Abstract
Cellular HIV-1 reservoirs that persist despite antiretroviral treatment are incompletely defined. We show that during suppressive antiretroviral therapy, CD4+ T memory stem cells (TSCM) harbor high per-cell levels of HIV-1 DNA, and make increasing contributions to the total viral CD4+ T cell reservoir over time. Moreover, phylogenetic studies suggested long-term persistence of viral quasispecies in CD4+ TSCM cells. Thus, HIV-1 may exploit stem cell characteristics of cellular immune memory to promote long-term viral persistence.
Antiretroviral combination therapy effectively suppresses HIV-1 replication, but replication-competent virus can persist in memory CD4+ T cells despite treatment 1–2. The memory CD4+ T cell compartment includes central-memory (TCM), effector-memory (TEM) and terminally-differentiated (TTD) cells, which most likely evolve through a sequential developmental program with progressive commitment to more differentiated cell types3–4. The presence of a more immature memory T cell population with stem cell-like properties has previously been hypothesized based on experimental animal studies5–9, and recently, small proportions of T cells with stem cell characteristics have been discovered in humans10–11, mice12 and non-human primates13. These cells, termed “T memory stem cells” (TSCM), seem to represent the earliest developmental stage of memory T cells, and can differentiate into large numbers of effector T cells, while maintaining their own pool size through homeostatic self-renewal. We hypothesized that HIV-1 can use CD4+ TSCM cells as a preferred niche for promoting long-term viral persistence.
To test this concept, we initially investigated the susceptibility of CD4+ TSCM cells to HIV-1 infection. These experiments demonstrated that CD4+ TSCM cells, phenotypically defined as described in previous studies10,14 and in Supplementary Figure 1, were approximately as susceptible as CD4+ TCM cells to infection with an R5-tropic HIV-1 isolate (Fig. 1a), although their surface expression of CCR5 was slightly lower (Supplementary Fig. 2a/b). In addition, CD4+ TSCM cells were highly susceptible to infection with a VSV-G pseudotyped HIV-1 construct (Fig. 1a, Supplementary Fig. 3), despite comparatively low expression of T cell activation makers (Supplementary Fig. 4). We also observed that HIV-1 RNA was readily detectable in CD4+ TSCM cells from untreated HIV-1 patients (Supplementary Fig. 2c). CD4+ TSCM cells had low sensitivity to cytopathic effects associated with HIV-1 infection (Supplementary Fig. 2d), and expressed reduced levels of the cell-intrinsic HIV-1 restriction factors TRIM5α, APOBEC3G and SAMHD1 (Supplementary Fig. 2e). Together, these data indicate that CD4+ TSCM cells are permissive to HIV-1 infection, and can serve as physiologic target cells for HIV-1.
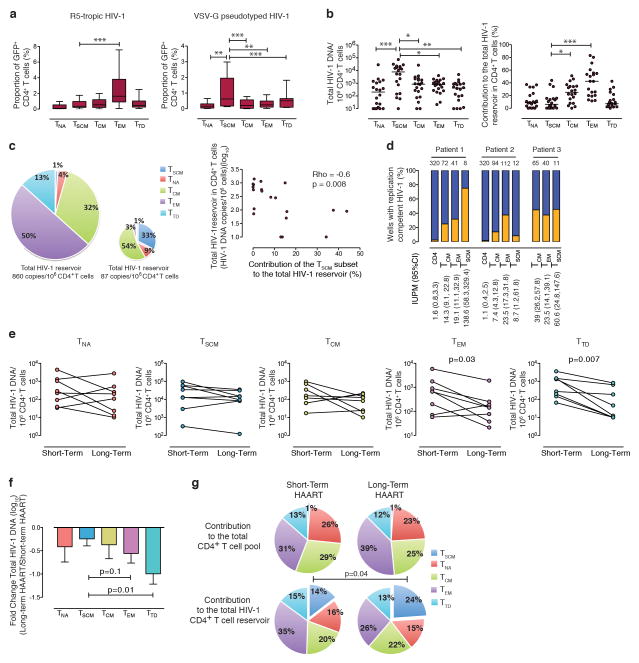
Figure 1. CD4+ TSCM cells represent a long-term reservoir for HIV-1 in HAART-treated patients.
(a): Proportion of HIV-1 infected cells after ex-vivo infection with GFP-encoding R5-tropic or VSV-G pseudotyped HIV-1 (R5: n=17, VSV-G: n=14). (b) Cell-associated HIV-1 DNA in sorted CD4+ T cell populations (left panel) and corresponding contributions to the total HIV-1 reservoir in CD4+ T cells from HAART-treated individuals (right panel). (c): Left panel: Representative pie charts reflecting the contribution of CD4+ TSCM cells to the total viral CD4+ T cell reservoir in two persons with large and small HIV-1 reservoirs in total CD4+ T cells, respectively. Right panel: Spearman correlation between contributions of CD4+ TSCM cells to the total HIV-1 CD4+ T cell reservoir, and corresponding size of the HIV-1 reservoir in total CD4+ T cells. (d): Reactivation of replication-competent HIV-1 from memory CD4+ T cell subsets. Orange bars reflect proportions of wells with detectable replication-competent HIV-1, blue bars indicate proportions of wells without detectable replication-competent HIV-1. Numbers above columns reflect total numbers of wells analyzed for each CD4+ T cell population; numbers below columns reflect estimated frequencies of cells with replication-competent HIV-1 per million cells (IUPM) based on limiting-dilution analysis. (e): Longitudinal evolution of HIV-1 DNA in CD4+ T cell subsets in n=8 study persons who initiated antiretroviral therapy in primary infection. (f): Pair-wise fold-differences in HIV-1 DNA measured after short-term and long-term antiretroviral therapy. Mean and standard error are shown. (g): Corresponding contribution of individual CD4+ T cell subsets to the total CD4+ T cell pool, and to the total HIV-1 CD4+ T cell HIV-1 reservoir after short-term and long-term antiretroviral therapy. Statistical significance was tested with Wilcoxon rank sum test. *,**, *** reflect p<0.05, p<0.01, p<0.001, respectively, after Bonferroni correction for multiple comparisons in panel a and b.
We next determined the levels of HIV-1 DNA in sorted CD4+ TSCM cells from HIV-1 infected patients, who had been treated with suppressive antiretroviral therapy for a median of 7 years (Supplementary Table). Proportions of CD4+ TSCM cells in these patients did not differ from an HIV-1 negative control cohort (Supplementary Fig. 5). In these HAART-treated patients, per-cell levels of HIV-1 DNA were highest in CD4+ TSCM cells, but their average contribution to the total viral CD4+ T cell reservoir was only approximately 8% (Fig. 1b). Interestingly, the contribution of CD4+ TSCM cells to the total viral reservoir in CD4+ T cells varied considerably among different HAART-treated patients, and was inversely associated with HIV-1 DNA levels in the entire CD4+ T cell compartment (Fig. 1c). Such a negative association was selectively observed for the CD4+ TSCM cell compartment (Supplementary Fig. 6) and resulted in a disproportionately increased contribution of CD4+ TSCM cells to the total viral reservoir in patients with a smaller viral reservoir in CD4+ TCM and TEM cells. This suggests that HIV-1-infected CD4+ TSCM cells represent one of the most stable and durable components of the viral CD4+ T cell reservoir that becomes increasingly visible when viral reservoirs in alternative CD4+ T cell subsets decline. HIV-1 DNA was also detectable in CD4+ TSCM cells from elite controllers15, although at significantly lower levels than in HAART-treated patients (Supplementary Fig. 7).
Since only a small proportion of CD4+ T cell-associated HIV-1 DNA encodes for replication-competent virus16, we performed viral outgrowth assays from three study subjects who had been on continuous suppressive antiretroviral combination therapy for a median of 28 months (range 14–42 months). These studies demonstrated that replication-competent virus was retrievable from CD4+ TSCM cells in all three cases, and that the estimated frequency of cells harboring replication-competent HIV-1 in CD4+ TSCM cells exceeded the corresponding frequencies in CD4+ TCM and TEM cells in two of the three patients (Fig. 1d). These findings indicate that HIV-1 DNA in CD4+ TSCM cells is functionally capable of resuming active viral gene expression.
Due to their stem cell-like properties, CD4+ TSCM cells may represent a privileged site for long-term viral persistence. To better investigate this, we longitudinally analyzed HIV-1 DNA in sorted CD4+ T cell subsets from eight individuals who started antiretroviral therapy in primary infection, and then remained on suppressive antiretroviral therapy without treatment interruptions. Using pair-wise comparisons between cell-associated HIV-1 DNA during earlier stages of antiretroviral therapy (median of 1 year, range: 10–14 months) and during later stages of treatment (median of nine years, range 7–11 years), we observed stable or mildly decreasing viral DNA in CD4+ TSCM cells; viral DNA decline in CD4+ TCM and CD4+ TNA cells was slightly more pronounced (Fig. 1e). In contrast, in the more short-lived CD4+ TEM and TTD cell populations, a significant reduction in per-cell levels of total HIV-1 DNA was noticed over time. Notably, among all CD4+ T cell subsets, the relative longitudinal decline in total HIV-1 DNA at per-cell levels was smallest in CD4+ TSCM cells, although differences between CD4+ TSCM cells and CD4+ TNA/TCM cells did not reach statistical significance in our small study cohort (Fig. 1f). Interestingly, we observed that CD4+ TSCM cells made a relatively small contribution to the total CD4+ T cell reservoir after the first year of suppressive antiretroviral therapy (Fig. 1g). Yet, after long-term antiretroviral treatment, there was a significant increase in the contribution of CD4+ TSCM cells to the total viral reservoir in CD4+ T cells, despite the fact that the numeric contribution of CD4+ TSCM cells to the total CD4+ T cell pool did not change. The contribution of CD4+ TCM cells to the total viral CD4+ T cell reservoir also slightly increased over time, but this did not reach the level of statistical significance. In contrast, the contribution of CD4+ TEM cells to the viral CD4+ T cell reservoir declined, despite a numerically increased proportion of TEM cells in the total CD4+ T cell pool (Fig. 1g). These data, although collected from a limited number of patients, suggest that CD4+ TSCM cells can support long-term viral persistence in patients treated with HAART.
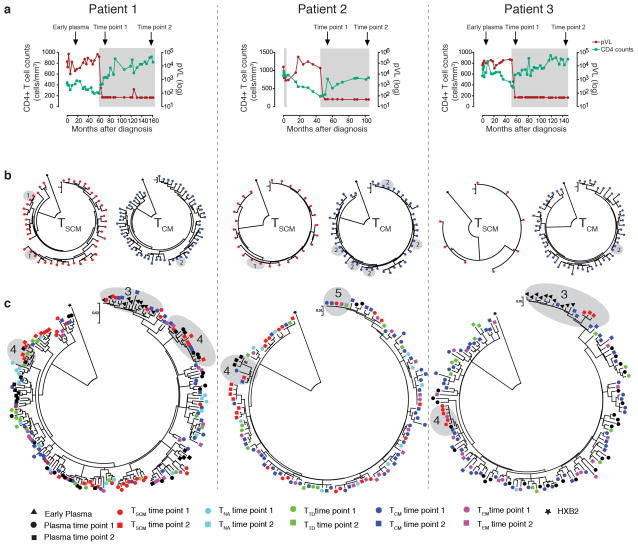
We subsequently performed proviral Env sequencing in DNA samples isolated from longitudinally sorted CD4+ T cell subsets of three HIV-1 patients who remained antiretroviral therapy-naïve during the initial years of disease, followed by continuous treatment with suppressive antiretroviral agents (Fig. 2a). We observed substantial variability between viral sequences from CD4+ TSCM cells collected at the beginning of antiretroviral therapy and again several years later, likely reflecting sampling of cells infected with different circulating viral strains during early disease stages (Fig. 2b). Yet, in CD4+ TSCM and CD4+ TCM cells (which were sampled in approximately 10–30-fold higher frequencies than CD4+ TSCM cells), we noticed several identical HIV-1 sequences in samples collected at the beginning of antiretroviral therapy and after 4–8 years of continuous treatment, consistent with long-term viral persistence in these CD4+ T cell subsets (Fig. 2b). Interestingly, we also observed that plasma sequences from early untreated disease stages were phylogenetically most closely related to HIV-1 DNA isolated from CD4+ TSCM and TCM cells collected 6–12 years later, suggesting that viral strains circulating in early infection can persist long-term upon infection of these memory CD4+ T cell subsets (Fig. 2c). In addition, pair-wise sequence comparisons revealed that the genetic distance between early HIV-1 RNA plasma sequences and HIV-1 DNA sequences from CD4+ T cell subsets collected during later stages of infection was lowest for CD4+ TSCM and CD4+ TCM cells (Supplementary Fig. 8). Sequences from CD4+ TSCM cells also showed frequent phylogenetic associations with contemporaneous and ensuing plasma sequences isolated during suppressive antiretroviral therapy, consistent with a possible interchange between viral strains in CD4+ TSCM cells and circulating viral species (Fig. 2c). Finally, we noted viral sequences from CD4+ TSCM cells that were identical to those from CD4+ TCM, TEM and TTD cells isolated several years later, supporting the role of CD4+ TSCM cells as precursor cells for more differentiated CD4+ T cell subsets (Fig. 2c). Although these phylogenetic studies were performed in a limited number of patients, they emphasize the role of CD4+ TSCM and TCM cells as a long-term reservoir for HIV-1.
Figure 2. Phylogenetic analysis of HIV-1 sequences isolated from CD4+ TSCM cells.
(a): Longitudinal evolution of CD4+ T cell counts and viral loads in the three study patients. Shaded areas reflect periods of antiretroviral treatment exposure. Arrows indicate time of CD4+ T cell and plasma sampling. (b): Phylogenetic analysis of HIV-1 sequences longitudinally amplified from sorted CD4+ TSCM and TCM cells at the beginning of antiretroviral treatment initiation, and after 4–8 years of continuous suppressive therapy in three study persons. Identical HIV-1 sequences in TSCM cells (1) and TCM cells (2) are highlighted by gray circles. (c): Circular phylogenetic trees of HIV-1 sequences amplified from indicated CD4+ T cell subsets and from plasma collected at indicated timepoints. Gray circles reflect: Phylogenetic relationships between HIV-1 DNA sequences from CD4+ TSCM cells and circulating HIV-1 viral RNA sequences isolated during early untreated disease (3) or during contemporaneous and ensuing timepoints (4). Identical HIV-1 sequences isolated from CD4+ TSCM, and from CD4+ TCM, TEM, TTD cells isolated at later timepoints, are also highlighted (5).
This study indicates that despite their small frequencies, CD4+ TSCM cells stand out among other memory CD4+ T cell subsets as the cell population in which long-term HIV-1 persistence is particularly evident, likely due to intrinsic cellular programs of these cells that maintain superior abilities to self-renew, resist apoptosis and survive for extremely long periods of time10,13. Interestingly, pharmaceutical inhibition of stem-cell specific molecular pathways is being investigated for targeting cancer stem cells17, and the specific targeting of cellular pathways responsible for stem cell-like properties of CD4+ TSCM cells may also have adjunct or additive effects on reducing persistence of HIV-1 infected CD4+ TSCM cells. Thus, optimism may be warranted that our increasing understanding of how stem cell-like properties of cellular immune memory maintain HIV-1 persistence despite HAART can be translated into improved clinical strategies for inducing HIV-1 eradication and cure18.
Patients and Methods
Patients
PBMC samples from HIV-infected individuals were used for this study according to protocols approved by the Institutional Review Board of Massachusetts General Hospital in Boston. All study participants gave written informed consent.
Cell sorting and flow cytometry
CD4+ TSCM cells and other CD4+ T cell subsets were isolated according to a previously described protocol14 with minor modifications. At least 100 Million PBMC were stained with monoclonal antibodies directed against CD4, CD3, CD45RA, CCR7, CD62L, CD122, CD95. After 20 minutes, CCR7+ CD45RA+ naïve CD4+ T cells, CCR7+ CD45RA− central-memory CD4+ T cells, CCR7− CD45RA− effector-memory CD4+ T cells, CCR7− CD45RA+ terminally-differentiated CD4+ T cells and CCR7+ CD45RA+ CD62L+ CD95+ CD122+ CD4+ T memory stem cells were live-sorted in a specifically designated biosafety cabinet (Baker Hood), using a FACS Aria cell sorter (BD Biosciences) at 70 pounds per square inch. Cell sorting was performed by the Ragon Institute Imaging Core Facility at Massachusetts General Hospital, and resulted in isolation of live lymphocytes with the defined phenotypic characteristics of >95% purity, as determined by three dedicated experiments in which sorted cells were subjected to repeat flow cytometric analysis (Supplementary Fig. 1b). For phenotypic characterization, cells were additionally stained with CCR5, CXCR4, CD38 or HLA-DR antibodies or Annexin V, and acquired on a LSRII flow cytometer (BD Biosciences). Data were analyzed using FlowJo software (Treestar).
Assessment of cell-associated HIV-1 DNA
Isolated CD4+ T cells were digested as previously described2 to extract cell lysates. We amplified total HIV-1 DNA with primers and probes previously described19. As a standard curve, we amplified serial dilutions of chronically infected 293T cells (kindly provided by Dr. Bushman, University of Pennsylvania). Proviral HIV-1 DNA copy numbers were calculated relative to CCR5 gene copy numbers according to standard procedures.
Analysis of cell-associated HIV-1 RNA
Cell-associated HIV-1 RNA in sorted CD4+ T cells was quantified by real-time RT-PCR, using primers and probes previously described20. HIV-1 RNA copy numbers were determined according to a standard HIV-1 RNA sample run in serial dilutions, and final results were expressed as the number of HIV-1 RNA copies per microgram of total RNA. The assay used had a detection threshold of 1 HIV-1 RNA copy/μg of total RNA.
Gene expression analysis
Expression of selected gene transcripts in individual CD4+ T cell subsets was analyzed by semiquantitative RT-PCR using Taqman gene expression assays with standardized primers/probes, and normalized to the expression of the housekeeping gene Actb (encoding β-actin) in each CD4+ T cell subset.
In vitro HIV-1 infection assays
Unselected PBMC from HIV-1 negative donors were cultured in RPMI medium supplemented with 10% FCS and 50 U/ml of rhIL-2. A total of 10×106 PBMCs were infected with a GFP-encoding VSV-G-pseudotyped virus (MOI=1, unless otherwise indicated) or a GFP-encoding R5-tropic viral strains (Ba-L, MOI=1, both isolates kindly provided by Dr. Littman, New York University). Cells were then washed twice with PBS and cultured at 200,000 cells/well in 96-well round-bottom plates for 5 days. On day 5, cells were stained with surface antibodies to identify individual CD4+ T cell subsets, washed and analyzed on a LSRII flow cytometer.
Analysis of HIV-1 replication products
HIV-1 reverse transcripts were amplified from cell lysates with primers hRU5-F2 and hRU5-R and probe hRU5-P (early RT products), or with primers GagF1 and GagR1 and probe P-HUS-103 (late RT products)21. Integrated HIV-1 DNA was detected using nested PCR with Alu-1/Alu-2 primers and HIV-1 LTR primer L-M667 for the first-round PCR and LTR primer AA55M, Lambda T primers, and MH603 probe for the second-round quantitative PCR, as described previously22. Serial dilutions of HIV-1 DNA from cell lysates of the HIV-1–infected cell line 293T (provided by F. Bushman, University of Pennsylvania, Philadelphia, PA, USA) were used for reference purposes. Proviral HIV-1 DNA copy numbers were calculated relative to the CCR5 gene previously quantified with the same standard curve. 2-LTR HIV-1 DNA was quantified as previously described23.
Viral outgrowth assays
Sorted CD4+ T cell populations were seeded at 10,000 cells/well (TSCM cells) or 20,000 cells/well (TCM and TEM cells) in round-bottom 96-well plates. Subsequently, cells were stimulated with PHA (2μg/ml), rh IL-2 (100 units/ml) and irradiated allogeneic PBMCs from HIV-negative healthy donors. CD8-depleted, PHA-stimulated PBMC from HIV-negative donors were added to each well on day 3 and again on day 7 and 14 of culture. Latently HIV-1 infected ACH-2 cells were run as positive control cells, and CD4+ cell-depleted PBMC samples from HAART-treated patients that were otherwise treated identically served as negative controls. The cultures were subjected to removal of 33% of the cell suspension every seven days and replenished with fresh rhIL-2-containing media. After 14–21 days, cell supernatant from each well was harvested and the number of wells containing infectious HIV-1 was assessed by incubation of the supernatant with TZM-bl cells, a permissive HeLa cell clone that contains HIV-1 Tat-responsive reporter genes for firefly luciferase under control of the HIV-1 LTR, permitting sensitive and accurate measurements of infection. Luciferase activity was quantified by luminescence and is directly proportional to the number of infectious virus particles present in the initial inoculum. Estimated frequencies of cells with replication-competent HIV-1 were calculated using limiting dilution analysis as described in24; all data were consistent with a single-hit Poisson distribution, as determined using a goodness-of-fit analysis based on a likelihood ratio test24.
Viral sequencing
Cell lysates from sorted T cell populations and plasma were used for HIV-1 envelope sequencing encompassing the V3 region. For plasma samples, 6 mL of plasma from each time point were ultracentrifuged at 170,000g for 30 min prior to proteinase K digestion and RNA isolation by acid guanidinium isothiocyanate. One-step RT-PCR reaction was then performed in triplicates using outer primers envA/LA1725. PCR products were used as a template to generate an amplicon by nested PCR with inner primers LA12 and LA1325. For V3 amplification from HIV-1 DNA in cell lysates, two-step nested PCR was performed with the same primer pairs. For amplification of HIV-1 RNA and DNA sequences, 2–4 separate reactions were conducted for each sample during first-round PCR; these PCR products were then pooled and used as templates for second-round PCR. Amplification products were inserted into TOPO cloning vectors, and used to transform competent bacteria. Individual bacterial colonies were amplified by overnight culture, and extracted DNA was ligated and directly sequenced by T7 or T3 primers on an ABI 3100 PRISM automated sequencer, without prior PCR-based amplification. Sequences were aligned with an HXB2 reference sequence using BioEdit V7.1.9. A neighbour-joining method, as implemented in MEGA426, was used to construct phylogenetic trees with phylogenetically informative HIV-1 nucleotide sequences. These sequences omit nucleotide mutations that occur only once and may therefore possibly be introduced by polymerase-induced errors during PCR27. Phylogenetically informative sites were identified as described before (http://indra.mullins.microbiol.washington.edu/DIVEIN/insites.html). This conservative approach may slightly underestimate nucleotide diversity relative to single-template amplification methods, but a direct comparison between HIV-1 sequences derived by PCR/cloning and single-genome amplification in a number of our samples demonstrated equivalent population structure (Supplementary Fig. 9), consistent with prior studies28. For comparison purposes, viral sequences were analyzed by single genome amplification according to a protocol described before29.
Statistics
Data are summarized as individual data plots with horizontal lines reflecting the median, or as box and whisker plots indicating the median, interquartile range, and minimum and maximum values. Spearman’s correlation coefficient was calculated to analyze correlations. Differences were tested for statistical significance using Wilcoxon rank sum tests, Mann-Whitney U test, Kruskal-Wallis or Fisher’s exact test, followed by Bonferroni correction or Dunn’s test for multiple comparisons where applicable.
Supplementary Material
Acknowledgments
This work was supported by the American Foundation for AIDS Research (grant 108302-51-RGRL to ML) and by the National Institutes of Health (grants AI098487 and AI106468 to ML, AI089339 to XGY, AI098480 to TJH, AI100699 to JZL). ML is a recipient of the Clinical Scientist Development Award from the Doris Duke Charitable Foundation (grant number 2009034). MJB is supported by a fellowship award from the European Molecular Biology Laboratory and by the Tosteson postdoctoral fellowship award from Massachusetts General Hospital. Patient blood sample collection was supported by the National Institutes of Health (grant AI074415), by the Mark and Lisa Schwartz Foundation and the Bill and Melinda Gates Foundation.
Footnotes
Author contributions
Research idea, study design and concept: MJB, XGY, ML
Writing of manuscript: MJB, ML
Performance of experiments: MJB, HS, EMG, JL, JZL, TJH,
Data analysis and interpretation: MJB, XGY, ML
Contribution of PBMC samples: FP, BDW, ESR
Technical assistance: CL, AS, KS
Biostatistical assistance RZ, ZO
Critical review of manuscript: JZL, TJH, BDW, ESR, XGY
References
- 1.Finzi D, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 2.Chomont N, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 4.Lanzavecchia A, Sallusto F. Progressive differentiation and selection of the fittest in the immune response. Nat Rev Immunol. 2002;2:982–987. doi: 10.1038/nri959. [DOI] [PubMed] [Google Scholar]
- 5.Fearon DT, Manders P, Wagner SD. Arrested differentiation, the self-renewing memory lymphocyte, and vaccination. Science. 2001;293:248–250. doi: 10.1126/science.1062589. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Joe G, Hexner E, Zhu J, Emerson SG. Host-reactive CD8+ memory stem cells in graft-versus-host disease. Nat Med. 2005;11:1299–1305. doi: 10.1038/nm1326. [DOI] [PubMed] [Google Scholar]
- 7.Papatriantafyllou M. T cell memory: the stem of T cell memory. Nat Rev Immunol. 2011;11:716. doi: 10.1038/nri3098. [DOI] [PubMed] [Google Scholar]
- 8.Ahmed R, Bevan MJ, Reiner SL, Fearon DT. The precursors of memory: models and controversies. Nat Rev Immunol. 2009;9:662–668. doi: 10.1038/nri2619. [DOI] [PubMed] [Google Scholar]
- 9.Emerson SG. T memory stem cells: looking for stem cells in an immune haystack. Trans Am Clin Climatol Assoc. 2008;119:289–293. discussion 293–284. [PMC free article] [PubMed] [Google Scholar]
- 10.Gattinoni L, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cieri N, et al. IL-7 and IL-15 instruct the generation of human memory stem T cells from naive precursors. Blood. 2013;121:573–584. doi: 10.1182/blood-2012-05-431718. [DOI] [PubMed] [Google Scholar]
- 12.Gattinoni L, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med. 2009;15:808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lugli E, et al. Superior T memory stem cell persistence supports long-lived T cell memory. J Clin Invest. 2013 doi: 10.1172/JCI66327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lugli E, et al. Identification, isolation and in vitro expansion of human and nonhuman primate T stem cell memory cells. Nat Protoc. 2012;8:33–42. doi: 10.1038/nprot.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol. 2013;13:487–498. doi: 10.1038/nri3478. [DOI] [PubMed] [Google Scholar]
- 16.Ho YC, et al. Replication-Competent Noninduced Proviruses in the Latent Reservoir Increase Barrier to HIV-1 Cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi-Yanaga F, Kahn M. Targeting Wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res. 2010;16:3153–3162. doi: 10.1158/1078-0432.CCR-09-2943. [DOI] [PubMed] [Google Scholar]
- 18.Katlama C, et al. Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. Lancet. 2013 doi: 10.1016/S0140-6736(13)60104-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liszewski MK, Yu JJ, O’Doherty U. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods. 2009;47:254–260. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasternak AO, et al. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J Clin Microbiol. 2008;46:2206–2211. doi: 10.1128/JCM.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mbisa JL, Delviks-Frankenberry KA, Thomas JA, Gorelick RJ, Pathak VK. Real-time PCR analysis of HIV-1 replication post-entry events. Methods Mol Biol. 2009;485:55–72. doi: 10.1007/978-1-59745-170-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brussel A, Delelis O, Sonigo P. Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. Methods Mol Biol. 2005;304:139–154. doi: 10.1385/1-59259-907-9:139. [DOI] [PubMed] [Google Scholar]
- 23.Buzon MJ, et al. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat Med. 2010;16:460–465. doi: 10.1038/nm.2111. [DOI] [PubMed] [Google Scholar]
- 24.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Sharkey M, et al. Episomal viral cDNAs identify a reservoir that fuels viral rebound after treatment interruption and that contributes to treatment failure. PLoS Pathog. 2011;7:e1001303. doi: 10.1371/journal.ppat.1001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 27.Herbeck JT, et al. Demographic processes affect HIV-1 evolution in primary infection before the onset of selective processes. J Virol. 2011;85:7523–7534. doi: 10.1128/JVI.02697-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordan MR, et al. Comparison of standard PCR/cloning to single genome sequencing for analysis of HIV-1 populations. J Virol Methods. 2010;168:114–120. doi: 10.1016/j.jviromet.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josefsson L, et al. Hematopoietic precursor cells isolated from patients on long-term suppressive HIV therapy did not contain HIV-1 DNA. J Infect Dis. 2012;206:28–34. doi: 10.1093/infdis/jis301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.