Abstract
Archaea have been isolated from the human colon, vagina, and oral cavity, but have not been established as causes of human disease. In this study, we reveal a relationship between the severity of periodontal disease and the relative abundance of archaeal small subunit ribosomal RNA genes (SSU rDNA) in the subgingival crevice by using quantitative PCR. Furthermore, the relative abundance of archaeal small subunit rDNA decreased at treated sites in association with clinical improvement. Archaea were harbored by 36% of periodontitis patients and were restricted to subgingival sites with periodontal disease. The presence of archaeal cells at these sites was confirmed by fluorescent in situ hybridization. The archaeal community at diseased sites was dominated by a Methanobrevibacter oralis-like phylotype and a distinct Methanobrevibacter subpopulation related to archaea that inhabit the gut of numerous animals. We hypothesize that methanogens participate in syntrophic relationships in the subgingival crevice that promote colonization by secondary fermenters during periodontitis. Because they are potential alternative syntrophic partners, our finding of larger Treponema populations sites without archaea provides further support for this hypothesis.
Methanogenic Archaea have been isolated from the human oral cavity (1), as well as the human gut (2) and vagina (3). Despite their detection among the human microbiota, an association has not been demonstrated between severity of disease and the relative abundance of members of the domain Archaea. (4). The oral disease, chronic periodontitis, is a polymicrobial infection afflicting 35% of U.S. adults (5). The disease may result in loss of teeth and has been implicated in endocarditis, atherosclerosis, stroke, and preterm delivery of low-birth-weight infants (6). Chronic periodontitis has been linked to multiple members of the domain Bacteria (7); however, no one member explains the majority of cases, nor the role of microbes in this disease. At the same time, there is little information on the relationship between Archaea and chronic periodontitis. Although methanogenic Archaea have been found in the mouth of patients with periodontitis, previous studies lacked fundamental controls and were not quantitative, precluding the establishment of a significant or clinically relevant association between Archaea and disease (1, 8-14). The present study establishes correlations between the presence of disease and the presence of archaeal DNA, the severity of periodontal disease and the relative abundance of archaeal DNA in subgingival plaque, and between disease resolution and diminished archaeal DNA abundance. Archaea were found to be restricted to a subset of human subjects and to be comprised of two distinct rDNA phylotypes within the genus Methanobrevibacter. These data have important implications for the etiology of a disease that is exceedingly common, and for the role of methanogens in the human microbial ecosystem.
Materials and Methods
Subject Enrollment. Subjects were enrolled at the University of California, San Francisco (UCSF), School of Dentistry in the Ratcliff Center for Clinical Research (Division of Periodontology). The use of human subjects in this investigation was approved by the Stanford University Administrative Panel on Human Subjects in Medical Research and the UCSF Committee on Human Research. Subjects were at least 25 years old, were missing no more than 14 teeth, had a clinical diagnosis of generally healthy gingiva or chronic periodontitis, and were free of other oral soft tissue disease. Periodontal status of each subject was determined by measuring clinical attachment loss (CAL) to the nearest millimeter at the mesiobuccal, buccal, distobuccal, mesiolingual, lingual, and distolingual sites around each tooth. Mean full-mouth CAL values were used to place patients in the following categories: healthy (mean CAL <0.6 mm), slight periodontitis (0.6 mm ≤ mean CAL <1.6 mm), moderate periodontitis (1.6 mm ≤ mean CAL <2.5 mm), and severe periodontitis (mean CAL ≥2.5 mm; Table 1). Subjects were excluded if they were diabetic, HIV-positive, pregnant, lactating, or had taken antibiotics in the previous 3 months, because these factors have been implicated in altering oral bacterial composition. Subjects completed a survey regarding age, gender, race, and habits of oral hygiene.
Table 1. Vital statistics for enrolled human subjects.
| Healthy | Slight | Moderate | Severe | |
|---|---|---|---|---|
| Mean CAL, mm* | 0.19 ± 0.01 | 1.59 | 1.98 ± 0.02 | 3.83 ± 0.04 |
| No. of patients | 8 | 1 | 12 | 37 |
| White | 8 | 0 | 10 | 18 |
| Black | 0 | 1 | 1 | 10 |
| Hispanic | 0 | 0 | 0 | 7 |
| Asian | 0 | 0 | 1 | 2 |
| Male | 3 | 1 | 2 | 25 |
| Female | 5 | 0 | 10 | 12 |
| Age† | 43.3 ± 13.6 | 34 | 42.6 ± 11.7 | 46.0 ± 12.2 |
Mean CAL = mean full-mouth CAL ± SE
Mean ± SD
Sample Collection. Subgingival plaque samples were collected from 6–12 periodontal pockets from each subject by using Hartzell R-1, R-2 curettes. Supragingival plaque was removed from tooth surfaces before sampling. Separate sterile curettes were used for each plaque sample. Sampling included both clinically healthy and diseased sites. Clinical assessments at each site included the presence or absence of bleeding on probing (BOP), probing depth (PD), and CAL. Clinical assessments and sample collections were performed by one researcher (G.C.A.). Each site was classified as healthy (no BOP, CAL ≤1 mm, PD ≤3 mm), having gingivitis (BOP, CAL ≤1 mm, PD ≤4 mm), slight periodontitis (BOP, CAL 2–3 mm, and PD ≥4 mm), moderate periodontitis (BOP, CAL 4–5 mm, and PD ≥4 mm), or severe periodontitis (BOP, CAL ≥6 mm, and PD ≥4 mm, Table 2). In addition, a sample was taken from the dorsum of the tongue with a sterile plastic spatula. Less than 1 mg of plaque material from each sampled site was placed in a 1-ml O-ring microcentrifuge tube containing 200 μl of γ-irradiated H2O. A 100-μl aliquot was moved to another tube for fluorescent in situ hybridization (FISH) after vortexing. The remaining aliquot was frozen immediately and kept at -80°C before further processing.
Table 2. Vital statistics for patient samples.
| Periodontitis patient sample sites
|
|||||||
|---|---|---|---|---|---|---|---|
| Tongue | Healthy | Gingivitis | Slight | Moderate | Severe | Healthy controls | |
| Samples | 20 | 38 | 9 | 28 | 34 | 96 | 29 |
| White | 9 | 20 | 2 | 12 | 16 | 45 | 29 |
| Black | 6 | 12 | 5 | 7 | 14 | 30 | 0 |
| Hispanic | 2 | 5 | 0 | 2 | 4 | 14 | 0 |
| Asian | 3 | 1 | 2 | 7 | 0 | 7 | 0 |
| Mean PD, mm* | NA | 3.24 ± 0.08 | 4.04 ± 0.15 | 4.71 ± 0.12 | 6.37 ± 0.08 | 8.90 ± 0.12 | 1.99 ± 0.10 |
| Mean CAL, mm† | NA | 0.45 ± 0.50 | 1.00 | 2.47 ± 0.06 | 4.69 ± 0.05 | 7.96 ± 0.11 | 0.04 ± 0.03 |
| Male | 12 | 20 | 7 | 12 | 14 | 66 | 11 |
| Female | 8 | 18 | 2 | 16 | 20 | 30 | 20 |
NA, not applicable.
Mean PD = probing (pocket) depth ± SE
Mean CAL ± SE
Nucleic Acids Extraction. Nucleic acids were extracted from each 100-μl plaque sample by adding an equal volume of 0.1% blue dextran (Sigma) and 2× volume of cell lysis buffer (100 mM Tris·HCl, pH 7.4/20 mM EDTA/5 M guanidine isothiocyanate/2% Triton X-100). Proteinase K (Sigma) was added to a final concentration of 250 μg/ml, and the sample was incubated at 65°C for 30 min. Samples were then agitated in a FastPrep FP120 instrument (Qbiogene, Carlsbad, CA) at 4.0 m/s for 30 s with 0.1 g each of 0.1-mm, 0.5-mm, and 1-mm diameter baked zirconia/silica beads (Biospec, Bartlesville, OK). An equal volume of 99% benzyl alcohol was added, and the sample was vortexed before centrifugation at 7,000 × g for 5 min. The aqueous phase was removed to a new microcentrifuge tube, and the nucleic acids were precipitated by the addition of 1/10 volume 3 M NaOAc, 2 volumes 100% EtOH, and centrifugation at 16,000 × g for 30 min. Nucleic acid pellets were washed with 70% EtOH, dried in a Speedvac (Thermo Savant, Holbrook, NY), and resuspended in 50 μlof γ-irradiated H2O. Samples that were subsequently PCR-amplified were incubated in 20 μg/ml RNaseA at 37°C for 30 min. Negative lysis controls, consisting of γ-irradiated water, were carried throughout the experiment.
PCR, Cloning, and Sequencing. Fragments of 16S rDNA from oral Archaea were PCR amplified by using broad-range archaeal primers SDArch0333aS15 (5′-TCCAGGCCCTACGGG-3′, modified from ref. 15) and either SDArch0958aA19 (5′-YCCGGCGTTGAMTCCAATT-3′; ref. 16) or SDArch1378aA20 (5′-TGTGTGCAAGGAGCAGGGAC-3′). Each 25-μl PCR consisted of 1 μlof extracted DNA, buffer (10 mM Tris·HCl, pH 8.5/1.5 mM MgCl2/50 mM KCl/200 μM of each dNTP/0.05% Triton X-100), 400 nM each primer, and 1.25 units of AmpliTaq (Applied Biosystems, Foster City, CA). Archaeal 16S rDNA genes were amplified under the following cycle conditions: 35 cycles of 94°C (30 s), 58°C (30 s), and 72°C (30 s) followed by a 3-min extension at 72°C. Primer specificity and sensitivity were determined by using cloned 16S rDNA genes from Halobacterium salinarum (pHs16S; ATCC 33171), Sulfolobus acidocaldarius (pSc; ATCC 33909), and Escherichia coli B/r (pEc) (17) (see Supporting Text, Table 3, and Figs. 5–7, which are published as supporting information on the PNAS web site). PCR products of the appropriate size from patient samples were cloned by using the TOPO-TA kit (Invitrogen) as per manufacturer's instructions and screened for the appropriately sized inserts. Inserts were sequenced on an ABI 377 sequencer (Applied Biosystems, Foster City, CA) using M13(-20)F and M13(-27)R primers for duplicate coverage with ABI PRISM BigDye Terminators v2.0 reagents (Applied Biosystems).
Phylogenetic Analysis. Initial alignment of amplified sequences was performed with the automated 16S rDNA sequence aligner of the arb software package (www.arb-home.de) against a database of 12,569 complete and partial rRNA sequences. Ambiguously and incorrectly aligned positions were aligned manually on the basis of conserved primary sequence and secondary structure. Identity matrices were generated from either 572 or 998 masked (unambiguously aligned) positions, depending on the primer pair used for sequence library construction. Following the proposition of Kroes et al. (18), sequences with ≥99% identity were considered as a single phylo-type. The phylogenetic associations of all representative sequences were determined by using a maximum-likelihood algorithm (19). These associations were confirmed by using a parsimony algorithm (19) and a neighbor-joining algorithm (20) of kimura two-parameter corrected evolutionary distances. Sequences were deposited in the GenBank database (accession numbers AY374553 and AY374554).
FISH. Polyribonucleotide probe (polyprobe) was generated from Microcon-100 (Millipore, Bedford, MA) purifed PCR amplicons of cloned SBGA-1 16S rRNA that contained a T7 RNA polymerase promoter by using a protocol modified from that of DeLong et al. (21). The polyprobe was transcribed by using a modification of the manufacturer's recommended protocol for RNAMaxx (Stratagene). Each 25-μl transcription-labeling reaction consisted of 1 μg of purified SBGA-1 amplicon, 1× transcription buffer, 30 mM DTT, 0.4 mM ATP, 0.4 mM GTP, 0.4 mM CTP, 0.8 mM UTP, 0.25 mM Cy3-UTP (Amersham Pharmacia), 200 units of T7 RNA polymerase, balance DEPC-H2O. Transcription reactions were incubated at 37°C for 2 h. DNA was removed from the sample by the addition of 1× DNase buffer (200 mM Tris·HCl, pH 8.4/20 mM MgCl2/500 mM KCl), 50 units of DNaseI (Invitrogen), and 100 units of RNaseOUT (Invitrogen) followed by a 15-min incubation at room temperature. The reaction was stopped by the addition of EDTA (pH 7.2) to 2 mM final concentration and heat inactivation at 65°C for 10 min. Polyprobe was purified with Microcon-100 columns and subsequently hydrolyzed by the addition of MgCl2 to 30 mM final concentration with incubation at 90°C for 10 min. Hydrolyzation was stopped by placing the reaction on ice and adding EDTA (pH 7.2) to a final concentration of 50 mM.
Subgingival plaque samples were vigorously vortexed for 5 min. Samples were fixed in 0.5× PBS (145 mM NaCl/8.7 mM Na2HPO4/1.5 mM NaH2PO4, pH 7.4) and 3.7% (wt/vol) formalin overnight at 4°C, transferred to 4-mm-diameter Teflon slides (Erie Scientific, Portsmouth, NH), and air dried. Samples were dehydrated by successive passage in 50%, 80%, and 90% EtOH for 3 min each. Ten microliters of hybridization buffer [0.9 M NaCl/20 mM Tris·HCl, pH 7.4/1% SDS/1 mg/ml poly(A)/50% formamide], and 50 ng of polyprobe was added to each well. Conditions that optimized polyprobe specificity were determined by using cloned artificial targets for FISH with a range (0–80%) of formamide concentrations, and by using E. coli transformed with vector but no SBGA-1 insert (22). Slides were incubated in the dark at 65°Cfor8hin chambers humidified with 0.9 M NaCl, 20 mM Tris·HCl. After hybridization, the slides were washed in 50 ml of wash solution (70 mM NaCl/20 mM Tris·HCl, pH 7.4/5 mM EDTA/0.01% SDS) for 2 h at 45°C in the dark. Samples were counterstained with 20 μl of 5 μM YOPRO-1 (Molecular Probes) for 15 min in the dark at room temperature. Slides were rinsed with water, air dried in the dark, and mounted with 1.7 μl of Vectashield (Vector Laboratories, Burlingame, CA). Micrographs were taken with a Bio-Rad MRC 1024ES laser scanning confocal imaging system mounted on a Nikon Eclipse TE300 microscope.
5′ Nuclease Assay. Archaeal, bacterial, and treponemal rRNA gene copies were quantified by using a 5′ nuclease assay and an ABI Prism 7900HT Sequence Detection System (Applied Biosystems) (see Figs. 6–8, which are published as supporting information on the PNAS web site). Each 10-μl reaction mix consisted of 1 μl of extracted DNA, 1× TaqMan Universal PCR master mix without AmpErase UNG (Applied Biosystems), 900 nM each primer, 200 nM probe, and 0.5 units of AmpliTaq Gold DNA polymerase (Applied Biosystems). The cycling conditions were 95°C for 10 min, followed by 50 cycles of 95°C for 30 s, 55°C for 30 s, 60°C for 45 s, 65°C for 15 s, and 72°C for 15 s. All probes were conjugated to a 6-carboxyfluorescein (FAM) reporter, 6-carboxy-tetramethylrhodamine (TAMRA) quencher, and HPLC purified. The threshold was set at 0.01, with the baseline measured from cycles 3 to 15. Total archaeal or bacterial gene copy number was estimated for each sample from a standard curve generated from a 10-fold serial dilution of pHs16S or pEc16S, respectively (Supp. Figs. 6 and 7). Archaeal small subunit (SSU) rDNA was quantified by using primers SDArch0333aS15 and SDArch0958aA19 and probe S*Univ0515aA19 (5′-FAM-TTACCGCGGCKGCTGGGACTAMRA-3′; ref. 23). Bacterial SSU rDNA was quantified by using primers SDBact0008aS20 (5′-AGAGTTTGATCCTGGCTCAG-3′; ref. 23) and S*Univ0515aA19 and probe SDBact0338aS18 (5′-FAM-GTCGCCTCCCGTAGGAGT-TAMRA-3′; ref. 24). Assay specificity and possible inhibition were tested with reactions by using standard serial dilutions spiked with 1.8 ng of human DNA, as well as Archaea-negative patient samples spiked with the archaeal standard curve dilutions. Archaeal rDNA abundance is reported as a percentage of total prokaryotic rRNA gene copy number [ar-chaeal gene copy number/(archaeal gene copy number + bacterial gene copy number)] to normalize for site-to-site difference in the total abundance of biomass. Treponemal SSU rDNA was quantified by using primers SGTrep0093aS19 (5′-TCTCCTAGAGYGGCGGACT-3′) and SGTrep0767aA20 (5′-TCCTGTTTGCTCCCCGCACY-3′) in conjunction with the SDBac0338aA18 probe. The treponemal standard curve was generated by using cloned rDNA from ZAS-9 (25). Treponemal rDNA abundance is reported as a percentage of total prokaryotic rRNA gene copy number.
Quantification and Statistics. As in previous work (26), we found a greater degree of interexperimental variation than intraexperimental variation in measurements of sequence-specific DNA abundance. A large portion of this variation was due to slight differences in the slope of the real-time PCR standard curve, which was then amplified in the conversion of log to absolute gene copy number. To minimize interexperimental variation, we constructed a composite standard curve that encompassed the standard curves from all individual experiments, similar to the procedure previously described (26). All samples and standards were analyzed in duplicate within each individual experiment. Samples with an intraexperimental coefficient of variation >1 were reanalyzed. Significant differences in archaeal, bacterial, and treponemal rRNA gene copy numbers between disease states were assessed by a two-tailed, unpaired t test. The hypothesis that the relative proportion of archaea increased with the degree of CAL was tested by one-way ANOVA. All errors are reported as standard error unless otherwise specified.
Results
The subjects enrolled in this study were classified as either possessing generally healthy gingiva or exhibiting various degrees of periodontitis (Table 1). The mean CAL, a measure of disease severity, in patients with severe periodontitis was significantly higher (P < 0.01) than in patients with moderate periodontitis. The mean CAL of patients with either severe or moderate periodontitis was significantly higher than (P < 0.01) the CAL of the healthy control population. We examined 205 subgingival plaque samples from healthy and diseased sites, as well as 20 tongue scrapings, from 50 periodontitis patients. In addition, we included 29 subgingival plaque samples and two tongue scrapings from eight healthy control subjects (Table 2). To determine the effect of conventional treatment on Archaea-positive sites, we examined 77 posttreatment plaque samples for Archaea from previously studied sites that had been treated with scaling and root planing plus routine maintenance care every 3 months for a 12- to 18-month period.
We developed a quantitative Archaea-specific SSU rDNA 5′ nuclease assay to assess the relationship between the abundance of archaeal phylotypes and disease severity. We also measured the abundance of Bacteria-specific SSU rDNA to normalize for variations in microbial biomass between samples, as described (26). The Archaea- and Bacteria-specific assays had lower detection limits of 100 and 1,000 gene copies, respectively (Figs. 6 and 7).
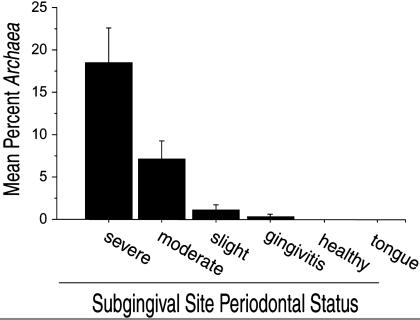
Archaeal SSU rDNA was not detected in any of the 31 samples from the healthy control population. Archaeal SSU rDNA was detected in 36% of the periodontitis patients. Archaeal SSU rDNA was detected in 76.6% of periodontitis sites but was not detected in samples from healthy sites or tongue scrapings from Archaea-positive periodontitis patients. There was a direct correlation between the relative abundance of archaeal SSU rDNA and the severity of disease within the Archaea-positive subset of patients (Fig. 1). Archaeal SSU rDNA accounted for 18.5 ± 4.2%, 7.2 ± 2.1%, 1.3 ± 0.7%, and 0.4 ± 0.3% of total prokaryotic (Bacteria plus Archaea) SSU rDNA in samples from sites with severe periodontitis, moderate periodontitis, slight periodontitis, and gingivitis, respectively. Although the abundance of bacterial SSU rDNA increased with the severity of periodontal disease (Fig. 2), the relative abundance of archaeal SSU rDNA in relation to total prokaryotic SSU rDNA was significantly higher (P < 0.05) in severe and moderate periodontitis sites compared to slight periodontitis sites within the Archaea-positive subset of patients. There was a significant relationship between the degree of CAL and the relative abundance of archaeal SSU rDNA (P < 0.0001) within the periodontitis patient subpopulation. There was no discernible relationship between ethnicity, gender, or age and the abundance of archaeal SSU rDNA; however, the data set may have been too small to resolve statistically significant relationships involving these parameters.
Fig. 1.
Relative abundance of archaeal SSU rDNA, expressed as the mean percent of total prokaryotic SSU rDNA in Archaea-positive patients. Error bars represent standard error.
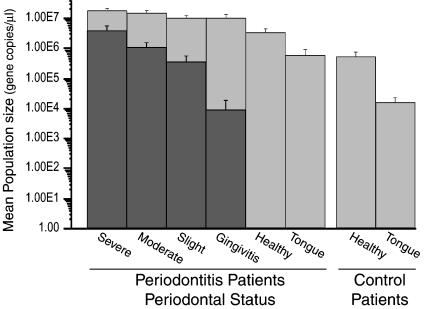
Fig. 2.
Mean bacterial (light gray) and archaeal (dark gray) rRNA gene copy number in periodontal health and disease. Gene copy number was determined by a quantitative 5′ nuclease assay. Error bars represent standard error.
As expected, bacterial rRNA gene copy numbers were significantly higher (P < 0.001) in severely and moderately diseased periodontitis sites compared to the healthy sites of periodontitis patients. Similarly, the mean bacterial rRNA gene copy numbers were significantly higher at slight periodontitis and gingivitis sites compared to the healthy sites of periodontitis patients (P < 0.015). The mean bacterial gene copy number was significantly lower (P < 0.005) in samples from the healthy control group (5.3 × 105 ± 2.4 × 105 gene copies per μl) compared to samples from healthy sites in periodontitis patients (3.5 × 106 ± 9.5 × 105 gene copies per μl; Fig. 2).
The analysis of 77 samples from six patients obtained 12–18 months after treatment, and the comparison of these results with those obtained from these sites before treatment, revealed a significant decrease in the relative abundance of archaeal SSU rDNA from a mean of 12.3 ± 4.6% to 0.0056 ± 0.0035% (P < 0.001). This decrease was accompanied by a drop in the patients' mean CAL from 3.8 ± 0.072 to 2.4 ± 0.19, indicating an improvement in disease status. The decrease in the relative abundance of archaeal SSU rDNA was caused by a decline at each sampled site and not caused by a reduction in prokaryotic biomass or an increase in bacterial 16S rDNA copy number, the latter of which remained nearly constant at 1.0 × 107 ± 1.8 × 106 copies before treatment and 1.5 × 107 ± 2.3 × 106 copies after treatment (P > 0.1).
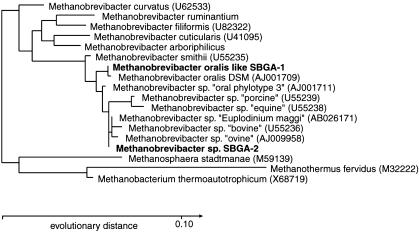
To investigate the diversity of Archaea in the human subgingival crevice, SSU rDNA was amplified with domain-specific primers and cloned independently from samples collected from six patients with periodontitis. For this purpose, we used the same archaeal primer set as that used in the 5′ nuclease assay, and a set that amplified a larger segment of the SSU rDNA; these primers were tested for both sensitivity and specificity (see Supporting Text). All 105 sequenced clones fell within the genus Methanobrevibacter of the Euryarchaea division. Phylogenetic analysis by both maximum-likelihood and maximum parsimony algorithms produced identical topologies (Fig. 3). Analysis using a neighbor-joining distance method produced a topology that differed from the other two analyses only in its placement of Methanobrevibacter cuticularis at the root of the clade containing Methanobrevibacter filiformis, Methanobrevibacter ruminantium, and Methanobrevibacter arboriphilius.
Fig. 3.
Phylogenetic relationships of Archaea in the subgingival crevice inferred from SSU rDNA analysis. Phylotypes identified in this study are shown in bold. GenBank accession numbers for database sequences are given in parentheses except for M. ruminantium and M. arboriphilicus, which are only available from the Ribosomal Database Project (38). This dendrogram was constructed from 572 homologous sequence positions (349–957, E. coli numbering) using a maximum-likelihood algorithm.
The clone libraries were dominated (81% of clones) by a phylotype (SBGA-1) with 99.8% identity to the 572 nucleotides of Methanobrevibacter oralis available from GenBank (Fig. 3). Using reverse primer SDArch1378aA20, we were able to extend the sequence from what was probably Methanobrevibacter oralis by an additional 436 nucleotides and demonstrate that this phylotype is clearly distinguishable from Methanobrevibacter smithii. Phylotype SBGA-1 shared 97.7% identity with M. smithii over 998 nucleotides (positions 349-1378, E. coli numbering). The remainder (19%) of the cloned sequences was composed of the phylotype SBGA-2. This phylotype shared 99.8% identity with a Methanobrevibacter sequence associated with the ciliate Euplodinium maggi, which inhabits the ovine rumen (27, 28). This phylotype was also closely related (99.5% identity) to, but distinct from, the human oral “phylotype 3” identified by Kulik et al. (1). Together these three phylotypes, along with phylotypes from a number of ruminants and swine, formed a clade that shared ancestry with M. oralis to the exclusion of M. smithii (Fig. 3). Although each of the phylotypes within this clade was distinguishable from the others, the nucleotide differences occurred in unpaired, nonhelical regions and may represent sequencing errors, interoperon variability, or different strains of a single species. Phylotype SBGA-2 shared 98.6% sequence identity with M. oralis, but was clearly distinct at eight positions [350 (T), 560 (T), 658 (C), 747 (G), 838 (G), 848 (C), 849 (T), 850 (C); E. coli numbering), some of which are predicted to form compensatory base-pairing structures (658–747 and 838–848).
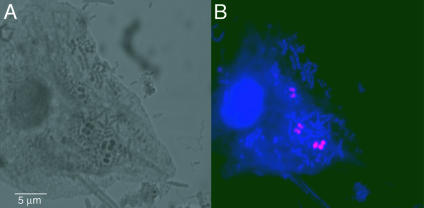
We used cloned SBGA-1 16S rDNA to generate an Archaea-specific RNA polyprobe for FISH that enabled us to characterize further the members of this domain in subgingival plaque samples. When we used cloned artificial targets for FISH, we found that hybridization in a solution of 50% formamide (vol/vol) at 65°C followed by a 45°C low-salt wash provided the optimal discrimination between SBGA-1 targets and nontarget sequences (22). These hybridization conditions were similar to those used previously with polyribonucleotide probes (21). We identified a population consisting primarily of diplococcobacilli (Fig. 4) with approximate dimensions of 0.9 μm × 0.9 μm. The morphology and dimensions were largely consistent with those of M. oralis (8), although the cell width observed in this study was approximately twice that previously reported.
Fig. 4.
Confocal micrographs of bacteria and archaea in subgingival plaque from a patient with severe periodontitis. (A) “Bright field” micrograph. (B) Fluorescent in situ hybridization micrograph of oral archaeum using SBGA-1 Cy-3-labeled polyprobe (pink) and counterstained with the nucleic acid dye YO-PRO-1 (blue).
Methanogenesis by M. oralis is a hydrogen-consuming process. In syntrophic relationships, this process facilitates the growth of hydrogen-producing organisms, which include some of the known oral bacterial pathogens. If syntrophy is an important feature of more severely diseased periodontal pockets, one might expect to find other syntrophic partners in methanogen-negative, diseased sites. Treponemes are a potential hydrogen competitor, and are a monophyletic group for which group-specific primers can be designed. Therefore, we determined the relative abundance of Treponema species rDNA within the same collection of plaque samples as used to investigate the relative abundance of archaeal rDNA (Fig. 8). We found that treponeme rDNA represented 12.4 ± 3.8% of the prokaryotic rDNA in a set of specimens from severe and moderate periodontitis sites that had no detected archaeal rDNA. In contrast, the relative abundance of treponemal rDNA at sites with archaeal rDNA was 6.2 ± 1.4%; this difference was statistically significant (P < 0.05). There was no significant difference in the relative abundance of treponemal rDNA found at sites with slight periodontitis or gingivitis, regardless of whether archaeal rDNA was detected or not; however, the number of sites with these disease classifications and detected archaea was small.
Discussion
Members of the domain Archaea are highly diverse in form and function, but curiously, disease causation is not among their demonstrated capabilities. We found that the relative abundance of archaeal SSU rDNA increased in relationship to the severity of periodontal disease within a cohort of patients. There was a corresponding decrease in the relative abundance of the archaeal SSU rDNA coinciding with an improvement in periodontal status after treatment. The etiology of a polymicrobial disease such as periodontitis is likely to be more complex than suggested by the traditional paradigm of disease involving a single virulent organism. Traditional approaches for establishing causation require the use of a relevant model system with the presumption that transfer of a purified microbial isolate will be sufficient to reproduce disease, as specified in Koch's postulates. Molecular criteria have been proposed for imputing a causative role when the putative factor cannot be easily isolated or purified (29).
Archaea were detected in only a subset of patients with severe disease. The assay used was capable of detecting amounts of Archaea representing as little as 0.001% of the prokaryotic population, suggesting that the methodology was not a limiting factor in detection. Two hypotheses, which are not mutually exclusive, may be advanced to explain the presence of oral methanogens in only a subset of periodontitis patients. The first hypothesis is that host genetics may predispose some individuals to colonization by oral methanogens. However, a comparison of the prevalence of oral and colonic methanogens found that all individuals harboring oral methanogens also harbored colonic methanogens, but not vice versa (9), suggesting that host genetics is not a sufficient explanation for the exclusion of methanogens from the oral cavity. An additional study of monozygotic and dizygotic twins found that host genetics did not play a significant factor in differences in breath methane emission, a hallmark of colonic methanogens (30).
The second hypothesis proposes niche exclusion of methanogens by other hydrogen-metabolizing microbes in some patients. Sulfate-reducing bacteria (SRB) are potential competitors that have been reported to be harbored by ≈64% of periodontitis patients, and their presence has been correlated with pocket depth (31). Under standard conditions, sulfate-reducing bacteria should out-compete methanogens (32), assuming that the availability of sulfate is not limited. However, if the interactions between subgingival SRB and methanogens are similar to those in the colon, then the two groups may coexist within the same environment (33, 34). Recent research has indicated that both may coexist in the oral cavity (13).
Members of the genus Treponema are also potential hydrogen competitors, and include a well-known periodontal pathogen, Treponema denticola. Previous work has demonstrated that T. denticola, like Porphyromonas gingivalis and Tannerella for-sythensis, is associated with severe periodontitis, as a member of the “red” polymicrobial disease complex (35). It has also recently been demonstrated that some Treponema species are capable of homoacetogenesis, a hydrogen-consuming process (36). We found that the relative abundance of treponemal rDNA was significantly lower in sites with archaeal rDNA than in sites without archaeal rDNA, suggesting that some Treponema species may compete with methanogens. Our results present the possibility that methanogens and treponemes may serve as alternative syntrophic partners with other members of the subgingival biofilm community, such as other members of the red complex. In this scenario, methanogenic Archaea indirectly promote periodontal disease in some patients by serving as a hydrogen sink, thereby permitting the proliferation of one or more pathogenic secondary fermenters to levels beyond that which would be possible in the absence of the archaea.
The apparent restricted diversity exhibited by the oral Archaea may reflect the adaptation of a small minority of organisms within this broad domain of life to this particular niche. The length and morphology of the cells labeled with the Cy-3 archaea-specific polyprobe were consistent with those of Methanobrevibacter oralis. However, the cells observed in this study were nearly twice the width of those previously reported, which may reflect differences in growth rate or nutritional status (8). Members of the genus Methanobrevibacter are strict anaerobes, and previous research has shown that mature subgingival plaque provides the highly reduced environment necessary for anaerobic growth (37). Although SBGA-1 was identified in all six of the patients from which archaeal clone libraries were created, phylotype SBGA-2 was recovered from only two of the six patients. Although this latter phylotype appears to be a minor constituent of the methanogenic population, the number of patients examined was not large enough to determine the true distribution of this phylotype.
We speculate that syntrophic interactions between Archaea and other members of the microbial flora may be an important feature of some polymicrobial diseases (4). The identity and role of the complementary syntrophic partner(s) should provide an important avenue for future research in eliciting the microbial mechanisms involved in chronic periodontitis and other polymicrobial diseases.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health/National Institute of Dental and Craniofacial Research Grant R01-DE13541 and an Ellison Medical Foundation grant (to D.A.R.) and the University Exploratory Research Program of Procter and Gamble (to P.W.L.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CAL, clinical attachment loss; FISH, fluorescence in situ hybridization; SSU, small subunit.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY374553 and AY374554).
References
- 1.Kulik, E. M., Sandmeier, H., Hinni, K. & Meyer, J. (2001) FEMS Microbiol. Lett. 196, 129-133. [DOI] [PubMed] [Google Scholar]
- 2.Miller, T. L. & Wolin, M. J. (1982) Arch. Microbiol. 131, 14-18. [DOI] [PubMed] [Google Scholar]
- 3.Belay, N., Mukhopadhyay, B., Conway de Macario, E., Galask, R. & Daniels, L. (1990) J. Clin. Microbiol. 28, 1666-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckburg, P. B., Lepp, P. W. & Relman, D. A. (2003) Infect. Immun. 71, 591-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albandar, J. M., Brunelle, J. A. & Kingman, A. (1999) J. Periodontol. 70, 13-29. [DOI] [PubMed] [Google Scholar]
- 6.Champagne, C. M., Madianos, P. N., Lieff, S., Murtha, A. P., Beck, J. D. & Offenbacher, S. (2000) J. Int. Acad. Periodontol. 2, 9-13. [PubMed] [Google Scholar]
- 7.Paster, B. J., Boches, S. K., Galvin, J. L., Ericson, R. E., Lau, C. N., Levanos, V. A., Sahasrabudhe, A. & Dewhirst, F. E. (2001) J. Bacteriol. 183, 3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrari, A., Brusa, T., Rutili, A., Canzi, E. & Biavati, B. (1994) Curr. Microbiol. 29, 7-12. [Google Scholar]
- 9.Brusa, T., Canzi, E., Allievi, L., Del Puppo, E. & Ferrari, A. (1993) Curr. Microbiol. 27, 261-265. [Google Scholar]
- 10.Brusa, T., Conca, R., Ferrara, A., Ferrari, A. & Pecchioni, A. (1987) J. Clin. Periodontol. 14, 470-471. [DOI] [PubMed] [Google Scholar]
- 11.Robichaux, M., Howell, M. & Boopathy, R. (2003) Curr. Microbiol. 47, 12-16. [DOI] [PubMed] [Google Scholar]
- 12.Brusa, T., Canzi, E., Conca, R., Ferrara, A., Ferrari, A. & Pecchioni, A. (1989) Ann. Microbiol. 39, 161-165. [Google Scholar]
- 13.Robichaux, M., Howell, M. & Boopathy, R. (2003) Curr. Microbiol. 46, 53-58. [DOI] [PubMed] [Google Scholar]
- 14.Belay, N., Johnson, R., Rajagopal, B. S., de Macario, E. C. & Daniels, L. (1988) Appl. Environ. Microbiol. 54, 600-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barns, S. M., Fundyga, R. E., Jeffries, M. W. & Pace, N. R. (1994) Proc. Natl. Acad. Sci. USA 91, 1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLong, E. F. (1992) Proc. Natl. Acad. Sci. USA 89, 5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brosius, J., Palmer, M. L., Kennedy, P. J. & Noller, H. F. (1978) Proc. Natl. Acad. Sci. USA 75, 4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroes, I., Lepp, P. W. & Relman, D. A. (1999) Proc. Natl. Acad. Sci. USA 96, 14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein, J. (1989) Cladistics 5, 164-166. [Google Scholar]
- 20.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406-425. [DOI] [PubMed] [Google Scholar]
- 21.DeLong, E. F., Taylor, L. T., Marsh, T. L. & Preston, C. M. (1999) Appl. Environ. Microbiol. 65, 5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouverney, C. C., Armitage, G. C. & Relman, D. A. (2003) Appl. Environ. Microbiol. 69, 6294-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane, D. J., Pace, B., Olsen, G. J., Stahl, D. A., Sogin, M. L. & Pace, N. R. (1985) Proc. Natl. Acad. Sci. USA 82, 6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giovannoni, S. J., DeLong, E. F., Olsen, G. J. & Pace, N. R. (1988) J. Bacteriol. 170, 720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lilburn, T. G., Kim, K. S., Ostrom, N. E., Byzek, K. R., Leadbetter, J. R. & Breznak, J. A. (2001) Science 292, 2495-2498. [DOI] [PubMed] [Google Scholar]
- 26.Brinig, M. M., Lepp, P. W., Ouverney, C. C., Armitage, G. C. & Relman, D. A. (2003) Appl. Environ. Microbiol. 69, 1687-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, T. L. & Lin, C. (2002) Int. J. Syst. Evol. Microbiol. 52, 819-822. [DOI] [PubMed] [Google Scholar]
- 28.Abraham, E. R. (1998) Nature 391, 577-580. [Google Scholar]
- 29.Fredericks, D. N. & Relman, D. A. (1996) Clin. Microbiol. Rev. 9, 18-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Florin, T. H., Zhu, G., Kirk, K. M. & Martin, N. G. (2000) Am. J. Gastroenterol. 95, 2872-2879. [DOI] [PubMed] [Google Scholar]
- 31.Langendijk, P. S., Hanssen, J. T. & Van der Hoeven, J. S. (2000) J. Clin. Periodontol. 27, 943-950. [DOI] [PubMed] [Google Scholar]
- 32.Gibson, G. R., Macfarlane, G. T. & Cummings, J. H. (1993) Gut 34, 437-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strocchi, A., Furne, J., Ellis, C. & Levitt, M. D. (1994) Gut 35, 1098-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pochart, P., Dore, J., Lemann, F., Goderel, I. & Rambaud, J. C. (1992) FEMS Microbiol. Lett. 77, 225-228. [DOI] [PubMed] [Google Scholar]
- 35.Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C. & Kent, R. L., Jr. (1998) J. Clin. Periodontol. 25, 134-144. [DOI] [PubMed] [Google Scholar]
- 36.Leadbetter, J. R., Schmidt, T. M., Graber, J. R. & Breznak, J. A. (1999) Science 283, 686-689. [DOI] [PubMed] [Google Scholar]
- 37.Kenney, E. B. & Ash, M. M., Jr. (1969) J. Periodontol. 40, 630-633. [DOI] [PubMed] [Google Scholar]
- 38.Cole, J. R., Chai, B., Marsh, T. L., Farris, R. J., Wang, Q., Kulam, S. A., Chandra, S., McGarrell, D. M., Schmidt, T. M., Garrity, G. M. & Tiedje, J. M. (2003) Nucleic Acids Res. 31, 442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.