Abstract
Telomerase is a ribonucleoprotein containing an essential telomerase RNA template and telomerase reverse transcriptase (TERT) that maintains telomeres. The dosage requirements for mammalian TERT in telomere length homeostasis are not known, but are of importance in cellular senescence, stem cell renewal, and cancer. Here, we characterize telomere maintenance and function upon successive breeding of mice deficient in mTert. These studies reveal a unique dosage requirement for telomere length maintenance by TERT; despite haploinsufficiency for the maintenance of long telomeres, mTert+/- mice retain minimal telomere DNA at all chromosome ends and do not exhibit the infertility typical of telomerase-deficient strains. Unlike the long (>50 kbp) average telomere lengths of wild-type laboratory mice, mTert+/- animals mice possess short telomere lengths similar to humans and wild-derived mice. Unexpectedly, mTert+/- mice are ersatz carriers for genetic instability, because their mating led to accelerated genetic instability and infertility in null progeny. Thus, limiting TERT levels play a key role in the maintenance of genome integrity, with important ramifications for the maintenance of short telomeres in human cancer and aging.
Keywords: telomerase reverse transcriptase, haploinsufficiency, genetic instability, telomere signal-free end
Telomerase contains two essential components: the telomerase reverse transcriptase (TERT), and the telomerase RNA, which provides the template for the reverse transcription of new telomere DNA by TERT (reviewed in ref. 1). Loss of function of telomerase is unusual in that it leads to a latent phenotype in which the phenotypic consequences of telomere attrition become evident only after telomere DNA has sufficiently eroded. Loss of either the telomerase RNA or TERT in unicellular organisms that constitutively express telomerase activity, such as fungi and protozoa, leads to an ever-shorter telomere phenotype that ultimately induces a growth arrest termed senescence (reviewed in ref. 1).
Some multicellular model organisms such as mice contain telomerase activity in most tissues (2–5). Other species, such as humans and plants, retain telomerase activity in undifferentiated germ-line or highly proliferating cells (reviewed in refs. 6 and 7). Loss of TERT in plants, or the telomerase RNA in mice, leads to heritable and progressive telomere shortening that results in loss of detectable telomere DNA, defects in cellular proliferation, and an increase in genetic instability and infertility (8–15). In some instances, shortened telomeres in telomerase RNA-deficient mice leads to an increase in tumor susceptibility, whereas in other circumstances tumor formation is reduced, illustrating the complex interrelationship between factors that affect genome integrity in the progression to cancer (16–19).
Genetic experiments in mice lacking the telomerase RNA established that the onset of genetic instability and infertility correlates with the presence of critically shortened chromosome ends (i.e., those that lack a detectable telomeric DNA signal), rather than short average telomere lengths per se (20, 21). Although the precise molecular definition of a critically short telomere remains elusive, senescent human primary fibroblasts and uncapped telomeres (through loss of the telomere binding factor TRF2) exhibit enrichment of several proteins associated with double-strand DNA breaks at the telomere (22–24). In budding yeast, shortened telomeres also mimic a DNA-damage response (25, 26). Loss of telomerase or telomere-end protection appears to alter the end-structure (perhaps by perturbing the telomere 3′ G-strand overhang or t loop), rendering telomeres more susceptible to nucleolytic attack and end-to-end fusion (reviewed in refs. 27–29). Use of the term “critically short” or “dysfunctional” is thus intended to convey that a telomere has lost some or all of these protective functions.
Although the suppressive effect of telomerase inhibition on human cancer cell growth has been well documented (reviewed in ref. 30), the physiological role of telomerase activity in human tissues is largely unknown. In human T lymphocytes and primary fibroblasts, telomeres shorten despite the presence of low levels of telomerase activity (31–33), and yet inhibition of telomerase drives these cells into premature senescence, suggesting that telomerase activity is important (33, 34). Recently, autosomal dominant forms of dyskeratosis congenita and aplastic anemia were identified that showed linkage disequilibrium with inactivating mutations in the telomerase RNA gene (35–39). X-linked mutations in human dyskerin also lead to destabilization of the telomerase RNA and telomere shortening (40). However, mice bearing a similar dyskerin mutation show slight telomere attrition only after four null generations, whereas dyskeratosis is observed as early as the first null generation (41). No human diseases have been reported in association with mutations in TERT, and the long-term consequences of partial or complete disruption of murine Tert are unknown. We show that mTert exhibits haploinsufficiency in adult mice, and we characterize a separation in dosage requirements for the maintenance of long versus short telomeres. The data suggest that mTert+/- mice will provide a model for probing the importance of low levels of telomerase activity in stem cell function and cancer.
Materials and Methods
Generation of mTert Null Mice and Mating Scheme. Production of mTert-deficient mice in a C57BL/6/129/SvJ (“mixed”) genetic background has been described (45). In brief, heterozygous mTert+/- founders were intercrossed to produce generation 1 (G1) mTert-/- animals (Fig. 1). Null progeny from separate mating events (to minimize inbreeding artifacts in successive generations; refs. 11 and 46) were mated to obtain G2 mTert-/- animals, etc., up to the eighth generation, at which point the mice became infertile (Fig. 1). Concurrently, mTert+/- founder animals were mated to wild-type C57BL/6 mice (The Jackson Laboratory) to obtain a more homogeneous genetic background, and the mTert+/- progeny were successively crossed to wild-type C57BL/6 mice for up to 10 generations (BC1–BC10) (Fig. 1). To obtain mTert null mice in a C57BL/6 background, BC6 mTert+/- mice were crossed to obtain mTert-/- mice (G1), which were bred to generate successive generations of mTert-/- progeny (up to G4). Animal genotype was determined by PCR amplification of genomic DNA using primers specific to either the wild-type locus or the disrupted allele and verified by Southern blotting as described (45). We confirmed that mTert mRNA levels were reduced by ≈50% in mTert+/- testes compared with wild-type animals, although we were unable to discern a 50% reduction of telomerase activity in mTert+/- versus mTert+/+ cell extracts (45, 47) (data not shown).
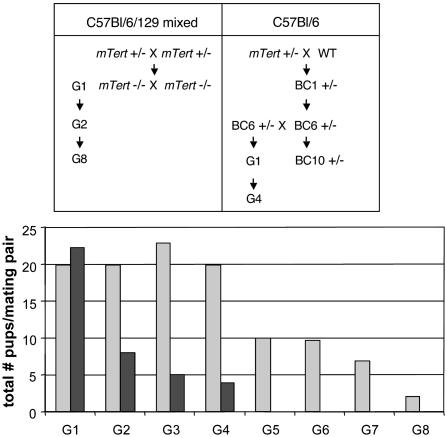
Fig. 1.
Mating scheme and litter sizes. (Upper) Schematic diagram of breeding strategy used to generate mTert-/- mice in a mixed (C57BL/6/129) or C57BL/6 genetic background. See Materials and Methods for details. (Lower) Total number of pups, divided by the number of mating pairs, produced in 6 months in mixed (light gray) and C57BL/6 (dark gray) mTert-/- mice. In the mixed background, the number of mating pairs analyzed were n = 2 (G1, G2, and G5), n = 3 (G3 and G4), n = 4 (G6), n = 6 (G7), and n = 9 (G8). In C57BL/6, the number of mating pairs analyzed were n = 10 (G1), n = 6 (G2), n = 11 (G3), and n = 7 (G4).
Telomere Length Measurements and Cytogenetic Analyses. Unless otherwise indicated, experiments were performed on activated splenocytes isolated from age-matched mice 8–12 weeks old. For the isolation of splenocytes, single-cell suspensions were plated onto dishes pretreated with 5 μg/ml anti-mouse CD3e (BD Biosciences), and cultured for 24 h in RPMI medium 1640 with l-glutamine (Invitrogen), 10% vol/vol FBS (Sigma), and 1% vol/vol 2-mercaptoethanol (Invitrogen). Cells were then grown for another 24 h in fresh media supplemented with 5 ng/ml IL-2 (BioSource), arrested in 0.1 μg/ml colcemid (Roche Diagnostics) for 2–6 h, harvested, and prepared for metaphase spreads as described (48).
Chromosomal abnormalities and fluorescent signal intensity of individual telomere ends were determined by using quantitative fluorescence in situ hybridization (Q-FISH) (48) with a telomeric Cy3-conjugated PNA probe (Applied Biosystems) on fixed cell preparations. Digital images were captured and processed as described (47), ensuring that the exposure times were in the linear or near-linear range for telomere fluorescence of samples containing a wild-type (C57BL/6) distribution of telomere lengths. Telomere fluorescence intensity was quantified by using tfl-telo software provided by P. Lansdorp (Terry Fox Laboratory, Vancouver) and expressed in arbitrary fluorescence units (11, 48, 49). The Q-FISH distribution profiles are not converted to values corresponding to kbp, because the use of plasmid standards containing <2.0 kbp of telomeric DNA negates accurate extrapolation to long average telomere lengths. Instead, in each Q-FISH experiment, at least one wild-type C57BL/6 animal (mean telomere length ≈50 kbp) was analyzed in parallel with the test samples, and comparisons are drawn to this control. Statistical analysis was performed by using a Student's t test to compare testes mass (two-tailed, nonpaired samples of unequal variance) and a χ2 test (1 degree of freedom) to compare the incidence of aneuploidy, telomere signal-free end (SFE), or end-to-end fusions between samples.
Results and Discussion
Reduced Fertility and Telomere Shortening in mTert-/- Mice in a Mixed C57BL/6/129 Background. We reported previously the isolation of founder animals heterozygous for mTert+/-, which were derived from chimeric C57BL/6 animals that contained mTert+/- 129/SvJ murine embryonic stem cells, thus generating a mixed C57BL/6/129 murine background (Fig. 1) (45). Founder mTert+/- animals were bred together to generate mTert-/- progeny, which exhibited a complete absence of mTert transcript and telomerase activity, and a progressive loss of telomere DNA after two generations (45). Another laboratory also independently generated mTert-/- mice, although telomere attrition was not reported (50). Here, we followed telomere lengths with continued breeding of mTert-/- mice in a mixed C57BL/6/129 genetic background, by using a cousin-mating strategy designed to minimize inbreeding artifacts in later generations (Fig. 1) (11, 46). Successive mating of mTert null mice in the mixed genetic background yielded normal numbers of offspring up to the fourth generation, after which a decrease in fertility became apparent (Fig. 1). Litter sizes decreased up to the eighth generation (G8), after which no offspring could be obtained.
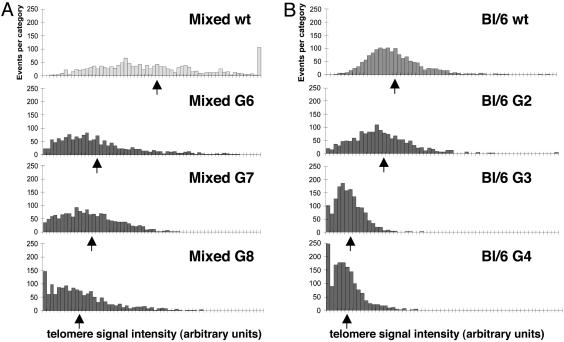
We then examined these later generation mTert-/- mice for similarities to mice lacking the telomerase RNA, including decreased testes mass, shortened telomeres, and an increase in telomere signal-free ends (10, 14). The average testes mass in mTert-/- G7 and G8 mice was significantly decreased compared to wild-type mice (Table 1) (Student's t test, P < 0.02). Using a quantitative measure of telomere length distribution, Q-FISH (see Materials and Methods), we also found a marked decline in telomere length in seventh and eighth generation mTert-/- mice compared with wild-type C57BL/6/129 mice (which exhibit a telomere length distribution intermediate between that of C57BL/6 and 129/SvJ strains; refs. 11 and 51–53) (Fig. 2A and Table 1). Finally, the incidence of telomere SFE rose with each increasing generation of mTert-/- mice, to statistically significant levels by G7 (5.5%, P < 0.05) and G8 (10.7%, P < 0.05) (Table 1). It should be noted that the lower limit of detection of telomeric signal by Q-FISH is not equivalent to zero telomeric DNA repeats (48). Although the incidence of SFE clearly correlated with the onset of infertility in mTert-/- mice at later generations, no statistically significant increase in end-to-end fusions and aneuploidy was observed, compared to wild-type control animals (Table 1). The failure to detect an increase in aneuploidy above background levels in activated splenocytes was also noted in mice deficient in the telomerase RNA; even at later generations, significant differences in genetic stability were apparent only in murine embryonic fibroblasts, or in activated splenocytes isolated from older animals (11, 13, 14). Thus, by several criteria, the terminal phenotype of mTert-/- mice is very similar to that of mice lacking the telomerase RNA. As described previously in Saccharomyces cerevisiae (54–56), we conclude that complete loss of an essential telomerase component, whether the telomerase RNA or TERT, yields a functionally equivalent phenotype in mammals.
Table 1. Summary of fertility and cytogenetic data in mTert+/+, mTert+/—, and mTert—/— mice.
| Generation | Average total offspring* (no. of mating pairs) | Testes mean ± SD,† mg | % SFE‡ (no. of mice) | End-to-end fusions per metaphase§ (no. of metaphases) | % Aneuploidy¶ |
|---|---|---|---|---|---|
| B6 wt | 26 (7) | 174 ± 22 (7) | 0.08 (4) | 0 (47) | 14.9 |
| B6 BC8+/— | 27 (2) | 178 ± 34 (3) | 0.21 (3) | 0.03 (37) | ND |
| B6 G1 | 22 (10) | ND | ND | ND | ND |
| B6 G2 | 8 (6) | 131 ± 52 (5) | 0.7 (2) | 0 (33) | ND |
| B6 G3 | 5 (11) | 151 ± 57 (7) | 4.0 (2) | 0 (36) | 39.0 |
| B6 G4 | 4 (7) | 97 ± 50 (6) | 15.4 (2) | 0 (42) | 29.0 |
| Mixed G6 | 9 (4) | 140, 189 (2) | 2.3 (2) | 0.18 (33) | ND |
| Mixed G7 | 7 (6) | 72 ± 10 (3) | 5.5 (3) | 0.09 (44) | 15.9 |
| Mixed G8 | 2 (9) | 76 ± 76 (7) | 10.7 (2) | 0 (34) | 53.0 |
B6, C57BL/6; mixed, C57BL/6/129; BC8+/—, mTert+/— backcrossed for eight generations; ND, not determined.
Animals aged 8—20 weeks were mated, the total number of pups produced during a 6-month period was recorded for each mating pair, and the average total number of pups for each generation was calculated
Expressed in milligrams. Number of mice analyzed is indicated in parentheses. n = 7 (B6 wt), 3 (BC8+/—), 5 (B6 G2), 7 (B6 G3), 6 (B6 G4), 3 (mixed G7), and 7 (mixed G8). Only two mixed G6 males were examined for testicular mass, therefore both values are listed
Expressed as percentage of all chromosome ends with no fluorescent signal. The number in parentheses is the number of mice measured per generation (400 chromosomes per mouse). Student's t tests revealed a statistically significant increase in SFE in B6 G3, B6 G4, mixed G7, and mixed G8 mice compared with wild-type control mice
Represents dicentric chromosomes only. Number in parentheses indicates the number of metaphases measured. Number of mice represented is the same as the % SFE column
Represents metaphases with greater or less than 40 chromosomes. Numbers of metaphases measured and mice represented is same as in the %SFE column
Fig. 2.
Telomere length analysis of mTert-/- mice in two different genetic backgrounds. (A) Q-FISH analysis from wild-type (wt), G6, G7, and G8 mTert-/- mice in a mixed C57BL/6/129 genetic background (Mixed). Each histogram represents 400 chromosomes from one mouse (at least two mice were analyzed from each generation, one representative mouse is shown). The arrows indicate the mean telomere length of the mouse represented. See Table 1 for quantification of cytogenetic data. (B) Q-FISH analysis from wild-type, G2, G3, and G4 mTert-/- mice in a C57BL/6 genetic background (Bl/6). Each histogram represents 400 chromosomes from one mouse (at least two mice were analyzed from each generation, one representative mouse is shown). The arrows indicate the mean telomere length of the mouse represented. See Table 1 for quantification of cytogenetic data. The x axes indicate telomere fluorescence signal intensity (in arbitrary units) for each chromosome end, with each tick mark representing 50 arbitrary units, and y axes indicate the number of ends (events) in each signal intensity category. Telomere SFEs are the events shown at “zero” arbitrary telomere fluorescence units on the x axis.
Accelerated Onset of Infertility of mTert-/- Mice in a C57BL/6 Pure Genetic Background. To confirm the terminal mTert-/- phenotype in a more homogeneous genetic background, we carried out successive crosses of mTert+/- founder animals with wild-type C57BL/6 mice for 10 generations (BC1–BC10) (Fig. 1). After six generations (BC6), mTert+/- mice were crossed together to obtain mTert null mice in a C57BL/6 background. These mice were also subjected to the same cousin-mating strategy as described above. Surprisingly, a dramatic reduction in litter size became apparent after the first mTert-/- generation in a C57BL/6 background (Fig. 1). By the fourth generation, few offspring were obtained and testes were significantly smaller (Table 1) (P < 0.02). In fact, G3 and G4 mTert-/- mice in a C57BL/6 background possessed an even greater degree of telomere shortening and increased incidence of SFE than G7 and G8 mTert-/- animals in a mixed C57BL/6/129 background (Fig. 2 and Table 1).
The acceleration of infertility and appearance of SFE in mTert-/- mice in a C57BL/6 background by at least four generations was not observed in C57BL/6 mice lacking the telomerase RNA (20). Hemann et al. (20) found that telomerase RNA-deficient C57BL/6 mice remained viable for the same number of generations as in a mixed C57BL/6/129 background (up to G6). In a separate laboratory, telomerase RNA-deficient C57BL/6 mice reached a terminal phenotype two generations earlier (G4), a difference that was attributed to shorter initial telomere lengths in the wild-type C57BL/6 animals (42). Thus, we found it difficult to justify the accelerated phenotype in mTert-/-BL/6 mice solely in terms of initial telomere length differences between the mixed and C57BL/6 genetic backgrounds. The contribution of genetic modifier effects unrelated to mTert also seemed unlikely, given the consistency of the phenotype in several independent crosses and the ability to rescue the terminal phenotype upon restoration of one functional allele of mTert (see below).
Mice Heterozygous for mTert Are Haploinsufficient for the Maintenance of Long Telomeres. The precocious sterility and onset of SFE in mTert null mice in the pure C57BL/6 background and our previous observation that mTert is haploinsufficient for the maintenance of long telomeres in murine embryonic stem (ES) cells (45, 47) prompted us to examine whether mice heterozygous for mTert may be similarly haploinsufficient. If this were the case, we predicted that telomere lengths in mTert+/- C57BL/6 animals would be shorter than wild-type C57BL/6 animals; hence, when mTert+/- mice were bred together, initial telomere lengths would be shorter still, thus contributing to an earlier onset of phenotypes related to critically shortened telomeres.
We performed Q-FISH analysis of mTert+/- animals and their +/+ littermates after successive crosses to C57BL/6 mice. Unfortunately, we were unable to obtain age-matched splenocytes for mTert+/- mice backcrossed to C57BL/6 mice for up to six generations. However, mTert+/- mice backcrossed for 7, 8, 9, and 10 generations (BC7–BC10) consistently possessed telomeres shorter than age-matched wild-type C57BL/6 mice and their mTert+/+ littermates (Fig. 3 and data not shown). In fact, mTert+/+ littermates also possessed shorter telomeres than wild-type C57BL/6 mice (Fig. 3, compare Top and Middle, and data not shown), an observation consistent with haploinsufficiency of mTert that cannot be completely corrected within one wild-type generation. These data demonstrate that telomere lengths in mTert+/- C57BL/6 mice have shortened beyond that of wild-type C57BL/6 mice. Thus, the initial telomere lengths in mTert-/- C57BL/6 mice are shorter than would be expected in the absence of mTert haploinsufficiency (Fig. 2 and data not shown). We conclude that the advanced onset of SFE and infertility in mTert-/- C57BL/6 animals is influenced by a shorter telomere length distribution in C57BL/6 mice compared with the mixed C57BL/6/129 genetic background, and even further telomere erosion that occurred during the successive mating of mTert+/- mice. In S. cerevisiae, there is already precedence for slight alterations in telomere length that have measurable effects upon long-term viability. For example, diploids heterozygous for more than one telomerase component show slightly shorter, but stable, telomere lengths compared with wild-type diploid yeast (55, 57). Resultant haploid progeny that are null for either telomerase component demonstrate a measurable acceleration in the terminal senescent phenotype by inheriting slightly shorter telomeres from their diploid parent (55, 57) (D. Edmonds, personal communication).
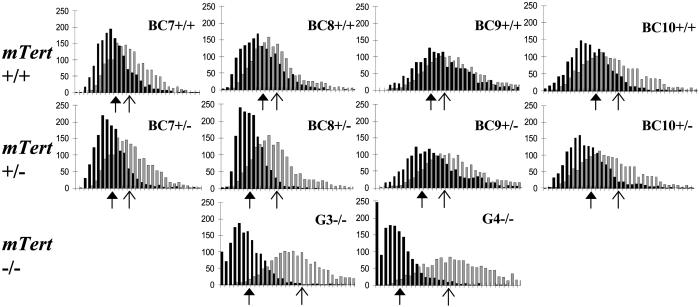
Fig. 3.
Telomere length analysis in backcrossed mTert+/- mice. (Top and Middle) Q-FISH analysis from 7th, 8th, 9th, and 10th generation mTert+/+ and mTert+/- littermates (black) from mTert+/- mice successively backcrossed to wild-type C57BL/6 mice (The Jackson Laboratory). In each case, a C57BL/6 mouse analyzed concurrently with each littermate pair is overlaid for comparison (gray). (Bottom) Q-FISH analysis from wildtype (gray) and G3 and G4 mTert-/- mice (black), reproduced from Fig. 2 for comparison. Note the absence of SFE in mTert+/- mice. For unknown reasons, all BC8+/- mice showed a slightly shorter average telomere length than BC9+/- and BC10+/- mice. Axes are labeled as in Fig. 2. The arrows indicate the mean telomere length of each mouse analyzed (solid arrows indicate mTert+/+, mTert+/- or mTert-/- mice; open arrows indicate wild-type C57BL/6 control mice).
Despite short average telomere lengths, mTert+/- mice remained fertile up to 10 generations (BC10), and did not exhibit testicular atrophy (Table 1 and data not shown). Comparison of mTert+/- and mTert-/- mice with similar average telomere lengths (e.g., BC9 mTert+/- mice and G3 mTert-/- mice) revealed an almost complete absence of SFE in mTert+/- animals (Fig. 3, Table 1, and data not shown). This observation parallels the selective maintenance of critically shortened telomeres that we observed in continually passaged mTert+/- ES cells (47). Although insufficient to maintain overall average telomere lengths, one functional allele of mTert appears nonetheless sufficient to maintain minimal telomere DNA at all chromosome ends. We estimate the average mean telomere length in later generation mTert+/- mice (BC8–10) to be <30 kbp (data not shown), which is roughly comparable to average telomere lengths in the human germ line (58) and wild-derived mouse strains (59).
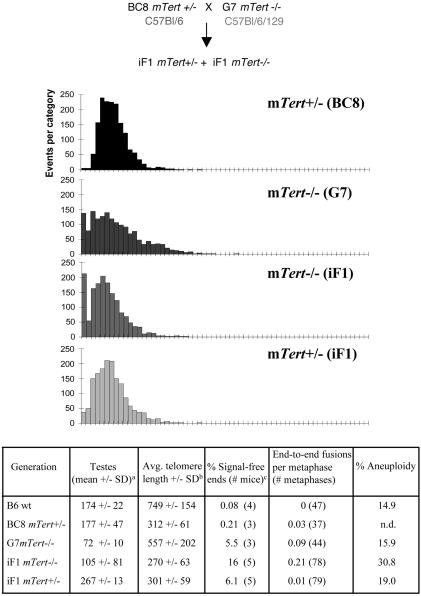
One Functional Allele of mTert Is Sufficient to Partially Rescue Telomere Instability. In mice lacking the telomerase RNA, the loss of detectable telomere DNA in later generations can be rescued by crosses that reintroduce one allele of the telomerase RNA (20, 21). Because later-generation mTert+/- mice appeared resilient to a complete loss of detectable telomeric DNA, we reasoned that one allele of mTert might be sufficient to rescue the telomere SFE observed in mTert-/- animals (Fig. 4). Later-generation mTert-/- G7 mice (with critically shortened telomeres) were mated to BC8 mTert+/- mice, and the phenotypes of the intergeneration F1 (iF1) littermates were compared. The iF1 mTert+/- progeny consistently possessed slightly longer telomeres compared to iF1 mTert-/- littermates (Fig. 4 and data not shown). In addition, iF1 mTert-/- littermates exhibited a decrease in testes mass (P = 0.07) and a statistically significant increase in end-to-end fusions and SFE (P < 0.01) compared to iF1 mTert+/- progeny. Perhaps the ability to discern a significant difference in end-to-end fusions in splenocytes derived from mTert+/- versus mTert-/- iF1 mice is facilitated by the ability to directly compare littermates. Similar results were also obtained in a separate mating analysis carried out in a C57BL/6 background (G3 mTert-/- mice mated to mTert+/- mice). These data suggest that limiting mTert is sufficient to maintain or rescue critically shortened telomeres, despite an inability to maintain the average telomere lengths typical for wild-type C57BL/6 mice.
Fig. 4.
Telomere length and cytogenetic analysis of mTert-/- and mTert+/- mice obtained from intergeneration crosses. (Top) Mating scheme for obtaining iF1 mTert-/- and mTert+/- mice (see text for details). (Middle) Q-FISH analysis of BC8 mTert+/- and G7 mTert-/- mice (the actual distributions shown are from littermates of the parents) and iF1 mTert-/- and iF1 mTert+/- littermates. Axes are labeled as in Fig. 2. (Bottom) Cytogenetic analysis of the mice shown above. See Table 1 for detailed explanation of each measurement. a, Expressed in milligrams. Data for B6 WT, BC8+/-, and G7-/- are reproduced from Table 1. The testes measured in iF1 animals (-/-, 3; +/-, 5) were taken from older, larger animals and are proportionate to total body mass.
It is unusual that one functional copy of mTert compromises some, but not all, aspects of its function in vivo. Unlike mTert+/- mice and ES cells, heterozygosity of the telomerase RNA does not lead to telomere shortening in ES cells (43) or a marked acceleration of the terminal phenotype in C57BL/6 mice (20, 21). Curiously, mice heterozygous for the telomerase RNA exhibit haploinsufficiency for lengthening of shorter telomeres in an interspecies cross of mouse strains with short (wild-derived) and long telomere lengths (laboratory inbred) (44). We have not determined whether heterozygosity of mTert is sufficient for lengthening of shorter telomeres in such an interspecies cross. However, the ability of mTert+/- mice to partially rescue critically shortened telomeres in a C57BL/6 background may portend an ability to lengthen the shortest telomeres when crossed with a wild-derived mouse. Another important distinction between dosage effects in mammalian telomerase is that, unlike young mTert+/- mice, humans harboring a mutation in the telomerase RNA do not remain asymptomatic; whether this disorder represents true haploinsufficiency remains to be verified (35, 36). Thus, additional experiments are required to determine whether direct parallels between the dosage requirements for the telomerase RNA and TERT can be drawn between humans and mice.
Two models that might explain the etiology of mTert haploinsufficiency can be considered. One model, termed the balance hypothesis (60), postulates that the dosage of a particular gene product evolved to be optimal in relation to other gene products, and that perturbation of this balance may have a deleterious effect on the remaining gene products (i.e., in some cases, the heterozygote possesses a more severe phenotype than the nullizygote) (60). A second hypothesis, articulated by Veitia (61), is that multicomponent processes have evolved a delicate equilibrium that may be dosage-sensitive to allow a more flexible response to external influences. This hypothesis implies that excess telomerase activity has been selected against; in fact, overexpression of murine TERT increases the incidence of skin neoplasias upon chemical treatment (62) and the incidence of breast carcinomas in aging mice (63). The fact that mTert+/- mice retain the ability to maintain critically short telomeres, but not longer ones, supports the notion of a delicate equilibrium in the accessibility of the telomere to telomerase. Short telomeres in humans and yeast have already been established as preferential substrates for telomerase extension, in part due to erosion of binding sites for inhibitory telomere-associated proteins, such as Rap1/hRAP1 and hTRF1 (22, 64–66). Thus, although half the dosage of mTert may be inadequate to overcome TRF1-mediated inhibition when telomeres are long, it may nonetheless be sufficient for telomere lengthening when telomeres are very short.
The mouse has proven to be an excellent model system for dissecting changes in genetic stability associated with haploinsufficiency. For example, in mice heterozygous for the Bloom's syndrome helicase (BLM) or p27, an increase in genetic instability is observed in heterozygous progeny without a concomitant loss of the remaining wild-type allele (67, 68). In other murine models of genetic instability, such as Msh2, Rb, and p18, the haploinsufficiency is subtle, and genetic instability and tumor susceptibility arise only upon close examination of mutation frequencies or challenge with carcinogens (69–72). In humans, other diseases associated with heritable changes in DNA have been found to exhibit genetic anticipation (i.e., earlier onset and increased penetrance with each generation) (73). Partial loss of telomerase function may predispose to an insidious haploinsufficiency that leads to genetic instability after a number of generations or upon environmental challenge (35–39), especially if telomerase activity is important for maintaining genome integrity in some tissues with increasing age (33, 34). The mTert+/- and mTert-/- mice thus provide a genetic model to compare two strains that both possess very short telomeres, and differ only in their ability to maintain critically short telomeres, for their predisposition to age-associated diseases and cancer. In addition, the mTert+/- mice might provide a more rapid means to assess the in vivo efficacy and/or potential toxicity of therapeutic inhibitors of telomerase.
Acknowledgments
We thank D. Durocher, C. Greider, G. Morin, P. Lansdorp, C. Price, M. Tyers, and members of the laboratory for fruitful discussion or critical comments on the manuscript. We also thank the participants of the Aspen Summer Physics Symposium (August 2003) for stimulating discussion on possible mechanisms of haploinsufficiency in eukaryotes. This work was supported by National Institutes of Health Grant AG16629-04 (to L.H.).
Abbreviations: TERT, telomerase reverse transcriptase; Q-FISH, quantitative fluorescence in situ hybridization; SFE, signal-free end; ES, embryonic stem; iF1, intergeneration F1.
References
- 1.Blackburn, E. H. (2001) Cell 106, 661-673. [DOI] [PubMed] [Google Scholar]
- 2.Venkatesan, R. N. & Price, C. (1998) Proc. Natl. Acad. Sci. USA 95, 14763-14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin-Rivera, L., Herrera, E., Albar, J. P. & Blasco, M. A. (1998) Proc. Natl. Acad. Sci. USA 95, 10471-10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prowse, K. R. & Greider, C. W. (1995) Proc. Natl. Acad. Sci. USA 92, 4818-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kishi, S., Uchiyama, J., Baughman, A. M., Goto, T., Lin, M. C. & Tsai, S. B. (2003) Exp. Gerontol. 38, 777-786. [DOI] [PubMed] [Google Scholar]
- 6.Greider, C. W. (1998) Proc. Natl. Acad. Sci. USA 95, 90-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riha, K. & Shippen, D. E. (2003) Chromosome Res. 11, 263-275. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald, M. S., Riha, K., Gao, F., Ren, S., McKnight, T. D. & Shippen, D. E. (1999) Proc. Natl. Acad. Sci. USA 96, 14813-14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riha, K., McKnight, T. D., Griffing, L. R. & Shippen, D. E. (2001) Science 291, 1797-1800. [DOI] [PubMed] [Google Scholar]
- 10.Hemann, M. T., Rudolph, K. L., Strong, M. A., DePinho, R. A., Chin, L. & Greider, C. W. (2001) Mol. Biol. Cell 12, 2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blasco, M. A., Lee, H. W., Hande, M. P., Samper, E., Lansdorp, P. M., DePinho, R. A. & Greider, C. W. (1997) Cell 91, 25-34. [DOI] [PubMed] [Google Scholar]
- 12.Chin, L., Artandi, S. E., Shen, Q., Tam, A., Lee, S. L., Gottlieb, G. J., Greider, C. W. & DePinho, R. A. (1999) Cell 97, 527-538. [DOI] [PubMed] [Google Scholar]
- 13.Rudolph, K. L., Chang, S., Lee, H. W., Blasco, M., Gottlieb, G. J., Greider, C. & DePinho, R. A. (1999) Cell 96, 701-712. [DOI] [PubMed] [Google Scholar]
- 14.Lee, H. W., Blasco, M. A., Gottlieb, G. J., Horner, J. W., Jr., Greider, C. W. & DePinho, R. A. (1998) Nature 392, 569-574. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg, R. A., Chin, L., Femino, A., Lee, K. H., Gottlieb, G. J., Singer, R. H., Greider, C. W. & DePinho, R. A. (1999) Cell 97, 515-525. [DOI] [PubMed] [Google Scholar]
- 16.Artandi, S. E., Chang, S., Lee, S. L., Alson, S., Gottlieb, G. J., Chin, L. & DePinho, R. A. (2000) Nature 406, 641-645. [DOI] [PubMed] [Google Scholar]
- 17.Wong, K. K., Chang, S., Weiler, S. R., Ganesan, S., Chaudhuri, J., Zhu, C., Artandi, S. E., Rudolph, K. L., Gottlieb, G. J., Chin, L., et al. (2000) Nat. Genet. 26, 85-88. [DOI] [PubMed] [Google Scholar]
- 18.Artandi, S. E. & DePinho, R. A. (2000) Curr. Opin. Genet. Dev. 10, 39-46. [DOI] [PubMed] [Google Scholar]
- 19.Farazi, P. A., Glickman, J., Jiang, S., Yu, A., Rudolph, K. L. & DePinho, R. A. (2003) Cancer Res. 63, 5021-5027. [PubMed] [Google Scholar]
- 20.Hemann, M. T., Strong, M. A., Hao, L. Y. & Greider, C. W. (2001) Cell 107, 67-77. [DOI] [PubMed] [Google Scholar]
- 21.Samper, E., Flores, J. M. & Blasco, M. A. (2001) EMBO Rep. 2, 800-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundblad, V. (2003) Nature 424, 926-927. [DOI] [PubMed] [Google Scholar]
- 23.Takai, H., Smogorzewska, A. & de Lange, T. (2003) Curr. Biol. 13, 1549-1556. [DOI] [PubMed] [Google Scholar]
- 24.d'Adda di Fagagna, F., Reaper, P. M., Clay-Farrace, L., Fiegler, H., Carr, P., von Zglinicki, T., Saretzki, G., Carter, N. P. & Jackson, S. P. (2003) Nature 425, 194-198. [DOI] [PubMed] [Google Scholar]
- 25.Nautiyal, S., DeRisi, J. L. & Blackburn, E. H. (2002) Proc. Natl. Acad. Sci. USA 99, 9316-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ijpma, A. S. & Greider, C. W. (2003) Mol. Biol. Cell 14, 987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertuch, A. A. & Lundblad, V. (2003) Genes Dev. 17, 2347-2350. [DOI] [PubMed] [Google Scholar]
- 28.Lydall, D. (2003) J. Cell Sci. 116, 4057-4065. [DOI] [PubMed] [Google Scholar]
- 29.Karlseder, J. (2003) Cancer Lett. 194, 189-197. [DOI] [PubMed] [Google Scholar]
- 30.Harrington, L. & Robinson, M. O. (2002) Oncogene 21, 592-597. [DOI] [PubMed] [Google Scholar]
- 31.Yui, J., Chiu, C. P. & Lansdorp, P. M. (1998) Blood 91, 3255-3262. [PubMed] [Google Scholar]
- 32.Vaziri, H., Dragowska, W., Allsopp, R. C., Thomas, T. E., Harley, C. B. & Lansdorp, P. M. (1994) Proc. Natl. Acad. Sci. USA 91, 9857-9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masutomi, K., Yu, E. Y., Khurts, S., Ben-Porath, I., Currier, J. L., Metz, G. B., Brooks, M. W., Kaneko, S., Murakami, S., DeCaprio, J. A., et al. (2003) Cell 114, 241-253. [DOI] [PubMed] [Google Scholar]
- 34.Roth, A., Yssel, H., Pene, J., Chavez, E. A., Schertzer, M., Lansdorp, P. M., Spits, H. & Luiten, R. M. (2003) Blood 102, 849-857. [DOI] [PubMed] [Google Scholar]
- 35.Vulliamy, T., Marrone, A., Goldman, F., Dearlove, A., Bessler, M., Mason, P. J. & Dokal, I. (2001) Nature 413, 432-435. [DOI] [PubMed] [Google Scholar]
- 36.Vulliamy, T. J., Knight, S. W., Mason, P. J. & Dokal, I. (2001) Blood Cells Mol. Dis. 27, 353-357. [DOI] [PubMed] [Google Scholar]
- 37.Vulliamy, T., Marrone, A., Dokal, I. & Mason, P. J. (2002) Lancet 359, 2168-2170. [DOI] [PubMed] [Google Scholar]
- 38.Comolli, L. R., Smirnov, I., Xu, L., Blackburn, E. H. & James, T. L. (2002) Proc. Natl. Acad. Sci. USA 99, 16998-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theimer, C. A., Finger, L. D., Trantirek, L. & Feigon, J. (2003) Proc. Natl. Acad. Sci. USA 100, 449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell, J. R., Wood, E. & Collins, K. (1999) Nature 402, 551-555. [DOI] [PubMed] [Google Scholar]
- 41.Ruggero, D., Grisendi, S., Piazza, F., Rego, E., Mari, F., Rao, P. H., Cordon-Cardo, C. & Pandolfi, P. P. (2003) Science 299, 259-262. [DOI] [PubMed] [Google Scholar]
- 42.Herrera, E., Samper, E., Martin-Caballero, J., Flores, J. M., Lee, H. W. & Blasco, M. A. (1999) EMBO J. 18, 2950-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niida, H., Shinkai, Y., Hande, M. P., Matsumoto, T., Takehara, S., Tachibana, M., Oshimura, M., Lansdorp, P. M. & Furuichi, Y. (2000) Mol. Cell Biol. 20, 4115-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hathcock, K. S., Hemann, M. T., Opperman, K. K., Strong, M. A., Greider, C. W. & Hodes, R. J. (2002) Proc. Natl. Acad. Sci. USA 99, 3591-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, Y., Snow, B. E., Hande, M. P., Yeung, D., Erdmann, N. J., Wakeham, A., Itie, A., Siderovski, D. P., Lansdorp, P. M., Robinson, M. O. & Harrington, L. (2000) Curr. Biol. 10, 1459-1462. [DOI] [PubMed] [Google Scholar]
- 46.Wright, S. (1921) Genetics 6, 111-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, Y., Kha, H., Ungrin, M., Robinson, M. O. & Harrington, L. (2002) Proc. Natl. Acad. Sci. USA 99, 3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zijlmans, J. M., Martens, U. M., Poon, S. S., Raap, A. K., Tanke, H. J., Ward, R. K. & Lansdorp, P. M. (1997) Proc. Natl. Acad. Sci. USA 94, 7423-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poon, S. S., Martens, U. M., Ward, R. K. & Lansdorp, P. M. (1999) Cytometry 36, 267-278. [DOI] [PubMed] [Google Scholar]
- 50.Yuan, X., Ishibashi, S., Hatakeyama, S., Saito, M., Nakayama, J., Nikaido, R., Haruyama, T., Watanabe, Y., Iwata, H., Iida, M., et al. (1999) Genes Cells 4, 563-572. [DOI] [PubMed] [Google Scholar]
- 51.Kipling, D. & Cooke, H. J. (1990) Nature 347, 400-402. [DOI] [PubMed] [Google Scholar]
- 52.Starling, J. A., Maule, J., Hastie, N. D. & Allshire, R. C. (1990) Nucleic Acids Res. 18, 6881-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hemann, M. T. & Greider, C. W. (1999) Nucleic Acids Res. 27, 3964-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singer, M. S. & Gottschling, D. E. (1994) Science 266, 404-409. [DOI] [PubMed] [Google Scholar]
- 55.Lendvay, T. S., Morris, D. K., Sah, J., Balasubramanian, B. & Lundblad, V. (1996) Genetics 144, 1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lingner, J., Hughes, T. R., Shevchenko, A., Mann, M., Lundblad, V. & Cech, T. R. (1997) Science 276, 561-567. [DOI] [PubMed] [Google Scholar]
- 57.Lingner, J., Cech, T. R., Hughes, T. R. & Lundblad, V. (1997) Proc. Natl. Acad. Sci. USA 94, 11190-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allsopp, R. C., Vaziri, H., Patterson, C., Goldstein, S., Younglai, E. V., Futcher, A. B., Greider, C. W. & Harley, C. B. (1992) Proc. Natl. Acad. Sci. USA 89, 10114-10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hemann, M. T. & Greider, C. W. (2000) Nucleic Acids Res. 28, 4474-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papp, B., Pal, C. & Hurst, L. D. (2003) Nature 424, 194-197. [DOI] [PubMed] [Google Scholar]
- 61.Veitia, R. A. (2003) J. Theor. Biol. 220, 19-25. [DOI] [PubMed] [Google Scholar]
- 62.Gonzalez-Suarez, E., Samper, E., Ramirez, A., Flores, J. M., Martin-Caballero, J., Jorcano, J. L. & Blasco, M. A. (2001) EMBO J. 20, 2619-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Artandi, S. E., Alson, S., Tietze, M. K., Sharpless, N. E., Ye, S., Greenberg, R. A., Castrillon, D. H., Horner, J. W., Weiler, S. R., Carrasco, R. D. & DePinho, R. A. (2002) Proc. Natl. Acad. Sci. USA 99, 8191-8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li, B. & de Lange, T. (2003) Mol. Biol. Cell. 14, 5060-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Lange, T. (1998) Cancer J. Sci. Am. 4, Suppl. 1, S22-S25. [PubMed] [Google Scholar]
- 66.Marcand, S., Wotton, D., Gilson, E. & Shore, D. (1997) Ciba Found. Symp. 211, 76-93; discussion 93-103. [DOI] [PubMed] [Google Scholar]
- 67.Goss, K. H., Risinger, M. A., Kordich, J. J., Sanz, M. M., Straughen, J. E., Slovek, L. E., Capobianco, A. J., German, J., Boivin, G. P. & Groden, J. (2002) Science 297, 2051-2053. [DOI] [PubMed] [Google Scholar]
- 68.Fero, M. L., Randel, E., Gurley, K. E., Roberts, J. M. & Kemp, C. J. (1998) Nature 396, 177-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bouffler, S. D., Hofland, N., Cox, R. & Fodde, R. (2000) Br. J. Cancer 83, 1291-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, S., Lloyd, R., Bowden, G., Glickman, B. W. & de Boer, J. G. (2002) Environ. Mol. Mutagen 40, 243-250. [DOI] [PubMed] [Google Scholar]
- 71.Bai, F., Pei, X. H., Godfrey, V. L. & Xiong, Y. (2003) Mol. Cell Biol. 23, 1269-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng, L., Flesken-Nikitin, A., Chen, P. L. & Lee, W. H. (2002) Cancer Res. 62, 2498-2502. [PubMed] [Google Scholar]
- 73.Lindblad, K. & Schalling, M. (1999) Semin. Neurol. 19, 289-299. [DOI] [PubMed] [Google Scholar]