Abstract
Chk1 both arrests replication forks and enhances repair of DNA damage by phosphorylation of downstream effectors. While there has been a distinguished effort in identifying effectors of Chk1 activity, there are still mechanisms of its activities that are yet to be identified. Metnase/SETMAR is a SET and transposase domain protein that promotes both DNA double strand break (DSB) repair and re-start of stalled replication forks. In this study, we show that Metnase is phosphorylated only on Ser495 (S495) in vivo in response to DNA damage by ionizing radiation. Chk1 is the major mediator of this phosphorylation event. We had previously shown that wild type (wt) Metnase associates with chromatin near an artificially induced DSB in an engineered cell system. However, an S495A Metnase mutant, which could not be phosphorylated by Chk1, had a defect in its DSB chromatin association. The S495A mutant also failed to support repair of an induced DSB when compared with wild type (wt) Metnase. Interestingly, the S495A mutant demonstrated increased restart of stalled replication forks compared to wt Metnase. Thus, S495 phosphorylation of Metnase differentiates between its two main functions, enhancing DSB repair and repressing replication fork restart. In summary, these data lend insight into the mechanism by which Chk1 enhances repair of DNA damage while at the same time repressing stalled replication fork restart.
Keywords: Chk1, Metnase, SETMAR, phosphorylation, DNA repair, replication fork, transposase, ionizing radiation
INTRODUCTION
Metnase, also known as SETMAR, is a SET [Su(var)3-9, Enhancer-of-zeste, Trithorax] and a transposase fusion protein present only in anthropoid primates, and not in other mammals (Cordaux et al., 2006; Jordan, 2006; Lee et al., 2005; Robertson, 1997). The Metnase SET domain di-methylates histone 3 lysine 36 (H3K36me2 Lee et al., 2005), while the transposase domain has most but not all of the classic transposase nuclease activities, including 5’-terminal inverted repeats (TIR)-specific DNA binding, DNA looping, assembly of paired end complex (PEC), and DNA cleavage (Liu et al., 2007).
We found that Metnase enhances non-homologous end joining and promotes genomic integration of foreign DNA, perhaps via its association with DNA Ligase IV (Lee et al., 2005, Hromas et al, 2008). Both the histone methylase and nuclease domains are essential for Metnase-mediated DSB repair and genomic integration in vivo (Bundock and Hooykaas, 2005; Lee et al, 2005). The SET domain is responsible for di-methylating H3K36 adjacent to induced DNA DSBs, and this di-methylation stabilized the Ku and MRN complex at the DSB, which explained Metnase’s enhancement of DSB repair (Fnu et al, 2011). Metnase is also essential for proper re-starting of stalled replication forks, perhaps by decatenating positively supercoiled DNA in front of the fork (De Haro et al, 2010; Williamson, et al 2008; Wray, et al; 2009; Roman et al, 2009). Interestingly, while Metnase is only present in primates (Lee et al, 2005), it can fit seamlessly into the murine NHEJ repair apparatus when over-expressed in these non-primate cells, and enhance DNA repair in that organism as well (Wray et al 2010).
Replication fork arrest upon DNA damage is regulated by a cascade of phosphorylation events, initiated upon sensing the DSB by ATM/ATR, with effectors of fork arrest and DNA repair as targets (Zhou and Elledge, 2000). Since Metnase is a novel component of the DSB repair pathway, we hypothesized that it was also regulated by phosphorylation. Therefore, in this study, we explored the phosphorylation of Metnase upon DNA damage, the origin of that phosphorylation, and the consequences of that phosphorylation. We discovered that Metnase is phosphorylated at S495 in response to DNA damage, mainly by Chk1, and dephosphorylated by PP2A. A Ser495Ala (S495A) mutant of Metnase was not able to be phosphorylated by Chk1, and had decreased localization to induced DSBs. This decreased DSB association resulted in decreased H3K36me2 formation in the DSB region. In addition, the S495A mutant demonstrated decreased re-ligation capacity of the induced DSB. However, the S495A mutant had an increased ability to re-start stalled replication forks. Thus, Chk1 phosphorylation differentiates between Metnase’s ability to repair DSBs and its ability to enhance replication fork recovery. These data provide a epistatic model for Metnase’s DSB repair function, and also shed light on the mechanism by which Chk1 mediates both of its major functions, cell cycle arrest and DNA repair.
RESULTS
Metnase is phosphorylated on S495 in response to DNA damage
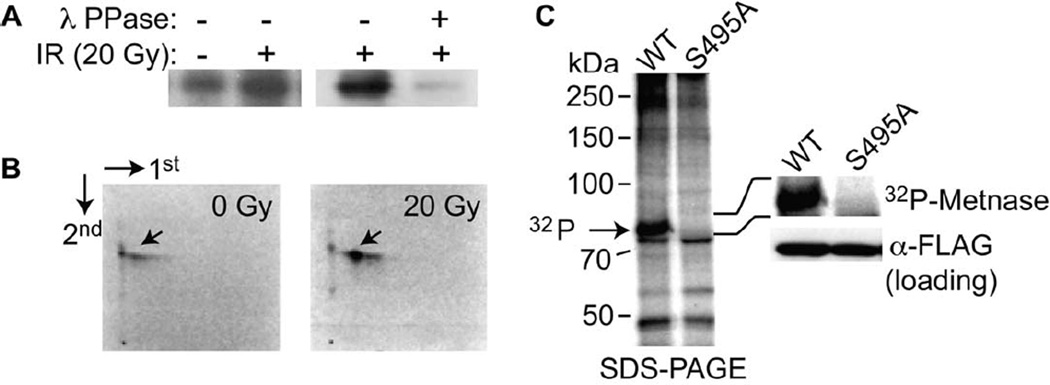
Since much of DNA replication and repair is regulated by phosphorylation, we investigated whether Metnase, which has roles in both of these functions, is also phosphorylated. Metnase possesses several sites that could potentially be phosphorylated by kinases known to be active in DNA replication and repair. There are two ATM/ATR consensus sites, 50-DIDPTQ-56 and 639-FVESQ-643 (Kim et al., 1999). In addition, the sequence surrounding S495 (489-YDNRRRSA-496) overlaps a Chk1/2 consensus target phosphorylation site (LxRxxS) with a p21-activated kinase (PAK) consensus site (O'Neill et al., 2002; Brzeska et al., 1997). It is also a possible site for Protein Kinase C (PKC). Each of these kinases can be activated by DNA damage, and would be candidate for phosphorylation of Metnase (Canman et al., 1998; Matsuoka et al., 1998; Roig and Traugh, 1999). We therefore examined phosphorylation of Metnase in vivo, after DNA damage. Inorganic 32P was indeed incorporated into Metnase in vivo, and this incorporation was markedly enhanced following DNA damage with ionizing radiation (IR) (Fig. 1A). Incorporation of 32P into Metnase was due to protein phosphorylation, since most of the incorporated 32P was removed following incubation with λ-protein phosphatase (Fig. 1A). There is a baseline level of Metnase phosphorylation observed in the absence of IR treatment (Fig. 1A, lane 1), which may be due to phosphorylation of Metnase from endogenous DNA damage during cell cycle progression (Kastan, 2001; Wilkes et al., 2003).
Figure 1. IR-induced phosphorylation of Metnase.
A. Cells over-expressing Flag-Metnase were incubated 32P-orthophosphoric acid for 1 hr, and then exposed to 20 gy ionizing radiation (IR). Metnase was immunoprecipitated using anti-Flag, electorphoretically separated, and autoradiographed. The loss of signal after λ-phosphatase implies a direct phosphorylation event. B. Two dimensional phosphopeptide mapping analysis of 32P-Metnase following IR treatment demonstrated only one species, and this species was induced following IR. This phosphopeptide was ioslated and analyzed for amino acid sequence (Supplemental Fig. 1). The phosphorylation occurred at S495. C. Phosphorylation analysis of Metnase and the S495A mutant in vivo after 20 gy IR. Wild-type (wt) and the S495A mutant of Metnase were immunoprecipitated after 32P labeling. Only the wt Metnase was phosphophorylated, shown in magnified form on the right of the panel.
To further dissect Metnase phosphorylation, we carried out two-dimensional phosphopeptide mapping analysis of Metnase after IR. Human 293 cells over-expressing V5-Metnase were metabolically labeled with 32P-orthophosphate, Metnase immunoprecipitated using anti-V5, and the immunoprecipitate analyzed by 2D gel electrophoresis for peptide(s) carrying 32P. A single phosphopeptide was identified, and whose presence was found to be significantly enhanced following IR (Fig. 1B). To determine Metnase’s phosphorylation site in vivo, Metnase was immunoprecipitated from human 293 cells over-expressing V5-Metnase following IR (20 Gy), and analyzed for phosphopeptide(s) using nano-LC/MS/MS. MS/MS analysis showed only one phosphopeptide, which perfectly matched to S495 (Fig. 1 B, Supplemental Fig. 1). To examine whether the S495 was the only site of phosphorylation on Metnase, we generated a S495A mutant of Metnase, and expressed it in 293 cells. The S495A Metnase species showed no phosphorylation in vivo when cells were incubated with 32P and irradiated (Fig. 1C). This result was consistent with the 2D gel mapping result, which also identified only one phosphopeptide following IR treatment.
Phosphorylation of S495A by Chk1
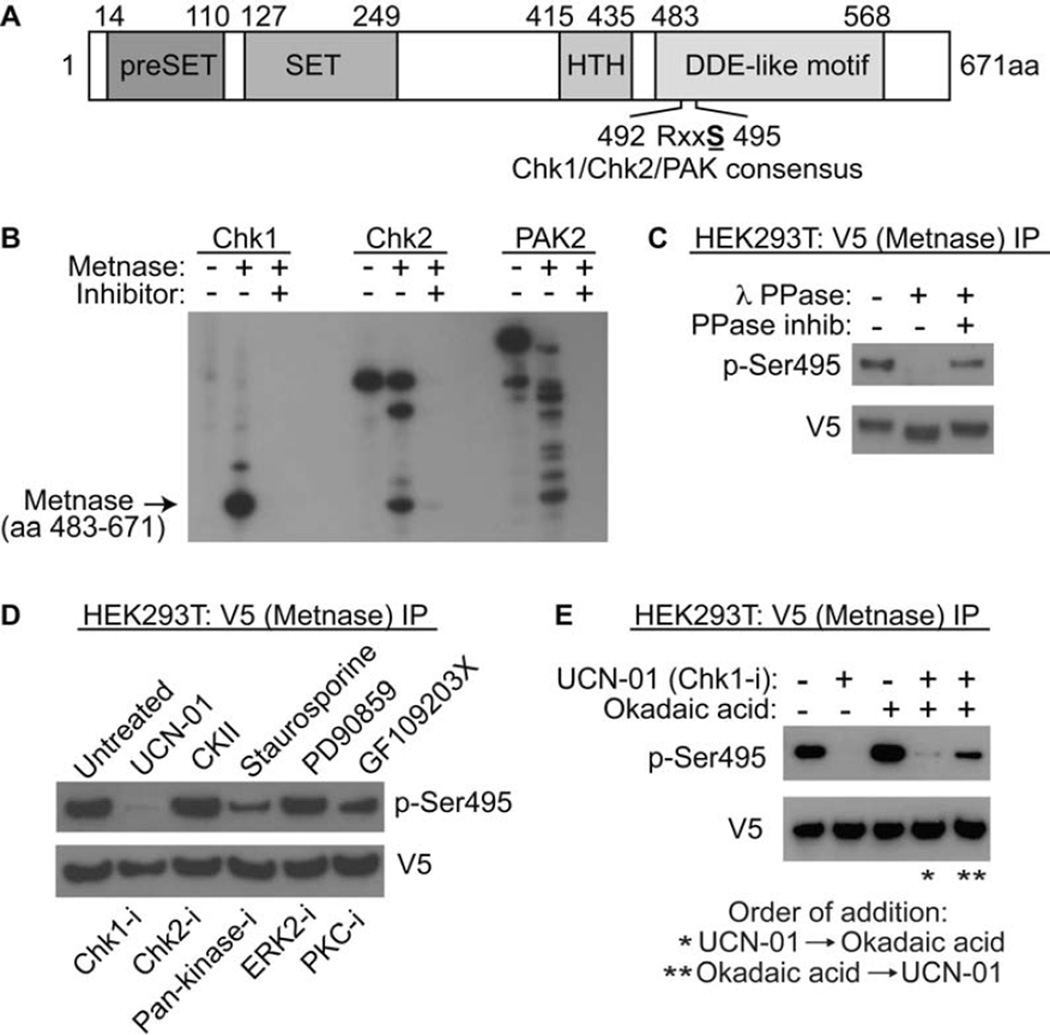
The S495 is a Chk1 and 2, and PAK consensus phosphorylation site (RxxS), as mentioned above. This site is in the DDE-like transposase nuclease domain, although not in the active site (Fig. 2A, Supplemental Fig. 2). We virtually modeled the 3-dimensional location of the S495 site within the Metnase tertiary structure (Supplemental Fig. 2). Previous structural analysis proposed that the DNA strand was located across the groove where S494 is located, as DNA approaches the catalytic triad at the active site (Goodwin et al 2010). Our model indicates that the region is quite flexible, but that phosphorylation could affect the mobility of this region. This in turn may influence DNA positioning as the strand approaches the catalytic triad. Thus, this model predicts that the nuclease function of Metnase could be altered by phosphorylation.
Figure 2. Phosphorylation of Metnase by Chk1 in vitro.
A. Schematic diagram of Metnase. The PreSET domain contains a cysteine- and histidine-rich putative Zn++ binding motif, and the SET domain has the histone methylase motif. The transposase nuclease domain contains the two conserved motifs, HTH and DDE-like (in Metnase this is DDN, Roman et al 2008). The S495 is within RxxS Chk1/2/Pak consensus sites. B. Chk1 phosphorylates Metnase in vitro much more efficiently than Chk2 and PAK2. In vitro phosphorylation of pure recombinant Metnase by pure recombinant Chk1, Chk2 and PAK2 kinases, with and without their inhibitors (UCN-01, CKII, and staurosporin, respectively), is shown. C. The anti-phosho-Ser495 (anti-pS495) antibody recognizes the phosphorylation of Metnase. V5-tagged wt Metnase was immunoprecipitated from human 293T cells using the V5 tag, and treated with 400U λ-protein phosphatase in the presence or absence of phosphatase inhibitors (as described in Methods). The immunoprecipitate was analyzed for the presence of phosphorylated S495 using anti-sera specific for the phoshorylated S495. The loss of signal in the presence of phosphatasae implies the direct phosphorylation of Metnase at S495. D. Metnase phosphorylation in vivo in the presence of kinase inhibitors, listed above the panel. Below the panel are the types of kinases inhibited. The Chk1 and to a lesser extent the pan-kinase inhibitor block the phosphporylation of S495 on Metnase as detected by western analysis using anti-pSer495 after immunoprecipitation of V5-tagged wt Metnase. The samples are as follows: 1- no treatment; 2- UCN-01 (Chk1 inhibitor); 3- CKII (Chk2 inhibitor); staurosporine (pan-kinase inhibitor, including PAK2 and Chk1); PD90859 (ERK2 inhibitor); GF109203X (PKC inhibitor). E. The PP2A inhibitor Okadaic acid reverses the ability of the Chk1 kinase inhibitor UCN-01 to inhibit phosphorylation of S495 on Metnase. After immunoprecipitation of V5-tagged wt Metnase from cells pre-treated with UCN-01, okadic acid, or both, the pS495 was detected using western analysis. Okadaic acid was effective only when it was added prior to UCN-01.
We examined whether Chk1, Chk2, or PAK2 could phosphorylate the S495 of Metnase using in vitro kinase assays. Pure recombinant Chk1, Chk2, and Pak2 were incubated with pure recombinant Metnase nuclease fragment (Roman et al, 2008), and 32P-ATP. Proteins were segregated by denaturing gel electrophoresis, and the gel autoradiographed. While Chk2 and PAK2 could both phosphoporylate Metnase to some extent, Chk1 had 8-fold greater kinase activity for Metnase than either of those (Fig. 2B). We then generated an antibody against phosphorylated S495 of Metnase (anti-pS495), and tested whether it could immunoprecipiate phosphorylated Metnase (Fig. 2C). V5-tagged Metnase was immunoprecipitated, and exposed to λ-phosphatase. The anti-pS495 recognized only the untreated, phosphorylated, form of Metnase (Fig. 2C). We then examined the ability of several kinase inhibitors to block the phosphorylation of Metnase (Fig. 2D). Since cell populations have a fraction of phosphorylated Metnase present constitutively, as noted above, perhaps from endogenous replication stress, we analyzed these kinase activities without DNA damage. The Chk1 inhibitor UCN-01 decreased the presence of phosphorylated Metnase significantly, with the pan-kinase inhibitor staurosporin partially inhibitory, but the other kinase inhibitors, including the PKC inhibitor G109203X, were not nearly as active.
Since PP2A has been reported to antagonize the effects of Chk1 (Leung-Pineda and Ryan 2006), we tested whether the PP1/PP2A inhibitor Okadaic acid could reverse the de-phosphorylation of Metnase (Fig. 2E). We found that okadaic acid could indeed increase the phosphorylation of Metnase, and overcome the decrease in phosphorylation seen with UCN-01. However, okadaic acid worked only when it was added prior to UCN-01, indicating a continuous dynamic between Metnase phosphorylation and de-phosphorylation within the cell.
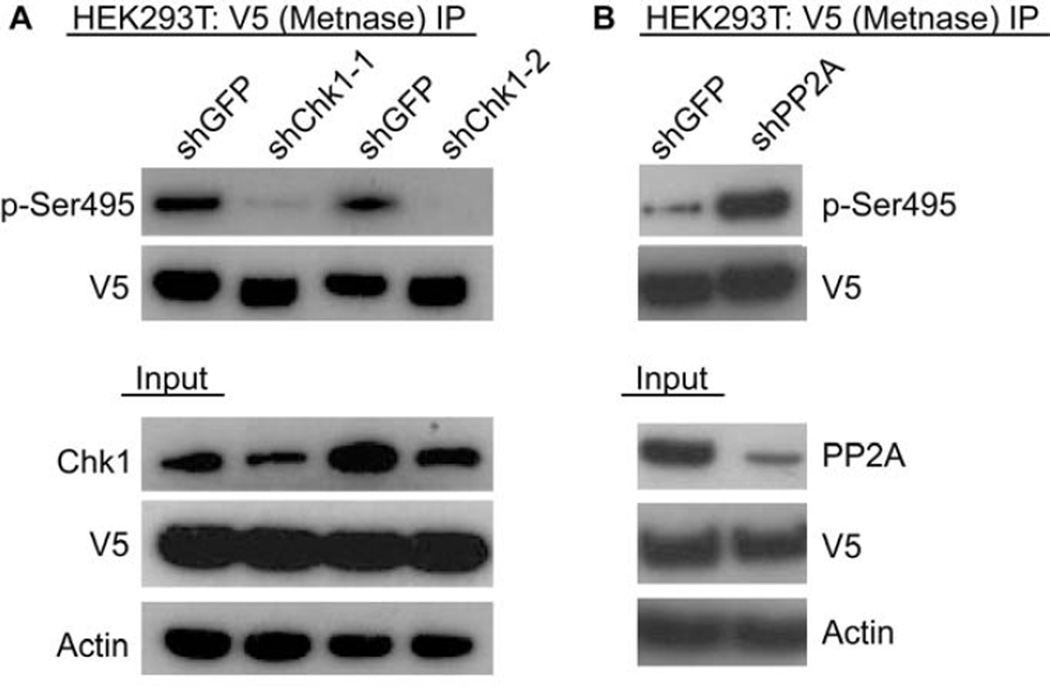
However, these inhibitors each have some off-targets effects, and it was possible that the decrease in S495 phosphorylation seen with UCN-01 could have been due to its minor activity against Chk2. Therefore, we used previously characterized shRNA to repress the expression of Chk1, and examined whether this altered the presence of pS495 (Fig. 3A). We found that even mildly decreasing Chk 1 expression nearly completely abrogated the phosphorylation of Metnase. In like manner, okadaic acid inhibits both PP1 and PP2A. We used shRNA to repress PP2A specifically, and found that it significantly increased the presence of phosphorylated S495 (Fig. 3B), implying that PP2A could specifically de-phosphorylate Metnase. When PP2A was repressed, then the activity of Chk1 was unopposed, and the phosphorylation of Metnase increased.
Figure 3. Cellular phosphorylation of Metnase by Chk1, and dephosphorylation by PP2A.
Repressing expression of Chk1 blocks Metnase S495 phosphorylation. V5-tagged Metnase was immunoprecipitated from cells with and without repression of Chk1 using specific siRNAs. The presence of phosphorylated Metnase was detected using western analysis with the anti-pS495. B. Repressing expression of PP2A enhances Metnase S495 phosphorylation. V5-tagged Metnase was immunoprecipitated from cells with and without siRNA repression of PP2A. The presence of phosphorylated Metnase was detected using western analysis with the anti-pS495.
Effect of Metnase phosphorylation on replication fork recovery and DNA repair
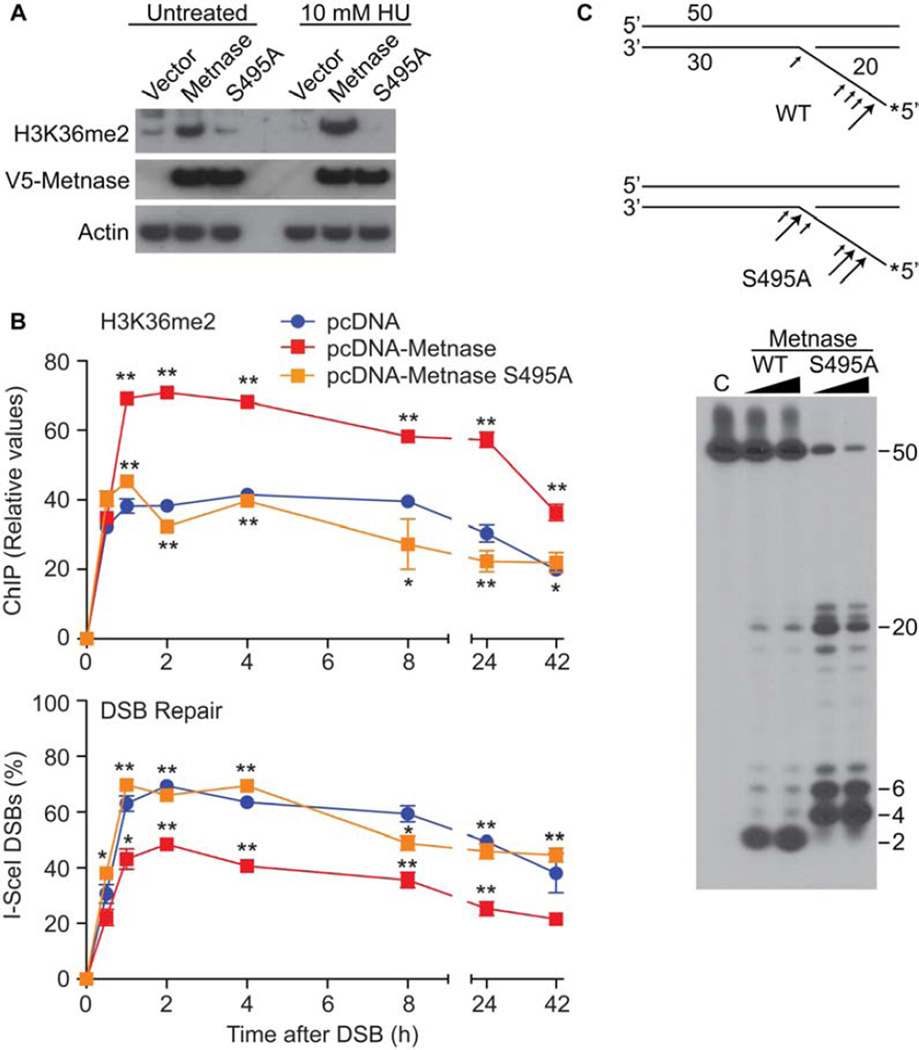
Metnase enhances DNA DSB repair via its histone methylase activity (Fnu et al, 2011). After a DNA DSB is formed, Metnase associates with chromatin adjacent to the DSB, and di-methylates H3K36. This di-methylation stabilizes NHEJ components such as the Ku complex at the DSB and increases the re-ligation of the DSB (Fnu et al, 2010). We isolated chromatin before and after ionizing radiation and hydroxyurea to measure its association with wt and S495A mutant Metnase by western blot (Fig. 4). We found that DNA damage with ionizing radiation increased the amount of total Metnase associated with chromatin moderately (2-fold). However, the S495A Metnase had little association with chromatin constitutively, and no increase after ionizing radiation (Fig. 4). It should be noted that the S495A species was expressed at lower levels than the wt Metnase, but not to the same extent as the decrease of the association of the S495A with chromatin. We then examined whether hydroxyurea could induce the phosphorylation of Metnase. Tagged Metnase was immunoprecipitated and then assessed for phosphorylation using the anti-pS495 (Fig. 4B). After 6 hours of hydroxyurea exposure, there was a 3-fold increase in the phosphorylation of Metnase. We also tested whether it was the phosphorylated Metnase species that associated with chromatin after IR (Fig. 4C). IR induces the association of phosphorylated Metnase with chromatin, while HU decreases this association. It should be noted that the endogenous levels of phosphorylated Metnase protein are lower in the IR samples, as they spend time at room temperature during the mock and test radiation exposure.
Figure 4. Phosphorylation of Metnase S495 promotes its association with chromatin after IR but not after replication stress.
A. Western analysis of chromatin-associated wild-type and S495A Metnase. Cells expressing FLAG-tagged Metnase or the S495A mutant were treated with 10 Gy IR, and chromatin was isolated at indicated times. Ku70 and histone H3 were used as nuclear protein loading controls. Expression of wild-type and S495A Metnase, analyzed by Western blot in total cell extracts, is shown to the right. B. V5-tagged wild-type or S495A Metnase were immunoprecipitated with anti-V5 and phospho-S495 Metnase was detected by Western blot (top panel). Middle and lower panels are V5-Metnase input and actin loading controls, respectively. C. Cells expressing FLAG-tagged Metnase were treated with 10 Gy IR, 10 mM HU, or mock treated, and phospho-S495 Metnase in chromatin-bound or unbound fractions was detected by Western blot. D. Chromatin association of wild-type and S495A Metnase as measured by coimmunoprecipitation analysis. FLAG-tagged wild-type or S495A Metnase were immunoprecipitated with anti-FLAG (middle panel) and their co-immunoprecipitation with histone H3 (top panel) was detected by Western blot. The lower panel shows input Metnase levels prior to immunoprecipitation. E. ChIP analysis of Metnase recruitment to a single DSB induced by I-SceI nuclease in HT1904 cells transfected with empty vector or over-expressing wild-thype or S495A Metnase (Fnu et al, 2011). ChIP was quantified by real-time PCR at indicated times after DSB induction. * P <0.05, ** P <0.01.
We next asked whether the S495A Metnase species could co-immunoprecipitate with histone 3, the target of Metnase’s histone methylase function (Fig. 4D). We found that the S495A Metnase interacted significantly less with histone 3 than the wt Metnase. We then used our previously described HT1904 cells in ChIP experiments to measure the relative concentration and rate at which wt versus S495A Metnase associated with a single Isce-I-induced DSB (Fnu et al, 2011). We found that transfecting wt Metnase increased the amount of Metnase detectable at this induced DSB, but that the S495A mutant failed to co-localize to the DSB region (Fig. 4E).
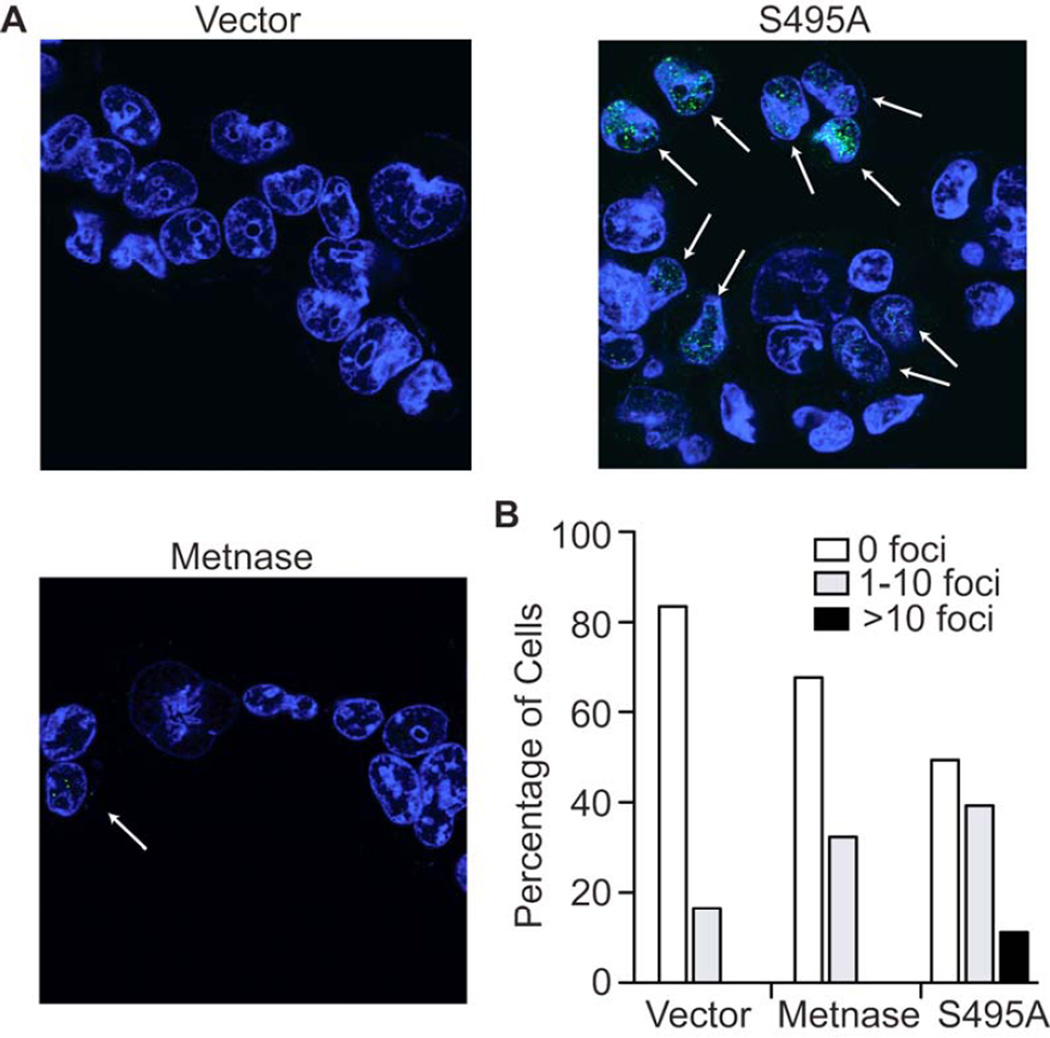
We then examined the role of the S495 phosphorylation on the activities of Metnase in 293T cells, which do not express Metnase by virtue of their transformation with T antigen (Williamson et al, 2008). Wt Metnase enhances restart of stalled forks after replication stress from hydroxyurea in these cells (De Haro et al, 2010). Recovery of replication forks after hydroxyurea exposure can be measured by the appearance of immunofluorescent BRDU foci, which designate regions of active incorporation of nucleotides (Helleday et al 2010). We found that the cells with the S495A Metnase had significantly more BRDU foci per cell than cells expressing wt Metnase after exposure and release from hydroxyurea for 15 minutes (Fig. 5A). There were 13% of 293T cells transduced with S495A mutant Metnase expressing greater than 10 foci per cell compared with no cells expressing wt Metnase (Fig. 5B). Thus, Metnase phosphorylation decreases its ability to enhance recovery of replication forks from stress, consistent with the known function of Chk1.
Figure 5. Expression of the S495A Metnase mutant increases replication fork recovery.
A. Confocal immunofluorescence of BRDU foci (green) in 293T cells expressing either wt or S495A Metnase species after recovery from exposure to hydroxyurea. The foci represent replication forks newly incorporating BRDU after hydroxyurea has been removed. B. Quantifying cells expressing BRDU foci demonstrates that cells with S495A Metnase species have significantly more foci than with wt Metnase. More than 500 cells were counted on at least 5 slides for each experimental arm.
We then assessed whether the S495A mutant could dimethylate H3K36, since it appeared that this mutant was defective in associating with an induced DSB. Metnase is universally expressed except in cells transformed by T antigen, for reasons that are not clear. However, such cells serve as a useful model to measure some Metnase functions. We found that in 293T cells the S495A mutant had a decreased ability to di-methylate H3K36 compared to wt Metnase by western analysis (Fig. 6A). We recently described a cell system, termed HT1904, in which a single DSB can be rapidly induced in the vast majority of cells using adenoviral expression of Isce-I (Fnu et al). Since the majority of cells have the DSB, this system can be used for Chromatin Immunoprecipitation (ChIP) analysis of chromatin components in real time, and repair of the DSB over that time course. ChIP analysis for the formation of H3K36me2 at a single induced DSB found that the S495A Metnase mutant was unable to induce H3K36me2 (Fig. 6B). More importantly, the S495A mutant was unable to re-ligate the induced DSB to the same extent as wt Metnase when measured by real time PCR across an Isce-I site (Fig. 6B).
Figure 6. Metnase phosphorylation regulates its biochemoical activities.
Metnase dimethylation of histone 3 lysine 35 (H3k36) and subsequent enhancement of DSB repair requires S495 phosphorylation. A. Western analysis of H3K36 dimethyltion (me2) by wt and S495A Metnase after DNA damage induced by hydroxyurea (HU). B. ChIP analysis (upper panel) over time after induction of a single DSB by ISCE-I for the adjacent presence of H3K36me2 with over-expression of wt or S495A Metnase (Fnu et al, 2011). The lower panel shows real time PCR assessment of the re-ligation of this induced single DSB over time. * P <0.05, ** p<0.01. C. Flap cleavage nuclease activities of pure isolated eukaryotic wt and S495A Metnase. The upper schematic diagrams the altered S495A flap nuclease activity. The size of the arrows correlates with the amount of endonucleolysis.
In addition, recombinant purified S495A Metnase protein, which in isolated form has no phosphorylated species (Supplemental Fig. 3B), had an altered nuclease activity, compared to pure recombinant wt Metnase, which is isolated from mammalian cells in a mixture of phosphorylated and non-phosphorylated forms (Fig. 6C). Cleavage products by the S495A species were larger in size, and were also more abundant. Finally, we had previously reported that Metnase interacts with TopoIIα and enhances its decatenation ability (Williamson et al, 2008, Wray et al, 2009). Therefore, we next examined whether the S495A mutant affected the ability of Metnase to enhance Topo IIα decatenation. We found that pure recombinant S495A protein was still able to promote TopoIIα decatenation (Supplemental Fig. 3).
DISCUSSION
Much of DNA replication and repair is regulated by effector protein phosphorylation (Bartek and Lukas 2003). Metnase is a recently described histone methylase and nuclease that plays a role in both replication and repair (De Haro et al 2010, Lee et al 2005). Since many of the effectors of these functions are regulated by phosphorylation, we examined whether Metnase was also regulated by phosphorylation. We used 32P labeling of intracellular Metnase, 2D phosphopeptide isolation, and mass spectroscopy to show that the major phosphorylation site of Metnase is S495. Several lines of evidence demonstrate that the S495A is the only Metnase phosphorylation site. First, mutating this site completely blocked incorporation of 32P orthophosphate into Metnase after DNA damage induction. Second, only one labeled phosphopeptide was seen in the 2D gel after DNA damage.
The S495 is within a Chk1 and 2 and a PAK consensus sequence. While each of these kinases could phosphorylate Metnase protein in vitro, Chk1 was many times more active. Inhibiting Chk1 with UCN-01 or repressing it with siRNA resulted in markedly decreased phosphorylation of Metnase, while other specific kinase inhibitors had only minimal effect. Even a modest knock-down of Chk1 has a dramatic on the loss of phosphorylation of Metnase, probably due to counter-balancing phosphastases that are still active (Leung-Pineda and ryan et al 2006). Indeed, consistent with this, we found that the Chk1 phosphatase PP2A mediated dephosphorylation of Metnase. Metnase may be undergoing a continuous, dynamic cycle of phosphorylation-dephosphorylation with Chk1 and PP2A. With decreased Chk1, there is more de-phosphorylation of Metnase from an increase in the activity of PP2A. A baseline level of phosphorylated Metnase was seen, not surprising since Chk1 is active even in unperturbed cells, likely by endogenous replication stress and DNA damage (Bartek and Lukas 2003).
The sequence where Metnase is phosphorylated is also a reasonable PKC site. However, we do not consider PKC to be a major kinase for Metnase for several reasons. First, Metnase is nuclear in location, and PKC is mainly cytoplasmic. Second, the PKC inhibitor GF109203X had little effect on the phosphorylation of Metnase. Third, while PKC is crucial to the apoptotic decisions a cell makes upon DNA DSB damage, it plays little known role in actual repair of DNA DSBs (Reyland).
These data indicated that the most important kinase phosphorylating Metnase is Chk1, although there is some redundancy. This redundancy could be important in activating the DNA repair activity of Metnase, as there may be situations where DNA is damaged, and Chk1 is not activated, but Chk2 and PAK are. Nonetheless, these data place Metnase within the epistatic cascade initiated by DNA damage, whether from exogenous or endogenous sources. These data also implicate a role for PP2A in replication fork recovery, as dephosphorylated Metnase appears to be more active in this role. This is consistent with previous reports that PP2A is required for deactivating Chk1 by dephosphorylation of S317, and for appropriate metabolism of RPA in the recovery of progression of stalled forks (Leung-Pineda and Ryan et al 2006; Feng and Wakemen et al 2009).
The phosphorylation of Metnase inhibits its role in replication fork recovery, and stimulates its role in DNA DSB repair. It is possible that the S495A mutant’s alteration of Metnase’s nuclease capability was due to altered structure of Metnase, from the introduction of this mutation, although our virtual modeling provided no evidence for that. The S495 is on the exterior of the Metnase protein, and would not be predicted to alter its structure. Thus, the altered nuclease capability of unphosphorylated S495A Metnase has several implications. First, it means that the nuclease activity of Metnase could have an important role in replication fork recovery, since this was increased in the S495A species. The dephosphorylated Metnase nuclease activity more closely resembles FEN-1, cutting close to the flap junction, than the wt Metnase activity. This altered nuclease capability may be more effective for fork restart as compared to end-joining of a DSB. Second, it also implies that the specific nuclease activity of the wt Metnase species is important for Metnase’s role in DSB repair, since this function is decreased in the S495A mutant. We have recently reported that Metnase is important for resecting overhangs from incompatible free DNA ends during non-homologous end joining repair (Beck et al, 2011). The nuclease activity of phosphorylated Metnase may be important in the processing of these free incompatible free DNA ends.
The decrease in cellular H3K36me2 seen with the S495A Metnase species may be due to the decreased chromatin association seen in this mutant. Indeed, the reduction in DSB repair seen with the S495A mutant implies that H3K36me2 formation and DSB repair are both enhanced by phosphorylation. It also implies that this repair is dependent on chromatin association of Metnase and subsequent H3K36me2 formation, as we previously demonstrated (Fnu et al 2011). In addition, it also suggests that the H3K36 di-methylation activity and chromatin association is not important for the ability of Metnase to enhance replication fork recovery after stress. The inability of the S495A mutant to associate with chromatin surrounding a DSB and mediate the re-ligation of this DSB indicates that phosphorylation of the S495 is a significant step in its role in DNA DSB repair. It also implies a mechanism for Metnase function in these roles: In this model the presence of DNA lesions activate Chk1, which phosphorylates Metnase. Phosphorylated Metnase is then recruited to chromatin adjacent to the DSB, and di-methylates H3K36 there, which would stabilize recruited NHEJ components, such as Ku70 and NBS1, and thereby enhance repair. Since the SET domain is not structurally altered by the S495 phosphorylation, it is likely that histone methylation is intact after phosphorylation. Thus, the decrease in H3K36me2 at the DSB region in the S495A species is likely from decreased localization of this mutant Metnase to the chromatin at that site. The ChIP data demonstrating the decreased presence of Metnase at the DSB over the time course of repair of an induced DSB corroborates this hypothesis.
The canonical role of Chk1 is to halt replication until DNA is repaired by phosphorylating cdc25a, which leads to its ubiquitinylation and degradation, and subsequent replication arrest (Donzelli and Draetta 2003). The phosphorylation of Metnase is another mechanism by which Chk1 can perform its essential roles, inhibiting replication recovery and promoting repair of DNA lesions. For Metnase to promote replication fork recovery after arrest, perhaps via its nuclease activity, it needs to be in the dephosphorylated state. However, phosphorylation splits Metnase activities, as phosphorylation enhances its localization to chromatin, and subsequent H3K36 dimethylation and DSB re-ligation. Thus, the phosphorylation of Metnase differentiates its two main functions, inhibiting the replication fork recovery assistance and enhancing DNA repair, the classic roles of Chk1. This is significant for the cell, as it would be detrimental to promote replication fork recovery before DNA damage is repaired. This would result in significant genomic instability or cell death. In summary, these data define Metnase as a novel Chk1 effector, mediating important downstream functions of the Chk1 cascade.
METHODS AND MATERIALS
Cell culture, proteins, and antibodies
Establishment of human 293 and 293T cells expressing wild type and mutant Metnase were carried out as previously described (Williamson et al, 2008). Cells were grown in Dulbecco's Modified Eagle Medium (DMEM) (Hyclone, Fisher Scientific, Pittsburgh, PA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and antibiotic-antimycotic reagent (penicillin G, 10 units/ml; streptomycin, 10 µg/ml; amphotericin B, 0.25 µg/ml) (Invitrogen, Carlsbad, CA). Purified His-Chk1, GST-Chk2 and GST-PAK2 kinases were obtained from Signal Chem (Richmond, British Columbia, CA). The following antibodies were used: FLAG, actin (Sigma, St. Louis, MO); PP2A (Millipore, Billerica, MA); H3K36me2 (Abcam, Cambridge, MA); Chk1, Chk2, PAK2, BrdU (Cell Signaling, Danvers, MA); V5 (Invitrogen, Carlsbad, CA); phosphorylated S495 Metnase (pS495, custom made, ProSci Inc, Poway, CA). Control and Metnase-specific siRNAs were used as described previously to manipulate Metnase levels (Lee et al., 2006, Fnu et al 2011).
Preparation of cell lysates and immunoblot analysis
Cells (1×105) were collected, washed with PBS, and lysed in a buffer containing 25 mM HEPES (pH 7.5), 0.3 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5% Triton X-100, 20 mM β-glycerolphosphate, 1 mM sodium vanadate, 1 mM DTT, protease inhibitor cocktails (Sigma). Cell lysates (50 µg) were loaded onto a SDS-PAGE and, following gel electrophoresis, proteins were transferred to a PVDF membrane (Millipore, Billerica, MA) and immunoblotted with primary antibody followed by peroxidase-coupled secondary antibody (Amersham, Piscataway, NJ) and an enhanced chemiluminescence (Amersham) reaction prior to visualization on Kodak-o-mat film.
In vivo orthophosphate labeling
Human 293 cells over-expressing Flag-Metnase were incubated with 1 mCi of 32P-orthophosphoric acid for 1 hr. Following IR treatment (20 Gy), cells were further incubated for 1 hr prior to harvest. Cell extracts (500 ug) were mixed with agarose-conjugated Flag-antibody beads for 1 hr for immunoprecipitation of Flag-Metnase. Samples were analyzed by 10% SDS-PAGE, dried and exposed to x-ray film.
Phosphopeptide mapping using nano-LC/MS/MS
Coomassie blue-stained gel pieces were placed in Eppendorf centrifuge tubes. The gel pieces were immersed in 100ul of 50% 0.1M ammonium bicarbonate and 50% acetonitrile (by volume) to completely covers the gel pieces. Samples were then incubated at 37°C for 30min. After incubation, the reagent was drawn and discarded. The gel pieces were dehydrated with 100 ul of pure acetonitrile for approximately 5min, and excess acetonitrile decanted. The gel pieces were rehydrated with 100ul of 10mM DTT (fleshly made) and incubated at 55° C for 45 min for reduction. After moved reduction solution, 100ul of 55mM iodoacetamide (fleshly made) was added to the gel pieces and incubated for 30 min at 37oC for alkylation. The gel pieces were dehydrated with 100 ul of pure acetonitrile for approximately 5 min. The excess acetonitrile was decanted and dried under vacuum for 15min. The gel pieces were then rehydrated with a trypsin solution (Promega V5280, 0.1–0.5 ug) and incubated overnight at 37°C. Following digestion, peptides were extracted with a 0.1% trifluoroacetic acid (TFA) for 30min at 37°C. Tryptic digests (10 ul) were injected onto a 75um×5 cm C-18 reverse-phase column, and peptides were eluted with a gradient from 5 to 45% acetonitrile developed over 30min at a flow rate of 250nl/min using Agilent 1100 series nanopump. The column was interfaced with a LTQ ion trap mass spectrometer (Thermo), and data were collected in the Data Dependent Neutral Loss MS3 mode. Neutral loss masses were 98 (+1), 49 (+2) and 32.7 (+3). MS/MS and MS/MS/MS spectra were searched against the IPI human protein database with SEQUEST algorithm.
Kinase and phosphatase assays
For the in vitro kinase assays, 100 ng of the purified kinase (Chk1, Chk2 or PAK2,) were incubated with 200 ng pure recombinant Metnase nuclease fragment (aa 433–671) in kinase buffer (25 mM MOPS, 12.5 mM α-glycerol-phosphate, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA, 0.25 mM DTT) with 250 uM γ-32P-ATP at 30°C for 30 minutes. Recombinant Metnase was generated as previously described (Roman et al, 2008). The reaction was stopped by the addition of loading dye and then separated by 4–12% gradient SDS-PAGE, the gel dried and exposed to x-ray film. Phosphatase reactions were carried out using immunoprecipitated V5-tagged Metnase protein with 400 U/ml λ-phosphatase (New England Biolabs, Ipswich, MA) in the presence or absence of phosphatase inhibitors (0.5 M EDTA, 0.5 M NaF, and 0.1 M NaVO4). Reactions were carried out at 37°C for 30 minutes and then separated by 4–12% gradient SDS-PAGE. Loss of phosphorylation was shown either by exposing the dried gel to x-ray film or immunoblotting with the phosphorylated serine495-specific antibody (anti-pS495). This anti-sera had been pre-cleared for reaction with Metnase unphosphorylated peptide. Kinase inhibition was carried out in vivo by incubating 293 cells with various inhibitors for 1 hr, then immunoblotting for the presence of phosphorylated S495 Metnase using the specific anti-p-ser495 antibody. Concentrations used: UCN-01 10 uM, Staurosporine 50 uM, CKII 20 uM, PD90859 10 uM, and GF109203x 10 uM, all for 1 hr. For phosphatase assays, Okadaic acid, a PP1 and 2A inhibitor, was incubated for 1 hr at 10 uM.
Chk1 and protein phosphatase 2A (PP2A) knock down
293T cells stably over-expressing wt Metnase were transiently transfected with shRNA against Chk1 and PP2A (SABiosciences, Frederick, MD) according to the manufacturer’s instructions. Forty-eight hours post-transfection cells were harvested, protein lysates were prepared. Wt Metnase was immunoprecipitated and immunoblotted with anti-pSer495 to determine the level of phosphorylation.
Nuclease Assays
Wt and S495A mutant Metnase protein were isolated as previously described (Roman et al, 2007). Nuclease activity of these proteins were carried out by incubation with γ-P32 radiolabeled pseudo-Y structures and autoradiography after separation of the cleaved products by urea denaturing gel electrophoresis as described (Roman et al, 2007).
BrdU immunofluorescence
293T cells stably expressing either the wt Metnase or the S495A Metnase mutant, along with the empty vector control stable cell line were seeded onto coverslips and allowed to adhere for 6 hours. After this time, 10 mM hydroxyurea (HU) was added for 18 hours. The HU was washed off and replaced with media containing 0.03 mg/ml BrdU. BrdU incorporation was stopped at 15 minutes, then fixed and processed for immunostaining using a BrdU-specific antibody (Cell Signaling) according to the manufacturer’s instructions (Helleday et al 2010). Images were collected using a Zeiss LSM-510 confocal microscopy. Five hundred cells were counted in each condition from at least 3 distinct slides.
Chromatin immunoprecipitation (ChIP) and real time PCR
ChIP analysis for the presence of wt and mutant Metnase, and H3K36me2, at the chromatin region adjacent to an induced DSB was performed as previously described (Fnu et al, 2011). In addition, assessment of the re-ligation of the induced DSB as a measure of end-joing DNA repair using real time PCR was also described previously (Fnu et al, 2011). Briefly, HT1904 cells containing a single allele Isce-I site in a puromycin resistance gene (puro) were transiently transfected with empty control vector (pcDNA3.1) or the Metnase expression vectors (pcDNA3.1-Metnase, pcDNA3.1-S495A). These cells were then infected with high titer adenovirus expressing Isce-I to induce a DSB in the puro gene. Cells were harvested at various time points for ChIP analysis of H3K36me2, or wt versus S495A mutant Metnase, associated with the DSB region, and real time PCR analysis of re-ligation of the Isce-I DSB site. Each experiment was performed twice in triplicate. Standard error of the mean is calculated and shown, as well as Student’s t-test for significance. All ChIP experimental values were obtained also by real time PCR normalized to the input DNA using amplification of GAPDH.
Assessment of chromatin-bound Metnase species
Immunoblot analysis of chromatin-bound Metnase was carried out as previously described (Lee et al., 2006). Human 293 cells were harvested one hour following IR treatment, and treated with cell lysis buffer I (25 mM Hepes, pH 7.5, 0.3 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5% Triton X-100, 0.5 mM DTT, and 1mM PMSF) and incubated for 90 min on ice. After centrifugation at 14,000 g for 30 min, pellets were extensively washed four times with cell lysis buffer I, resuspended in SDS–PAGE loading buffer, and analyzed by a 10% SDS-PAGE. Gels were transferred to PVDF membrane, probed with anti-FLAG (for Metnase), -Ku70, or -histone H3 antibody (monoclonal mouse IgG, Oncogene Sciences) followed by HRP-conjugated secondary antibody. Proteins were visualized by using the ECL system (Amersham).
Modeling of Metnase
The S495 phosphorylation site was modeled in a 3-dimensional structure of Metnase, The 3-dimensional structures of Metnase were generated using PyMOL v. 0.99 X-ray crystallographic structures of Metnase (3F2K, 3K9J, 3K9K) and the MOS-1 Mariner transposase (2F7T) (Arnold and Keifer 2009, Goodwin 2010). The Metnase structure from the SWISS-MODEL Repository was superimposed in DeepView/SwissPDBViewer v.4.0.1 using the alpha carbons.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. J-S You at the IU Proteomics Core facilities for MS/MS phosphopeptide analysis, and Dr. H. Chun for providing purified ATM kinase. This research was supported by grants from NIH (CA92111 to SHL), the IU Cancer Center, and the Walther Oncology Center. BDB was supported by NIH predoctoral training grant (NIDDK 1 T32 DK 0719-15). The authors acknowledge K. Valerie for the kind gift of the adenovirus I-SceI system, the support of NIH R01 GM084020 and NIH R01 CA100862 (J. N.), an APRC supplement CA100862 (J.N. and R.H.), NIH R01 CA102283 (R.H.), NIH R01 HL075783 (R.H.), NIH R01 CA139429 (R.H.), and the Leukemia and Lymphoma Society SCOR 7388-06 (R.H.).
Footnotes
CONTRIBUTIONS: R.H. conceived of and analyzed experiments, and wrote the manuscript. S.F., E.A.W., Y.-J.L., S.-J.P., B.D.B., and J.-S. Y. performed and analyzed the experiments. A.L. performed the modeling analysis. J.A.N. analyzed experiments, and wrote the manuscript. S.-H.L. designed experiments, analyzed data, and wrote the manuscript.
DISCLOSURES: R.H. serves as a scientific advisor to Northlake Pharmaceuticals. No other author has any disclosure.
REFERENCES
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Nuclear localization signals (NLS) Crit Rev Eukaryot Gene Expr. 1993;3:193–227. [PubMed] [Google Scholar]
- Arnold K, Kiefer F, Kopp J, Battey JN, Podvinec M, Westbrook JD, Berman HM, Bordoli L, Schwede T. The Protein Model Portal. J. Struct. Funct. Genomics. 2009;10:1–8. doi: 10.1007/s10969-008-9048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck BD, Lee SS, Williamson E, Hromas RA, Lee SH. Biochemical characterization of metnase's endonuclease activity and its role in NHEJ repair. Biochemistry. 50:4360–4370. doi: 10.1021/bi200333k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska H, Knaus UG, Wang ZY, Bokoch GM, Korn ED. p21-activated kinase has substrate specificity similar to Acanthamoeba myosin I heavy chain kinase and activates Acanthamoeba myosin I. Proc Natl Acad Sci U S A. 1997;94:1092–1095. doi: 10.1073/pnas.94.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, Hooykaas P. An Arabidopsis hAT-like transposase is essential for plant development. Nature. 2005;436:282–284. doi: 10.1038/nature03667. [DOI] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci U S A. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, Virshup DM, Firulli AB. Phosphopeptide mapping of proteins ectopically expressed in tissue culture cell lines. Biol Proced Online. 2004;6:16–22. doi: 10.1251/bpo69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fnu S, Williamson EA, DeHaro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Yamada C, Tsurumi T, Hanaoka F, Matsuzawa K, Inagaki M. Cell cycle- and chromatin binding state-dependent phosphorylation of human MCM heterohexameric complexes. A role for cdc2 kinase. J Biol Chem. 1998;273:17095–17101. doi: 10.1074/jbc.273.27.17095. [DOI] [PubMed] [Google Scholar]
- Goodwin KD, He H, Imasaki T, Lee S-H, Georgiadis MM. Crystal Structure of the Human Hsmar1-Derived Transposase Domain in the DNA Repair Enzyme Metnase. Biochemistry. 2010;49:5705–5713. doi: 10.1021/bi100171x. [DOI] [PubMed] [Google Scholar]
- Haren L, Ton-Hoang B, Chandler M. Integrating DNA: transposases and retroviral integrases. Annu Rev Microbiol. 1999;53:245–281. doi: 10.1146/annurev.micro.53.1.245. [DOI] [PubMed] [Google Scholar]
- Jordan IK. Evolutionary tinkering with transposable elements. Proc Natl Acad Sci U S A. 2006;103:7941–7942. doi: 10.1073/pnas.0602656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB. Cell cycle. Checking two steps. Nature. 2001;410:766–767. doi: 10.1038/35071218. [DOI] [PubMed] [Google Scholar]
- Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, Kwan L, Nickoloff JA, Hromas R. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc Natl Acad Sci U S A. 2005;102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Park SJ, Ciccone SL, Kim CR, Lee SH. An in vivo analysis of MMC-induced DNA damage and its repair. Carcinogenesis. 2006;27:446–453. doi: 10.1093/carcin/bgi254. [DOI] [PubMed] [Google Scholar]
- Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Leis J, Johnson S, Collins LS, Traugh JA. Effects of phosphorylation of avian retrovirus nucleocapsid protein pp12 on binding of viral RNA. J Biol Chem. 1984;259:7726–7732. [PubMed] [Google Scholar]
- Liu D, Bischerour J, Siddique A, Buisine N, Bigot Y, Chalmers R. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol Cell Biol. 2007;27:1125–1132. doi: 10.1128/MCB.01899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Mirzoeva OK, Wells J, Olivares H, Williams BR, Zinkel RA, Farnham PJ, Petrini JH. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol Cell Biol. 2001;21:6006–6016. doi: 10.1128/MCB.21.17.6006-6016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JH. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murti KG, He DC, Brinkley BR, Scott R, Lee SH. Dynamics of human replication protein A subunit distribution and partitioning in the cell cycle. Exp Cell Res. 1996;223:279–289. doi: 10.1006/excr.1996.0083. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Taniguchi T, Ranganathan V, New HV, Moreau LA, Stotsky M, Mathew CG, Kastan MB, Weaver DT, D'Andrea AD. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat Cell Biol. 2002;4:913–920. doi: 10.1038/ncb879. [DOI] [PubMed] [Google Scholar]
- O'Neill T, Giarratani L, Chen P, Iyer L, Lee CH, Bobiak M, Kanai F, Zhou BB, Chung JH, Rathbun GA. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J Biol Chem. 2002;277:16102–16115. doi: 10.1074/jbc.M111705200. [DOI] [PubMed] [Google Scholar]
- Park SJ, Ciccone SL, Freie B, Kurimasa A, Chen DJ, Li GC, Clapp DW, Lee SH. A positive role for the Ku complex in DNA replication following strand break damage in mammals. J Biol Chem. 2004;279:6046–6055. doi: 10.1074/jbc.M311054200. [DOI] [PubMed] [Google Scholar]
- Park SJ, Lee YJ, Beck BD, Lee SH. A positive involvement of RecQL4 in UV-induced S-phase arrest. DNA Cell Biol. 2006;25:696–703. doi: 10.1089/dna.2006.25.696. [DOI] [PubMed] [Google Scholar]
- Reyland ME. Protein kinase C isoforms: Multi-functional regulators of cell life and death. Front Biosci. 2009;14:2386–2399. doi: 10.2741/3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HMaZ, KL Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene. 1997;205:203–217. doi: 10.1016/s0378-1119(97)00472-1. [DOI] [PubMed] [Google Scholar]
- Roig J, Traugh JA. p21-activated protein kinase gamma-PAK is activated by ionizing radiation and other DNA-damaging agents. Similarities and differences to alpha-PAK. J Biol Chem. 1999;274:31119–31122. doi: 10.1074/jbc.274.44.31119. [DOI] [PubMed] [Google Scholar]
- Tahara SM, Traugh JA. Cyclic Nucleotide-independent protein kinases from rabbit reticulocytes. Identification and characterization of a protein kinase activated by proteolysis. J Biol Chem. 1981;256:11558–11564. [PubMed] [Google Scholar]
- Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol Cell Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venere M, Mochan TA, Halazonetis TD. Chk2 leaves the PML depot. Nat Cell Biol. 2002;4:E255–E256. doi: 10.1038/ncb1102-e255. [DOI] [PubMed] [Google Scholar]
- Wang H, Zeng ZC, Bui TA, DiBiase SJ, Qin W, Xia F, Powell SN, Iliakis G. Nonhomologous end-joining of ionizing radiation-induced DNA double-stranded breaks in human tumor cells deficient in BRCA1 or BRCA2. Cancer Res. 2001;61:270–277. [PubMed] [Google Scholar]
- Weinmann AS, Bartley SM, Zhang T, Zhang MQ, Farnham PJ. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol Cell Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS, Farnham PJ. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- Wilkes MC, Murphy SJ, Garamszegi N, Leof EB. Cell-type-specific activation of PAK2 by transforming growth factor beta independent of Smad2 and Smad3. Mol Cell Biol. 2003;23:8878–8889. doi: 10.1128/MCB.23.23.8878-8889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.