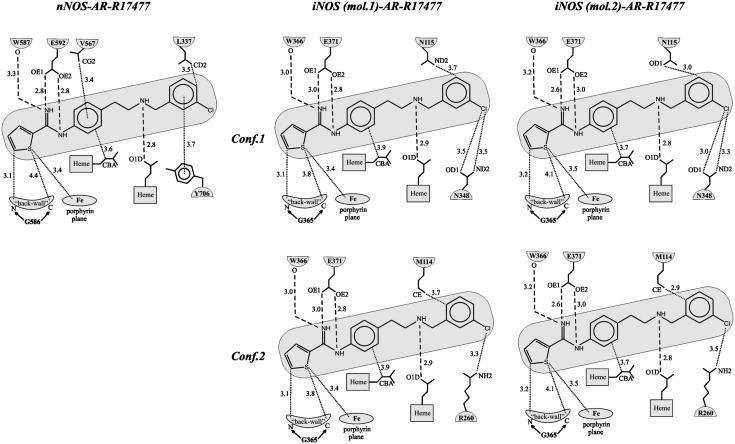
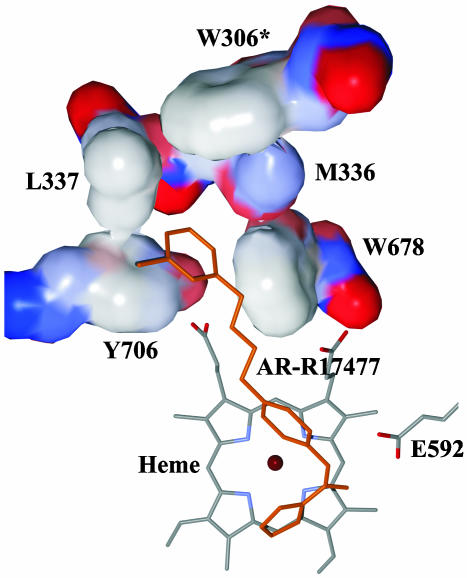
The AR-R17477 interactions are shown schematically in Fig. 2. None of the inhibitor atoms makes direct contact to the second molecule of the biological NOSoxy dimer. Analogously to the guanidinium group of the natural substrate l-Arg, the nitrogen atoms of the thiophecarboxamidine group form hydrogen bonds with the E592/E371 side chain and the main chain oxygen atom of W587/W366 (nNOS/iNOS). The thiophene group lines up on the “back wall” of the active site (14) formed by the residues F584, S586, G586, and W587 (nNOS) and F363, N364, G365, and W366 (iNOS), respectively, making van der Waals contacts with G586/G365. The sulfur atom forms van der Waals contacts with the heme porphyrin, with the distance between the sulfur and the porphyrin plane being 3.4–3.5 Å (Fig. 2). The phenyl group next to the thiophecarboxamidine is located atop the porphyrin edge containing the propionic groups. The aromatic ring is tilted with respect to the heme plane, making an angle of 54°. This orientation may be caused by the stabilizing van der Waals interactions of the phenyl group with the aliphatic part of the heme propionic group (Fig. 2) and the side chain of V567 in case of nNOS. The latter interaction is missing in iNOS because of a slight shift and different orientation of the corresponding V346. The aliphatic link connecting the phenyl and the chlorophenyl groups occupies the space between the propionic groups of the heme so that the amino group forms a hydrogen bond with the O1D oxygen of the propionic carboxyl. Up to this point, the interactions of AR-R17477 are very similar in the nNOSoxy and iNOSoxy complexes. However, the interactions of the remaining part of the molecule including the chlorophenyl group are quite different. In the nNOSoxy complex, the chlorophenyl group fits nicely into a hydrophobic pocket created by side chains of Y706, W678, M336, and L337 and topped off by the relatively distant W306 of the second molecule of the nNOSoxy biological dimer (Fig. 3). Here, the chlorophenyl forms good stacking interactions with the aromatic ring of Y706 and the chlorine atom is located in a large cavity of the substrate access channel where it faces no steric clashes. L337 adjoins the flexible loop region comprising residues P338–T350. Filling in the hydrophobic pocket by the chlorophenyl group may contribute to the stabilization of the flexible loop region, which could explain the significant improvement of the nNOSoxy-AR-R17477 crystal quality compared to crystals of other nNOSoxy complexes including the complex with the natural substrate.
Fig. 2.
Schematic representation of the interactions of AR-R17477 and nNOSoxy and iNOSoxy (two molecules in asymmetric unit, two conformations each). Dashed lines correspond to hydrogen bonds, dotted lines correspond to van der Waals interactions.
Fig. 3.
Hydrophobic pocket accommodating the chlorophenyl aromatic ring of AR-R17477 bound to rat nNOSoxy. For the residues Y706, L337, M336, W678, and W306 of the second molecule in nNOSoxy biological dimer (W306*), the van der Waals surface is shown. The color scheme of the surface corresponds to the atom charge (blue, positive; red, negative; white, neutral).
In case of iNOSoxy, the interactions of the chlorophenyl group change dramatically because of the replacement of a leucine in nNOS (L337) to asparagine in iNOS (N115) (Figs. 2 and 4). To avoid a steric clash with the polar N115 side chain, the chlorophenyl moves aside, resulting in two conformations (Figs. 1 and 2). In one conformation (conf.1 in Figs. 1, 2, and 4), the chlorophenyl group still remains in the same region of the substrate access channel, but the stacking interaction with the tyrosine is broken and the whole group is shifted toward N348, so that the chlorine atom makes close van der Waals contacts with the oxygen and nitrogen atoms of the N348 side chain. However, this still does not prevent close contacts between the oxygen of the N115 side chain and part of the chlorophenyl aromatic ring (Fig. 2). In the other conformation (conf.2), the chlorophenyl group occupies a different region of the substrate access channel where the chlorine atom is located closer to R260. Also, in this case there remains one close contact between the chlorophenyl aromatic ring and the terminal carbon of the M114 side chain. Apart from the fact that these two conformations are obviously less favorable from steric and energetic points of view compared to the conformation in nNOS, the L337/N115 replacement seems to disturb the hydrogen bond between the amino group of the aliphatic link and the O1D oxygen of the propionic group of the heme. This gives the nitrogen of the amino group more flexibility, which is reflected in the lack of electron density for this atom in iNOSoxy (Fig. 1).
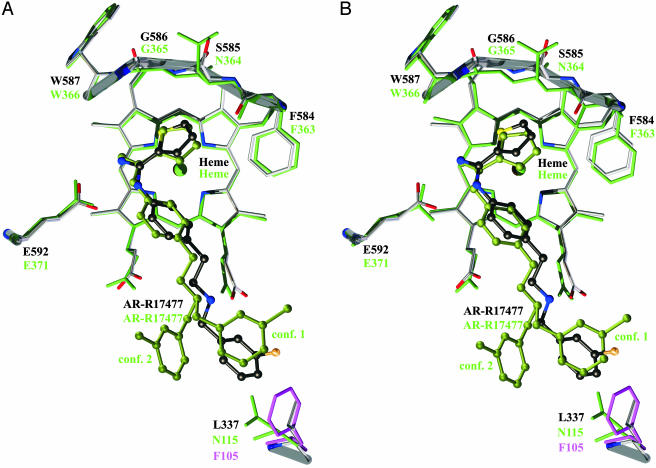
Fig. 4.
Superposition of mouse iNOSoxy molecules 1 (A) and 2 (B) in asymmetric unit (green) and rat nNOSoxy (gray) complexed with AR-R17477. F105 of human eNOS is shown in magenta. The thick transparent rods outline the main chain atoms of the backwall of nNOSoxy (also in Fig. 5).
Site-Directed Mutagenesis Experiments. The analysis of the crystal structures suggests that the major component of the structural basis of isoform-specific binding of AR-R17477 is the interaction with the substrate access channel residues L337 and N115 in nNOS and iNOS, respectively. We have tested this hypothesis by directed mutagenesis (Table 1). First, we have swapped these residues in rat nNOS and mouse iNOS, creating the mutants rat nNOS L337N and mouse iNOSoxy N115L. Also, the corresponding residue in human iNOS enzyme is a threonine, and we have replaced this residue in human iNOSoxy with a leucine as in rat nNOS. The result is the mutant human iNOSoxy T121L. Finally, we have introduced in nNOS the residue found at that same position in human eNOS, which is F105. The mutant that we have generated is therefore rat nNOSoxy L337F. As can be seen from Table 1, the affinity of AR-R17477 for nNOS and iNOS correlates with the original characteristics of the protein from which this residue was derived. In particular, the affinity increases significantly when replacing the mouse or human iNOS amino acid with the rat nNOS leucine residue. In contrast, the affinity becomes very poor when a phenylalanine is present at this location, as in human eNOSoxy or the mutant L337F rat nNOSoxy. The analysis of the H2O2-dependent Nω-hydroxy-l-Arg (l-NOHA) oxidation by the mutants shows that mouse N115L iNOSoxy and human T121L iNOSoxy behave like rat nNOSoxy. The turnover number for l-NOHA oxidation decreases to 35 mol/min per mol of enzyme similar to wild-type nNOSox compared to 70 mol/min per mol of enzyme for wild-type iNOS (19, 22). The Kd for l-arginine binding is similar to wild type (0.5–1.0 mM), but the kinetics of binding (as measured by the appearance of Soret peak at 395 nm; ref. 21) changed quite significantly; for N115L and T121L, the rate is 0.2 mOD/min, which is very close to the rat nNOSoxy l-arginine binding rate of 0.32 mOD/min, compared to the wild-type iNOSoxy rate of 0.032 mOD/min. This change in phenotype of the iNOS mutants supports the importance of access channel residues in determining the isoform-specific ligand interaction and their influence on the dynamics of ligand binding coupled with catalytic activity. The binding analysis (not shown) indicates that both the oxidized and the reduced forms of NOS in complex with AR-R17477 cannot bind cyanide (CN–), an O2 mimic, or carbonmonoxide (CO). Comparison of the crystal structures of the mouse iNOSoxy·l–Arg·CN– (20) and iNOSoxy·AR–R17477 complexes (see supporting information) supports this conclusion. The thiophene group blocks the heme active site so that neither oxygen nor the water molecule that has been suggested to participate in proton transfer during the first step of the NOS hydroxylation reaction (20) can bind. This makes AR-R17477 a desirable and pharmacologically potent competitive inhibitor, because it inhibits not only NO production but also superoxide generation in nNOS, a common phenomenon observed with other l-arginine analog inhibitors because of uncoupled NADPH oxidation.
AR-R17477 and Human Isozymes. AR-R17477 also inhibits the human NOS isoforms (Table 1). Superposition of the structures of human eNOSoxy and rat nNOSoxy shows that human eNOS F105 corresponding to rat nNOS L337 points into the substrate access channel and would leave no space for the chlorophenyl group (Fig. 4). This explains the low affinities of eNOS and rat nNOSoxy L337F for AR-R17477. The situation is structurally similar to mouse iNOSoxy (Fig. 4), in line with the finding that the corresponding IC50 values are also similar (Table 1). Thus, the structural observation being in perfect agreement with the inhibition values (Table 1) implies that the source of isoform selectivity for AR-R17477 is provided by the residues L337/N115/F105 (rat nNOS/mouse iNOS/human eNOS) in the substrate access channel that participate in the formation of the hydrophobic binding pocket of the chlorophenyl group. Its positioning influences the interactions of the amino group and depends on the size and hydrophobicity of the binding pocket.
From this point of view, AR-R17477 may not be so selective within the human isoforms as it is for rat nNOS against mouse iNOS and human eNOS. The polar side chains of H342 and T121 in human nNOS and iNOS, respectively, may alter the hydrophobic properties of the pocket, making interactions of the chlorophenyl group unfavorable. Moreover, the superposition of the structures of human iNOSoxy (PDB ID 4NOS), human eNOSoxy (PDB ID 3NOS), and a homology model of the human nNOSoxy-AR-R17477 complex (Fig. 5) shows that the side chain of H342 overlaps well enough with the phenyl ring of F105 in human eNOSoxy and may, therefore, cause steric hindrance and the same “push out” effect. Although the side chain of a threonine is shorter than the one of a phenylalanine or histidine, the same situation is observed in human iNOSoxy, where the side chain of T121 overlaps with the side chains of F105 (human eNOSoxy) and H342 (homology model of human nNOSoxy) because of a different conformation of the loop containing T121.
Fig. 5.
Superposition of the hydrophobic pocket residues in the substrate access channel of human iNOSoxy (green), human eNOSoxy (magenta), and the homology model of human nNOSoxy (gray). The reference structure of AR-R17477 from rat nNOSoxy-AR-R17477 is shown in dark-gray (model 1). The QM/MM optimized models of modified AR-R17477 are shown in black (model 2 containing a chloropyridine ring) and orange (model 3 containing a hydroxypyridine ring). In models 2 and 3, the pyridine nitrogen can form a hydrogen bond with the side chains of both H342 (human nNOS) and T121 (human iNOS). The phenyl ring of F105 (human eNOS) does not allow such an interaction and should preserve the “push-out” effect described in the text. The red balls represent the water molecules from the human iNOSoxy crystal structure that overlap with AR-R17477 atoms in this region.
These features make the design of isoform-specific inhibitors exploiting the substrate access channel of human isozymes less straightforward, without, however, excluding it. For example, the replacement of a carbon in meta-position relative to the chlorine and methyl in the aromatic ring of the chlorophenylmethyl group by a nitrogen can allow (with a minor conformational adjustment) the resulting chloropyridine ring to form a hydrogen bond with either H342 of human nNOS or T121 of human iNOS (Fig. 5). This hydrogen bond can be formed when the conformation of such a derivative of AR-R17477 is relaxed (which is supported by QM/MM PM3 calculations) and the hydrogen bond between the amino group of the aliphatic link and O1D oxygen of the heme propionic group is not disturbed. The stacking interaction between the aromatic ring of the chloropyridine and the tyrosine can be preserved, although in this case the stacking will be rather tilted like the stacking of the phenyl group of AR-R17477 and the porphyrin of the heme. This conformation can be further stabilized if the chlorine atom is replaced by a hydroxy group, which can form another hydrogen bond with N574/N354 (human nNOS/human iNOS). The models of chloropyridine and hydroxy-pyridine derivatives of AR-R17477 optimized by QM/MM PM3 calculations for human iNOSoxy structure revealed the same interaction pattern as those optimized for human nNOSoxy homology model (Fig. 5). Thus, the above consideration is applicable to both human nNOS and iNOS, but not to human eNOS, where the presence of nitrogen in chloropyridine aromatic ring will not compensate the “push out” effect of F105. This difference can be used to achieve the selectivity for human nNOS and iNOS against human eNOS. It is not clear whether the replacement of the chlorine atom by a hydroxy group would lead to increase or decrease of the selectivity, which means that both ways should be tried. It is interesting to note that the water molecules in the vicinity of T121 in the human iNOSoxy crystal structure superimpose well with the chlorine, methyl carbon, and the carbon in the para-position relative to the chlorine of the chlorophenylmethyl group of AR-R17477 in complex with rat nNOS, making a kind of pharmacophore for the chlorophenylmethyl group (Fig. 5). This observation may be useful when designing possible modifications of this part of AR-R17477.
Another part of AR-R17477 that can be modified to increase selectivity between human nNOS and iNOS is the thiophene group. It has been shown (14) that the “back wall” of the active site, made up of β-strand S15, differs significantly in mouse iNOSoxy and rat nNOSoxy, the latter having a larger active site (Fig. 4). This feature contributes to rat nNOS's specificity for Nω-propyl-l-Arg over mouse iNOS, and must also be present in the human isoforms. Replacement of the thiophene in AR-R17477 by large or inflexible groups making optimal contacts to the back-wall residues in nNOS would lead to less favorable interaction of these groups with the back wall of iNOS, resulting in increase of nNOS selectivity.
Conclusions
The crystal structures of AR-R17477-NOSoxy complexes presented here reveal a distant hydrophobic pocket that is a very promising source of isoform specificity because of unique residues in the substrate access channel. This finding provides an important guideline for rational design of nNOS-specific inhibitors. Although AR-R17477 should be modified to increase selectivity in human isozymes, such a modification seems to be possible even without significant modification of the compound. The structural data on the AR-R17477-NOSoxy complexes together with available structures of human isoforms make it possible to start a rational design of inhibitors whose selectivity is based on the isoform-unique residues in the substrate access channel. Structural analysis of the AR-R17477-NOSoxy complexes coupled with information provided by the distribution of water molecules in the vicinity of the substrate access channel allows us to reduce the structural field of search for such compounds significantly and to increase the probability of success. The modification of the thiophene group may help us to use the differences in the positioning of β-strand S15 that makes up the back wall of the heme cavity, which can provide additional source of selectivity between nNOS and iNOS isoforms.
Supplementary Material
Acknowledgments
We thank Chris Veale from Astra Zeneca for the generous gift of AR-R17477, Elisabeth Hartmann and Georg Holtermann for excellent technical help, Roger S. Goody for hospitality and support, and the Deutsche Forschungsgemeinschaft (I.S.) for generous financial support. D.K.G. is very grateful to Dr. J. B. Weinberg for continuing support and sharing laboratory space for this study, and to the American Heart Foundation for financial support (Grant 0365338U).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NOS, NO synthase; eNOS, endothelial NOS; iNOS, inducible NOS; nNOS, neuronal NOS; AR-R17477, N-(4-(2-((3-chlorophenylmethyl) amino) ethyl) phenyl)-2-thiophecarboxamidine dihydrochloride; QM/MM, quantum mechanics/molecular mechanics.
Data deposition: The coordinates and structure factor amplitudes reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1VAG and 1VAF).
References
- 1.Alderton, W. K., Cooper, C. E. & Knowles, R. G. (2001) Biochem. J. 357, 593–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raman, C. S., Li, H., Martasek, P., Kral, V., Masters, B. S. & Poulos, T. L. (1998) Cell 95, 939–950. [DOI] [PubMed] [Google Scholar]
- 3.Bec, N., Gorren, A. C., Voelker, C., Mayer, B. & Lange, R. (1998) J. Biol. Chem. 273, 13502–13508. [DOI] [PubMed] [Google Scholar]
- 4.Ignarro, L. J., Buga, G. M., Wood, K. S., Byrns, R. E. & Chaudhuri, G. (1987) Proc. Natl. Acad. Sci. USA 84, 9265–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garthwaite, J., Charles, S. L. & Chess-Williams, R. (1988) Nature 336, 385–388. [DOI] [PubMed] [Google Scholar]
- 6.Moncada, S., Palmer, R. M. & Higgs, E. A. (1991) Pharmacol. Rev. 43, 109–142. [PubMed] [Google Scholar]
- 7.Babu, B. R. & Griffith, O. W. (1998) Curr. Opin. Chem Biol. 2, 491–500. [DOI] [PubMed] [Google Scholar]
- 8.Crane, B. R., Arvai, A. S., Gachhui, R., Wu, C., Ghosh, D. K., Getzoff, E. D., Stuehr, D. J. & Tainer, J. A. (1997) Science 278, 425–431. [DOI] [PubMed] [Google Scholar]
- 9.Crane, B. R., Arvai, A. S., Ghosh, D. K., Wu, C., Getzoff, E. D., Stuehr, D. J. & Tainer, J. A. (1998) Science 279, 2121–2126. [DOI] [PubMed] [Google Scholar]
- 10.Fischmann, T. O., Hruza, A., Niu, X. D., Fossetta, J. D., Lunn, C. A., Dolphin, E., Prongay, A. J., Reichert, P., Lundell, D. J., Narula, S. K., et al. (1999) Nat. Struct. Biol. 6, 233–242. [DOI] [PubMed] [Google Scholar]
- 11.Li, H., Shimizu, H., Flinspach, M., Jamal, J., Yang, W., Xian, M., Cai, T., Wen, E. Z., Jia, Q., Wang, P. G., et al. (2002) Biochemistry 41, 13868–13875. [DOI] [PubMed] [Google Scholar]
- 12.Garvey, E. P., Oplinger, J. A., Furfine, E. S., Kiff, R. J., Laszlo, F., Whittle, B. J. & Knowles, R. G. (1997) J. Biol. Chem. 272, 4959–4963. [DOI] [PubMed] [Google Scholar]
- 13.Zhang, H. Q., Fast, W., Marletta, M. A., Martasek, P. & Silverman, R. B. (1997) J. Med. Chem 40, 3869–3870. [DOI] [PubMed] [Google Scholar]
- 14.Fedorov, R., Hartmann, E., Ghosh, D. K. & Schlichting, I. (2003) J. Biol. Chem. 278, 45818–45825. [DOI] [PubMed] [Google Scholar]
- 15.Raman, C. S., Li, H., Martasek, P., Babu, B. R., Griffith, O. W., Masters, B. S. & Poulos, T. L. (2001) J. Biol. Chem. 276, 26486–26491. [DOI] [PubMed] [Google Scholar]
- 16.Reif, D. W., McCarthy, D. J., Cregan, E. & Macdonald, J. E. (2000) Free Radical Biol. Med. 28, 1470–1477. [DOI] [PubMed] [Google Scholar]
- 17.Harukini, I., Traystman, R. J. & Kirsch, J. R. (1999) Crit. Care Med. 27, 2508–2511. [DOI] [PubMed] [Google Scholar]
- 18.O'Neill, M. J., Murray, T. K., McCarty, D. R., Hicks, C. A., Dell, C. P., Patrick, K. E., Ward, M. A., Osborne, D. J., Wiernski, T. R., Roman, C. R., et al. (2000) Brain Res. 871, 234–244. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh, D. K., Rashid, M. B., Crane, B., Taskar, V., Mast, M., Misukonis, M. A., Weinberg, J. B. & Eissa, N. T. (2001) Proc. Natl. Acad. Sci. USA 98, 10392–10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fedorov, R., Ghosh, D. K. & Schlichting, I. (2003) Arch. Biochem. Biophys. 409, 25–31. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh, D. K., Wu, C., Pitters, E., Moloney, M., Werner, E. R., Mayer, B. & Stuehr, D. J. (1997) Biochemistry 36, 10609–10619. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh, D. K., Crane, B. R., Ghosh, S., Wolan, D., Gachhui, R., Crooks, C., Presta, A., Tainer, J. A., Getzoff, E. D. & Stuehr, D. J. (1999) EMBO J. 18, 6260–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuehr, D. J. & Ikeda-Saito, M. (1992) J. Biol. Chem. 267, 20547–20550. [PubMed] [Google Scholar]
- 24.McMillan, K. & Masters, B. S. (1993) Biochemistry 32, 9875–9880. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch, W. (1993) J. Appl. Crystallogr. 26, 795–800. [Google Scholar]
- 26.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S. et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 27.Sheldrick, G. M. & Schneider, T. R. (1997) Methods Enzymol. 277, 319–343. [PubMed] [Google Scholar]
- 28.Taylor, W. R. & Orengo, C. A. (1989) J. Mol. Biol. 208, 1–22. [DOI] [PubMed] [Google Scholar]
- 29.Stewart, J. J. P. (1989) J. Comput. Chem. 10, 209–220. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.