Abstract
Loss-of-function mutations in the murine dominant white spotting/c-kit locus affect a diverse array of biological processes and cell lineages and cause a range of phenotypes, including severe anemia, defective pigmentation, sterility, mast cell deficits, a lack of interstitial cells of Cajal, spatial learning memory deficits, and defects in peripheral nerve regeneration. Here we show that tyrosine residues 567 and 569 in the juxtamembrane (Jx) domain of the murine Kit receptor tyrosine kinase are crucial for the function of Kit in melanogenesis and mast cell development, but are dispensable for the normal development of erythroid, interstitial cells of Cajal and germ cells. Furthermore, adult mice lacking both tyrosines exhibit splenomegaly, dysregulation of B-cell and megakaryocyte development, and enlarged stomachs. Analysis of signal transduction events induced by the mutant receptors after ligand stimulation indicates that Jx tyrosine mutations diminish receptor autophosphorylation and selectively attenuate activation of extracellular signal-regulated kinase/mitogen-activated protein kinases. Together, these observations demonstrate that the Jx domain of Kit plays a cell-type specific regulatory role in vivo and illustrate how engineered mutations in Kit can be used to understand the complex biological and molecular events that result from activating a receptor tyrosine kinase.
The Kit receptor tyrosine kinase (RTK) is centrally involved in the development of multiple cell lineages, including hematopoietic and germ cells, melanocytes, and the interstitial cells of Cajal (ICC) (1–4). Insights into the roles of this receptor and its cognate ligand, stem cell factor (SCF), in these developmental processes have been greatly facilitated by the large series of naturally occurring mutations in the murine genes that encode these molecules, the dominant white spotting (W) and steel (Sl) loci, respectively. Thus, mice with loss-of-functions in either the W or Sl loci are anemic and exhibit white spotting, sterility, and a concomitant loss of the ICC and intestinal pacemaker activity.
Ligand binding to the Kit RTK induces receptor dimerization and autophosphorylation of specific tyrosine residues (5, 6). These phosphorylation events create docking sites for specific Src homology 2 (SH2) domain-containing proteins, which in turn control various intracellular signaling pathways (7). Recruitment of particular targets is mediated by the ability of their SH2 domains to recognize specific phosphotyrosine (pTyr)-containing motifs on the activated receptor (8). Numerous signaling molecules have been identified as binding partners for specific pTyr residues on activated Kit, including the p85 subunit of phosphatidylinositol 3′ kinase (by means of tyrosine 719), phospholipase Cγ (by means of tyrosine 728), and the Grb2 and Grb7 adapter proteins (by means of tyrosine residues 702 and 934) (6). Additionally, signaling molecules, including Src family kinases and the protein tyrosine phosphatases Shp-1 and Shp-2, have been shown to associate with a dual tyrosine motif in the juxtamembrane (Jx) region of Kit (tyrosine residues 567 and 569) (6). Although mutations in Kit tyrosine residues have been shown to affect downstream signaling pathways, such as the mitogen-activated protein kinase (MAPK) and Akt pathways, the biological significance of most of these biochemical interactions remains unclear.
The pleiotropic nature of the W and Sl phenotypes makes the SCF/Kit pathway an ideal model for dissecting the in vivo role of the multiple signaling pathways that emanate from RTKs. For example, by introducing a specific tyrosine to phenylalanine mutation at tyrosine 719 in the Kit RTK, two groups have demonstrated that the resultant homozygous mutant mice are normal, except that homozygous mutant male mice are sterile because of decreased proliferation and increased apoptosis of spermatogonial cells (9, 10). Similar approaches with the Met and fibroblast growth factor RTKs have also revealed specific developmental defects, depending on which signaling pathway is perturbed through the loss of individual tyrosines in these RTKs (11, 12).
Amino acid substitutions or deletions in the Jx region of a number of RTKs, including Kit, Fms, and Flt3, can lead to dysregulation of tyrosine kinase activity and are associated with oncogenic transformation (13–15). In particular, oncogenic variants of Kit associated with human and murine mast cell leukemia carry either amino acid substitutions or deletions in the Jx domain (16, 17), and the majority of Kit variants associated with human gastrointestinal stromal tumors (GISTs) have activating mutations in the Jx region (13, 18, 19). Recent analysis has suggested that the Jx regions of RTKs, such as Eph receptors and Kit, have a dual role (20–22). In the autoinhibited state, the Jx region represses the activity of the kinase domain, but after stimulation, this inhibition is relieved and Jx pTyr sites can bind SH2-containing targets. The importance of the Kit Jx tyrosine residues is underscored by recent mass spectrometric analysis demonstrating that tyrosine residues 567 and 569 are significant autophosphorylation sites in the activated receptor (23).
In this study, we have explored the possible in vivo biological functions of the two tyrosine residues at positions 567 and 569 in the Jx domain of the murine Kit RTK by site-directed mutagenesis in vivo. We demonstrate that these tyrosine residues, acting in concert, are essential for melanogenesis, mast cell development, and gastrointestinal organogenesis but are not required for erythropoiesis, spermatogenesis and oogenesis, or development of the ICC. Homozygous mutant mice also exhibit profound dysregulation of adult B cell development and megakaryopoiesis, revealing previously uncharacterized regulatory functions of the Kit RTK in adult hematopoiesis. Furthermore, we show that Jx domain tyrosine mutations result in differential levels of receptor autophosphorylation and activation of extracellular signal-regulated kinase (Erk)-MAPK. Taken together, these findings demonstrate that the Jx domain of Kit plays a cell-type specific regulatory role in vivo.
Materials and Methods
In Vitro Progenitor Colony Assay. Progenitor cell assays of bone marrow cells were performed as described (24–27). For colony-forming unit (CFU)-pre-B, cells (1 × 104) were cultured in semisolid media with recombinant murine (rm)SCF (4 or 100 ng/ml) (R & D Systems) and rmIL-7 (100 ng/ml) (R & D Systems), and colonies were scored on day 12 of culture. CFU-megakaryocytic was assessed in collagen-based media (MegaCult-C, StemCell Technologies, Vancouver). For CFU-mast, 1 × 105 bone marrow cells were cultured for 7–10 days with rmSCF (50 ng/ml). Erythroid CFU and erythroid burst-forming unit assays were performed with MethoCult M3434 (StemCell Technologies) and scored on day-2 and day-7 to day-9 cultures, respectively.
Cytometry. Single-cell suspensions were prepared from tissues of 5- to 7-week-old mice and stained with FITC-, phycoerythrin- or allophycocyanin-conjugated antibodies (Pharmingen) reactive to B220, sIgM, sIgD, CD43, CD4, CD8, Gr-1, Ter119, or CD41. For examination of peritoneal mast cells, peritoneal cells were obtained by gentle lavage of the peritoneal cavity with 5 ml of PBS with 2% FBS and were treated with anti-2,4-dinitrophenyl-IgE mAb (Sigma), followed by FITC-conjugated anti-IgE mAb and allophycocyanin-conjugated anti-Kit mAb (Pharmingen) as described (28, 29). All samples were analyzed by flow cytometry with a FACSCalibur (Becton Dickinson).
Immunoprecipitation and Western Blotting. Site-direct mutagenesis of murine Kit cDNA was performed by using Altered Sites in Vitro Mutagenesis System (Promega) as described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Mutant Kit receptor cDNAs were subcloned into the mammalian expression vector pcDNA3 (Invitrogen) and introduced into subconfluent human embryonic kidney 293 cells by using polyethyleneimine-based transfection. Cells were serum starved for 18 h in DMEM before stimulation with 100 ng/ml rmSCF (R & D Systems) for 10 min at 37°C, where indicated. Cleared cell extracts were precipitated with anti-Kit antibodies (M-14, Santa Cruz Biotechnology). Antibodies used for Western blotting of Kit immunoprecipitates and total cell extracts include anti-pTyr 4G10 (Upstate Biotechnology), anti-Kit (Cell Signaling Technology, Beverly, MA), and anti-p85 (Upstate Biotechnology). Phospho-specific antibodies for tyrosine 719 on Kit and MAPK and Akt (Ser 473) were purchased from Cell Signaling Technology.
Results
Derivation of Kit567F, Kit569F, and KitFF Mice. We made use of Cre/loxP technology to create embryonic stem cells and mice in which tyrosine residues 567 and 569 in exon 11 of murine c-kit were mutated alone or in combination to phenylalanine by using the exon knock-in replacement strategy shown in Fig. 6, which is published as supporting information on the PNAS web site.
Heterozygous mice of all three genotypes (Kit+/567F, Kit+/569F, and Kit+/FF) were indistinguishable from wild-type Kit+/+ littermates, except for the appearance of white spotting, which is described below. In contrast, KitFF/FF mice appeared emaciated, and their gain of weight was delayed relative to wild-type or heterozygous mice in the KitFF line. Most homozygous KitFF mice had died by 3 months of age, although the median life span of homozygous KitFF mice was around weaning (i.e., 3–4 weeks after birth). Approximately 20% of homozygous Kit569F mice (15 of 75) died within 12 h after birth, and an additional 10% of these Kit569F/569F mice died neonatally. The remaining Kit569F/569F mice survived for at least 1 year. Interestingly, 10% of heterozygous Kit569F mice (13 of 124) also died during the neonatal stage, although most Kit+/569F mice survived for at least 1 year. No lethality was associated with heterozygous or homozygous Kit567F mice or with Kit567F/Kit569F mice. In the F2 generation, heterozygous and homozygous mutant mice with either a single (Kit567F or Kit569F) or double (KitFF) tyrosine to phenylalanine mutation were born at the expected Mendelian frequencies.
Melanocyte and Mast Cell Defects in KitFF Mutant Mice. The first obvious phenotype of homozygous mutant KitFF mice was extensive white spotting (Fig. 1A and Table 1, which is published as supporting information on the PNAS web site) with >90% of the body surface area white except around the ear. The coat color within pigmented areas was completely normal agouti or black, depending on the agouti background inherited. The eyes were dark in all cases. White spots were observed in 10–15% of homozygous Kit567F and Kit569F mutant mice and in 2–5% of heterozygous Kit567F and Kit569F mutant mice (Table 1). Microscopic examination of skin sections from the mutant mice confirmed the absence of melanin pigment in the hair shafts and of melanocytes in the hair bulbs in the regions where the coat was white (data not shown).
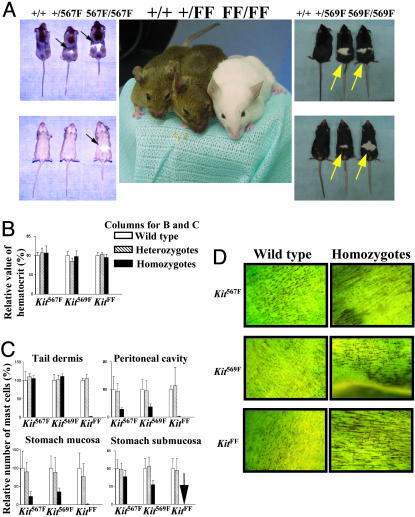
Fig. 1.
Examination of W phenotypes in Kit mutants. (A) White spotting on Kit567F, Kit569F, and KitFF mutant mice. Note KitFF homozygous mouse is completely white, although it has black eyes. Arrows indicate white spots. (B) The relative hematocrit values from heterozygotes and homozygotes of Kit567F, Kit569F, and KitFF mutants are expressed as a percentage of hematocrit value from wild type of the Kit mutants. (C) The relative numbers of mast cells present in each anatomical tissue from heterozygotes and homozygotes of Kit567F, Kit569F, and KitFF mutants are expressed as a percentage of the number of mast cells present in wild type of the Kit mutants. Arrow indicates that no toludine blue-stained cells were detected. (D) Supravital methylene blue staining of the adult small intestine. Homozygous small intestine from Kit567F, Kit569F, and KitFF mutants display the normal network of ICC. (Magnification: ×5.2.)
Heterozygous and homozygous Kit567F, Kit569F, and KitFF mutant mice were fully fertile (Table 2, which is published as supporting information on the PNAS web site) and revealed no differences in their hematocrit values and development of the network of ICC (Fig. 1 B and D). In contrast, loss of mast cells was detected in the tail dermis, the peritoneal cavity, and the stomach of KitFF mutants (Fig. 1C). Homozygous Kit567F and Kit569F mice also exhibited reduced number of mast cells in the peritoneal cavity and the stomach. Interestingly, we observed normal numbers of mast-cell precursors (CFU-mast) and it was possible to derive mast cells from the bone marrow of each Kit homozygous mutant (unpublished observations).
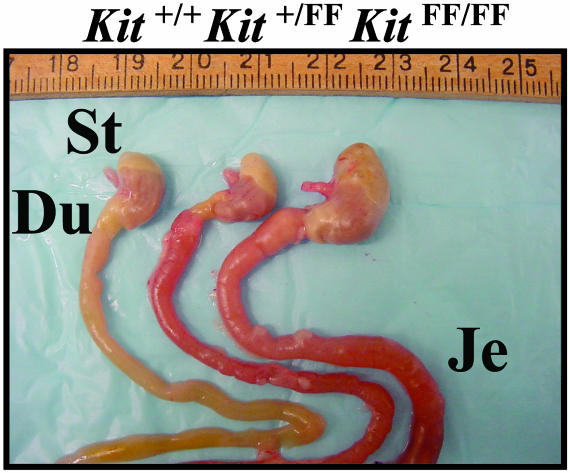
Mega-Stomach, -Duodenum, and -Jejunum in KitFF/FF Mice. Autopsy revealed gross distension of the stomach, the duodenum, and the jejunum in KitFF/FF mice (Fig. 2), although this phenotype was not observed in homozygous Kit567F, Kit569F, or W mutant mice (data not shown). Dissection of the entire gastrointestinal tract of homozygous KitFF mice revealed no narrow portion of the proximal stomach, duodenum, and jejunum, indicating that the distended gastrointestinal tract was not caused by an obstructing lesion.
Fig. 2.
Gross morphological abnormality of gastrointestinal tracts of KitFF/FF mice. Dissection of gastrointestinal tracts from wild-type (left), Kit+/FF (center), and KitFF/FF mice (right). Distension of the entire stomach (St), duodenum (Du), and jejunum (Je) is evident in the KitFF/FF mice.
We next examined the organization of the cells in the stomach and the intestine of KitFF/FF mice to determine the histological basis for this gross abnormality. Despite extensive histological analysis, we did not observe any significant defects of myenteric ganglion cells or of cellular organization of the stomach, intestine, or colon (data not shown).
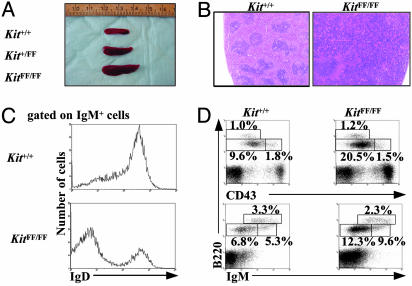
Splenomegaly and B Cell Dysregulation in Homozygous KitFF Mice. An unexpected phenotype observed in all of the KitFF/FF mice (but not homozygous Kit567F/567F, Kit569F/569F, or W mutant mice) was greatly enlarged spleens (splenomegaly; Fig. 3A and data not shown). To understand the cellular basis for this phenotype, we performed histological analysis of the spleens of wild-type and KitFF/FF mice. In contrast to the normal splenic architecture of wild-type mice (well defined lymphoid white and red pulps) (Fig. 3B and data not shown), the splenic architecture in KitFF/FF mice was grossly abnormal, with enlarged regions of white pulp and characteristic discrete lymphoid follicles. Closer microscopic examination of the spleens of KitFF/FF mice also revealed a marked increase in cells that were morphologically identical to megakaryocytes (data not shown).
Fig. 3.
Enlarged spleen and alterations of B lymphopoiesis and megakaryopoiesis in KitFF/FF mice. (A) Gross anatomy of spleens from Kit+/+, Kit+/FF, and KitFF/FF mice. (B) Hematoxylin and eosin staining of spleen sections from KitFF/FF mice showing enlarged white pulp. (Magnification: ×50.) (C) Reduced expression of IgD is detected on IgM+ B cells in the KitFF/FF spleens. (D) B cell precursors are accumulated in the bone marrow of KitFF/FF mice. Representative two-color fluorescence plots show expression of B220 and CD43 (Upper) and B220 and IgM (Lower) on bone marrow cells. Percentages represent the fraction of the total gated live cells within the indicated boxes.
Consistent with the enlarged spleens of KitFF/FF mice, total cellularity of their spleens was increased compared with that of controls (Table 3, which is published as supporting information on the PNAS web site). We next investigated the cellular basis for this splenomegaly by examining and characterizing splenic cell populations by their cell surface phenotypes. The spleens of KitFF/FF mice contained reduced proportions of CD4+ cells, whereas the absolute numbers of CD4+ and CD8+ cells were almost equivalent amongst Kit+/+, Kit+/FF, and KitFF/FF spleens. The increased cellularity of KitFF/FF spleens could be largely attributed to the marked increase in the numbers of B220+IgM+ B cells, consistent with the enlarged white pulps (Table 3). The B cell populations in these mice expressed reduced levels of IgD on their cell surface (Fig. 3C and Table 3). CD41+ megakaryocytes were also markedly increased in the spleens and the bone marrow of KitFF/FF mice, consistent with the histological observations on KitFF/FF spleens (Table 3 and data not shown). Together, these data suggest that the enlarged spleens of KitFF/FF mice are caused by abnormally high numbers of B cells and megakaryocytes.
The reduction of IgD expression levels on B cells suggests that the B cells in the spleens of KitFF/FF mice are blocked at an immature stage of lineage development, resulting in the accumulation of immature B cells. To investigate this possibility, we characterized expression levels of other cell surface markers on bone marrow cells from these mice. Total cellularity of KitFF/FF bone marrow was normal, as was the numbers of Gr-1+ granulocytes and Ter119+ erythrocytes (Table 3). Interestingly, KitFF/FF mice had significantly increased numbers of B220lowIgM- and B220lowCD43- pre-B cells in the bone marrow, whereas the absolute numbers of B220lowIgM+ immature B cells and B220lowCD43+ pro-B cells in KitFF/FF bone marrow were the same as on Kit+/+ and Kit+/FF mice (Fig. 3D and Table 4, which is published as supporting information on the PNAS web site). These data suggest that KitFF/FF mice have a defect in the differentiations of pre-B cells to immature B cells.
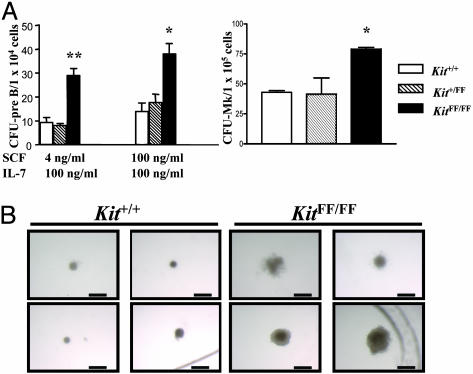
To determine whether the alterations in B lymphopoiesis and megakaryopoiesis arose at the progenitor cell level, bone marrow cells from Kit+/+, Kit+/FF, and KitFF/FF mice were cultured in semisolid media with various cytokines. Colony numbers of CFU-pre-B cells were dramatically increased in KitFF/FF bone marrow cells, compared with Kit+/+ bone marrow cells (Fig. 4A). Moreover, the lymphoid colonies derived from KitFF/FF bone marrow cells were larger (Fig. 4B). Similarly, the numbers of CFU-megakaryocytic from KitFF/FF bone marrow were increased 2-fold relative to that from wild-type bone marrow (Fig. 4A). There were no significant differences in the number of erythroid CFU, erythroid burst-forming units, or CFU-mast between controls and KitFF/FF bone marrow populations (data not shown). Bone marrow progenitor cells from these mice were not factor (SCF)-independent, nor did we observe any hypersensitivity to IL-7 of SCF (data not shown).
Fig. 4.
Hematopoietic progenitor activities in bone marrow cells. (A) In vitro colony assay was performed with bone marrow cells from Kit+/+, Kit+/FF, and KitFF/FF mice. The means and SEM of the number of colonies per 1 × 104 CFU-pre-B or 1 × 105 CFU-megakaryocytic (CFU-Mk) are shown from triplicate cultures. *, P < 0.05; **, P < 0.01. (B) Enlarged size of pre-B colonies formed from KitFF/FF bone marrow cells. Typical appearance of pre-B colonies in response to IL-7 (100 ng/ml) and SCF (100 ng/ml) in semisolid medium. (Scale bar: 500 μm.)
The splenomegaly and block in B cell development in KitFF/FF mice raised the possibility that these mutations might predispose to B lymphopoiesis. To date, we have not observed any lymphomas in these mice, although most animals were dead by 3 months, probably as the results of their enlarged stomachs.
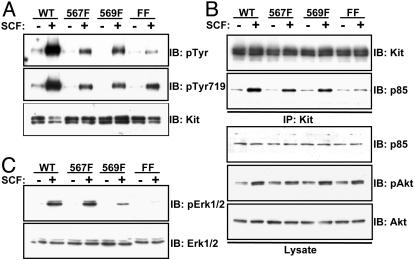
The Jx Tyrosine Mutations Cause Differences in Downstream Signaling. Substitution of tyrosines 567 and/or 569 with phenylalanine might be anticipated to interfere with Kit activation by removing the phosphoregulatory Jx switch, but could also modify downstream signaling by preventing the recruitment of SH2-containing targets. Given that mice bearing mutations in tyrosine residues 567 and/or 569 do not show a complete phenotypic overlap with Kit loss-of-function mutations (1–4), we reasoned that these mutations might indeed interfere with specific growth factor-induced signaling pathways. To investigate any potential differences in signaling by the mutated receptors, we examined their ability to be activated by SCF treatment and to stimulate intracellular signaling pathways. To first determine whether the mutant Kit receptors were autophosphorylated in response to Kit ligand stimulation, we expressed wild-type Kit and the mutated Kit receptors (567F, 569F, FF) in 293 cells and examined the phosphorylation status of each receptor after SCF stimulation. All mutant Kit receptors were autophosphorylated in response to SCF in 293 cells (Fig. 5A), indicating that substitution of the Jx tyrosines does not lead to a complete failure of kinase activation, as observed for Eph receptors (20, 21, 30). However, tyrosine phosphorylation of the 567F and 569F single mutant receptors was reduced compared with wild-type and phosphorylation of the FF double mutant receptor was even further decreased. These findings may be partially explained by the fact that these tyrosine residues are themselves autophosphorylation sites (23, 31). All of the mutant Kit receptors were autophosphorylated on tyrosine 719 in the kinase insert region after SCF stimulation, as judged by immunoblotting with phospho-specific antibodies (Fig. 5A). However, tyrosine 719 phosphorylation was reduced in the Jx mutants, as compared with wild types (Fig. 5A). Together, these results are consistent with a role for the Jx tyrosines in Kit activation but show that the mutant Kit receptors retain inducible kinase activity and the potential to activate downstream targets, including those mediated through tyrosine residue 719.
Fig. 5.
Jx tyrosine residues of Kit regulate MAPK but not Akt activation. (A) Autophosphorylation levels of mutant Kit receptors. HEK 293 cells were transiently transfected with Kit or Kit mutants and stimulated with SCF (100 ng/ml) where indicated. Cell extracts were immunoprecipitated with anti-Kit antibodies and subjected to immunoblotting with anti-pTyr (4G10) (Top) or anti-pTyr719 (Middle). (Bottom) Total Kit protein levels in transfected cell extracts. (B) Recruitment of p85 and activation of Akt by mutant Kit receptors. Cells were treated as described in A, and Kit immunoprecipitates were Western blotted by using anti-p85 antibodies to detect bound p85 (Upper). Total p85 protein levels in cell extracts are shown. Lysates from treated cells were also subjected to Western blot analysis by using an anti-phospho-Akt antibody. Total Akt levels are shown in Lower. (C) Activation of Erk signaling by mutant Kit receptors. Lysates from A were subjected to Western blot analysis by using an anti-phospho-Erk1/2 antibody (Top). The membrane was subsequently reprobed with anti-Erk1/2 antibodies to demonstrate the total Erk protein in each lane (Lower).
Because tyrosine residue 719 has been demonstrated to be the binding site for the p85 subunit of phosphatidylinositol 3′ kinase (6), we next examined whether the mutant Kit receptors could recruit p85 and stimulate activation of the downstream phosphatidylinositol 3′ kinase target Akt. As predicted, all mutant receptors were associated with p85 in response to SCF stimulation (Fig. 5B). The reduced binding of p85 to the FF double mutant observed here may be a consequence of decreased Src-mediated p85 recruitment to tyrosine residues 567 and 569 (32). Despite reduced association of p85 with the FF mutant, this receptor stimulated phosphorylation of Akt at levels comparable with those of the wild type and 567F and 569F single mutants (Fig. 5B). Collectively, these findings demonstrate that the mutant Kit receptors are functionally active and can induce signaling responses after growth-factor stimulation.
In an attempt to identify potential differences in downstream signaling pathways induced by the mutant Kit receptors in response to SCF stimulation, we analyzed the ability of the 567F, 569F, and FF receptors to activate the Erk-MAPK pathway. Interestingly, although 567F could stimulate phosphorylation of Erk1 and Erk2 similar to WT, 569F induced only a weak increase in Erk phosphorylation, and that induced by FF was barely detectable (Fig. 5C).
Discussion
Decades of elegant analysis of the large series of mutant alleles of the W and Sl loci have previously shown that this complementary gene pair is essential for hematopoiesis, melanogenesis, and gametogenesis (1). With the molecular cloning of both loci, it became possible to ask where these genes are normally expressed during development and in the adult (33). These expression studies suggested possible new roles for the Kit RTK in the central and peripheral nervous systems and in the gut region; indeed, functional analyses subsequently showed that the Kit pathway is involved in long-term spatial memory (34), peripheral nerve regeneration (35), and the development of the ICC and intestinal pacemaker activity (4, 36).
In this study, we have generated several alleles of the W/c-kit locus by a gene-targeting knock-in strategy. All of the naturally occurring W alleles are loss-of-function mutations that affect the levels of Kit tyrosine kinase activity as the result of either structural modifications in the kinase domain itself (23, 37) or regulatory modifications that down-regulate the levels of Kit RTK expression (37–39). Thus, the loss-of-function mutations in the W series of alleles presumably down-modulate all of the signaling pathways that normally emanate from the activated Kit RTK. In contrast, the three mutant alleles generated here lie outside the kinase domain itself in two highly conserved tyrosine residues located just below the plasma membrane (40). These mutations might therefore uncouple a specific pathway(s) activated by tyrosine phosphorylation of residues 567 and 569 from the other pathways regulated by the Kit RTK. In contrast to Eph receptors, such as EphA4, for which substitution of the Jx tyrosines with phenylalanine results in constitutive autoinhibition of the kinase domain and a kinase inactive phenotype in vivo (30), the Kit FF mutant receptor retains partial signaling activity. These data suggest that the precise molecular mechanisms by which the Kit and Eph receptor Jx regions regulate kinase activity are likely to be distinct.
Indeed, analysis of the phenotypes of mice homozygous for the KitFF mutation revealed a previously uncharacterized combination of affected cell lineages: melanocytes, mast cells, megakaryocytes, and B cells. Erythropoiesis, gametogenesis, and development of the ICC appeared to be normal in these mutant animals. The ability of the mutant receptors described here to become tyrosine phosphorylated on the p85 binding site of Kit, tyrosine 719, and subsequently activate Akt are consistent with the absence of fertility defects in Kit567F, Kit569F, and KitFF mutant mice compared with those observed in mice lacking tyrosine 719 (9, 10). Moreover, the residual autophosphorylation of the FF mutant receptor raises the possibility that the normal erythropoiesis in these mice may be due to the ability of the FF mutant receptor to recruit additional signaling molecules required for erythropoiesis.
It is interesting to compare the B cell phenotype described here with the phenotypes of mice lacking Shp-1 or Lnk, two SH2 domain-containing proteins known to modulate the Kit RTK by binding to the activated receptor. The SH2 domain-containing Shp-1 tyrosine phosphatase negatively regulates a number of signaling pathways controlled by tyrosine kinases, including those regulated by the Kit RTK. In vitro studies have shown that Shp-1 binds to and dephosphorylates the activated Kit RTK (41) through the interaction of Shp-1 with tyrosine 569 of Kit (42). The conclusion that Shp-1 is an important attenuator of Kit signaling is further supported by the finding that the hyperproliferation of multiple hematopoietic cell lineages in mice with mutations at the motheaten (me) locus are the result of loss-of-function mutations in Shp-1 (43, 44). Furthermore, analysis of mice carrying both the mild Wv mutation in combination with the me or mev mutations also revealed genetic interactions between these two loci, including amelioration of some of both the W and me phenotypes (45, 46).
Thus, the mutations we have generated here may be a partial phenocopy of the me mutation, delinking Kit RTK activation from its attenuation by Shp-1. However, enhanced numbers of mast cells in the skin and inflammatory lung disease, both of which are observed in me mice, were not detected in Kit569F mutant mice (data not shown). Furthermore, we only observed splenomegaly when both tyrosines 567 and 569 were mutated. Thus, disruption of the Kit/Shp-1 interaction cannot alone explain the results described here.
The SH2 domain-containing adaptor protein Lnk is another protein that interacts with the activated Kit RTK (27, 47). Mice lacking Lnk as the result of gene targeting in embryonic stem cells are viable, but display splenomegaly, enhanced B cell precursors, and hypersensitivity of hematopoietic progenitor cells to SCF, the Kit ligand (24, 27, 47). These observations, together with findings that Lnk is tyrosine-phosphorylated by Kit in a mast cell line (27, 47), suggest that Lnk may be a relevant Kit-activated attenuator of hematopoietic and lymphoid cell lineages.
W mutant mice do not demonstrate deficits in B cell development. However, several lines of evidence suggest an essential role of the Kit signaling pathway in lymphopoiesis. First, SCF, the Kit ligand, stimulates bone marrow-derived B cell progenitors from adult mice synergistically with IL-7 (48, 49). Second, the inability to detect lymphoid defects in adult W mice is likely caused by the early death of mice with null or severe loss-of-function W mutations, likely because of the severe macrocytic anemia (50). Thus, animals lacking Kit die before they manifest any lymphoid defect. However, W/W mice overexpressing erythropoietin survive to adulthood, and display defects in both B cell and T cell development (50). The data presented here confirm and extend these observations and demonstrate that the Jx domain of Kit plays a key role in regulating Kit activity in B lymphoid progenitor cells. Our studies have also implicated the Jx domain of Kit in regulating megakaryocyte development, a cell lineage known also to be affected by mutations at the W locus (51).
Activating mutations in the Jx domain of the Kit RTK are found in mastocytosis and GISTs in human (13, 17). GISTs are now thought to derive from the ICC (13). Recently, Sommer et al. (52) introduced a targeted deletion of valine 558 in the Jx domain into the mouse germ-line. The resulting heterozygous animals died of a GIST-like disease, associated with elevated mast cell numbers. The enlarged stomachs, duodenum, and jejunum in KitFF/FF mice raise the intriguing possibility that these mutations might also predispose to either obstructive or paralytic ileus caused by a GIST-like malignancy or a defect in electrical activity associated with pacemaker function, respectively. However, no malignancies were observed in the gastrointestinal tracts of KitFF/FF mice, and we were unable to observe any cellular abnormalities in myenteric ganglion cells or cellular organization in ICC, the stomach, intestine, or colon of KitFF/FF mice.
Mutation of both Jx tyrosine residues in Kit leads to extensive white spotting and reduced numbers of mast cells in the periphery of mutant mice. Interestingly, these phenotypes are also observed in mutant mice lacking only one of the two tyrosine residues, although the defects are less severe. These differences in phenotypic severity raise the intriguing possibility that the Jx tyrosines may cooperate to activate a common downstream signal transduction pathway to allow migration and/or proliferation of melanocytes and mast cells. Accordingly, we have found that the FF mutant receptor is significantly compromised in its ability to stimulate phosphorylation of Erk-MAPK, and this effect is also seen to some extent with the 569F mutant receptor. Consistent with these findings, Hong et al. (32) have recently demonstrated that tyrosine residues 567 and 569, in the absence of all other tyrosine residues on Kit, are sufficient to activate MAPK signaling. Overlapping defects in Kit ligand-induced Erk phosphorylation and mast cell development are also observed in mice lacking either the Gab2 adapter protein (29) or p85 (28). Moreover, mice bearing a mutant Kit receptor unable to recruit p85 directly also display reduced numbers of peritoneal mast cells (10). The ability of the Jx domain tyrosine residues to indirectly recruit p85 through association with Src family kinases at these sites (32) suggests that assembly of a signaling complex of Src, p85, and Gab2 on the activated Kit receptor may be required to stimulate Erk activation and subsequent development of specific cell lineages, such as mast cells.
In summary, by using a gene targeting approach, we have derived a series of W/c-kit mutations within the highly conserved Jx domain. Analysis of the resulting homozygous mutant mice has revealed a previously uncharacterized combination of phenotypes, adding to the already rich array of cell lineages and developmental processes controlled by the Kit/SCF signaling pathway. Clearly, further studies of these and other mutant alleles of Kit will provide an ideal experimental system in which to understand the complex biological events and molecular processes that are regulated by a single cell surface receptor.
Supplementary Material
Acknowledgments
We thank A. Nagy for providing pCAGGS-NLS-Cre, S. Tondat for embryonic stem cell aggregation, K. So for histological sections, S. Zhao for flow cytometric analysis, R. F. Paulson for helpful discussions, and the staff and the faculty of the Samuel Lunenfeld Research Institute for their dedicated support. This work was supported by grants from the National Cancer Institute of Canada (to A.B and T.P.), the Ontario Research and Development Challenge Fund Fellowship (to Y.K.), the National Cancer Institute of Canada/Terry Fox Run Research Fellowship (to N.J.), the Canadian Institutes of Health Research Fellowship (to M.K.), the Canadian Institutes of Health Research Scholarship (to J.B.C.), and the Canadian Institutes of Health Research Fellowship and Uehara Memorial Foundation Fellowship (to K.T.) J.L.W. is an International Investigator of the Howard Hughes Medical Institute and an Investigator of the Canadian Institutes of Health Research. T.P. is a Distinguished Scientist of the Canadian Institutes of Health Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: W, white spotting; ICC, interstitial cells of Cajal; Jx, juxtamembrane; pTyr, phosphotyrosine; MAPK, mitogen-activated protein kinase; Erk, extracellular signal-regulated kinase; RTK, receptor tyrosine kinase; SCF, stem cell factor; Sl, steel; SH2, Src homology-2; GIST, gastrointestinal stromal tumor; me, motheaten; CFU, colony-forming unit.
References
- 1.Russell, E. S. (1979) Adv. Genet. 20, 357-459. [PubMed] [Google Scholar]
- 2.Bernstein, A., Forrester, L., Reith, A. D., Dubreuil, P. & Rottapel, R. (1991) Semin. Hematol. 28, 138-142. [PubMed] [Google Scholar]
- 3.Besmer, P. (1991) Curr. Opin. Cell Biol. 3, 939-946. [DOI] [PubMed] [Google Scholar]
- 4.Huizinga, J. D., Thuneberg, L., Klüppel, M., Malysz, J., Mikkelsen, H. B. & Bernstein, A. (1995) Nature 373, 347-349. [DOI] [PubMed] [Google Scholar]
- 5.Ullrich, A. & Schlessinger, J. (1990) Cell 61, 203-212. [DOI] [PubMed] [Google Scholar]
- 6.Scheijen, B. & Griffin, J. D. (2002) Oncogene 21, 3314-3333. [DOI] [PubMed] [Google Scholar]
- 7.Pawson, T. & Nash, P. (2003) Science 300, 445-452. [DOI] [PubMed] [Google Scholar]
- 8.Pawson, T. & Scott, J. D. (1997) Science 278, 2075-2080. [DOI] [PubMed] [Google Scholar]
- 9.Blume-Jensen, P., Jiang, G., Hyman, R., Lee, K. F., O'Gorman, S. & Hunter, T. (2000) Nat. Genet. 24, 157-162. [DOI] [PubMed] [Google Scholar]
- 10.Kissel, H., Timokhina, I., Hardy, M. P., Rothschild, G., Tajima, Y., Soares, V., Angeles, M., Whitlow, S. R., Manova, K. & Besmer, P. (2000) EMBO J. 19, 1312-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maina, F., Casagranda, F., Audero, E., Simeone, A., Comoglio, P. M., Klein, R. & Ponzetto, C. (1996) Cell 87, 531-542. [DOI] [PubMed] [Google Scholar]
- 12.Partanen, J., Schwartz, L. & Rossant, J. (1998) Genes Dev. 12, 2332-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirota, S., Isozaki, K., Moriyama, Y., Hashimoto, K., Nishida, T., Ishiguro, S., Kawano, K., Hanada, M., Kurata, A., Takeda, M., et al. (1998) Science 279, 577-580. [DOI] [PubMed] [Google Scholar]
- 14.Myles, G. M., Brandt, C. S., Carlberg, K. & Rohrschneider, L. R. (1994) Mol. Cell. Biol. 14, 4843-4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokota, S., Kiyoi, H., Nakao, M., Iwai, T., Misawa, S., Okuda, T., Sonoda, Y., Abe, T., Kahsima, K., Matsuo, Y. & Naoe, T. (1997) Leukemia 11, 1605-1609. [DOI] [PubMed] [Google Scholar]
- 16.Furitsu, T., Tsujimura, T., Tono, T., Ikeda, H., Kitayama, H., Koshimizu, U., Sugahara, H., Butterfield, J. H., Ashman, L. K., Kanayama, Y., et al. (1993) J. Clin. Invest. 92, 1736-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsujimura, T., Morimoto, M., Hashimoto, K., Moriyama, Y., Kitayama, H., Matsuzawa, Y., Kitamura, Y. & Kanakura, Y. (1996) Blood 87, 273-283. [PubMed] [Google Scholar]
- 18.Nakahara, M., Isozaki, K., Hirota, S., Miyagawa, J., Hase-Sawada, N., Taniguchi, M., Nishida, T., Kanayama, S., Kitamura, Y., Shinomura, Y. & Matsuzawa, Y. (1998) Gastroenterology 115, 1090-1095. [DOI] [PubMed] [Google Scholar]
- 19.Andersson, J., Sjogren, H., Meis-Kindblom, J. M., Stenman, G., Aman, P. & Kindblom, L. G. (2002) Am. J. Pathol. 160, 15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binns, K. L., Taylor, P. P., Sicheri, F., Pawson, T. & Holland, S. J. (2000) Mol. Cell. Biol. 20, 4791-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wybenga-Groot, L. E., Baskin, B., Ong, S. H., Tong, J., Pawson, T. & Sicheri, F. (2001) Cell 106, 745-757. [DOI] [PubMed] [Google Scholar]
- 22.Chan, P. M., Ilangumaran, S., La Rose, J., Chakrabartty, A. & Rottapel, R. (2003) Mol. Cell. Biol. 23, 3067-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mol, C. D., Lim, K. B., Sridhar, V., Zou, H., Chien, E. Y., Sang, B. C., Nowakowski, J., Kassel, D. B., Cronin, C. N. & McRee, D. E. (2003) J. Biol. Chem. 278, 31461-31464. [DOI] [PubMed] [Google Scholar]
- 24.Takaki, S., Sauer, K., Iritani, B. M., Chien, S., Ebihara, Y., Tsuji, K., Takatsu, K. & Perlmutter, R. M. (2000) Immunity 13, 599-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieran, M. W., Perkins, A. C., Orkin, S. H. & Zon, L. I. (1996) Proc. Natl. Acad. Sci. USA 93, 9126-9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura, Y., Hart, A., Hirashima, M., Wang, C., Holmyard, D., Pittman, J., Pang, X. L., Jackson, C. W. & Bernstein, A. (2002) J. Exp. Med. 195, 941-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Velazquez, L., Cheng, A. M., Fleming, H. E., Furlonger, C., Vesely, S., Bernstein, A., Paige, C. J. & Pawson, T. (2002) J. Exp. Med. 195, 1599-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukao, T., Yamada, T., Tanabe, M., Terauchi, Y., Ota, T., Takayama, T., Asano, T., Takeuchi, T., Kadowaki, T., Hata Ji, J. & Koyasu, S. (2002) Nat. Immun. 3, 295-304. [DOI] [PubMed] [Google Scholar]
- 29.Nishida, K., Wang, L., Morii, E., Park, S. J., Narimatsu, M., Itoh, S., Yamasaki, S., Fujishima, M., Ishihara, K., Hibi, M., et al. (2002) Blood 99, 1866-1869. [DOI] [PubMed] [Google Scholar]
- 30.Kullander, K., Mather, N. K., Diella, F., Dottori, M., Boyd, A. W. & Klein, R. (2001) Neuron 29, 73-84. [DOI] [PubMed] [Google Scholar]
- 31.Lennartsson, J., Blume-Jensen, P., Hermanson, M., Pontén, E., Carlberg, M. & Rönnstrand, L. (1999) Oncogene 18, 5546-5553. [DOI] [PubMed] [Google Scholar]
- 32.Hong, L., Munugalavadla, V. & Kapur, R. (2004) Mol. Cell. Biol. 24, 1401-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Motro, B., van der Kooy, D., Rossant, J., Reith, A. & Bernstein, A. (1991) Development (Cambridge, U.K.) 113, 1207-1221. [DOI] [PubMed] [Google Scholar]
- 34.Motro, B., Wojtowicz, J. M., Bernstein, A. & van der Kooy, D. (1996) Proc. Natl. Acad. Sci. USA 93, 1808-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lourenssen, S., Motro, B., Bernstein, A. & Diamond, J. (2000) NeuroReport 11, 1159-1165. [DOI] [PubMed] [Google Scholar]
- 36.Klüppel, M., Huizinga, J. D., Malysz, J. & Bernstein, A. (1998) Dev. Dyn. 211, 60-71. [DOI] [PubMed] [Google Scholar]
- 37.Reith, A. D., Rottapel, R., Giddens, E., Brady, C., Forrester, L. & Bernstein, A. (1990) Genes Dev. 4, 390-400. [DOI] [PubMed] [Google Scholar]
- 38.Geissler, E. N., Ryan, M. A. & Housman, D. E. (1988) Cell 55, 185-192. [DOI] [PubMed] [Google Scholar]
- 39.Klüppel, M., Nagle, D. L., Bucan, M. & Bernstein, A. (1997) Development (Cambridge, U.K.) 124, 65-77. [DOI] [PubMed] [Google Scholar]
- 40.Mori, S., Ronnstrand, L., Yokote, K., Engstrom, A., Courtneidge, S. A., Claesson-Welsh, L. & Heldin, C. H. (1993) EMBO J. 12, 2257-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi, T. & Ihle, J. N. (1993) Mol. Cell. Biol. 13, 3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozlowski, M., Larose, L., Lee, F., Le, D. M., Rottapel, R. & Siminovitch, K. A. (1998) Mol. Cell. Biol. 18, 2089-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsui, H. W., Siminovitch, K. A., de Souza, L. & Tsui, F. W. (1993) Nat. Genet. 4, 124-129. [DOI] [PubMed] [Google Scholar]
- 44.Kozlowski, M., Mlinaric-Rascan, I., Feng, G. S., Shen, R., Pawson, T. & Siminovitch, K. A. (1993) J. Exp. Med. 178, 2157-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paulson, R. F., Vesely, S., Siminovitch, K. A. & Bernstein, A. (1996) Nat. Genet. 13, 309-315. [DOI] [PubMed] [Google Scholar]
- 46.Lorenz, U., Bergemann, A. D., Steinberg, H. N., Flanagan, J. G., Li, X., Galli, S. J. & Neel, B. G. (1996) J. Exp. Med. 184, 1111-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takaki, S., Morita, H., Tezuka, Y. & Takatsu, K. (2002) J. Exp. Med. 195, 151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin, F. H., Suggs, S. V., Langley, K. E., Lu, H. S., Ting, J., Okino, K. H., Morris, C. F., McNiece, I. K., Jacobsen, F. W., Mendiaz, E. A., et al. (1990) Cell 63, 203-211. [DOI] [PubMed] [Google Scholar]
- 49.McNiece, I. K., Langley, K. E. & Zsebo, K. M. (1991) J. Immunol. 146, 3785-3790. [PubMed] [Google Scholar]
- 50.Waskow, C., Paul, S., Haller, C., Gassmann, M. & Rodewald, H. (2002) Immunity 17, 277-288. [DOI] [PubMed] [Google Scholar]
- 51.Chervenick, P. A. & Boggs, D. R. (1969) J. Cell. Physiol. 73, 25-30. [DOI] [PubMed] [Google Scholar]
- 52.Sommer, G., Agosti, V., Ehlers, I., Rossi, F., Corbacioglu, S., Farkas, J., Moore, M., Manova, K., Antonescu, C. R. & Besmer, P. (2003) Proc. Natl. Acad. Sci. USA 100, 6706-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.