Abstract
Acetyl-CoA carboxylases (ACCs) are crucial for the metabolism of fatty acids, making these enzymes important targets for the development of therapeutics against obesity, diabetes, and other diseases. The carboxyltransferase (CT) domain of ACC is the site of action of commercial herbicides, such as haloxyfop, diclofop, and sethoxydim. We have determined the crystal structures at up to 2.5-Å resolution of the CT domain of yeast ACC in complex with the herbicide haloxyfop or diclofop. The inhibitors are bound in the active site, at the interface of the dimer of the CT domain. Unexpectedly, inhibitor binding requires large conformational changes for several residues in this interface, which create a highly conserved hydrophobic pocket that extends deeply into the core of the dimer. Two residues that affect herbicide sensitivity are located in this binding site, and mutation of these residues disrupts the structure of the domain. Other residues in the binding site are strictly conserved among the CT domains.
Obesity has become a serious health problem worldwide over the past few decades, due in part to increased food intake in conjunction with reduced physical activity by the general population (1–3). In the United States, ≈30% of the population is obese, and another 35% of the population is overweight. Moreover, obesity is associated with a variety of serious human diseases, especially type 2 diabetes, cardiovascular diseases, depression, and cancer. There is clearly a critical need for therapeutic agents that can regulate body weight and obesity.
Acetyl-CoA carboxylases (ACCs) are crucial for the metabolism of fatty acids. They catalyze the production of malonyl-CoA from acetyl-CoA and CO2, a reaction that also requires the hydrolysis of ATP (4–7). Two isoforms of this enzyme have been identified in mammals. ACC1, a cytosolic enzyme, controls the first and the committed step in the biosynthesis of long-chain fatty acids (5). In comparison, ACC2 is associated with the mitochondrial membrane, and its malonyl-CoA product potently inhibits the shuttle that transports long-chain acyl-CoAs from the cytosol to the mitochondria for oxidation (8, 9). Mice lacking ACC2 have elevated fatty acid oxidation and reduced body fat and body weight, establishing ACC2 as a target for anti-obesity and anti-diabetes agents (6, 10).
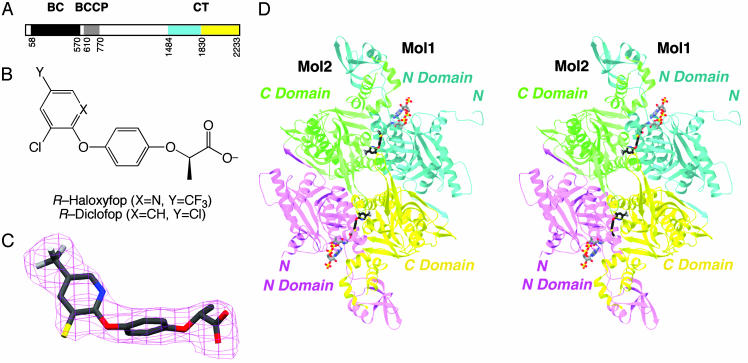
ACCs catalyze the carboxylation of acetyl-CoA in two steps: ATP-dependent carboxylation of a biotin group covalently linked to a lysine residue in the biotin carboxyl carrier protein (BCCP), and then the transfer of this activated carboxyl group to acetyl-CoA. In mammals, yeast, and most other eukaryotes, ACC is a large, multidomain enzyme (Fig. 1A), with a biotin carboxylase (BC) domain that catalyzes the first step and a carboxyltransferase (CT) domain that catalyzes the second step of the reaction. Biotin is covalently linked to the BCCP domain in the enzyme (Fig. 1 A).
Fig. 1.
Crystal structure of CT domain in complex with haloxyfop. (A) Domain organization of yeast ACC. The N and C subdomains of CT are colored in cyan and yellow, respectively. (B) Chemical structures of the herbicides (R)-haloxyfop and (R)-diclofop. (C) Final 2Fo – Fc electron density at 2.8-Å resolution for haloxyfop, contoured at 1σ. (D) Schematic stereodrawing of the structure of yeast CT domain dimer in complex with haloxyfop. The N domains of the two monomers are colored in cyan and magenta, and the C domains are colored in yellow and green. The inhibitor is shown in stick models, in black for carbon atoms. The CoA molecule is shown for reference (11), in gray. C was produced with setor (28), and D was produced with ribbons (29).
The CT domain contains ≈800 residues (90 kDa), and constitutes approximately the C-terminal one-third of the eukaryotic, multidomain ACCs (Fig. 1 A). The amino acid sequences of this domain are highly conserved; for example there is 52% sequence identity between the CT domains of yeast ACC and human ACC2. Therefore, structural information on the yeast CT domain should be applicable to the human CT domains. We have recently reported the crystal structures of the CT domain of yeast ACC and its complex with CoA (11). The structure contains two subdomains, N and C domains (Fig. 1 A), and the active site is located at the interface of a dimer of the enzyme.
The CT domain is the site of action of two different classes of widely used commercial herbicides (12–15), as represented by haloxyfop and diclofop (FOPs, Fig. 1B) and sethoxydim (DIMs). These compounds are potent inhibitors of ACCs from sensitive plants and kill them by shutting down fatty acid biosynthesis. This observation confirms that an inhibitor of the CT domain is sufficient to block the function of ACC, and it establishes this domain as a valid target for the development of inhibitors against these enzymes, especially the human ACCs for the treatment of obesity and diabetes.
However, the molecular mechanism for the inhibitory action of the herbicides is currently not known. The herbicides were the only known potent inhibitors of ACCs until the recent report of potent inhibitors against mammalian ACCs (16). The herbicides represent a lead for the discovery of new inhibitors against these enzymes. Therefore, elucidating the mechanism of action of these compounds will also provide a starting point for the design and development of inhibitors against the human ACCs.
Materials and Methods
Protein Production and Crystallization. The cloning, expression, purification, and crystallization of the CT domain (residues 1429–2233) of yeast ACC followed protocols as described earlier (11). On the basis of that first crystal structure, we designed additional bacterial expression constructs for this domain, and found that the construct covering residues 1476–2233 produces a large amount of soluble protein in Escherichia coli. This protein can be purified by following the same protocol, and it readily produces large crystals of the free enzyme. The reservoir solution contains 0.1 M sodium citrate (pH 5.5), 200 mM NaCl, 8% (wt/vol) polyethylene glycol (PEG) 8000, and 10% (vol/vol) glycerol. The protein is at 10 mg/ml concentration. These free enzyme crystals were cryoprotected by the introduction of 25% (vol/vol) ethylene glycol and flash-frozen in liquid propane for data collection at 100 K.
To prepare the haloxyfop complex, crystals of the free enzyme (covering residues 1429–2233) were soaked with various concentrations of the herbicide for different lengths of time. Noting the poor affinity of the inhibitor, initial attempts used high concentration of the compound (5 mM and higher); however, this procedure invariably led to dissolution of the crystal or loss of x-ray diffraction. Good-quality diffraction was maintained after soaking a crystal for 1 h at 1 mM haloxyfop. The crystal was flash-frozen in liquid propane.
To prepare the diclofop complex, CT domain (residues 1476–2233) was crystallized from a solution containing 2 mM diclofop. The reservoir solution contains 0.1 M sodium citrate (pH 5.5), 200 mM NaCl, 8% (wt/vol) PEG 8000, and 10% (vol/vol) glycerol.
Structure Determination. X-ray diffraction data were collected at the X4A beamline of the National Synchrotron Light Source (NSLS). The diffraction images were processed with the hkl package (17). The wild-type free enzyme, the haloxyfop complex, and the L1705I/V1967I mutant crystals belong to space group C2 and are isomorphous with each other, as well as with the free enzyme structure that we reported earlier (11). The unit cell parameters for the free enzyme crystal are a = 246.8 Å, b = 125.2 Å, c = 145.5 Å, and β = 94.1°. The structure refinement was carried out with the program cns (18). Clear electron density for the herbicide was observed from the crystallographic analysis (Fig. 1C). The atomic model was built with the program o (19). The crystallographic information is summarized in Table 1.
Table 1. Summary of crystallographic information.
| Haloxyfop complex | Diclofop complex | Free enzyme | L17051/V19671 | |
|---|---|---|---|---|
| Maximum resolution, Å | 2.8 | 2.5 | 2.5 | 2.6 |
| No. of observations | 285,486 | 584,662 | 420,147 | 315,564 |
| Rmerge,* % | 9.3 (28.8) | 7.6 (31.7) | 6.9 (31.4) | 5.0 (25.6) |
| Resolution range for refinement | 27-2.8 | 30-2.5 | 27-2.5 | 28-2.6 |
| No. of reflections | 96,167 | 88,661 | 130,501 | 117,132 |
| Completeness, % | 88 (74) | 96 (89) | 86 (64) | 87 (69) |
| R factor,† % | 21.8 (27.9) | 21.7 (26.1) | 21.9 (26.7) | 21.2 (27.0) |
| Free R factor,† % rms deviation | 25.2 (30.9) | 24.8 (29.5) | 25.0 (29.0) | 23.7 (28.9) |
| Bond lengths, Å | 0.009 | 0.007 | 0.007 | 0.007 |
| Bond angles, ° | 1.2 | 1.2 | 1.2 | 1.1 |
Rmerge = Σh Σt|Ihi — <Ih>|/Σh Σt Ihi. The numbers in parentheses are for the highest-resolution shell.
R = Σh|Fho — Fhc|/ΣhFho.
Crystals of the diclofop complex are in a new crystal form. They belong to space group P3221, with cell parameters of a = b = 136.8 Å and c = 244.4 Å. There is one dimer of the CT domain in the asymmetric unit. The structure was solved by the molecular replacement method with the program como (20), using the structure of the haloxyfop complex as the search model.
Mutagenesis and Kinetic Assays. The L1705I and V1967I single-site mutants as well as the L1705I/V1967I double mutant were made with the QuikChange kit (Stratagene), from the expression construct that covers residues 1476–2233. The mutants were sequenced, expressed in E. coli, and purified by following the same protocol as that for the wild-type CT domain. The catalytic activity of the CT domain was assayed by following protocols described earlier (11, 21). For inhibition studies, the activity of the enzyme in the presence of 0, 0.5, or 2 mM haloxyfop was determined (Fig. 6, which is published as supporting information on the PNAS web site). The concentration of the malonyl-CoA substrate was kept close to the expected Km, 75 μM for the wild-type enzyme and 750 μM for the mutant proteins. The wild-type enzyme was at 2.5 μM, whereas the mutant proteins were at 20 μM.
Results and Discussion
The Overall Structures. The crystal structure of the CT domain of yeast ACC in complex with haloxyfop (Fig. 1B) has been determined at 2.8-Å resolution (Table 1). The free enzyme crystal was soaked in a solution containing 1 mM haloxyfop. Our earlier kinetic experiments showed that haloxyfop has a Ki of about 0.25 mM against this CT domain (11). Nonetheless, the crystallographic analysis clearly revealed the presence of haloxyfop in the structure, with well defined electron density (Fig. 1C).
To assess whether there are conformational changes in the enzyme upon inhibitor binding, we determined the structure at 2.5-Å resolution of the free enzyme of the CT domain (Table 1). The free enzyme structure that we reported earlier was based on a crystal that was grown in the presence of acetyl-CoA (11). For the current structure, acetyl-CoA was not included in the crystallization solution. In addition, we used a new expression construct to prepare the protein samples for this crystal. This construct covers residues 1476–2233 of yeast ACC, removing about 50 residues at the N terminus (1429–1475) that were disordered in the earlier structure (11). This protein sample readily produces large crystals of the CT domain.
The enzyme·diclofop complex was prepared by cocrystallization, using the new CT domain protein sample covering residues 1476–2233. These cocrystals are in a different crystal form compared with the crystals of the free enzyme and the haloxyfop complex, and the structure was determined by the molecular replacement method (20) (Table 1).
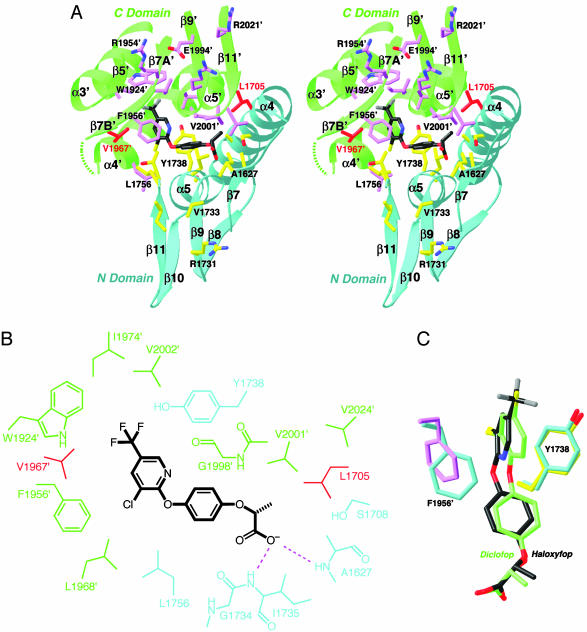
Binding Mode of Haloxyfop. Haloxyfop is bound in the active site region, at the interface between the N domain of one monomer and the C domain of the other monomer of the dimer (Fig. 1D). The pyridyl ring of the inhibitor is sandwiched between the side chains of Tyr-1738 and Phe-1956′ (primed residue numbers indicate the C domain of the other monomer), showing π–π interactions (Fig. 2A). The trifluoromethyl group is positioned over the plane of the Trp-1924′ side chain, as well as near the side chains of Val-1967′, Ile-1974′, and Val-2002′. The phenyl ring in the center of the inhibitor is situated between the Gly-1734–Ile-1735 and Gly-1997′–Gly-1998′ amide bonds (Fig. 2B). One of the carboxylate oxygen atoms of the inhibitor is hydrogen-bonded to the main-chain amides of Ala-1627 and Ile-1735 (Fig. 2B), whereas the other is exposed to the solvent.
Fig. 2.
The binding mode of haloxyfop. (A) Stereographic drawing showing the binding site for haloxyfop. The N domain of one monomer is colored in cyan, and the C domain of the other monomer is in green. The side chains of residues in the binding site are shown in yellow and magenta, respectively. The dashed segment indicates the disordered residues 1959′–1964′. The drawing was produced with ribbons (29). (B) Schematic drawing of the interactions between haloxyfop and the CT domain. (C) Overlay of the binding mode of haloxyfop (in black) and diclofop (in green). The conformations of residues Tyr-1738 and Phe-1956′ in the haloxyfop (yellow and magenta) and diclofop (cyan) complexes are also shown.
The methyl group of haloxyfop has van der Waals interactions with the side chains of Ala-1627 and Leu-1705 (Fig. 2B). In contrast, this methyl group in the S stereoisomer of haloxyfop will clash with one of the carboxylate oxygens of the inhibitor (Fig. 2 A), explaining the selectivity for the R stereoisomer of this class of compounds (12, 22).
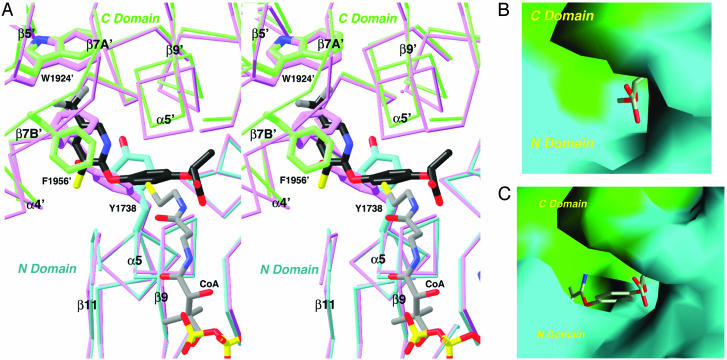
Large Conformational Changes in the Enzyme upon Haloxyfop Binding. There are significant conformational changes in the active site of the enzyme upon herbicide binding. Most importantly, the side chains of Tyr-1738 and Phe-1956′ assume new positions in the inhibitor complex to become π-stacked with the pyridyl ring of haloxyfop (Fig. 3A). These two side chains help cover the hydrophobic core of the dimer interface in the free enzyme. Their positions in the free enzyme actually clash with the position of haloxyfop (Fig. 3A), and the herbicide binding pocket does not exist in the free enzyme (Fig. 3B). With the conformational changes, a binding pocket is created on the surface of the CT domain and the trifluoromethylpyridyl group of the inhibitor is inserted deeply into the hydrophobic core of the dimer (Fig. 3C).
Fig. 3.
Conformational change in the CT domain upon inhibitor binding. (A) Stereographic structural overlay of the CT domain free enzyme (in magenta) and the haloxyfop complex (in cyan and green for the N and C domains) near the inhibitor binding site. The binding mode of CoA (11) is also shown. The poorer structural overlap in the C domain is due to the change in the dimer organization. (B) Molecular surface of the active site of the free enzyme. The model of haloxyfop is included for reference. Most of the inhibitor is in steric clash with the enzyme. (C) Molecular surface of the binding site in the haloxyfop complex. For both B and C, residues 1759–1772 and 2026′–2098′ have been removed to give a better view of the binding site. A was produced with ribbons (29), and B and C were produced with grasp (30).
Whereas the conformational change at Tyr-1738 is limited to its side chain, extensive structural differences are observed near the Phe-1956′ residue (Fig. 3A). The side chain of this residue rotates by ≈120° around the χ1 torsion angle, and its main chain moves by ≈2 Å such that the residue does not clash with the inhibitor (Fig. 3A). This movement triggers a conformational change for an entire segment (residues 1955′–1967′) of the CT domain, and residues 1959′–1964′ in this segment (helix α4A′, Fig. 4) are disordered in the herbicide complex (Fig. 2 A).
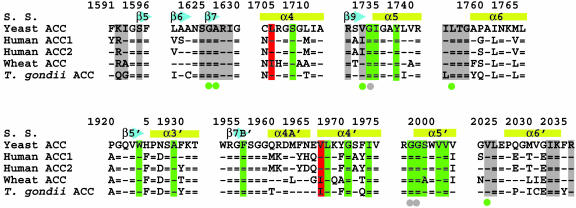
Fig. 4.
Sequence conservation in the binding site. Sequence alignment of residues in the haloxyfop (in green) and CoA (in gray) binding pockets. The two residues that confer herbicide resistance, Leu-1705 and Val-1967, are highlighted in red. A dash represents a residue that is identical to that in yeast ACC, whereas an equals sign represents a residue that is strictly conserved among ACCs. S. S., secondary structure.
Inhibitor binding at the dimer interface also causes a change in the organization of the dimer (Fig. 3A). With one monomer of the dimer in superposition, a rotation of ≈2.5° is needed to bring the second monomer into overlap.
It is unlikely that the herbicide binding pocket can exist during the catalytic cycle of ACCs. Moreover, herbicide binding is incompatible with the binding or the conformation of the acetyl- and malonyl-CoA substrates for catalysis (Fig. 3A), consistent with our kinetic data demonstrating that haloxyfop is a competitive inhibitor with respect to malonyl-CoA (11). Previous studies with wheat ACC showed that the herbicides are nearly competitive with respect to the substrate acetyl-CoA (23). At the same time, the distance between the thiol group of CoA and the exposed carboxylate oxygen of haloxyfop is only 3 Å (Fig. 3A), explaining earlier observations that the CoA ester of this herbicide is a more potent inhibitor of ACC (10, 24, 25).
The formation of this binding site requires conformational variability for several residues in the active site of the enzyme (Fig. 3A). The structure of the CT domain as observed here in the inhibitor complex is unlikely to be stable on its own, because of the significant exposure of the hydrophobic core of the dimer. Factors that regulate the conformational dynamics of residues in this dimer interface may affect the inhibitor sensitivity of the CT domain. This could be one mechanism for the herbicide sensitivity of the plant ACCs (see below).
Diclofop Has a Similar Binding Mode. The binding mode of the herbicide haloxyfop as well as the conformational changes in the monomer and dimer of the CT domain are confirmed by the structure of the complex with diclofop (Fig. 1B), at 2.5-Å resolution (Table 1). This complex was produced by cocrystallization, and the crystals are in a different space group as compared with the free enzyme and the haloxyfop complex. Nonetheless, the structure of the diclofop complex is the same as that of the haloxyfop complex. This conservation confirms that the structural changes observed for the haloxyfop complex are unlikely to be biased by crystal packing interactions.
The carboxyl groups of the two inhibitors have essentially the same binding mode (Fig. 2C). The aromatic rings of diclofop show small but recognizable differences in their positions as compared with those of haloxyfop, which may be linked to the conformational change in the side chain of Phe-1956′ (Fig. 2C). In comparison, the other residues in the binding site, including Tyr-1738, have the same conformation in the two complexes. The α4A′ helix is disordered in the cocrystals with diclofop as well. Moreover, the dimer organization in the diclofop complex is the same as the haloxyfop complex, as it also has the 2.5° rotation of the second monomer relative to the first monomer.
Residues That Confer Resistance to Herbicides Are in the Binding Pocket. Most of the residues that interact with the herbicides are either strictly or highly conserved among all of the CT domains (Fig. 4). Only two residues in the binding site show appreciable variation among the different CT domains, Leu-1705 and Val-1967′ (Fig. 4). Remarkably, it is exactly the variation/mutation of these two residues that can confer resistance to the herbicides in plants (14, 15).
The residue that is equivalent to Leu-1705 in the CT domains of wheat and other sensitive ACCs is Ile, and the Ile → Leu mutation, a subtle change in the side chain of this residue, renders the enzyme resistant to both haloxyfop and sethoxydim (14). The residue that is equivalent to Val-1967 in sensitive plants is Ile, and the Ile → Asn mutation makes the plants resistant to the FOPs, but not the DIMs. The Ile → Val mutation may also confer resistance to haloxyfop, although it does not affect the sensitivity to clodinafop (15).
To assess whether the Leu-1705 and Val-1967 residues are the sole determinants of herbicide sensitivity by the CT domains, we created the L1705I/V1967I double mutant as well as the L1705I and V1967I single mutants of the yeast CT domain. These mutant proteins are still dimeric in solution as determined by light-scattering experiments (data not shown). Kinetic studies show that the mutations have only minimal impact on the sensitivity of the CT domain to haloxyfop (Fig. 6), with Ki remaining in the 0.5 mM range. Interestingly, the IC50 values of herbicides against resistant plant ACCs are also in the 0.1–0.5 mM range (15), suggesting that the yeast CT domain behaves like a resistant plant CT domain. It is unlikely that the binding mode of herbicides to ACCs from sensitive plants is significantly different from that observed here in complex with the yeast CT domain, although a definitive answer to this question will have to await structural studies on the plant enzymes.
The mutations did cause roughly 100-fold reduction in the catalytic activity of the enzyme, including a 10-fold increase in the Km for malonyl-CoA (11). Our crystal structure of the L1705I/V1967I double mutant, at 2.6-Å resolution, shows that large segments of the enzyme are disordered (Table 1 and Fig. 7, which is published as supporting information on the PNAS web site), which may have impeded substrate binding and catalysis.
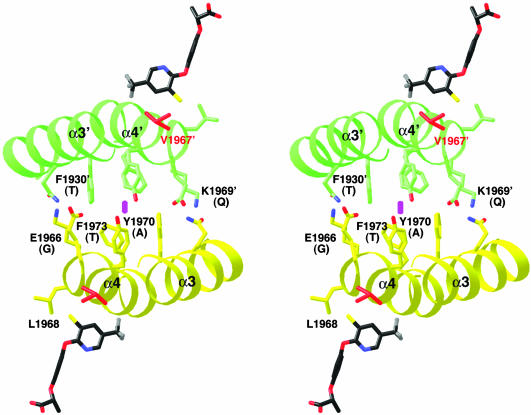
These experiments demonstrate that the Leu-1705 and Val-1967 residues are not the sole determinants of herbicide sensitivity by the yeast CT domain. Moreover, the apicoplast ACC from the parasite Toxoplasma gondii has a Leu residue at position 1705 (Fig. 4), but is still sensitive to the FOPs (26, 27). Therefore, herbicide sensitivity may be determined by these two residues together with other features in the dimer interface of the sensitive ACCs. For example, the pyridyl group of haloxyfop contacts one face of the α3′ and α4′ helices (Fig. 2 A), whereas the other face contacts its symmetry-mate across the twofold axis of the CT dimer (Fig. 5). Remarkably, the residues in this region of the dimer interface show significant variations between the plant and other ACCs (Fig. 4), with several large, aromatic residues replaced by small residues in the plant enzymes (Fig. 5). This variation may lead to a change in the dimer organization of the plant CT domains, which could be beneficial for herbicide binding. We observed a 2.5° rotation in the dimer interface of yeast CT domain upon haloxyfop binding, and it could be possible that the CT dimers of plant ACCs are preorganized for herbicide binding.
Fig. 5.
Differences between yeast and plant ACCs in the dimer interface of the CT domain. In this stereographic drawing, the α3 and α4 helices of one monomer are shown in yellow, and those of the other monomer are shown in green. The side chains in the dimer interface are shown and labeled. The equivalent residues in the plant ACCs are shown in parentheses. The haloxyfop molecules are shown for reference. The twofold axis of the dimer is indicated by the magenta oval.
Compounds that strongly inhibit both isoforms of rat ACC have recently been reported, representing the first known potent inhibitors of the mammalian enzymes (16). Kinetic studies suggest that they probably also function by interfering with the CT activity, confirming that the CT domain is a valid target for inhibiting the human ACCs (16). Moreover, these compounds can both inhibit de novo fatty acid biosynthesis and stimulate fatty acid oxidation, which may be clinically more efficacious (16). Our structures of the inhibitor complexes of the CT domain reveal a large conformational change in the active site of the enzyme, which produces a highly conserved and highly hydrophobic binding pocket that leads deeply into the dimer interface. This structural information should prove especially useful in the design and optimization of inhibitors against the CT domains of these important therapeutic targets.
Supplementary Material
Acknowledgments
We thank Randy Abramowitz and Xiaochun Yang for setting up the X4A beamline; Reza Khayat, Gerwald Jogl, Javed Khan, and Corey Mandel for help with data collection at the synchrotron source; and Yang Shen and Jiang Li for helpful discussions. This research was supported in part by National Institutes of Health Grant DK67238 (to L.T.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ACC, acetyl-CoA carboxylase; CT, carboxyltransferase.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1UYR, 1UYS, 1UYT, and 1UYV).
References
- 1.Friedman, J. M. (2003) Science 299, 856–858. [DOI] [PubMed] [Google Scholar]
- 2.Hill, J. O., Wyatt, H. R., Reed, G. W. & Peters, J. C. (2003) Science 299, 853–855. [DOI] [PubMed] [Google Scholar]
- 3.Pi-Sunyer, X. (2003) Science 299, 859–860. [DOI] [PubMed] [Google Scholar]
- 4.Alberts, A. W. & Vagelos, P. R. (1972) in The Enzymes, ed. Boyer, P. D. (Academic, New York), Vol. 6, pp. 37–82. [Google Scholar]
- 5.Wakil, S. J., Stoops, J. K. & Joshi, V. C. (1983) Annu. Rev. Biochem. 52, 537–579. [DOI] [PubMed] [Google Scholar]
- 6.Abu-Elheiga, L., Matzuk, M. M., Abo-Hashema, K. A. H. & Wakil, S. J. (2001) Science 291, 2613–2616. [DOI] [PubMed] [Google Scholar]
- 7.Cronan, J. E., Jr., & Waldrop, G. L. (2002) Prog. Lipid Res. 41, 407–435. [DOI] [PubMed] [Google Scholar]
- 8.McGarry, J. D. & Brown, N. F. (1997) Eur. J. Biochem. 244, 1–14. [DOI] [PubMed] [Google Scholar]
- 9.Ramsay, R. R., Gandour, R. D. & van der Leij, F. R. (2001) Biochim. Biophys. Acta 1546, 21–43. [DOI] [PubMed] [Google Scholar]
- 10.Lenhard, J. M. & Gottschalk, W. K. (2002) Adv. Drug Delivery Rev. 54, 1199–1212. [DOI] [PubMed] [Google Scholar]
- 11.Zhang, H., Yang, Z., Shen, Y. & Tong, L. (2003) Science 299, 2064–2067. [DOI] [PubMed] [Google Scholar]
- 12.Gronwald, J. W. (1991) Weed Sci. 39, 435–449. [Google Scholar]
- 13.Devine, M. D. & Shukla, A. (2000) Crop Protection 19, 881–889. [Google Scholar]
- 14.Zagnitko, O., Jelenska, J., Tevzadze, G., Haselkorn, R. & Gornicki, P. (2001) Proc. Natl. Acad. Sci. USA 98, 6617–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delye, C., Zhang, X.-Q., Chalopin, C., Michel, S. & Powles, S. B. (2003) Plant Physiol. 132, 1716–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harwood, H. J., Jr., Petras, S. F., Shelly, L. D., Zaccaro, L. M., Perry, D. A., Makowski, M. R., Hargrove, D. M., Martin, K. A., Tracey, W. R., Chapman, J. G., et al. (2003) J. Biol. Chem. 278, 37099–37111. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 18.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A. (1978) J. Appl. Crystallogr. 11, 268–272. [Google Scholar]
- 20.Jogl, G., Tao, X., Xu, Y. & Tong, L. (2001) Acta Crystallogr. D 57, 1127–1134. [DOI] [PubMed] [Google Scholar]
- 21.Guchhait, R. B., Polakis, S. E., Dimroth, P., Stoll, E., Moss, J. & Lane, M. D. (1974) J. Biol. Chem. 249, 6633–6645. [PubMed] [Google Scholar]
- 22.Rendina, A. R., Felts, J. M., Beaudoin, J. D., Craig-Kennard, A. C., Look, L. L., Paraskos, S. L. & Hagenah, J. A. (1988) Arch. Biochem. Biophys. 265, 219–225. [DOI] [PubMed] [Google Scholar]
- 23.Rendina, A. R., Craig-Kennard, A. C., Beaudoin, J. D. & Breen, M. K. (1990) J. Agric. Food Chem. 38, 1282–1287. [Google Scholar]
- 24.Kemal, C. & Casida, J. E. (1992) Life Sci. 50, 533–540. [DOI] [PubMed] [Google Scholar]
- 25.Taylor, W. S., Hixon, M., Chi, H., Marsilii, E. & Rendina, A. R. (1995) Pestic. Sci. 43, 177–180. [Google Scholar]
- 26.Jelenska, J., Sirikhachornkit, A., Haselkorn, R. & Gornicki, P. (2002) J. Biol. Chem. 277, 23208–23215. [DOI] [PubMed] [Google Scholar]
- 27.Zuther, E., Johnson, J. J., Haselkorn, R., McLeod, R. & Gornicki, P. (1999) Proc. Natl. Acad. Sci. USA 96, 13387–13392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans, S. V. (1993) J. Mol. Graphics 11, 134–138. [DOI] [PubMed] [Google Scholar]
- 29.Carson, M. (1987) J. Mol. Graphics 5, 103–106. [Google Scholar]
- 30.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281–296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.