Abstract
Genome-scale metabolic models are built using information from an organism's annotated genome and, correspondingly, information on reactions catalyzed by the set of metabolic enzymes encoded by the genome. These models have been successfully applied to guide metabolic engineering to increase production of metabolites of industrial interest. Congruity between simulated and experimental metabolic behavior is influenced by the accuracy of the representation of the metabolic network in the model. In the interest of applying the consensus model of Saccharomyces cerevisiae metabolism for increased productivity of triglycerides, we manually evaluated the representation of fatty acid, glycerophospholipid, and glycerolipid metabolism in the consensus model (Yeast v6.0). These areas of metabolism were chosen due to their tightly interconnected nature to triglyceride synthesis. Manual curation was facilitated by custom MATLAB functions that return information contained in the model for reactions associated with genes and metabolites within the stated areas of metabolism. Through manual curation, we have identified inconsistencies between information contained in the model and literature knowledge. These inconsistencies include incorrect gene-reaction associations, improper definition of substrates/products in reactions, inappropriate assignments of reaction directionality, nonfunctional β-oxidation pathways, and missing reactions relevant to the synthesis and degradation of triglycerides. Suggestions to amend these inconsistencies in the Yeast v6.0 model can be implemented through a MATLAB script provided in the Supplementary Materials, Supplementary Data S1 (Supplementary Data are available online at www.liebertpub.com/ind).
Introduction
Biodiesel can serve as a renewable alternative to petroleum-derived diesel. The current major feedstock for commercial production of biodiesel in the United States is soybean oil.1 However, the use of soybean oil poses several issues such as the necessity for arable cropland, competition with food uses, and high feedstock costs. Alternative feedstocks grown on marginal lands or generated from waste or agricultural and forest residues could address these issues. The microbial conversion of these alternative feedstocks into lipids has been demonstrated to be technically feasible.2 Nonetheless, high lipid productivity will be essential for commercial production of biodiesel that is economically and environmentally sustainable.
Metabolic engineering can serve to increase productivity by driving more metabolic flux into lipids. A challenge in developing effective schemes for metabolic engineering is predicting the effects of genetic manipulations a priori. This is due to the complexities of the metabolic network (eg, redundancy, high level of interconnectedness, presence of alternative metabolic routes, and control mechanisms). Thus, it is imperative that we move toward a systems approach to understanding the workings of cells. Genome-scale metabolic models can be used to predict or describe flux distributions throughout the entirety of a cell's known metabolic network. By extension, these models can also be used to predict the outcome of genetic perturbations and to determine optimal engineering strategies.3 An accurate representation of the metabolic network is crucial for simulation of cellular behavior. The endeavor to create an accurate representation is often an iterative process that requires repeated expansion or correction of the model based on comparison of model predictions to experimental data and/or the discovery of errors in the model.4 In this paper, we manually evaluated the representation of fatty acid, glycerolipid, and glycerophospholipid metabolism in the consensus model of yeast metabolism (Yeast v6.0).5 These areas of metabolism were chosen due to the connection and coordination between glycerophospholipid and glycerolipid (eg, triglyceride) metabolism and the function of fatty acids as precursors to both lipids. The description of these metabolic pathways in the model was compared to the knowledge described in the scientific literature. Through this comparison, we identified biologically inconsistent representations (eg, substrate specificity, compartmentalization of reactions, reaction directionality), missing connections between metabolites, and limitations in the lumped representation of metabolites. We have compiled a collection of suggested changes to the Yeast v6.0 model to contribute to the continuing community effort to develop a genome-scale reconstruction of yeast metabolism.
Materials and Methods
Yeast v6.0 Model
The Systems Biology Markup Language (SBML) file for the Yeast v6.0 model was downloaded from http://yeast.sourceforge.net and is included in the Supplementary Materials section, Supplementary Data S2, (Supplementary Data are available online at www.liebertpub.com/ind). This SBML file was read into MATLAB R2009a (MathWorks, Natick, MA) using the readCbModel function in the Constraints Based Reconstruction and Analysis (COBRA) Toolbox 2.0.5 via libSBML 5.6.0 and the SBMLToolbox 4.1.0.6–8
Curation Process
To facilitate model investigation, MATLAB functions were developed to explore the reactions, metabolites, and genes contained in the model. The following custom functions were frequently utilized during the curation process: metInfo, rxnInfo, and cgpr. These functions extract information of interest from the COBRA-format model and display this information in a human-readable format. The function metInfo displays the names of all the reactions with which a metabolite is involved; rxnInfo displays the genes associated with a reaction, lower/upper flux bounds, and the metabolites involved in the reaction. The sequential use of metInfo and rxnInfo allows one to leapfrog throughout the model. For instance, rxnInfo can be used to gain information on a particular reaction, and metInfo can be used to learn more about the metabolites in the reaction by generating a list of other reactions in which the metabolite participates. This cycle can be repeated for the newly generated list of reactions. The function cgpr, which stands for common gene name-protein-reaction relationship, displays information on the reactions associated with a gene. The common gene name utilized for this function should be consistent with the standard name specified in the Saccharomyces Genome Database (www.yeastgenome.org). All three of these functions are provided in the Supplementary Materials (Supplementary Data S3, S4, and S5).
Examination of the reconstruction of fatty acid, glycerolipid, and glycerophospholipid metabolism was initiated by first defining the set of genes involved in this area of metabolism. A recent review article served as a source for generating this list of genes.9 The cgpr function was utilized to compare the information contained in the model with current knowledge from scientific literature for each of the genes of interest. To ensure consideration of reactions not properly annotated with the appropriate gene(s) in the model, the model was also queried using metabolites as the search basis using the functions metInfo and rxnInfo.
Discrepancies between the model and literature evidence were addressed by revising the COBRA-format Yeast v6.0 model using functions contained in the COBRA toolbox. The function changeGeneAssociation was used to amend false gene-reaction relationships in the model. changeRxnBounds was used to change the lower and/or upper bound constraints for reaction flux, and addReaction was used to add reactions for catalytic activity and for transport of metabolites between compartments. removeRxns was typically used in conjunction with addReaction to remove the previous representation of a reaction and to replace with an updated representation. The collection of all changes made to the COBRA-format model was documented in a MATLAB code that, when executed, applies all the changes (Supplementary Data S1).
Determining Blocked Reactions and Essential Genes
Blocked reactions and essential genes in the model were determined in silico using an updated version of the testYeastModel MATLAB function included in the Yeast 5 paper (Supplementary Data S6).10 Blocked reactions were identified using flux variability analysis to search for reactions incapable of carrying fluxes under any media conditions (ie, the flux bounds on all exchange reactions are relaxed to allow for the unconstrained uptake of all the extracellular metabolites included in the model). Essential genes were predicted by screening all possible gene knockout strains for inability to produce biomass at or above the growth rate threshold of 10−6/hr in glucose minimal media; this simulation was performed using flux balance analysis. The solver used for both flux balance analysis and flux variability analysis was Gurobi 5.0.2 (Gurobi Optimization, Houston, TX). The list of in silico essential genes was compared to the list of in vivo essential genes compiled in testYeast.
Results and Discussion
Revised Representation of Fatty Acid Metabolism
Changes to representation of fatty acid synthesis
Fatty acids serve as building blocks for membrane lipids and as stores of chemical energy. The profile of fatty acids found in S. cerevisiae is mostly dominated by C16:0, C16:1, C18:0, and C18:1 fatty acids; minor amounts of C14:0 and very long-chain fatty acids, such as C26:0, are also present.9 Cellular fatty acids can be obtained through uptake from media, de novo synthesis, and lipid turnover.11 In this section, changes made to the Yeast v6.0 model that are relevant to de novo fatty acid synthesis are discussed.
S. cerevisiae has two fatty acid synthase (FAS) systems to generate fatty acids through successive additions of two carbons to a starting acetyl moiety. Cytoplasmic FAS—a complex composed of two subunits, Fas1p and Fas2p—releases C16:0 and C18:0 fatty acids esterified to coenzyme A.12 In contrast, mitochondrial fatty acid synthesis is achieved by sequential action of discrete, individual enzymes and generates C8:0 fatty acids bound to an acyl carrier protein (ACP).13 The enzymes involved in this process are acyl-carrier protein (Acp1p), malonyl-CoA:ACP transferase (Mct1p), β-ketoacyl-ACP synthase (Cem1p), 3-oxoacyl-ACP reductase (Oar1p), 3-hydroxyacyl-thioester dehydratase (Htd2p), and enoyl-ACP reductase (Etr1p).13 Although there is evidence to suggest in vitro catalytic capacity for longer fatty acids, in vivo evidence and understanding of the roles of mitochondrial FAS products with chain lengths greater than eight carbons are lacking.13
The Yeast v6.0 model depicts cytoplasmic fatty acid synthesis with a lumped reaction for the extension of acetyl-CoA (C2:0) to eight carbons and individual reactions for the addition of two carbons going from eight carbons up to 18 carbons in length (Fig. 1a). This representation is misleading since the reaction intermediates are shuttled within the complex and are not released into the cytosol in vivo. Cytoplasmic FAS is also modeled as being capable of utilizing and producing both free fatty acids and fatty acids esterified to coenzyme A. The erroneous inclusion of free fatty acids as products in the model may have arisen from comparison to mammalian cells.11 To amend both issues, the aforementioned reactions could be replaced with two reactions describing the net equation for generation of the dominant products of cytoplasmic FAS [ie, palmitoyl-CoA (C16:0) and stearoyl-CoA (C18:0)] from acetyl-CoA (Fig. 1b).
Fig. 1.
Curation of cytoplasmic and mitochondrial fatty acid synthesis (FAS) in the Yeast v6.0 model. (a) Reactions in the Yeast v6.0 model relevant to cytoplasmic and mitochondrial FAS. Dashed arrows indicate the net outcome of two reactions (ie, transport and hydrolysis). Mitochondrial FAS reactions leading to production of myristoleate, palmitoleate, and linoleic acid are blocked reactions, as indicated by X's in the figure. (b) Proposed representation of cytoplasmic and mitochondrial FAS. For the sake of space, the repeated reaction sequence of CEM1, OAR1, HTD2, and ETR1 for butanoyl-ACP (C4:0) to octanoyl-ACP is omitted, as indicated by the dashed arrow. The reactions for mitochondrial FAS are blocked reactions, as indicated by X's.
In the Yeast v6.0 model, mitochondrial fatty acid synthesis is depicted similarly to cytoplasmic fatty acid synthesis, which allows for the mitochondrial system to create products of the same carbon lengths as the cytoplasmic system (Fig. 1a). Acyl-ACPs from mitochondrial FAS are portrayed as being able to be transported to the cytoplasm where they can be hydrolyzed to free fatty acids through action of Fas1p and Fas2p. Yeast v6.0 also includes reactions to generate unsaturated acyl chains of 14, 16, and 18 carbon lengths through mitochondrial FAS. Given that the utility and destination of mitochondrial FAS products besides octanoyl-ACP are unknown, the reactions for longer acyl-chains may be considered inappropriate (Fig. 1b).13 Octanoyl-ACP serves as a precursor for lipoic acid, an essential cofactor in oxidative decarboxylation reactions.11 However, it should be noted that gaps remain in the reconstructed pathway for de novo synthesis of lipoic acid.
Changes to representation of fatty acid elongation
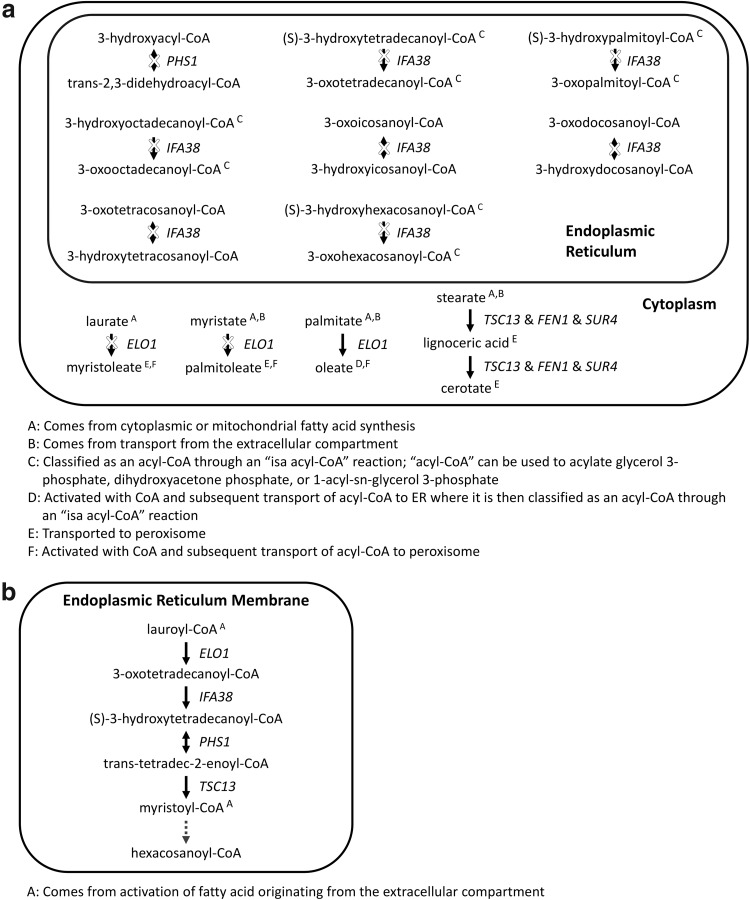
Another set of enzymes is needed to produce acyl chains longer than 18 carbons. These very long-chain fatty acids are synthesized through reaction mechanisms similar to fatty acid synthesis; the difference is that medium to very long-chain acyl-CoA's serve as substrates for addition of two-carbon units. Fatty acid elongation is catalyzed by the sequential action of the following endoplasmic reticulum (ER) membrane-localized enzymes: elongases Elo1p, Fen1p, or Sur4p; β-ketoacyl-CoA reductase, Ifa38p; β-hydroxyacyl-CoA dehydratase, Phs1p; and enoyl-CoA reductase Tsc13p.14–19
The Yeast v6.0 model describes fatty acid elongation in a biologically inconsistent manner. The model currently has seven reactions depicting the activity of Ifa38p on various metabolites; a reaction describing the activity of Phs1p on the generic metabolite 3-hydroxyacyl-CoA; three reactions for the elongation and desaturation of free fatty acids from C12:0 to C14:1, C14:0 to C16:1, and C16:0 to C18:1 that are associated with ELO1; and two reactions for the elongation of free fatty acids from C18:0 to C24:0 and C24:0 to C26:0 that are associated with TSC13, FEN1, and SUR4 (Fig. 2a). The current representation does not capture the interplay between the different enzymes involved in fatty acid elongation. For instance, the individual reactions for IFA38 and for PHS1 are not connected to the rest of fatty acid elongation since the other reactions consider net reactions as opposed to including intermediate steps. Evaluation of the net reactions reveals several inaccuracies. The gene associations for these reactions are incomplete since β-ketoacyl-CoA reductase and β-hydroxyacyl-CoA dehydratase are also needed to catalyze the net reaction. Another error is that the elongation process should utilize and generate fatty acids esterified to coenzyme A, not free fatty acids. In addition to the two issues discussed above, Elo1p is not able to introduce a double bond to the fatty acid chain.
Fig. 2.
Curation of fatty acid elongation in the Yeast v6.0 model. (a) Reactions in the Yeast v6.0 model relevant to fatty acid elongation. As indicated by X's, all of the reactions associated with PHS1 or IFA38 are blocked, as are two of the three reactions associated with ELO1. (b) Proposed representation of fatty acid elongation. For the sake of space, the repeated reaction sequence of (ELO1, FEN1, or SUR4), IFA38, PHS1, and TSC13 for myristoyl-CoA (C14:0) to hexacosanoyl-CoA (C26:0) is omitted, as indicated by the dashed arrow. For this omission, the embedded intermediates of palmitoyl-CoA (C16:0) and stearoyl-CoA (C18:0) are assumed to be also derived from activation of fatty acid originating from the extracellular compartment or from cytoplasmic fatty acid synthesis.
One solution to these issues is to remove the desaturation reactions associated with ELO1, expand upon all the steps involved in the elongation process with each individual gene associated with the appropriate reaction, and use acyl-CoA species instead of free fatty acids (Fig. 2b). This modification removes dead-ends in the model by improving metabolite connections and ensuring that the products of one reaction serve as the reactants in another reaction. The expansion of the individual reaction steps allows for the description of the acyl-CoA specificity of the different elongase enzymes. The elongases have different reactant preferences and also differ in the length of the acyl-chain produced by the elongation cycle.17,20 The proposed modification assumes that Elo1p acts on C12:0 and C14:0 acyl-CoA; Fen1p acts on even-length acyl-CoA's from C16:0 to C22:0; and Sur4p acts on even-length acyl-CoA's from C18:0 to C24:0. Overall, the expanded representation allows for resolution of the individual reactions associated with each gene.
Changes to representation of fatty acid desaturation
The production of C16:1 and C18:1 fatty acids is catalyzed by the Δ9-desaturase Ole1p, which catalyzes the insertion of a double bond between carbons 9 and 10. Ole1p acts on palmitoyl-CoA (C16:0) to form palmitoleoyl-CoA (C16:1) and on stearoyl-CoA (C18:0) to form oleoyl-CoA (C18:1).21 As described in prior sections, reactions that produce monounsaturated fatty acids through mitochondrial fatty acid synthesis or fatty acid elongation are removed from the model after applying the suggested changes for those processes. This modification restricts de novo synthesis of C16:1 and C18:1Δ fatty acids to occur only through Ole1p.9 The two reactions for OLE1 in the Yeast v6.0 model also have reconstruction errors. The introduction of a double bond in the fatty acid chain requires reducing equivalents from NADH.22 NADH/NAD+ cofactors for these reactions are not included in the Yeast v6.0 model. In addition, the product of desaturation of palmitoyl-CoA should be corrected to be palmitoleoyl-CoA, instead of hexadec-2-enoyl-CoA. Although both palmitoleoyl-CoA and hexadec-2-enoyl-CoA contain C16:1 fatty acids, the location of the double bond differs between the two metabolites.
Changes to repair blocked reactions in β-oxidation
Fatty acids can be broken down through a process called β-oxidation. Each round of β-oxidation removes two carbons from the fatty acid chain in the form of acetyl-CoA. This acetyl-CoA can be utilized for energy production through the citric acid cycle and for carbohydrate biosynthesis through the glyoxylate cycle.23 For S. cerevisiae, β-oxidation occurs only in peroxisomes.23 Since the peroxisomal membrane is impermeable to NAD(H) and acetyl-CoA, there must exist mechanisms to regenerate NAD+ for continued β-oxidation and to transport acetyl-CoA to mitochondria for energy production.24 Attention to this issue in curation of the model led to corrections that allow for simulated growth on fatty acids as sole carbon sources, which is reflective of in vivo metabolic capacity of S. cerevisiae. A visual comparison of the differences in depiction of β-oxidation before and after our curation is shown in Fig. 3.
Fig. 3.
Curation of β-oxidation in the Yeast v6.0 model. (a) Reactions in the Yeast v6.0 model relevant to β-oxidation of saturated and unsaturated fatty acids. All of the reactions shown are blocked, as indicated by X's. For the sake of space, the repeated reaction sequence of POX1, FOX2, and POT1 for stearoyl-CoA (C18:0) to octanoyl-CoA (C8:0) is omitted, as indicated by the dashed arrow. (b) Proposed representation of β-oxidation of saturated and unsaturated fatty acids. The metabolites relevant to β-oxidation of oleoyl-CoA are shown in parentheses and are written below the metabolites relevant to β-oxidation of palmitoleoyl-CoA. For the sake of space, dashed arrows are used to condense the combined action of the enzymes of classical β-oxidation (ie, Pox1p, Fox2p, and Pot1p) into a singular illustrated reaction.
During β-oxidation, NAD+ is needed for the step catalyzed by FOX2-encoded 3-hydroxyacyl-CoA dehydrogenase.23 The NAD+ used for β-oxidation is able to be regenerated through action of MDH3-encoded malate dehydrogenase, which reduces oxaloacetate into malate in an NADH-dependent manner.24 The Yeast v6.0 model has this reaction constrained strictly in the direction of conversion of malate to oxaloacetate. This direction constraint therefore prevents NAD+ from being able to be regenerated in the peroxisome. This can be remedied by making the malate dehydrogenase reaction reversible.
Since the acetyl-CoA end-product of β-oxidation is unable to diffuse across the peroxisomal membrane, S. cerevisiae relies on two different pathways to utilize this metabolite.24 One pathway is through the glyoxylate cycle, which yields succinate from two molecules of acetyl-CoA. Another pathway is through the carnitine shuttle. The carnitine acetyl-CoA transferase, Cat2p, is involved in transfer of acetyl units from the peroxisome to the mitochondria. Acetyl-CoA is converted to acetylcarnitine in the peroxisome. Acetylcarnitine can be transported to the mitochondria and subsequently the acetyl group can be transferred to a molecule of free coenzyme A for further metabolism in the tricarboxylic acid cycle (TCA) cycle. The Yeast v6.0 model represents both peroxisomal and mitochondrial carnitine acetyl-CoA transferase reactions in the same way (ie, (R)-carnitine+acetyl-CoA→coenzyme A+O-acetylcarnitine). This representation does not allow for mitochondrial production of acetyl-CoA from acetylcarnitine. Therefore, the mitochondrial CAT2 reaction can be revised by switching the products/reactants.
Revised Representation of Glycerolipid and Glycerophospholipid Metabolism
Addition of new genes
Following manual curation, 15 additional genes were identified for inclusion in the Yeast v6.0 model (Table 1). Eleven of these genes introduce the potential for new catalytic activity. The addition of AYR1 to the model ultimately allows for the formation of phosphatidate from the substrate of dihydroxyacetone phosphate (Fig. 4).25 Phosphatidate is a key intermediate in the synthesis of glycerophospholipids and triglycerides. With the inclusion of AYR1, the two different pathways for phosphatidate biosynthesis (ie, glycerol 3-phosphate pathway and dihydroxyacetone phosphate pathway) can both be accounted for in the model.
Table 1.
List of New Genes To Be Added to the Yeast v6.0 Model
| NEW GENE ADDITIONS THAT CATALYZE REACTIONS CURRENTLY NOT INCLUDED IN THE MODEL | |
|---|---|
| AYR1 | NADPH-dependent 1-acyl dihydroxyacetone phosphate reductase |
| CLD1 | Mitochondrial cardiolipin-specific phospholipase |
| CST26 | Protein that incorporates stearic acid into phosphatidylinositol |
| DGK1 | Diacylglycerol kinase |
| GCY1 | Putative NADP(+) coupled glycerol dehydrogenase |
| GEP4 | Mitochondrial phosphatidylglycerophosphatase |
| IDP2 | Cytosolic NADP-specific isocitrate dehydrogenase |
| LRO1 | Acyltransferase that catalyzes diacylglycerol esterification |
| PAH1 | Mg2+-dependent phosphatidate (PA) phosphatase |
| PHM8 | Lysophosphatidate (LPA) phosphatase |
| YJU3 | Monoglyceride lipase |
| NEW GENE ADDITIONS THAT CATALYZE REACTIONS ALREADY PRESENT IN THE MODEL | |
|---|---|
| FRQ1 | N-myristoylated calcium-binding protein that may have a role in intracellular signaling through its regulation of the phosphatidylinositol 4-kinase Pik1p |
| LDH1 | Exhibits active esterase plus weak triacylglycerol lipase activities |
| VAC14 | Protein involved in regulated synthesis of PtdIns(3,5)P(2); interacts with Fig4p; activator of Fab1p |
| VPS15 | Functions as a membrane-associated complex with Vps34p; active form recruits Vps34p to the Golgi membrane |
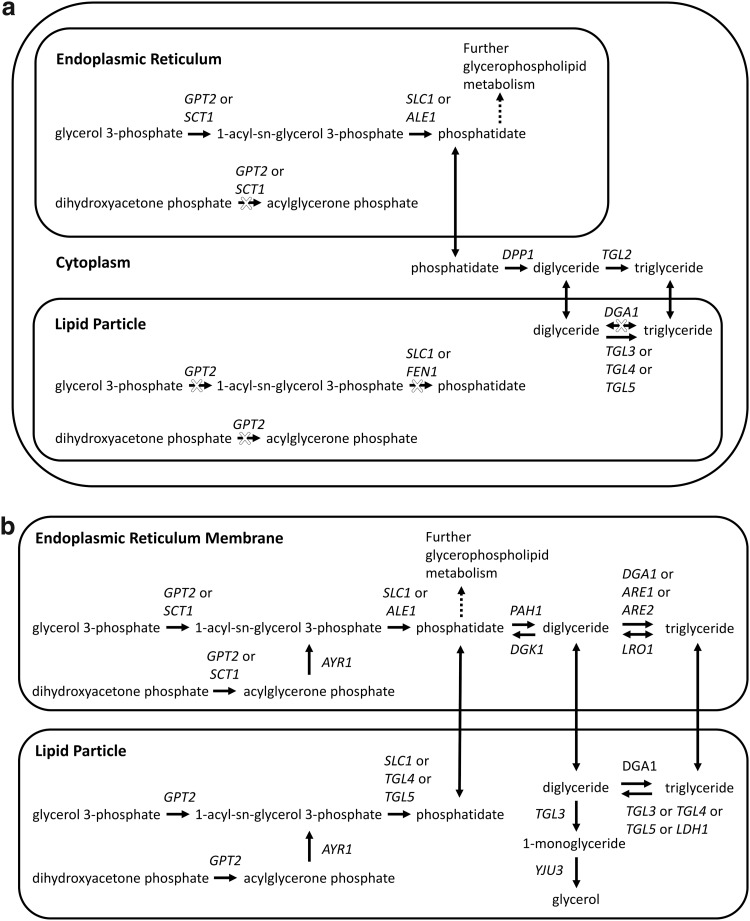
Fig. 4.
Curation of glycerolipid metabolism in the Yeast v6.0 model. (a) Reactions in the Yeast v6.0 model relevant to glycerolipid metabolism. Blocked reactions are indicated by X's. (b) Proposed representation of glycerolipid metabolism. The proposed modification expands each of the classes shown in this figure into its constituent species (not shown). For instance, the term phosphatidate in the figure refers to a collection of individual species [eg, phosphatidate (1–16:0, 2–16:1), phosphatidate (1–16:1, 2–16:1), etc].
The genes PAH1 and LRO1, which are relevant to triglyceride synthesis, can also be added to the model. The enzyme encoded by PAH1 dephosphorylates phosphatidate to yield diglyceride, which can be utilized in the synthesis of triglyceride or the glycerophospholipids phosphatidylethanolamine and phosphatidylcholine.26 The Yeast v6.0 model associates the conversion of phosphatidate to diglyceride with the gene DPP1 (Fig. 4a). Although Dpp1p does act upon phosphatidate to yield diglyceride, this activity is believed to be involved in lipid signaling through the regulation of the amounts of phosphatidate and diacylglycerol pyrophosphate present in the vacuolar membrane, as opposed to generating diglyceride for de novo synthesis of glycerophospholipids and triglyceride.27 Thus, the model can be amended to have Pah1p produce diglyceride at the ER membrane for the synthesis of triglyceride, phosphatidylethanolamine, and phosphatidylcholine (Fig. 4b). The phosphatidate phosphatase activity associated with Dpp1p can be relocated to the vacuolar membrane in the model and kept separate from the de novo synthesis pathways.
The synthesis of triglyceride from diglyceride can occur through either a mechanism using acyl-CoA or glycerophospholipids as acyl donors. The four enzymes that account for all triglyceride synthesis are Dga1p, Lro1p, Are1p, and Are2p. Dga1p catalyzes the majority of the acyl-CoA-dependent diglyceride acyltransferase activity, whereas Are1p and Are2p provide only minor activity.28–30 Lro1p catalyzes the transfer of the acyl group at the sn-2 position of phosphatidylethanolamine or phosphatidylcholine to diglyceride.31,32 The Yeast v6.0 model associates the conversion of diglyceride to triglyceride with the genes DGA1, TGL2, TGL3, TGL4, and TGL5 (Fig. 4a), instead of the genes DGA1, LRO1, ARE1, and ARE2 (Fig. 4b). The association of the triacylglycerol lipase (TGL) genes with this activity is erroneous since these genes enable the breakdown, not synthesis, of triglyceride. Thus, the reaction directionality for the TGL genes should be changed so that it goes irreversibly towards triglyceride hydrolysis. The genes associated with triglyceride lipase activity should also be amended to be TGL3, TGL4, TGL5, and LDH1 (Fig. 4b). TGL2 is not included in this association since triglyceride has not been observed at the mitochondria, which is where Tgl2p is localized.33,34 LDH1 is a new gene to the model and is added based on recent evidence.35
Additional genes and reactions connected to triglyceride breakdown should be included in the model (Fig. 4b). The inclusion of DGK1 allows for utilization of the diglyceride generated from triglyceride hydrolysis. DGK1 encodes for diacylglycerol kinase, which converts diglyceride into phosphatidate for use in glycerophospholipid synthesis.36 The pathways for triglyceride degradation in the model should also be supplemented with reactions for the complete hydrolysis of triglyceride into glycerol and free fatty acids. This can be accomplished by the addition of TGL3-encoded diglyceride lipase activity and YJU3-encoded monoglyceride lipase activity to the model.37,38
Expansion of species
Glycerolipids and glycerophospholipids can vary by the fatty acyl chain attached to the glycerol backbone of the molecule. The acyl chains can differ in chain length and/or the number of double bonds. These differences are influential in altering the physical properties of the biological membrane with respect to membrane thickness, intrinsic curvature, and fluidity.39 These physical properties can in turn affect the membrane's permeability, the activity of membrane-associated enzymes, and membrane fusion and fission.39 Furthermore, adjustment of the acyl chain composition is utilized by the cell to adapt to different conditions and has also been proposed as a means to increase tolerance to stress conditions.40,41 Theoretically, the number of glycerolipid and glycerophospholipid species that can exist is extremely large due to the vast permutations possible for different acyl chains positioned along the glycerol backbone. Thus, a modeling strategy to represent this structural diversity is needed and is explored herein.
Most of the models prior to the Yeast consensus model (ie, iFF708, iLL672, iMM904, and iND750) represent the glycerolipid and glycerophospholipid classes as defined composites of specific species.5,42–45 For instance, in iND750, the glycerophospholipid class phosphatidate is given a singular defined molecular formula of C3540H6544O800P100 in the model, which describes 100 molecules of phosphatidate; reactions using this metabolite have the stoichiometric coefficient scaled by 1/100. This formula arises from the stoichiometry of the various acyl-CoA's used in the two reactions describing the successive acylation of glycerol-3-phosphate. As an example, the first acylation is described in the iND750 model as:
Glycerol 3-phosphate+0.02 Decanoyl-CoA (n-C10:0CoA)+0.06 Dodecanoyl-CoA (n-C12:0CoA)+0.17 Hexadecenoyl-CoA (n-C16:1CoA)+0.09 Octadecynoyl-CoA (n-C18:2CoA)+0.24 Octadecenoyl-CoA (n-C18:1CoA)+0.27 Palmitoyl-CoA (n-C16:0CoA)+0.05 Stearoyl-CoA (n-C18:0CoA)+0.1 Tetradecanoyl-CoA (n-C14:0CoA) → 0.01 1-Acyl-sn-glycerol 3-phosphate+Coenzyme A
This reaction in iND750 is an abstraction that depicts the creation of 1-acyl-sn-glycerol 3-phosphate through fixed fractional contribution of various acyl-CoA species in acylating the sn-1 position of glycerol 3-phosphate. The advantages of this approach are its succinctness and its incorporation of information on reaction specificity through different stoichiometries for each species. Conversely, this approach implements rigid stoichiometries and imposes a requirement for all the species listed in the reaction equation. Thus, this approach does not capture the flexibility of the cell's lipidome and the possibility for remodeling of individual lipids because of its restrictive molecular formulas.
In contrast, the approach taken by the Yeast consensus model is to categorize individual specific species into general classes using “isa reactions” (eg., isa acyl-CoA: hexacosanoyl-CoA→acyl-CoA) and to use the term subsequently for the general class throughout metabolic reactions in the model. For example, the reaction described above for iND750 is represented in the Yeast v6.0 model as:
Glycerol 3-phosphate+Acyl-CoA→1-Acyl-sn-glycerol 3-phosphate+Coenzyme A+2 H+
The use of isa acyl-CoA reactions in defining the term acyl-CoA allows for logical “OR” operation in which any of the categorized individual acyl-CoA species can satisfy the acyl-CoA requirement in the reaction. The 1-acyl-sn-glycerol 3-phosphate produced in this reaction is consequently a generic term in the model since it is created using the generic acyl-CoA term.
The disadvantage of strictly using generic terms defined by isa reactions is that information on individual reaction specificity is lost. For instance, if a particular reaction only uses C16:0- and C16:1-CoA, this specificity cannot be indicated using the acyl-CoA generic term. Furthermore, species that would not be biochemically utilized in the reaction could be utilized in computer simulations due to this generic term. Another disadvantage is backward incompatibility in going from the generic term back to specific species. An illustration of this is the breakdown of triglyceride to yield free fatty acids. With the usage of the generic term fatty acid, the exact molecular formula is unknown, and thus the potential number of β-oxidation cycles that can occur cannot be determined.
Another approach to modeling the diversity of lipid species is to expand the general classes into their corresponding individual species and to utilize the appropriate species, as opposed to general class, in the equations for metabolic reactions. This approach has recently been applied to sphingolipid metabolism in the Yeast consensus model and has been continued in this paper for glycerolipid and glycerophospholipid metabolism.46 The general classes of glycerolipids and glycerophospholipids can be expanded to clarify the specific acyl group(s) attached to the glycerol backbone. This expansion can consider the dominant acyl species found in S. cerevisiae, ie, C16:0, C16:1, C18:0, and C18:1.9 An additional assumption can be applied to restrict the acyl chain found at the sn-2 position of the glycerol backbone to be either C16:1 or C18:1.47 As an example, phosphatidylcholine can be expanded out to eight different species: phosphatidylcholine (1–16:0, 2–16:1), (1–16:1, 2–16:1), (1–18:0, 2–16:1), (1–18:1, 2–16:1), (1–16:0, 2–18:1), (1–16:1, 2–18:1), (1–18:0, 2–18:1), and (1–18:1, 2–18:1). A major detriment of this representation is that it greatly increases the number of reactions, which may be deemed unnecessary for the modeling scope of certain users. However, incorporation of this level of detail will provide a platform for users interested in accounting for distribution and function of distinct molecular lipid species.
Compartmentalization
Multiple compartments, each housing specific enzymes, contribute to lipid synthesis in S. cerevisiae.9 The catalogue of lipids in various membranes is a result of both local metabolism and membrane trafficking between different compartments. For instance, most of the steps in the synthesis of phosphatidylcholine through the CDP-diacylglycerol pathway occur at the ER membrane. However, the major source of the intermediate phosphatidylethanolamine comes from activity at the mitochondrial membrane. Consequently, lipid molecules must be transferred between different membranes in order to fulfill a sequence of metabolic reactions.
The Yeast v6.0 model classifies reactions and species into 16 compartments: boundary, cell envelope, cytoplasm, ER, ER membrane, extracellular, Golgi, Golgi membrane, lipid particle, mitochondrion, mitochondrial membrane, nucleus, peroxisome, peroxisomal membrane, vacuole, and vacuolar membrane. In classifying reactions involved in lipid metabolism, the Yeast v6.0 model opts not to localize the reactions to membrane compartments (eg, localized to ER instead of ER membrane). Reactions in glycerolipid and glycerophospholipid metabolism could be updated to indicate if they occur at the membrane. The utilization of membrane compartments in the model provides more resolution on reaction localization. This change also requires the addition of transport reactions to allow transport of water soluble lipid precursors from the cytoplasm to the membrane and to allow reversible transfer of lipids between membrane compartments. The addition of these transport reactions can be based on modeling decisions to fill gaps within the model. As the interplay of organelles in lipid metabolism is further elucidated, these modeling decisions may be readdressed.
Blocked Reactions
Blocked reactions are defined as reactions that are incapable of carrying flux while still satisfying the constraints of flux bounds and the steady-state assumption. Upon implementing all of the suggested changes to the Yeast v6.0 model (Supplementary Data S1), the absolute number of blocked reactions actually increases by eight, but the relative number of blocked reactions decreases from 39.2% to 21.3% of all reactions (Table 2). This contrast arises from the large increase in the number of reactions in the model due to the expansion of classes into the individual species that can participate in various reactions. This expansion consequently biased the metric of percent blocked reactions to be more of an indicator of the topology of fatty acid, glycerolipid, and glycerophospholipid metabolism. Nevertheless, comparison between the set of blocked reactions for the model before and after modifications can allow assessment of gaps that were filled in the network and of areas of metabolism that can serve as a focus for future model development.
Table 2.
Statistics for the Yeast v6.0 Model Before and After Implementing the Changes Suggested Through Manual Curation
| YEAST v6.0 MODEL | BEFORE CHANGES | AFTER CHANGES |
|---|---|---|
| Genes | 901 | 916 |
| Reactions | 1,882 | 3,493 |
| Metabolites | 1,454 | 2,218 |
| Blocked Reactions | 737 (39.2%) | 745 (21.3%) |
The major source of reductions in the number of blocked reactions was due to changes relevant to β-oxidation (Fig. 3). These changes not only allowed for fatty acids to be broken down in the peroxisome but also allowed for the acetyl-CoA produced from β-oxidation to serve as a carbon and energy source for cell growth. Flux can now be carried through peroxisomal reactions utilizing acetyl-CoA—ie, carnitine acetyl-CoA transferase, citrate synthase, and malate synthase. Carnitine acetyl-CoA transferase mediates transfer of the acetyl unit from the peroxisome to the mitochondria, where it can then feed into the TCA cycle for further oxidation and energy release.23 The two glyoxylate cycle enzymes citrate synthase and malate synthase use acetyl-CoA to generate citrate and malate, respectively. These metabolites can be exported to the cytosol where they can undergo further transformation and can ultimately be used for synthesis of carbohydrates through gluconeogenesis.23 Given the increase in utilizable pathways in the model, the model now has the capacity to simulate growth on fatty acids as the sole carbon source.
One area of metabolism that was originally capable of carrying flux in simulations but was blocked following changes to the model was mitochondrial fatty acid synthesis (Fig. 1). In the original model, certain acyl-ACP species created in the mitochondria could be transported to the cytoplasm where they can then be hydrolyzed to free fatty acids. Removal of the acyl-ACP transport and hydrolysis reactions, as was done in the model changes, leads to blocked mitochondrial fatty acid synthesis. The removal of these reactions was justified by the observation that the mitochondrial FAS system is unable to compensate for the loss of cytoplasmic fatty acid synthesis in fas1 or fas2 mutants.48 The blocked nature of mitochondrial fatty acid synthesis in the modified model does not stem from gaps within the pathway but rather lack of utility of its end products. Although mutants in mitochondrial fatty acid synthesis are viable in vivo, these strains display deficiency in mitochondrial respiration.13 This respiratory deficiency has a straightforward explanation: lack of lipoic acid, which is an essential cofactor for α-keto acid dehydrogenase complexes and is produced from octanoyl-ACP from mitochondrial fatty acid synthesis.13 The dysfunction in respiration is currently not captured by either the original model or the modified model, which can be attributed to several issues. The synthesis and attachment of the lipoic acid moiety to the glycine decarboxylase, pyruvate dehydrogenase, and α-ketoglutarate dehydrogenase enzyme complexes are not represented in the original Yeast v6.0 model or in the modified model. Furthermore, in the reaction mechanisms for the enzyme complexes, the lipoic acid moiety undergoes transformations but is ultimately regenerated. Thus, the net reactions do not include any terms for lipoic acid, which obviates the need for this metabolite and consequently yields a discrepancy between simulation and in vivo requirements.
Additional pathways, of which the end products lack utility, are also blocked in both the original and revised Yeast v6.0 model. Two examples are cardiolipin metabolism and phosphoinositide metabolism. Cardiolipin plays a role in multiple mitochondrial processes such as transport of proteins into the mitochondria, mitochondrial energy production, regulation of apoptosis, and membrane fusion.49 Phosphoinositides are involved in many functions such as signaling, recruitment of proteins to specific membranes, regulation of cell wall maintenance/synthesis, and vesicle-mediated membrane trafficking.50 Although reactions for the synthesis of cardiolipin and phosphoinositides are contained in the model, their involvement in the aforementioned processes is beyond the scope of both the original and revised model. The comprehensive modeling of metabolism, regulation, signaling, and other cellular processes involves complex and, in many cases, unknown interactions; this presents a great challenge for modeling.3 As it stands, modeling only purely metabolic reactions for these metabolites yields blocked reactions in the pathways. Since the revised model expands upon the individual species of the lipid class, there is more of a penalty in the number of blocked reactions for the revised model compared to the original model, which uses generic classes.
Predictions on Gene Essentiality
Analysis of gene essentiality has been one standard measure for the predictive ability of genome-scale metabolic models.4 In this test, the effect of single gene deletions is assessed on the basis of whether removal of reactions associated with each gene in the model blocks the ability for biomass production. The results of single gene deletions for both the original and modified Yeast v6.0 model using a simulated aerobic glucose-limited defined media are summarized in Table 3. Although there are individual differences in predictions of essential and lethal gene deletions, the aggregate statistics remain consistent between the original and modified model. This is to be expected, since manual curation focused on only a small portion of metabolism. Both models are able to match the in vivo phenotype of the single-gene deletion mutants for approximately 87% of the genes contained in the model. Both models have better prediction accuracies for viable gene deletions than lethal gene deletions, with approximately 98% of viable gene deletions correctly predicted versus 52% of lethal gene deletions correctly predicted.
Table 3.
Accuracy of Gene Essentiality Predictions for the Yeast v6.0 Model Before and After Implementing the Changes Suggested Through Manual Curation
| YEAST v6.0 MODEL | BEFORE CHANGES | AFTER CHANGES |
|---|---|---|
| True Positivea (TP) | 674 | 691 |
| True Negativeb (TN) | 114 | 114 |
| False Positive (FP) | 102 | 103 |
| False Negative (FN) | 11 | 8 |
| Sensitivityc | 98.4% | 98.9% |
| Specificityd | 52.8% | 52.5% |
| Overall accuracye | 87.5% | 87.9% |
Positive=viable gene deletion
Negative=lethal gene deletion
Sensitivity=TP/(TP+FN)
Specificity=TN/(TN+FP)
Overall accuracy=(TP+TN)/(TP+TN+FP+FN)
Whereas there is minimal change in relative accuracy between the original and modified model, focusing on individual differences in predictions of gene essentiality allows for assessment of how the changes made to the model affect simulation results and also provides further insight into the limitations of the models. The addition of 15 new genes to the model led to 14 new true positive predictions and one new false positive prediction. This false positive prediction was for the gene FRQ1, the product of which recruits PIK1-encoded phosphatidylinositol 4-kinase to the Golgi membrane and stimulates Pik1p activity.50 The false positive prediction (ie, deletion of FRQ1 was predicted to be viable despite in vivo lethality) can be attributed to the lack of utility of phosphoinositides in the model (see “Blocked Reactions”). Thus, the synthesis of phosphoinositides is treated as inessential in the model. This leads to additional false positives in both the original and modified model for genes that catalyze the synthesis of phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate (ie, STT4, PIK1, and MSS4). A simple workaround is to include these species in the pseudo-reaction representing biomass production, therefore requiring synthesis of these species to form biomass. For this approach, it should be noted that the separate pools of phosphatidylinositol 4-phosphate (PI 4-P) in the cell have their own distinct roles in cell functioning; absence of STT4, which generates PI 4-P at the plasma membrane, cannot be compensated by overproduction of PIK1, which generates PI 4-P at the Golgi membrane, and vice versa.50
Several of the genes involved in metabolism of very-long chain fatty acids had different predictions of gene essentiality between the original and modified model. Single-gene deletions of FAT1, FEN1, and SUR4 were correctly predicted as being viable in the modified model, while incorrectly predicted as lethal in the original model. FAT1-encoded acyl-CoA synthetase catalyzes the esterification of very long chain fatty acids with coenzyme A to form very long chain acyl-CoA's.51 Another means of generating very long chain acyl-CoA's is through fatty acid elongation (see “Changes to Representation of Fatty Acid Elongation”). FAT1 is incorrectly predicted as essential in the original model since it depicts fatty acid elongation as producing free fatty acids instead of acyl-CoA's. Therefore, the very long chain acyl-CoA's needed to synthesize sphingolipids is only able to be produced through FAT1-encoded acyl-CoA synthetase in the original model.
The depiction of fatty acid elongation in the original model also leads to its false negative predictions for FEN1 and SUR4. Both FEN1 and SUR4 encode for elongases that have partially overlapping ranges of acyl-CoA substrates and function; mutants with deletion of either FEN1 or SUR4 are viable, whereas deletion of both genes is lethal.14 The original model contained two lumped reactions for fatty acid elongation, which had gene associations of “TSC13 and FEN1 and SUR4.” This logical relationship employs the “AND” condition, which requires all the genes in the association to be present for fatty acid elongation to occur. In contrast, the modified model has individual reactions for the action of elongases on different acyl-CoA substrates and assigns the appropriate gene relationship based on substrate specificity of each elongase. As a result, the modified model accurately predicts the viability of single gene deletions of FEN1 or SUR4 and the lethality of simultaneous deletion of both genes.
For the fatty acid elongation system, one gene deletion that the original model is able to predict correctly as viable while the modified model predicts falsely as lethal is IFA38. However, this correct prediction in the original model is not due to accuracy in its representation of the functioning of IFA38-encoded β-ketoacyl-CoA reductase. All of the reactions in the original model that are associated with IFA38 are blocked reactions due to the lack of connection to the rest of fatty acid elongation (see “Changes to Representation of Fatty Acid Elongation”). Thus, all the reactions associated with IFA38 are inconsequential to simulation results using the original model and, therefore, deletion of IFA38 is predicted as viable. In comparison, the reactions associated with IFA38 in the modified model can carry metabolic flux and are also needed for fatty acid elongation to occur. The synthesis of very long chain fatty acids is essential in vivo, and the viability of ifa38Δ suggests capacity for residual β-ketoacyl-CoA reductase activity.16 It has been hypothesized that Ayr1p is responsible for this residual activity based on its homology to IFA38 and the synthetic lethality of ifa38Δayr1Δ.16 However, overexpression of AYR1 does not suppress the slow growth of ifa38Δ mutants, and in vitro data generated in another study do not support any role of Ayr1p in fatty acid elongation in ifa38Δ mutants.16,20 Based on a lack of direct evidence, the gene AYR1 was not associated with fatty acid elongation in the modified model. This modeling decision meant that the only gene product capable of catalyzing the β-ketoacyl-CoA reductase activity for fatty acid elongation in the modified model was that of IFA38 and, consequently, the deletion of IFA38 is falsely predicted as lethal for the modified model.
In comparing the original and modified model, there are also instances in which the original model is able to predict correctly the single gene deletion as lethal, whereas the modified model incorrectly predicts it as viable. Nevertheless, the reason for why the gene is essential for simulation of biomass production in the original model does not match with the true role of the gene in vivo. This is the case for the genes RIM2 and PET8. RIM2 encodes for a transporter that imports (deoxy)pyrimidine nucleoside triphosphates into the mitochondria in exchange for intra-mitochondrially generated (deoxy)pyrimidine nucleoside monophosphates.52 The imported (deoxy)pyrimidine nucleoside triphosphates are essential for synthesis of mitochondrial DNA and RNA. Neither the original nor modified model captures the role of RIM2 in providing precursors for mitochondrial DNA and RNA metabolism. Instead, the original model requires RIM2 for exchange of the mitochondrial nucleotides cytidine monophosphate (CMP) and cytidine triphosphate (CTP) for synthesis of the glycerophospholipids phosphatidylinositol, phosphatidylserine, and CDP-diacylglycerol. The modified model does not require RIM2 for this purpose because the reactions for PIS1-encoded phosphatidylinositol synthase and CHO1-encoded phosphatidylserine synthase are localized to the ER membrane, while CDS1-encoded CDP-diacylglycerol synthase is localized to both the ER membrane and mitochondrial membrane. It should be noted that the original model does have these enzymes localized to other organelles besides the mitochondria; however, the reactions in these alternate compartments are blocked and flux can only be carried through the mitochondrial reactions in the original model.
The correct prediction of PET8 as an essential gene using the original model and the wrong prediction using the modified model can be attributed to similar circumstances as those described previously for RIM2. The physiological role of Pet8p is to transport S-adenosylmethionine into the mitochondria, where it is utilized as a cofactor in biotin and lipoic acid synthesis and also as a methyl group donor for methylation of DNA, RNA, and protein.53 The involvement of S-adenosylmethionine in these processes is not accounted for in either the original or modified model. The reason for the essentiality of PET8 in the original model is for the transport of S-adenosylmethionine into the mitochondria for the methylation of phosphatidylethanolamine to phosphatidylcholine. The modified model, in contrast, has these methylation reactions localized to the ER membrane and therefore does not require mitochondrial S-adenosylmethionine for this purpose.
A direct consequence of the expansion of glycerolipid and glycerophospholipid species in the modified model was the correct prediction that ole1Δ mutants require unsaturated fatty acids for growth, which the original model could not foresee.54 OLE1 encodes for Δ9-desaturase, which is required for production of unsaturated fatty acids.21 The inability of the original model to predict the unsaturated fatty acid auxotrophy of ole1Δ mutants stems from its usage of isa reactions, which treat species as functionally equivalent. For instance, the isa acyl-CoA reaction categorizes various individual acyl-CoA species (eg, palmitoyl-CoA, palmitoleoyl-CoA, etc.) into the generic term of acyl-CoA. Thus, any of the categorized acyl-CoA species can fulfill the requirement for the acyl-CoA used in lipid metabolism. In contrast, the modified model employs the assumption that the acyl chain found at the sn-2 position of the glycerol backbone of glycerolipids and glycerophospholipids is either C16:1 or C18:1. This assumption imposes a specific requirement for unsaturated acids that must be met either through synthesis with Ole1p or through supplementation of the media.
Conclusions
Through the process of manual curation, we have identified inconsistencies between information contained in the Yeast consensus model (Yeast v6.0) and literature knowledge for fatty acid, glycerolipid, and glycerophospholipid metabolism. These inconsistencies include instances of incorrect gene-reaction associations, improper definition of substrates/products in reactions, and inappropriate assignments of reaction directionality. In addition to correcting these inconsistencies, the addition of 15 new genes and the introduction of increased specificity in representation of glycerolipid and glycerophospholipid metabolism through the expansion of lipid classes and denotation of membrane reactions are proposed. A complete consideration of triglyceride metabolism should include pathways for both its synthesis and degradation. Through the addition of new genes and reactions the proposed modifications account for the synthesis of the key intermediate phosphatidate from dihydroxyacetone phosphate; for the conversion of diglyceride generated from hydrolysis of triglyceride into phosphatidate; and for the complete breakdown of triglyceride into glycerol and free fatty acids.
One consequence of the proposed changes to the Yeast v6.0 model is the newly acquired ability to simulate growth on fatty acids as the sole carbon source. This functionality is enabled through the amendment of blocked reactions in β-oxidation. Analysis of the blocked reactions in the Yeast v6.0 model both before and after implementing the suggested changes revealed gaps reflecting the challenge in interlinking metabolic reactions to complex biological processes such as signaling, and the unaccounted-for roles of metabolites such as cofactors. The presence of these blocked reactions ultimately leads to an incomplete picture that does not capture the function of genes associated with the blocked reactions. This is especially consequential for the predictive accuracy of the effect of gene deletions. In this respect, deletions of genes associated with blocked reactions are predicted as viable. The high percentage of blocked reactions is not unique to the Yeast consensus model; the iMM904 and iND750 models of yeast metabolism have respectively 31% and 41% of all reactions blocked.10 Thus, with the various models, the accuracy of predictions for viability of gene deletions can be highly influenced by blocked reactions instead of a basis rooted in biological reality.
The suggested changes for Yeast v6.0 have been submitted to Dr. Kieran Smallbone, University of Manchester, UK, who maintains the Yeast metabolic network reconstruction, and have since been incorporated to generate Yeast v7.0. Prior to the publication of this paper, another genome-scale model of S. cerevisiae metabolism called iTO977 has been published. iTO977 merges the earlier models of iIN800 and Yeast v1.0 and then improves and expands on this through gap-filling methods and the introduction of additional genes, reactions, and metabolites based on literature evidence and database searches.5,55,56 The process presented in this paper for analysis of the representation of fatty acid, glycerolipid, and glycerophospholipid metabolism was repeated for iTO977, and a discussion of the differences between Yeast v6.0 and iTO977 is provided in the Supplementary Materials (Supplementary Data S7). Overall, there were instances in which iTO977 had the same issues as the Yeast v6.0 model, where iTO977 was in agreement with the proposed modifications, and where iTO977 had its own unique issues.
The modeling of the entirety of an organism's metabolic network is highly complex. The databases upon which these networks are built can contain errors, even in well-known pathways such as the TCA cycle.57 These inaccuracies can become embedded in the model unless efforts are taken to inspect the representation of metabolism. One approach to achieve this goal is manual curation of sub-portions of the model defined by pathways of interest. Although this requires significant time and effort, the benefit of such manual curation is an increased confidence and also awareness of limitations in the examined portion of the model.
An accurate representation of the metabolic network is a key component to ensure that application of the model yields results reflective of actual biochemistry. Genome-scale metabolic models have been used to aid the interpretation of high-throughput data, to guide metabolic engineering, and to generate testable hypotheses on cellular behavior.3 Our own particular interest is in using this framework to explore the molecular organization of lipid biosynthesis and how this synthesis influences the metabolic fluxes and pools of critical metabolites, such as phosphatidate, phosphatidylinositol, diglyceride, and triglyceride, which play important roles in signaling and response to environmental stress. Yeast adapt to environmental stresses such as product inhibition, nutrient limitation, elevated temperatures, and high osmolarity through manipulation of lipid yield and membrane composition. A better understanding of key lipid biosynthetic pathways is important for industrial biotechnologists seeking not only to increase lipid yields but also to push the environmental limits of microbial conversion systems.
Supplementary Material
Acknowledgments
The authors would like to thank ML Gaspar, MA Aregullin, and SA Jesch for their insights on lipid metabolism, BD Heavner for engaging discussions on manual curation and model evaluation, EC Evans for constructive comments on the manuscript, and K. Smallbone for his work in incorporating the changes into the consensus model. We also thank Hnin Aung's funding sources: US Department of Transportation, Federal Grant #DTOS59-07-G-00052; US Department of Agriculture, Award #2010-38502-21900; and the Spencer Family Fund; and the National Institutes of Health Grant GM-19629 to SAH.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.US Environmental Protection Agency. Biofuels and the environment: First triennial report to Congress; 2011. EPA/600/R-10/183F. [Google Scholar]
- 2.Kosa M. Ragauskas AJ. Lipids from heterotrophic microbes: Advances in metabolism research. Trends Biotechnol. 2011;29(2):53–61. doi: 10.1016/j.tibtech.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Oberhardt MA. Palsson BO. Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5 doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiele I. Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5(1):93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herrgard MJ. Swainston N. Dobson P, et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol. 2008;26(10):1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schellenberger J. Que R. Fleming RMT, et al. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox v2.0. Nat Protoc. 2011;6(9):1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bornstein BJ. Keating SM. Jouraku A. Hucka M. LibSBML: An API library for SBML. Bioinformatics. 2008;24(6):880–881. doi: 10.1093/bioinformatics/btn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keating SM. Bornstein BJ. Finney A. Hucka M. SBMLToolbox: An SBML toolbox for MATLAB users. Bioinformatics. 2006;22(10):1275–1277. doi: 10.1093/bioinformatics/btl111. [DOI] [PubMed] [Google Scholar]
- 9.Henry SA. Kohlwein SD. Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190(2):317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heavner B. Smallbone K. Barker B, et al. Yeast 5—an expanded reconstruction of the Saccharomyces cerevisiae metabolic network. BMC Syst Biol. 2012;6(1):55. doi: 10.1186/1752-0509-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tehlivets O. Scheuringer K. Kohlwein SD. Fatty acid synthesis and elongation in yeast. BBA-Mol Cell Biol L. 2007;1771(3):255–270. doi: 10.1016/j.bbalip.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Lomakin IB. Xiong Y. Steitz TA. The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell. 2007;129(2):319–332. doi: 10.1016/j.cell.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Hiltunen JK. Schonauer MS. Autio KJ, et al. Mitochondrial fatty acid synthesis type II: More than just fatty acids. J Biol Chem. 2009;284(14):9011–9015. doi: 10.1074/jbc.R800068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh C-S. Toke DA. Mandala S. Martin CE. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272(28):17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 15.Toke DA. Martin CE. Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J Biol Chem. 1996;271(31):18413–18422. doi: 10.1074/jbc.271.31.18413. [DOI] [PubMed] [Google Scholar]
- 16.Han G. Gable K. Kohlwein SD, et al. The Saccharomyces cerevisiae YBR159w gene encodes the 3-ketoreductase of the microsomal fatty acid elongase. J Biol Chem. 2002;277(38):35440–35449. doi: 10.1074/jbc.M205620200. [DOI] [PubMed] [Google Scholar]
- 17.Denic V. Weissman JS. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell. 2007;130(4):663–677. doi: 10.1016/j.cell.2007.06.031. [DOI] [PubMed] [Google Scholar]
- 18.Kohlwein SD. Eder S. Oh C-S, et al. Tsc13p Is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21(1):109–125. doi: 10.1128/MCB.21.1.109-125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul S. Gable K. Dunn TM. A six-membrane-spanning topology for yeast and Arabidopsis Tsc13p, the enoyl reductases of the microsomal fatty acid elongating system. J Biol Chem. 2007;282(26):19237–19246. doi: 10.1074/jbc.M701774200. [DOI] [PubMed] [Google Scholar]
- 20.Rössler H. Rieck C. Delong T, et al. Functional differentiation and selective inactivation of multiple Saccharomyces cerevisiae genes involved in very-long-chain fatty acid synthesis. Mol Genet Genomics. 2003;269(2):290–298. doi: 10.1007/s00438-003-0836-0. [DOI] [PubMed] [Google Scholar]
- 21.Stukey JE. McDonough VM. Martin CE. The OLE1 gene of Saccharomyces cerevisiae encodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem. 1990;265(33):20144–20149. [PubMed] [Google Scholar]
- 22.Martin CE. Oh C-S. Jiang Y. Regulation of long chain unsaturated fatty acid synthesis in yeast. BBA-Mol Cell Biol L. 2007;1771(3):271–285. doi: 10.1016/j.bbalip.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Hiltunen JK. Mursula AM. Rottensteiner H, et al. The biochemistry of peroxisomal β-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2003;27(1):35–64. doi: 10.1016/S0168-6445(03)00017-2. [DOI] [PubMed] [Google Scholar]
- 24.van Roermund CW. Elgersma Y. Singh N, et al. The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J. 1995;14(14):3480–3486. doi: 10.1002/j.1460-2075.1995.tb07354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Athenstaedt K. Daum G. 1-Acyldihydroxyacetone-phosphate reductase (Ayr1p) of the yeast Saccharomyces cerevisiae encoded by the open reading frame yil124w is a major component of lipid particles. J Biol Chem. 2000;275(1):235–240. doi: 10.1074/jbc.275.1.235. [DOI] [PubMed] [Google Scholar]
- 26.Han G-S. Wu W-I. Carman GM. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281(14):9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carman GM. Han G-S. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci. 2006;31(12):694–699. doi: 10.1016/j.tibs.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oelkers P. Cromley D. Padamsee M, et al. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem. 2002;277(11):8877–8881. doi: 10.1074/jbc.M111646200. [DOI] [PubMed] [Google Scholar]
- 29.Sandager L. Gustavsson MH. Ståhl U, et al. Storage lipid synthesis is non-essential in yeast. J Biol Chem. 2002;277(8):6478–6482. doi: 10.1074/jbc.M109109200. [DOI] [PubMed] [Google Scholar]
- 30.Sorger D. Daum G. Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl-glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J Bacteriol. 2002;184(2):519–524. doi: 10.1128/JB.184.2.519-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oelkers P. Tinkelenberg A. Erdeniz N, et al. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J Biol Chem. 275(21):15609–15612. doi: 10.1074/jbc.C000144200. 200. [DOI] [PubMed] [Google Scholar]
- 32.Dahlqvist A. Ståhl U. Lenman M, et al. Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci. 2000;97(12):6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ham HJ. Rho HJ. Shin SK. Yoon H-J. The TGL2 gene of Saccharomyces cerevisiae encodes an active acylglycerol lipase located in the mitochondria. J Biol Chem. 2010;285(5):3005–3013. doi: 10.1074/jbc.M109.046946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grillitsch K. Daum G. Triacylglycerol lipases of the yeast. Front Biol. 2011;6(3):219–230. [Google Scholar]
- 35.Debelyy MO. Thoms S. Connerth M, et al. Involvement of the Saccharomyces cerevisiae hydrolase Ldh1p in lipid homeostasis. Eukaryot Cell. 2011;10(6):776–781. doi: 10.1128/EC.05040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fakas S. Konstantinou C. Carman GM. DGK1-encoded diacylglycerol kinase activity is required for phospholipid synthesis during growth resumption from stationary phase in Saccharomyces cerevisiae. J Biol Chem. 2010;286(2):1464–1474. doi: 10.1074/jbc.M110.194308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurat CF. Natter K. Petschnigg J, et al. Obese yeast: Triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem. 2006;281(1):491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- 38.Heier C. Taschler U. Rengachari S, et al. Identification of Yju3p as functional orthologue of mammalian monoglyceride lipase in the yeast Saccharomyces cerevisiae. BBA-Mol Cell Biol L. 2010;1801(9):1063–1071. doi: 10.1016/j.bbalip.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Kroon AIPM. Metabolism of phosphatidylcholine and its implications for lipid acyl chain composition in Saccharomyces cerevisiae. BBA-Mol Cell Biol L. 2007;1771(3):343–352. doi: 10.1016/j.bbalip.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 40.You KM. Rosenfield C-L. Knipple DC. Ethanol tolerance in the yeast Saccharomyces cerevisiae Is dependent on cellular oleic acid content. Appl Environ Microbiol. 2003;69(3):1499–1503. doi: 10.1128/AEM.69.3.1499-1503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodríguez-Vargas S. Sánchez-García A. Martínez-Rivas JM, et al. Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl Environ Microbiol. 2007;73(1):110–116. doi: 10.1128/AEM.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Förster J. Famili I. Fu P, et al. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13(2):244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuepfer L. Sauer U. Blank LM. Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res. 2005;15(10):1421–1430. doi: 10.1101/gr.3992505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mo M. Palsson B. Herrgard M. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol. 2009;3(1):37. doi: 10.1186/1752-0509-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duarte NC. Herrgård MJ. Palsson BØ. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 2004;14(7):1298–1309. doi: 10.1101/gr.2250904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heavner BD. Henry SA. Walker LP. Evaluating sphingolipid biochemistry in the consensus reconstruction of yeast metabolism. Ind Biotechnol. 2012;8(2):72–78. [Google Scholar]
- 47.Wagner S. Paltauf F. Generation of glycerophospholipid molecular species in the yeast Saccharomyces cerevisiae. Fatty acid pattern of phospholipid classes and selective acyl turnover at sn-1 and sn-2 positions. Yeast. 1994;10(11):1429–1437. doi: 10.1002/yea.320101106. [DOI] [PubMed] [Google Scholar]
- 48.Schweizer E. Hofmann J. Microbial type I fatty acid synthases (FAS): Major players in a network of cellular FAS systems. Microbiol Mol Biol Rev. 2004;68(3):501–517. doi: 10.1128/MMBR.68.3.501-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osman C. Voelker DR. Langer T. Making heads or tails of phospholipids in mitochondria. J Cell Biol. 2011;192(1):7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strahl T. Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. BBA-Mol Cell Biol L. 2007;1771(3):353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi J-Y. Martin CE. The Saccharomyces cerevisiae FAT1 gene encodes an acyl-CoA synthetase that is required for maintenance of very long chain fatty acid levels. J Biol Chem. 1999;274(8):4671–4683. doi: 10.1074/jbc.274.8.4671. [DOI] [PubMed] [Google Scholar]
- 52.Marobbio CMT. Di noia MA. Palmieri F. Identification of a mitochondrial transporter for pyrimidine nucleotides in Saccharomyces cerevisiae: Bacterial expression, reconstitution and functional characterization. Biochem J. 2006;393(2):441–446. doi: 10.1042/BJ20051284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marobbio CMT. Agrimi G. Lasorsa FM. Palmieri F. Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 2003;22(22):5975–5982. doi: 10.1093/emboj/cdg574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stukey JE. McDonough VM. Martin CE. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J Biol Chem. 1989;264(28):16537–16544. [PubMed] [Google Scholar]
- 55.Osterlund T. Nookaew I. Bordel S. Nielsen J. Mapping condition-dependent regulation of metabolism in yeast through genome-scale modeling. BMC Syst Biol. 2013;7(1):36. doi: 10.1186/1752-0509-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nookaew I. Jewett M. Meechai A, et al. The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: A scaffold to query lipid metabolism. BMC Syst Biol. 2008;2(1):71. doi: 10.1186/1752-0509-2-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stobbe MD. Houten SM. van Kampen AHC, et al. Improving the description of metabolic networks: The TCA cycle as example. FASEB J. 2012;26(9):3625–3636. doi: 10.1096/fj.11-203091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.