Abstract
The epidermal growth factor receptor (EGFR) signaling network is activated in most solid tumors, and small-molecule drugs targeting this network are increasingly available. However, often only specific combinations of inhibitors are effective. Therefore, the prediction of potent combinatorial treatments is a major challenge in targeted cancer therapy. In this study, we demonstrate how a model-based evaluation of signaling data can assist in finding the most suitable treatment combination. We generated a perturbation data set by monitoring the response of RAS/PI3K signaling to combined stimulations and inhibitions in a panel of colorectal cancer cell lines, which we analyzed using mathematical models. We detected that a negative feedback involving EGFR mediates strong cross talk from ERK to AKT. Consequently, when inhibiting MAPK, AKT activity is increased in an EGFR-dependent manner. Using the model, we predict that in contrast to single inhibition, combined inactivation of MEK and EGFR could inactivate both endpoints of RAS, ERK and AKT. We further could demonstrate that this combination blocked cell growth in BRAF- as well as KRAS-mutated tumor cells, which we confirmed using a xenograft model.
Keywords: cancer, EGFR signaling, mathematical modeling, modular response analysis, signal transduction
Introduction
The signal transduction network downstream of the epidermal growth factor receptor (EGFR) has received much attention, as a majority of human cancers shows mutations leading to hyperactivation of the network (Hanahan and Weinberg, 2011). Based on detailed mechanistic understanding of the network, a large number of targeted therapies has been developed (Herbst et al, 2004; Roberts and Der, 2007; Prenen et al, 2010). However, despite positive treatment responses in some patients, a large fraction of patients do not benefit even if molecular markers such as KRAS or BRAF mutation status are used to stratify patient groups (Karapetis et al, 2008; Walther et al, 2009; Roth et al, 2010).
One reason for the somewhat disappointing response rate to these therapies is that they have been developed using the concept of linear signaling pathways downstream of the receptor. However, the EGFR signal is propagated through a complex network (Bublil and Yarden, 2007), involving cross talks to parallel pathways (Porter and Vaillancourt, 1998) and strong feedback loops on different levels (Blüthgen and Legewie, 2008; Legewie et al, 2008; Cirit et al, 2010; Avraham and Yarden, 2011). Quantitative analysis of these regulatory principles suggested that strong feedbacks can neutralize drug treatment (Friday et al, 2008; Cirit et al, 2010; Sturm et al, 2010; Fritsche-Guenther et al, 2011).
Mathematical modeling of signaling networks can help to understand the behavior of these complex networks, and can be used to simulate the effect of drugs in such a network. The structure of these mathematical models can be directly deduced from pathway maps (Oda et al, 2005). Detailed mechanistic models based on Ordinary differential equations (ODE) have been developed for the EGFR signaling network (Kholodenko et al, 1999; Schoeberl et al, 2002; Nelander et al, 2008). However, for such detailed models the parameterization remains a major challenge. More coarse-grain modeling approaches, such as logical models or non-mechanistic statistical models require less data for parameterization (Kreeger et al, 2009; Morris et al, 2011; Saez-Rodriguez et al, 2011, 2009; Tentner et al, 2012). These approaches allow qualitative predictions, but typically fail to deal with feedback loops or do not provide mechanistic insights. The approach we chose for this study is termed modular response analysis (MRA), which resides between the qualitative nature of Boolean models and detailed mechanistic models. It provides a framework to calculate the response of a linear approximation of an ordinary differential equation model to a perturbation (Bruggeman et al, 2002; Kholodenko et al, 2002), and has been developed to discover and parameterize networks from systematic perturbation studies (Santos et al, 2007; Stelniec-Klotz et al, 2012). The parameters of an MRA model are so-called local response coefficients that quantify how strong a change in activity of one node directly affects the activity of another node. These models then allow to quantitatively analyze feedback regulation, feedforward loops as well as cross talks, which is of major interest as these network motifs have major effects on drug sensitivity and network behavior (Friday et al, 2008; Cirit et al, 2010; Sturm et al, 2010; Fritsche-Guenther et al, 2011).
In this work, we exposed a panel of colon cancer cell lines to different stimuli and pharmaceutical inhibitors, and measured key signaling molecules in a medium-throughput approach. The data generated by this approach were then used to parameterize MRA-based mathematical models, which generated quantitative maps of the wiring between signaling molecules. We focused our efforts on RAS-mediated signal transduction pathways, as they are currently in the strategic focus of targeted therapeutics in solid cancers. We were able to identify feedbacks and cross talks of therapeutic relevance. Our model predicted that EGFR-directed therapeutics might be effective even in tumors carrying a mutation in RAS, if they are provided in combination with RAF or MEK inhibitors. We confirmed our predictions by phenotypic assays and a xenograft model.
Results
A pipeline to model signal transduction networks in cancer cell line panels
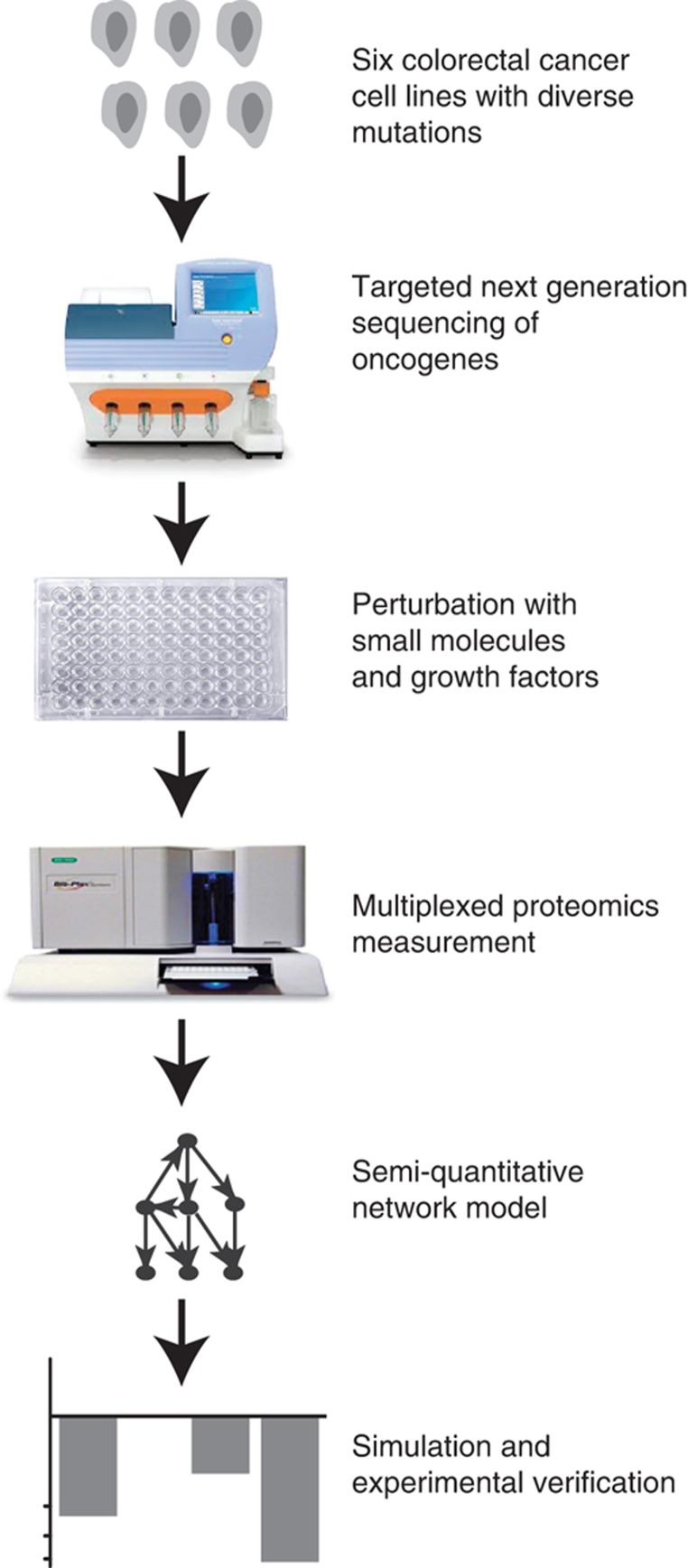
We developed a combined experimental and theoretical approach to dissect signaling networks in cancer cell lines to generate predictive mathematical models for their signal transduction pathways. The workflow of our pipeline was as follows (Figure 1): we selected a panel of six colon cancer cells that were genotyped for common oncogenes using targeted next-generation sequencing. Subsequently, the signaling network in these cell lines was perturbed using small molecule inhibitors and growth factors, and combinations thereof. Before and after perturbation, phosphorylation of key signaling molecules was quantified. This high-dimensional data set was then used to parameterize mathematical models of the signaling events. These models were simulated to predict effects of inhibition and potential combinatorial treatment.
Figure 1.
General outline of the study. A panel of six colon cancer cell lines was chosen, and profiled by sequencing selected cancer-related genes. The cells were then systematically perturbed with four kinase inhibitors and two ligands of growth receptors, and phosphorylation of key signaling proteins was measured using the Luminex platform. These data were used for parameterizing a mathematical model, which was then used to predict combinatorial treatments.
Network quantification by a systematic perturbation screen
We first chose six colon cancer cell lines for our systematic perturbation screen, LIM1215, HCT116, SW403, SW480, HT29 and RKO. We used targeted sequencing of 46 genes to verify that these cells represent a panel that reflects the genetic diversity of colon cancer (see Table 1 and Supplementary Table S1).
Table 1. Mutation spectrum of the cell line panel.
| Gene symbol | LIM1215 | HCT116 | SW403 | SW480 | HT29 | RKO | TCGA patients |
|---|---|---|---|---|---|---|---|
| ABL1 | P309A | Y257C | _ | ||||
| APC | K1462R | A1457T/K1462R | _ | ||||
| BRAF | V600E* | V600E* | 13 | ||||
| CTNNB1 | T41AH | _ | |||||
| FGFR3 | S400R | S400R | _ | ||||
| KRAS | A146T | G13D* | G12V*,H | G12V*,H | 6,8,28 | ||
| PIK3CA | H1047R* | H1047R* | 4 | ||||
| SMAD4 | Q311X*,H | _ | |||||
| SMO | V404M | _ | |||||
| STK11 | G58S | G58S | _ | ||||
| TP53 | R273HH | R273HH | R273H*,H | 26 |
Shown are non-silent mutation results from sequencing of known mutated regions in 46 cancer-related genes. SNPs reported by Cancer Genome Project are marked with asterisk (*), and H indicates homozygous mutations. The number of patients found in The Cancer Genome Atlas to harbor this particular mutation (in the order of appearance) is shown under TCGA patients.
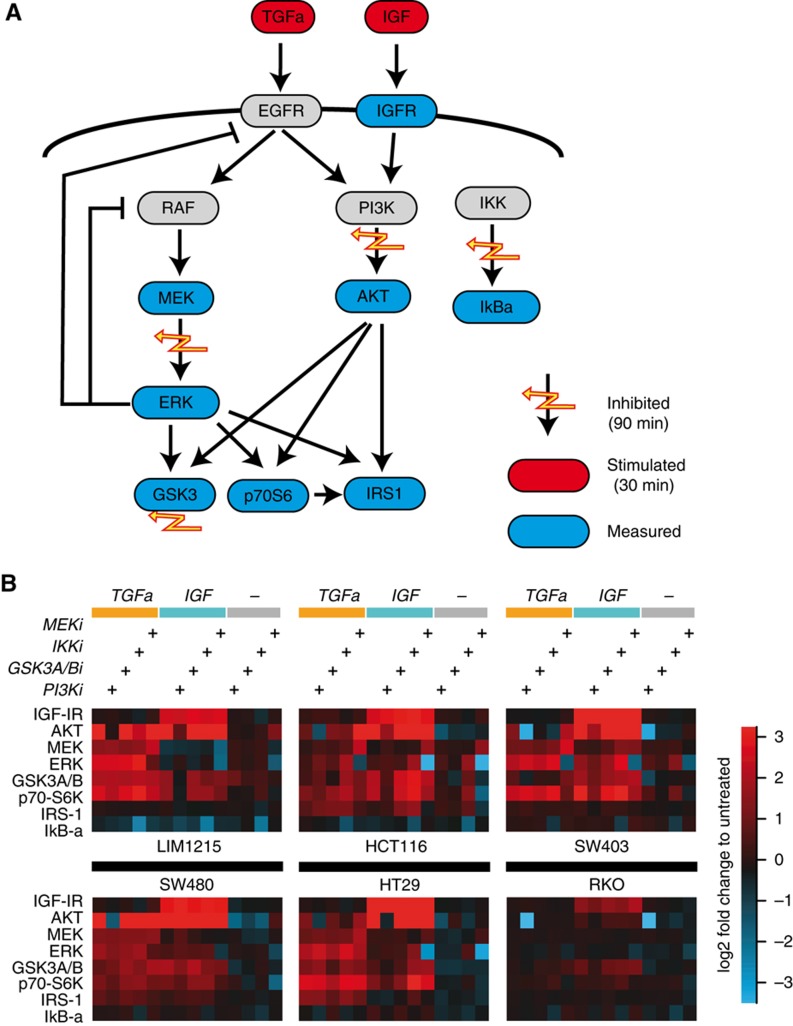
To generate a semi-quantitative database for further mathematical modeling, we decided to measure phosphorylation of selected signaling molecules for combinatorial perturbations. In line with previous approaches (Nelander et al, 2008; Saez-Rodriguez et al, 2009, 2011; Morris et al, 2011), we used a pair-wise design of perturbations, where each inhibition is combined with each stimulus. Specifically, we stimulated the cells with two growth factors, TGFa and IGF, activating the EGF receptor and the IGF-receptor, respectively (shown in red in Figure 2A). The cells have been pre-incubated for 1 h with pharmacological inhibitors against the kinases MEK, PI3K, IKK or GSK-3a/b (flashes in Figure 2A). As signaling typically displays a strong transient response followed by a long-term plateau, we performed time-series experiments to determine optimal time points for the experiments (see Supplementary Figure S1). We find that peak transients are within the first 10 min after stimulation, and the response to TGFa as well as IGF has reached a plateau at 30 min after stimulation. Thus, we chose the 30-min time point for further experiments, as the interpretation of MRA requires the signaling network to be approximately in steady state. We then used the Luminex proteomics platform to measure the phosphorylation of eight key signaling proteins (AKTS473, ERK2T185/Y187, MEK1S217/S221, p70S6KT421/S424, IGF-IRY1131, GSK-3a/bS21/S9, IkB-aS32/S36, and IRS-1S636/S639) that are within or in close proximity to the stimulated pathways and the inhibited kinases (shown in blue in Figure 2A).
Figure 2.
Generation of systematic perturbation data. (A) Perturbations consisted of two ligands (red nodes), and four pharmacological inhibitors (yellow flashes). These were applied alone and in inhibitor-ligand combinations for the indicated time points. Then eight phosphorylation signals were measured (blue nodes). (B) Log2 fold change of phosphorylation in response to the perturbations for the six indicated cell lines. Displayed response range was limited to ±3.5 (approx. 10-fold). Source data for this figure is available on the online supplementary information page.
The resulting data sets are shown in Figure 2B as log2 fold changes compared with unperturbed controls. When comparing the six different cell lines, we found strong response patterns to the perturbation in all cells, with the exception of RKO cells that show hardly any response and which we therefore excluded from further analysis.
All other cell lines, while clearly responding to the inhibitors and stimuli, showed subtle differences in their response patterns. For example, IRS-1 phosphorylation changed strongly in HCT116 cells at certain treatments, but was not altered in SW403 cells. To further infer qualitative and quantitative differences in the interactions between the signaling nodes and to predict effects of combinatorial treatments, we decided to analyze the data using a mathematical model.
A model-based approach to analyze the perturbation data set
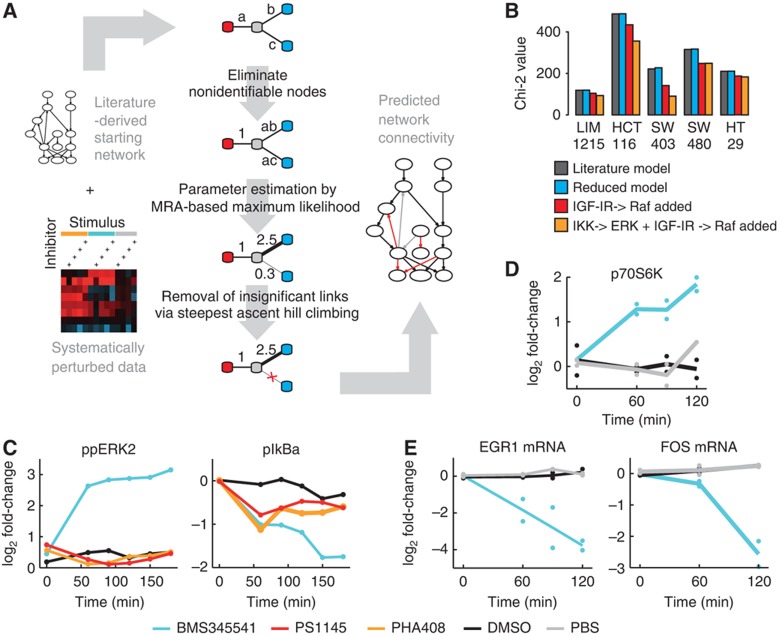
We developed an algorithm that can unambiguously determine network structure and parameters from the data set based on MRA. The algorithm extends our previously developed methodology (Stelniec-Klotz et al, 2012) by considering multiple perturbations and unobserved nodes. First, the model estimates the response coefficients and inhibitor strengths for a literature-based starting network. Then edges are iteratively removed if they are not supported by the data (tested by a likelihood ratio test), until no further edge can be removed (Figure 3A).
Figure 3.
EGFR modeling required network alteration that led to the discovery of a new effect of BMS345541 on ERK. (A) Scheme of the modeling pipeline from the starting network to its parameterization. Systematic perturbation data and a starting network serve as input. The core-fitting routine consists of three steps, and is illustrated by a four-node example network. First, non-identifiable parameters are detected and reparameterized. Second, the identifiable parameter combinations are fitted to the experimental data. Third, connections not significantly contributing to the fit are removed and subsequently the remaining parameters are refitted. The resulting network contains information about the strength, direction and sign of its connections. (B) Chi-squared values of the models trained on data of five cell lines. Dark bars indicate values for the initial model. Blue, red and yellow bars show values after model reduction of literature network, with a link from IGFR to RAF, or when additionally including a link from IKK to ERK, respectively. (C, D) Log2 fold change in time-series experiments after treatment with IKK inhibitors or solvent controls to investigate the IKK–ERK relationship in HCT116 cells. (C) IKK inhibitor BMS345541 treatment results in an increased phospho-ERK level, whereas treatment with IKK inhibitors PHA408, PS1145 and solvent control (DMSO) results in no increase. The inhibition of IkBa phosphorylation is comparable for all three inhibitors after 1 h (Luminex, n=1). (D) Phosphorylation of p70S6K, a cytoplasmic target of ERK, increases after 60 min of BMS345541 treatment (western blot, n=2) when compared with DMSO and PBS control. (E) Expression of ERK target genes EGR1 and FOS decreases after BMS345541 treatment (qrt–PCR, n=2). Source data for this figure is available on the online supplementary information page.
One of the major obstacles in comparing the networks quantitatively between cells was to find a parameterization of the model that can be uniquely determined from the data. For example, the individual response coefficients along the path from EGFR to the activation of AKT cannot be determined individually, as the only node measured is the phosphorylation of AKT (Figure 2A). Thus, we had to re-parameterize the MRA model. Elegant rule-based re-parameterization algorithms (Saez-Rodriguez et al, 2009) could not be applied, as they fail for more complex scenarios, where paths branch, feedbacks exist and inhibitors are placed within the path. We therefore developed an algorithmic approach that replaces unidentifiable parameters by an identifiable combination of parameters. These new parameters can then be reliably estimated from the data set and compared between different cell lines (see Materials and methods, Supplementary Method 1, and Supplementary Figure S2).
Model fit requires alteration of the network model
When we applied our modeling procedure to the perturbation data sets of the five selected cell lines, we obtained fits with Chi-squared values between 116 and 470 (Figure 3B, dark bars). Our algorithm then pruned the network and could remove several links in each cell line without significant increase in the Chi-squared values (Figure 3B, blue bars), indicating that these links are not important in mediating the perturbed signals (see an example for the stepwise reduction in Supplementary Figure S2).
When we compared the model predictions with the data, we found that our literature model failed to explain an increase of MEK and ERK phosphorylation after IGF treatment that was visible in multiple cells. When checking back with literature, we found that IGF-1 can lead to activated RAS by triggering the activation chain via IGF-IR, IRS-1 and Grb-2 (Florini et al, 1996). Hence, we included a link from IGF to RAF, which improved model performance for all five cell lines significantly (red bars in Figure 3A).
Additionally, our model could not account for an increase of phospho-ERK level upon treatment with IKK inhibitor BMS345541, which was particularly pronounced in HCT116 and SW403. Interestingly, this increase in ERK phosphorylation did not coincide with an increase in MEK levels, thus the effect of IKK inhibition on ERK cannot be explained via upstream components in the pathway. We thus decided to include this potential new interaction in the model and to repeat the model selection procedure. The inclusion of a link from IKK to ERK improved the fit of the model considerably for HCT116 and SW403 cells (see decrease of Chi-squared value in Figure 3B orange bars) and showed no improvement for the other cell lines.
The observed increase in ERK phosphorylation after treatment with IKK inhibitors may be either due to unspecific effects of the chosen IKK inhibitor or due to direct or indirect regulation of ERK by IKK or its downstream kinases. We therefore decided to characterize this interaction further. We found that treatment with the IKK inhibitor BMS345541 resulted in an increase of phosphorylation of ERK within 1 h (Figure 3C, left panel), but treatment with two other IKK inhibitors had no effect on ERK phosphorylation, although they also blocked phosphorylation of IkBa with similar strength at this time point (Figure 3C, right panel). From that we concluded that the interaction is likely to be a hitherto unknown side effect of the inhibitor. Interestingly, while phosphorylation of ERK and its cytoplasmic target p70S6K are increased in response to BMS345541 (Figure 3D), ERK’s nuclear activity seems to be decreased, as expression of typical immediate-early target genes of ERK such as EGR1 and FOS is strongly reduced (Figure 3E). This suggests that although ERK phosphorylation is increased, ERK activity seems to be redirected toward cytoplasmic targets, and treatment with BMS345541 may result in a repression of typical genetic programs stimulated by ERK. In line with this hypothesis, we found that treatment with moderate concentrations of BMS345541 results in impaired proliferation of HCT116 cells (Supplementary Figure S3). Thus, the modeling procedure identified an unknown side effect of BMS345541 on ERK, and consequently we excluded the data of BMS345541 from further analysis.
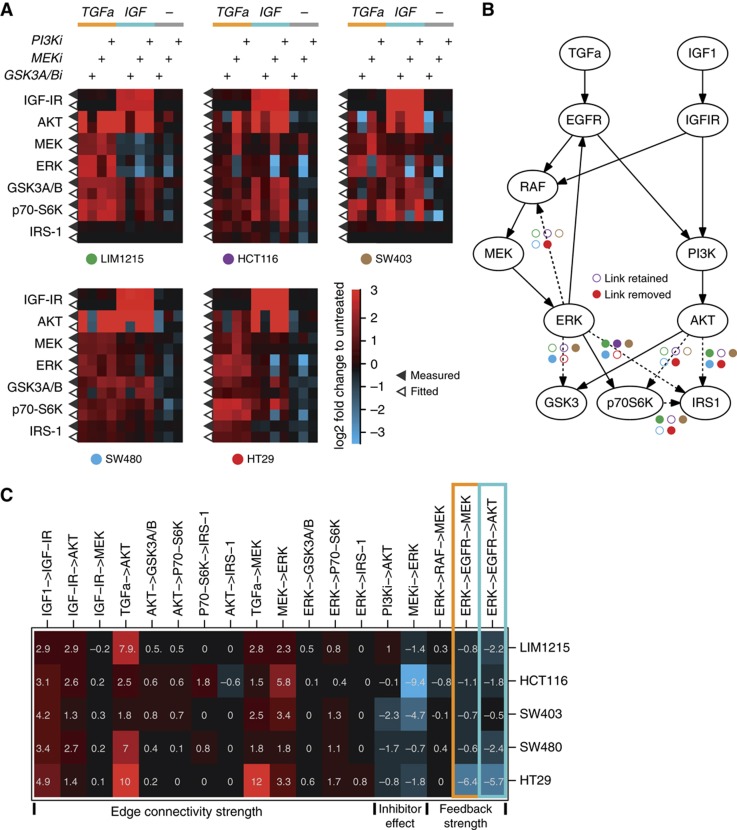
After these alterations in the network structure, the resulting model could mimic most of the responses in quantitative detail. Figure 4A shows the experimental measurement side-by-side with the corresponding model fit. For each signaling node, measured phosphorylation is displayed in the upper row (indicated by filled triangle) and the model simulation in the lower row (open triangle). By running the procedure on the data with simulated noise added, we confirm that the procedure robustly identifies the parameters (Supplementary Figure S4). In addition, we confirmed by 100 runs of our algorithm that for each data set the model structure and parameterization remained identical, irrespective of initial parameterization.
Figure 4.
Model fit uncoveres differences in network topology and signaling strength. (A) Heat maps for the five cell lines showing log2 fold changes of phosphorylation (filled triangles) and the corresponding model simulation (open triangles). (B) Network structure derived from the model fit. Dashed lines indicate edges that are removed in those cell lines marked by filled circles (same colors and layout as in A). (C) Model-deduced parameter values representing signaling strength for identifiable paths, inhibitors and feedback/cross talk expressed as log2 response coefficients. Orange and blue boxes indicate feedback from ERK via EGFR to MEK, and cross talk from ERK via EGFR to AKT.
Model fit uncovers differences in network structure
The topology of the final models is displayed in Figure 4B. Edges that our modeling procedure has removed in at least one cell line are depicted as dashed lines, with color-coded circles indicating the cell line. One interesting difference in the topology of the networks resides in the feedback from ERK to RAF, which has been removed in HT29 cells. This is in line with previous findings that BRAFV600E mutation disables MAPK feedback regulation via RAFs (Friday et al, 2008; Sturm et al, 2010; Fritsche-Guenther et al, 2011). Furthermore, the topology that connects the output kinases to ERK and AKT differs. For example, the phosphorylation site measured for IRS1 is not connected to ERK and AKT in LIM1215, SW480 and SW403, and only connected to ERK in HT29 cells. Taken together, the model-fitting procedure allowed to identify qualitative differences in signal transduction networks in our cell line panel, some of which can be related to specific mutations.
Quantitative differences between signaling networks
In addition to qualitative differences in the underlying network topology, we were also interested to study how signaling networks in these cells differ in quantitative aspects. Thus, we decided to inspect differences in the parameter sets. Overall, the models had on an average 15 parameters (between 14 for SW403 and 17 for HCT116). As these parameters typically correspond to complicated combinations of response coefficients, the most intuitive way to inspect these parameter combinations was by recomputing these parameter combinations such that they correspond to paths in the network. The values of such parameter combinations are visualized in Figure 4C. Interestingly, many paths that correspond to the intracellular logic, such as the response coefficient from MEK to ERK or from ERK to p70S6K have comparable values in all cell line models. This suggests that certain quantitative aspects of signaling are comparable in these cells, despite their heterogeneous genetic background. On the other hand, there are also strong differences between the cells, most of which relate to external perturbations. For example, TGFa does have a strong effect on MEK phosphorylation in HT29 cell, but can only weakly activate MEK in HCT116 cells.
All cells show negative response coefficients for feedback regulation from ERK via EGFR back to MEK (orange box in Figure 4C), with strongest negative values in HT29 cells.
An EGFR-dependent feedback causes cross talk between MAPK and AKT signaling
An interesting aspect of this ERK-EGFR feedback is that it connects ERK signaling with AKT activity in an EGFR-dependent manner, as ERK inhibits the EGF receptor that also stimulates AKT signaling. Thus, inhibition in the ERK pathway can lead to an activation of AKT if ligands for the EGFR receptors are present. This effect has been previously described as a mechanism of drug resistance in tumor cells with BRAF mutation (Prahallad et al, 2012). When we inspected the response coefficient of the path from ERK to AKT via EGFR, we found high negative numbers for HT29, the BRAF-mutated cell line (blue box in Figure 4C). However, the model fit unveiled that the feedback is present in all cell lines, and causes strong cross talk from ERK to AKT in all cell lines, including those harboring different KRAS mutations (blue box in Figure 4C, Table I).
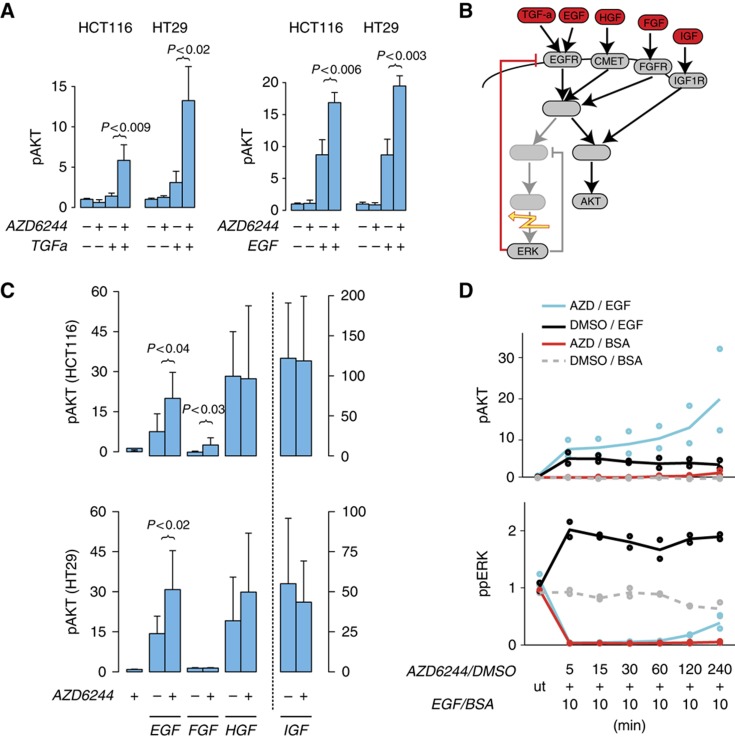
To confirm the EGFR-dependent cross talk between ERK and AKT, we performed independent experiments in HT29 and HCT116 cells, as examples for cells with BRAF and RAS mutations. Figure 5A shows that AKT phosphorylation is only slightly increased when a MEK inhibitor is applied alone, and that AKT can be only weakly stimulated with TGFa. In line with the cross talk hypothesis, we found that AKT phosphorylation was significantly increased (P<0.05) by a factor of 2–3 when cells were pre-treated with the MEK inhibitor, confirming that the cross talk operates irrespective of mutations in BRAF or RAS. We also observed a similar effect when we stimulate with EGF for 10 min, another ligand of the EGFR (Figure 5A).
Figure 5.
ERK–AKT cross talk is mediated by EGFR and independent of RAS or RAF mutation. (A) Increase of AKT phosphorylation in HCT116 (KRAS mutation) and HT29 (BRAF mutation) cells incubated with MEK inhibitor AZD6244 (0.1 μM) or its solvent control (DMSO) for 1 h before application of TGFa or BSA for 30 min. Effect can be also produced by 10 min EGF treatment in the presence of AZD6244 (1 μM). (B) Schematic view of the proposed ERK–AKT cross talk via the EGFR feedback (red), predicting no effect of MEK inhibition (flash) on stimulation of other growth receptors. (C) Increase of Akt phosphorylation compared with solvent control (DMSO) with combinatorial treatment of AZD6244 (5 μM) and different growth factors (10 min). (D) Response of phospho-ERK and AKT to 10 min EGF treatment for different preincubation times of AZD6244 (1 μM) in HCT116 cells. BSA and DMSO are solvent controls for EGF and AZD6244, respectively. All data measured using Luminex assays and shown as fold change; significant deviations are indicated. Brackets indicate significant one-sided t-test, P<0.05. Error bars indicate s.d. of n⩾3 samples. Source data for this figure is available on the online supplementary information page.
We then aimed to investigate whether the negative feedback directly acts at the level of the EGF receptor, or more generally desensitizes growth factor receptors, for example by acting on shared adaptor molecules (Dhillon et al, 2007). To disentangle the network that mediates the feedback, we stimulated cells with the growth factors HGF, FGF and IGF, which do not signal via EGFR (Figure 5B). In contrast to EGF, pre-inhibiting either cell line with AZD6244 had no significant effect on the response to HGF. Similarly, it had no effect on the response to FGF in HT29 cells, and only a weak effect in HCT116 cells. When using IGF as a control that stimulates primarily AKT, pre-treatment with MEK inhibitors had no effect on AKT phosphorylation. This suggests that the feedback common to HCT116 and HT29 acts on the EGFR and not on downstream adaptors that are shared by many growth factor receptors.
We next aimed to define the timescale on which the feedback operates. We treated HCT116 cells with the MEK inhibitor for various times and then stimulated the pathway with EGF for 10 min and measured AKT and ERK phosphorylation (Figure 5D). Interestingly, already with a total inhibition time of 15 min, we observed an increase of AKT phosphorylation, which increased further for longer pre-inhibition times, suggesting that the feedback operates as an integral feedback. Notably, after about 2 h, also non-EGF-treated but MEK-inhibited cells show a slight increase of AKT levels, indicating an EGFR-independent cross talk operating on a longer timescale. Furthermore, the EGFR-dependent feedback shows that EGF treatment can partially restore the pre-inhibitor phospho-ERK level after ∼4 h of inhibition.
EGFR inhibition prevents AKT activation by MEK inhibitors irrespective of mutation status
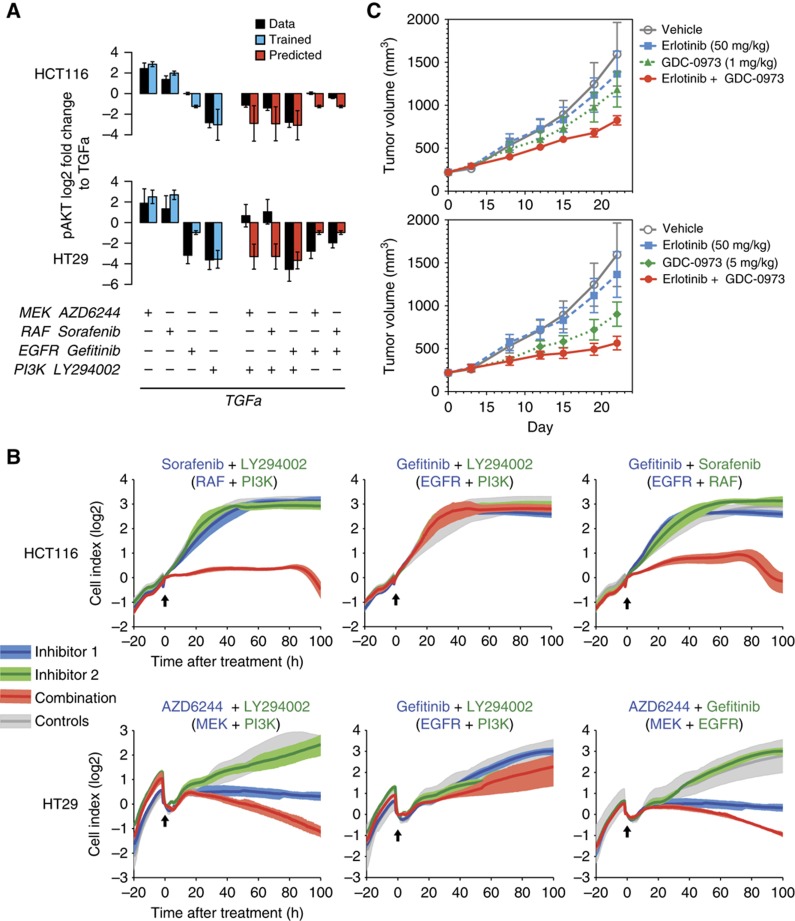
We then used our model to predict combinatorial treatments that reduce ERK activity without activating AKT. Our initial model was trained using only two inhibitors directly in the EGFR pathway, thus we decided to retrain the model for further inhibitors against EGFR (gefitinib) and RAF (sorafenib), see Figure 6A blue bars and Supplementary Figure S5. We then used the model to predict treatments of inhibitors of RAF or MEK combined with EGFR or PI3K plus the combination of the latter two (red bars in Figure 6A). For all these combinatorial perturbations, the model predicted a reduction in AKT activity when compared with TGFa treatment only. Measurement of phospho-AKT confirmed the predictions, except for combinations of MEK/RAF inhibitors and PI3K inhibitors in HT29 cells (black bars in Figure 6A).
Figure 6.
Model-derived combinatorial treatment options unveil effective combinations. (A) Model prediction of selected double inhibitions (red bars) and Luminex measurements (dark bars) for phosphorylation of AKT in HCT116 and HT29 cells in the presence of TGFa in log2 scale. Fits for single perturbations are shown on the left as blue bars. Error bars for modeled results represent s.d. of 100 noised simulations (see Supplementary Methods) (B) Cell proliferation (Xcelligence) of HCT116 and HT29 cells in response to single and double inhibition and control. Arrows mark time of treatment, and areas represent the maximal range of the growth curves, with dark lines reflecting the mean (n>=3). For panels A and B, concentration of AZD6244 was 1 μM. (C) Tumor growth of coloretal cancer cell DLD-1 cells transplanted into nude mice, which received a daily oral gavage of one of two different concentrations of MEK inhibitor GDC-0973, EGFR inhibitor erlotinib alone or combined. Error bars represent s.e.m. with n=10. Source data for this figure is available on the online supplementary information page.
EGFR inhibition together with inhibition in MAPK signaling prevents growth of tumor cells irrespective of mutation status
We next asked whether a combination therapy of MAPK inactivation together with an inhibitor against EGFR also stops proliferation of tumor cells. To assess this, we measured cell growth of HCT116 and HT29 cells with those four inhibitors alone, their vehicle control DSMO, and in selected combinations. Inhibition of the EGF receptor as well as inhibition of PI3K alone had no effect on proliferation in HT29 and HCT116 (Figure 6B), and also combined application of both inhibitors did not alter proliferation either. This is in line with the notion that the oncogenes KRAS and BRAF drive proliferation in these cells via MAPK signaling and do not require the EGFR to generate the pro-proliferative signal. We next investigated how manipulations downstream of their driver mutations alter proliferation in combination with PI3K inhibition. To inhibit the MAPK signaling pathway, we decided to use two different inhibitors in the two cell lines. Based on the differences in ERK–RAF feedback (Sturm et al, 2010; Fritsche-Guenther et al, 2011), we decided to inhibit MAPK activity with the RAF inhibitor sorafenib in HCT116 cells and the MEK inhibitor AZD6244 in HT29 cells. This decision is further supported by the corresponding measurements of the double inhibition in Figure 6A, where those combinations showed the strongest effects.
Application of sorafenib alone had no effect on growth of our RAS/PI3K mutant cell line model HCT116, suggesting that growth may depend on multiple redundant pathways. However, in combination with a PI3K inhibitor, the cells stop growing, indicating that MAPK and AKT signaling redundantly control cell growth. As our model predicts that a combination of RAF and EGFR inhibitor prevents AKT activation, we tested the EGFR inhibitor gefitinib together with sorafenib. In line with the model, this combinatorial treatment synergistically reduced growth as strong as the combination with PI3K inhibition.
In BRAF mutant HT29 cells, MEK inhibition blocked growth, and PI3K inhibition had no effect, confirming that HT29 cells depend solely on MAPK signaling for growth. The combination of MEK inhibitor and PI3K inhibitor led to a decrease of the cell index. Our model predicts that EGFR would also synergize with MEK inhibition to block AKT activation. In line with this prediction, EGFR inhibition had no effect when provided alone, but caused a decrease in cell index when cells were treated in combination with a MEK inhibitor.
EGFR synergizes with MEK inhibitors in vivo
We sought to substantiate the effects of co-targeting of MEK and EGFR in the DLD-1 colorectal xenograft model that harbors both KRASG13D and PIK3CAE545K mutations. DLD-1 tumors were established in nude mice and treated with erlotinib (dosed at a clinically relevant dose of 50 mg/kg), GDC-0973 (a potent MEK-1/2 allosteric inhibitor, dosed both at 1 and 5 mg/kg), or the combination of erlotinib+GDC-0973 at both dose levels. While erlotinib and the lower dose of GDC-0973 (1 mg/kg) were ineffective as single agents resulting in 1 and 22% tumor growth inhibition (%TGI), respectively, the combination demonstrated superior combination efficacy over either single agent alone with 36% TGI (Figure 6C, top panel). The 5 mg/kg dose of GDC-0973 demonstrated better single-agent activity with 42% TGI, however again combination with erlotinib demonstrated superior tumor inhibition to either drug used alone with 60% TGI (Figure 6C, bottom panel). The combination of these drugs was well tolerated in mice with minimal weight loss (Supplementary Figure S6).
Taken together, the results suggest that the EGF receptor is not required for maintaining the cellular phenotype in cells with RAS and RAF mutations. However, once MAPK signaling is blocked, the EGFR increases AKT activity and thus regains its key role in the network that was previously lost due to downstream mutations.
Discussion
A huge body of detailed mechanistic understanding about signaling processes has been accumulated within the last decades (Wang et al, 2002; Oda et al, 2005). Still it remains challenging to understand how a signaling network reacts when targeted therapies are applied because of strong cross talk between pathways and feedback loops (Kim et al, 2007; Friday et al, 2008; Sturm et al, 2010; Fritsche-Guenther et al, 2011; Kholodenko et al, 2012). In this study, we addressed this problem by quantifying MAPK/AKT signaling networks in a panel of colorectal cancer cell lines with differing genetic background. We developed an experimental/computational pipeline that compiles mathematical models for individual cell lines based on systematic perturbation. Our modeling approach resides between detailed mechanistic models (Schoeberl et al, 2002) and more coarse-grain logical models (Saez-Rodriguez et al, 2011, 2009) or even purely statistical descriptions (Tentner et al, 2012). The complexity of the model was chosen such that it could be parameterized with the limited perturbation data set, but allowed to reveal network features such as feedback loops that simpler representations cannot account for. MRA generally requires that the response of the system to perturbations can be modeled using linear equations, and the system is close to steady state. Thus, the modeling procedure can be used to interpret perturbation screens, but parameters should be interpreted in a phenomenological rather than in a precise mechanistic way. Consequently, the procedure is helpful to interpret perturbation data and generate new hypotheses that can be subsequently tested, such as in a previous study of transient EGF/NGF signaling (Santos et al, 2007). In our study, the majority of parameters could be well estimated from the data. However, if there are strong uncertainties in the parameters, methods such as MCMC (Hastings, 1970) or the profile likelihood (Raue et al, 2009) method can be readily applied to model parameters or structural uncertainties.
When we compared the signaling maps between the cell lines, we observed that the core signaling network was quantitatively very similar in all cells, although these cells were heterogeneous in their genetic constellation. This suggests that it is instrumental to build generic models of signaling despite genetic heterogeneity. Nevertheless, certain quantitative aspects differed strongly between cells, such as the strength of the response toward stimulation of the EGFR. We also found qualitative differences between cell lines, such as the loss of the ERK–RAF feedback in HT29 cells, which can be traced back to the BRAF V600E mutation (Friday et al, 2008). Similar studies on larger cell line collectives may unveil further differences in network wiring due to the underlying mutations.
Despite the diversity of mutations in the EGFR signaling network, we found a conserved strong feedback from ERK to EGFR in all five cell lines. At least in HCT116 cells, this feedback was detectable within 15 min but its strength increased further on timescales of hours, suggesting that this feedback operates as an integral feedback, which may have a role for signaling homeostasis (Yi et al, 2000; Prahallad et al, 2012). Different mechanisms of feedback regulation of the EGFR are known, which may all contribute. Transcriptional feedbacks such as MIG-6 (Yoon et al, 2012) or the Sprouty family (Mason et al, 2006; Halilovic et al, 2010) cannot account for the fast feedback regulation, but may be involved in later phases. Adaptors shared between receptors, such as SOS (Douville and Downward, 1997; Shankaran and Wiley, 2010) or Gab1 (Yu et al, 2002), are fast enough but most likely not the main feedback players, as the cross talk was only strong when EGFR was stimulated, but was not or only weakly present when HGF, FGF or IGF were applied. Thus, the strongest target of this feedback is most likely the EGFR itself. Pancreatic cancer cells exhibited increased phosphorylation of the EGFR at Y1068, Y1045 and Y845 when MEK was inhibited (Gan et al, 2010). Phosphorylation of T669 by ERK affects EGFR turnover (Birtwistle et al, 2007). Knockdown of CDC25A, known to be regulated by ERK (Wang et al, 2002), did also lead to increased phosphorylation of EGFR at Y1068 (Prahallad et al, 2012), suggesting that ERK changes CDC25A activity and by this regulates phosphorylation of EGFR (Wang et al, 2005).
A consequence of this feedback for targeted inhibition is that it leads to activation of AKT upon inhibition within MAPK signaling. Such feedback-mediated cross talk has been noted in many different tumors such as breast cancer (Mirzoeva et al, 2009; Lu et al, 2011), prostate cancer (Gan et al, 2010), melanoma (Gopal et al, 2010), gastric cancer (Yoon et al, 2009) and in colorectal cancer (Prahallad et al, 2012). Increased phosphorylation of AKT may cause drug resistance, as AKT activity stimulates survival and migration. Furthermore, AKT shares a complex network of transcription factors with ERK (Stelniec-Klotz et al, 2012) and both paths converge on key proteins important for cellular function such as cyclin-D1 for growth control (Halilovic et al, 2010) and Bad for apoptotic regulation (She et al, 2005), explaining the need to switch off both pathways.
Our model allowed devising combinatorial therapies that block ERK activation and at the same time prevent rise in AKT activity. While PI3K inhibitors may be used to block AKT signaling, EGFR inhibition can also prevent strong AKT activation, irrespective of whether RAF, RAS or PI3K was mutated. In line with this prediction, we found approximately the same synergistic reduction of growth for PI3K/MAPK combination than for EGFR/MAPK inhibition in vitro in two cell line models. Therefore, while upregulation of AKT by MAPK inhibition can be successfully blocked by PI3K or mTOR inhibition (Balmanno et al, 2009; Mirzoeva et al, 2009; Aksamitiene et al, 2010) in colorectal cancer models, upstream inhibition of EGFR may be similarly potent with possibly less side effects. Using a RAS/PI3K mutant colon cancer xenograft model, we confirmed that combinatorial therapy with inhibitors against MEK and EGFR is also an efficient therapy in vivo. This extends previous findings showing that BRAF inhibitors synergize with EGFR inhibition in a colorectal cancer xenograft model with BRAF mutation (Prahallad et al, 2012). For RAS-mutated tumors, so far no targeted therapy is available (Baines et al, 2011; Ward et al, 2012), and a mutation in RAS precludes EGFR-directed interventions. Our results suggest that RAS-mutated tumor cells can be successfully treated by EGFR inhibitors if provided together with MEK or RAF inhibitors.
While our model can predict successful combinatorial treatments, it has limitations. It cannot account for combinations that require sequential application of drugs (Lee et al, 2012), and fails to capture resistance due to tumor–stroma interactions (Sebens and Schafer, 2012). It is likely that in other cell types different combinations of drugs may be more successful, as the role of specific feedbacks can be different in different cell types, and can even switch between positive and negative effects depending on receptor expression (Birtwistle et al, 2007).
Tumor evolution is one of the major causes for eventual relapse (Iwasa et al, 2006). For example, relapse after long-term treatment with the anti-EGFR agents Panitumumab in KRAS wild-type colorectal carcinoma (Amado et al, 2008; Karapetis et al, 2008) is often caused by selection for mutations downstream of EGFR (Diaz et al, 2012; Misale et al, 2012). The combinatorial treatment predicted by our model may thus be advisable even for EGFR-addicted tumors, as it will counteract selection for additional mutations in RAS or RAF.
In colon cells, the EGFR receptor is very potent, but depending on tissue and mutational status, other receptors may be more important in stimulating the network. For example, c-Met, that is feedback regulated and signals to both ERK and AKT, was shown to mediate resistance to BRAF inhibition in melanoma (Wilson et al, 2012), suggesting c-Met as the most effective co-target for this particular situation. Hence, our results further highlight that downstream mutations such as in RAS and BRAF do not necessarily invalidate upstream drug treatment if provided in combination with a suitable downstream inhibitor.
Materials and methods
Model construction and evaluation
The general modeling approach is depicted in Figure 3A. First, we derived a starting network from the literature. We then developed a modeling framework that extends our previous algorithm (Stelniec-Klotz et al, 2012) to estimate a quantitative map of the signaling network using an approach derived from MRA (Kholodenko, 2000; Bruggeman et al, 2002; Friday et al, 2008; Cirit et al, 2010; Sturm et al, 2010; Fritsche-Guenther et al, 2011). Briefly, MRA links the log-fold change after perturbation (called global response coefficient R) to a matrix corresponding to link strengths (local response matrix r) by inversion. We adjust the equation linking R and r for two qualitatively different perturbations (stimulation and inhibition). Stimulations are modeled as entries in the perturbation vector and inhibitions impair the signal flow of these stimulations. In addition, we allowed inhibitors to affect the base level of the signaling of the inhibited node, which is incorporated in the perturbation vector as stimulus. As the experimental design allowed measuring and perturbing only selected nodes, many parameters were not identifiable. We developed an algorithm using Gaussian elimination that reparameterized the model such that all parameters became identifiable. These new parameters were then determined by maximizing the likelihood using a Levenberg–Marquardt optimization algorithm using the best fit of 10 000 random initializations of parameters. We verified that this initial parameter scan results in unique parameters, as running the procedure 100 times on the same data, each time initializing the random number generator with a different number resulted in identical parameter sets. This network was then pruned by using a greedy hill climbing approach in which we removed edges that after model refitting did not significantly decrease the goodness of fit (Likelihood ratio test, with significance threshold of 0.05). The implementation of this algorithm was essentially as described earlier (Stelniec-Klotz et al, 2012) but extended by the non-identifiability analysis and combinatorial perturbations. To extend the model for further inhibitors, we performed maximum likelihood estimates for the novel parameters, while keeping the others constant. Error bars for predictions were generated by 100 times fitting the model to data with added noise according to the error model. The program was written in the programming language C++, and symbolic matrix operations were conducted with the GiNaC library version 1.6 ( http://www.ginac.de/). An extensive description of the algorithm and results of a stepwise reduction can be found in Supplementary Method 1 and Supplementary Figure S2, respectively.
Cells and cell culture
Human colorectal cancer cell lines SW480, SW403, HCT116, RKO and HT29 were obtained from the ATCC (American Type Culture Collection, UK). LIM1215 were kindly provided by Professor John Mariadason (Ludwig Institute for Cancer Research Austin Hospital, Melbourne), Australia. All cell lines were maintained in DMEM (Dulbecco’s modified Eagle’s medium, Lonza) supplemented with 10% fetal calf serum, 1% ultraglutamine and 1% penicillin/streptomycin and incubated in a humidified atmosphere of 5% CO2 in air at 37 °C.
Reagents
The following inhibitors where used in the various assays: MEK inhibitors AZD6244 (0.1 μM unless otherwise specified, Selleck Chemicals LLC) and GDC-0973 (Genentech), RAF inhibitor sorafenib (10 μM, LC Laboratories), PI3K inhibitor LY294002 (10 μM, Axxora Deutschland), EGFR inhibitors gefitinib (10 μM, Cayman Chemicals) and erlotinib (Genentech), GSK3ß inhibitor SB216763 (5 μM, Sigma Aldrich), IKK-α/β inhibitor BMS345541 (25 μM, Sigma Aldrich), and IKK-ß inhibitors PS1145 (10 μM, AxonMedChem) and PHA408 (100 nM, AxonMedChem). The solvent control was DMSO (equal μM to each inhibitor).
We used the following ligands (all Peprotech): TGF-α (0.01 μg/ml), IGF-1 (0.1 μg/ml), EGF (0.025 μg/ml), FGF2 (0.005 μg/ml) and HGF (0.05 μg/ml) with 0.01% BSA in PBS as solvent.
Luminex assays
After treatment and incubation, lysates were collected according to supplier’s protocol and analyzed with the Bio-Plex Protein Array system (Bio-Rad, Hercules, CA) using beads specific for P-AKT (S473), P-ERK1/2 (Thr202/Tyr204/Thr185/Tyr187), P-ERK2 (Thr185/Tyr187), P-GSK3α/ß (S21/S6), P-IGF-1R (Tyr1131), P-IRS1 (S636/S639), P-IKBα (S32/S36), P-MEK1 (S217/S221), and P-p70S6K (Thr421/S424) according to the manufacturer’s instructions. Briefly, samples were washed with PBS and lysed with cell lysis buffer (Bio-Rad). Lysate protein concentration was determined with BCA (bicinchoninic acid) method. The beads and detection antibodies were diluted 1:5 or 1:3. For data acquisition, the Bio-Plex Manager software was used.
RNA isolation and quantitative RT–PCR analysis
RNA was isolated from HCT116 cells treated with BMS345541 using the RNeasy-mini-kit (Qiagen) according to the supplier’s protocol. To obtain cDNA from RNA, the high-capacity cDNA reverse transcription kit (Applied Biosystems) was used. Synthesis of double-stranded DNA during the PCR cycles was visualized with TaqMan gene expression assays FAM-dye labeled (gene of interest: EGR1 (Hs_00152928_m1), cFos (Hs_00170630_m1)) or VIC-dye labeled for loading control PGK1 (Hs_943178_g1) and TaqMan gene expression master mix (Applied Biosystems). Quantitative real-time PCR (qrt–PCR) analysis was performed using a StepOnePlus 96-well format Light-Cycler apparatus (Applied Biosystems). Experiments were run and analyzed with the StepOne 2.0 software. The data were analyzed quantitatively by measuring the threshold cycles (CT). CT values where normalized first by the CT of the internal control PGK1 (−ΔCT) and second by the ΔCT value of the zero time point (−ΔΔ CT), which are interpreted as log2 fold changes.
Immunoblotting
Protein extracts of cells were prepared as described for Bioplex analysis. Blotting procedure and materials were as previously described (Fritsche-Guenther et al, 2011; Stelniec-Klotz et al, 2012). The following primary antibodies were used: rabbit anti-human P-p70S6 (Thr389, Cell Signaling Technology, 1:500) or mouse anti-human GAPDH (Ambion, 1:12 500). Membranes were scanned using Li-COR Odyssey. The signals were quantified using Odyssey software.
Tumor and body weight measurements
The DLD-1 colorectal tumor cell line was inoculated subcutaneously on the flank of female athymic nude (nu/nu) mice (Harlan Laboratories) and tumors were allowed to establish to a size of 125–250 mm3. Animals were grouped out into six treatment groups (n=10 per group) based upon tumor volume and treatment was initiated with vehicles (MCT (methylcellulose+0.2% Tween 80) and 15% HBCD (15% hydroxypropyl-beta-cyclodextrin)), erlotinib (50 mg/kg, daily oral gavage formulated in 15% hydroxypropyl-beta-cyclodextrin), GDC-0973 (1 or 5 mg/kg, daily oral gavage formulated in MCT), or the combination of erlotinib+GDC-0973 at both doses. Tumor volumes were determined using digital calipers (Fred V. Fowler Company, Inc.) using the formula (L × W × W)/2. %TGI was calculated as the percentage of the area under the fitted curve (AUC) for the respective dose group per day in relation to the vehicle, such that %TGI=100 × 1—(AUCtreatment/day)/(AUCvehicle/day). Curve fitting was applied to log2-transformed individual tumour volume data using a linear mixed-effects model using the R package nlme, version 3.1–97 in R v2.12.0. Body weights and gross observations on animal welfare were also recorded throughout the study. All experimental procedures conformed to the guiding principles of the American Physiology Society and were approved by Genentech’s Institutional Animal Care and Use Committee.
Proliferation assay
For proliferation studies in real time, the XCelligence RTCA SP instrument was used. HCT116 and HT29 cells were plated 24 h before treatment with inhibitors or controls. Over the time of measurement, the system records the cell index (ci), which reflects the cell attachment to the electrodes and is proportional to the number of cells. Each treatment was measured at least in triplicates. As the initial cell number is variable and growth of the cell population is exponential, averaging the growth curves is not reasonable. Instead, we calculated the logarithmised and normalized cell index log2(ci(t)/ci(t of inhibitor application)) and took the average of these values.
Sequencing and detection of single-nucleotide variations (SNVs)
We performed targeted sequencing by the Ion AmpliSeq Cancer Panel 1.0 and the Ion PGM sequencer covering 13.7-kb-coding sequence from 46 cancer-relevant genes (Supplementary Table ST2) and 739 COSMIC-annotated SNP positions. Genomic DNA was extracted from pelleted CRC cell lines. The extracted DNA was quantified and the quality estimated by the Qubit 2.0 Fluorometer (Qubit dsDNA HS Assay Kit, Life Technologies) and Bioanalyzer 2100 (High Sensitivity DNA Kit, Agilent). The amplicon library was prepared according to the manufacturer’s protocols using 12–30 ng of DNA and loaded on an Ion 314 Chip (Life Technologies) to yield at least 10 Mb. To increase the efficiency, we spun the chip for 2 min before and after turning around the chip by 180° for each loading. IonTorrent Suite, Version 2.1 Variant Caller (Life Technologies) was used for variant calling, with positions with minor allele frequencies below 10% were defined homozygotic. SNVs were filtered as described in supplement.
Supplementary Material
Supplementary figures S1-S10, Supplementary methods 1 and 2
Model parameter
Acknowledgments
This study has been largely funded by the German Ministry for Education and Research (BMBF), grants Colonet (to CS, NB, RS) and FORSYS (to NB), by DFG grants SFB618 (A3 to NB, RS, A1 to CS) and SPP1395 (InKomBio to NB). Establishment of the deep sequencing platform was supported by the German Cancer Aid funding program ‘Centers of Excellence in Oncology’. We acknowledge skilled technical support by Cornelia Gieseler. We thank Ralf Steuer for critically reading the manuscript.
Author contributions: NB and CS conceived the study, AS and RF-G performed perturbation studies, DS performed sequencing, LB conducted xenograft experiments, BK and NB developed algorithm and performed mathematical modeling, RS supervised sequencing and cell culture, MM supervised xenograft experiments, FW and PD performed data analysis, NB, BK, CS, RS and YY discussed results, BK and NB wrote the manuscript with input from all authors.
Footnotes
LB was an employee, YY and MM are employees of Genentech, Inc., a member of the Roche group, and may have equity interest in Roche. The remaining authors declare that they have no conflict of interest.
References
- Aksamitiene E, Kholodenko BN, Kolch W, Hoek JB, Kiyatkin A (2010) PI3K/Akt-sensitive MEK-independent compensatory circuit of ERK activation in ER-positive PI3K-mutant T47D breast cancer cells. Cell Signal 22: 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD (2008) Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26: 1626–1634 [DOI] [PubMed] [Google Scholar]
- Avraham R, Yarden Y (2011) Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol 12: 104–117 [DOI] [PubMed] [Google Scholar]
- Baines AT, Xu D, Der CJ (2011) Inhibition of Ras for cancer treatment: the search continues. Future Med Chem 3: 1787–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmanno K, Chell SD, Gillings AS, Hayat S, Cook SJ (2009) Intrinsic resistance to the MEK1/2 inhibitor AZD6244 (ARRY-142886) is associated with weak ERK1/2 signalling and/or strong PI3K signalling in colorectal cancer cell lines. Int J Cancer 125: 2332–2341 [DOI] [PubMed] [Google Scholar]
- Birtwistle MR, Hatakeyama M, Yumoto N, Ogunnaike BA, Hoek JB, Kholodenko BN (2007) Ligand-dependent responses of the ErbB signaling network: experimental and modeling analyses. Mol Syst Biol 3: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüthgen N, Legewie S (2008) Systems analysis of MAPK signal transduction. Essays Biochem 45: 95–107 [DOI] [PubMed] [Google Scholar]
- Bruggeman FJ, Westerhoff HV, Hoek JB, Kholodenko BN (2002) Modular response analysis of cellular regulatory networks. J Theor Biol 218: 507–520 [PubMed] [Google Scholar]
- Bublil EM, Yarden Y (2007) The EGF receptor family: spearheading a merger of signaling and therapeutics. Current Opin Cell Biol 19: 124–134 [DOI] [PubMed] [Google Scholar]
- Cirit M, Wang C-C, Haugh JM (2010) Systematic quantification of negative feedback mechanisms in the extracellular signal-regulated kinase (ERK) signaling network. J Biol Chem 285: 36736–36744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon AS, Hagan S, Rath O, Kolch W (2007) MAP kinase signalling pathways in cancer. Oncogene 26: 3279–3290 [DOI] [PubMed] [Google Scholar]
- Diaz LA, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, Kinzler KW, Oliner KS, Vogelstein B (2012) The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486: 537–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douville E, Downward J (1997) EGF induced SOS phosphorylation in PC12 cells involves P90 RSK-2. Oncogene 15: 373–383 [DOI] [PubMed] [Google Scholar]
- Florini JR, Ewton DZ, Coolican SA (1996) Growth hormone and the insulin-like growth factor system in myogenesis. Endocr Rev 17: 481–517 [DOI] [PubMed] [Google Scholar]
- Friday BB, Yu C, Dy GK, Smith PD, Wang L, Thibodeau SN, Adjei AA (2008) BRAF V600E disrupts AZD6244-induced abrogation of negative feedback pathways between extracellular signal-regulated kinase and Raf proteins. Cancer Res 68: 6145–6153 [DOI] [PubMed] [Google Scholar]
- Fritsche-Guenther R, Witzel F, Sieber A, Herr R, Schmidt N, Braun S, Brummer T, Sers C, Blüthgen N (2011) Strong negative feedback from Erk to Raf confers robustness to MAPK signalling. Mol Syst Biol 7: 489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Shi C, Inge L, Hibner M, Balducci J, Huang Y (2010) Differential roles of ERK and Akt pathways in regulation of EGFR-mediated signaling and motility in prostate cancer cells. Oncogene 29: 4947–4958 [DOI] [PubMed] [Google Scholar]
- Gopal YNV, Deng W, Woodman SE, Komurov K, Ram P, Smith PD, Davies MA (2010) Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res 70: 8736–8747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halilovic E, She QB, Ye Q, Pagliarini R, Sellers WR, Solit DB, Rosen N (2010) PIK3CA mutation uncouples tumor growth and cyclin D1 regulation from MEK/ERK and mutant KRAS signaling. Cancer Res 70: 6804–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Hastings WK (1970) Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57: 97–109 [Google Scholar]
- Herbst RS, Fukuoka M, Baselga J (2004) Gefitinib—a novel targeted approach to treating cancer. Nat Rev Cancer 4: 956–965 [DOI] [PubMed] [Google Scholar]
- Iwasa Y, Nowak MA, Michor F (2006) Evolution of resistance during clonal expansion. Genetics 172: 2557–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au H-J, Langer C, Moore MJ, Zalcberg JR (2008) K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359: 1757–1765 [DOI] [PubMed] [Google Scholar]
- Kholodenko B, Yaffe MB, Kolch W (2012) Computational approaches for analyzing information flow in biological networks. Sci Signal 5: re1. [DOI] [PubMed] [Google Scholar]
- Kholodenko BN (2000) Negative feedback and ultrasensitivity can bring about oscillations in the mitogen-activated protein kinase cascades. Eur J Biochem 267: 1583–1588 [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Demin OV, Moehren G, Hoek JB (1999) Quantification of short term signaling by the epidermal growth factor receptor. J Biol Chem 274: 30169–30181 [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Kiyatkin A, Bruggeman FJ, Sontag E, Westerhoff HV, Hoek JB (2002) Untangling the wires: a strategy to trace functional interactions in signaling and gene networks. Proc Natl Acad Sci USA 99: 12841–12846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Rath O, Kolch W, Cho K-H (2007) A hidden oncogenic positive feedback loop caused by crosstalk between Wnt and ERK pathways. Oncogene 26: 4571–4579 [DOI] [PubMed] [Google Scholar]
- Kreeger PK, Mandhana R, Alford SK, Haigis KM, Lauffenburger DA (2009) RAS mutations affect tumor necrosis factor-induced apoptosis in colon carcinoma cells via ERK-modulatory negative and positive feedback circuits along with non-ERK pathway effects. Cancer Res 69: 8191–8199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB (2012) Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell 149: 780–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legewie S, Herzel H, Westerhoff HV, Blüthgen N (2008) Recurrent design patterns in the feedback regulation of the mammalian signalling network. Mol Syst Biol 4: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Muller M, Smith D, Dutta B, Komurov K, Iadevaia S, Ruths D, Tseng J-T, Yu S, Yu Q, Nakhleh L, Balazsi G, Donnelly J, Schurdak M, Morgan-Lappe S, Fesik S, Ram PT, Mills GB (2011) Kinome siRNA-phosphoproteomic screen identifies networks regulating AKT signalling. Oncogene 30: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Morrison DJ, Basson MA, Licht JD (2006) Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol 16: 45–54 [DOI] [PubMed] [Google Scholar]
- Mirzoeva OK, Das D, Heiser LM, Bhattacharya S, Siwak D, Gendelman R, Bayani N, Wang NJ, Neve RM, Guan Y, Hu Z, Knight Z, Feiler HS, Gascard P, Parvin B, Spellman PT, Shokat KM, Wyrobek AJ, Bissell MJ, McCormick F et al. (2009) Basal subtype and MAPK/ERK kinase (MEK)-phosphoinositide 3-kinase feedback signaling determine susceptibility of breast cancer cells to MEK inhibition. Cancer Res 69: 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, Bencardino K, Cercek A, Chen C-T, Veronese S, Zanon C, Sartore-Bianchi A, Gambacorta M, Gallicchio M, Vakiani E, Boscaro V et al. (2012) Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MK, Saez-Rodriguez J, Clarke DC, Sorger PK, Lauffenburger DA (2011) Training signaling pathway maps to biochemical data with constrained fuzzy logic: quantitative analysis of liver cell responses to inflammatory stimuli. PLoS Comput Biol 7: e1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelander S, Wang W, Nilsson B, She Q-B, Pratilas C, Rosen N, Gennemark P, Sander C (2008) Models from experiments: combinatorial drug perturbations of cancer cells. Mol Syst Biol 4: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Matsuoka Y, Funahashi A, Kitano H (2005) A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol 1: 0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter AC, Vaillancourt RR (1998) Tyrosine kinase receptor-activated signal transduction pathways which lead to oncogenesis. Oncogene 17: 1343–1352 [DOI] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R (2012) Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483: 100–103 [DOI] [PubMed] [Google Scholar]
- Prenen H, Tejpar S, Cutsem EV (2010) New strategies for treatment of KRAS mutant metastatic colorectal cancer. Clinl Cancer Res 16: 2921–2926 [DOI] [PubMed] [Google Scholar]
- Raue A, Kreutz C, Maiwald T, Bachmann J, Schilling M, Klingmüller U, Timmer J (2009) Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics 25: 1923–1929 [DOI] [PubMed] [Google Scholar]
- Roberts PJ, Der CJ (2007) Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 26: 3291–3310 [DOI] [PubMed] [Google Scholar]
- Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, Aranda E, Nordlinger B, Cisar L, Labianca R, Cunningham D, Van Cutsem E, Bosman F (2010) Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol 28: 466–474 [DOI] [PubMed] [Google Scholar]
- Saez-Rodriguez J, Alexopoulos LG, Epperlein J, Samaga R, Lauffenburger DA, Klamt S, Sorger PK (2009) Discrete logic modelling as a means to link protein signalling networks with functional analysis of mammalian signal transduction. Mol Syst Biol 5: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Rodriguez J, Alexopoulos LG, Zhang M, Morris MK, Lauffenburger DA, Sorger PK (2011) Comparing signaling networks between normal and transformed hepatocytes using discrete logical models. Cancer Res 71: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SDM, Verveer PJ, Bastiaens PIH (2007) Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol 9: 324–330 [DOI] [PubMed] [Google Scholar]
- Schoeberl B, Eichler-Jonsson C, Gilles ED, Müller G (2002) Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat Biotechnol 20: 370–375 [DOI] [PubMed] [Google Scholar]
- Sebens S, Schafer H (2012) The tumor stroma as mediator of drug resistance—a potential target to improve cancer therapy? Curr Pharm Biotechnol 13: 2259–2272 [DOI] [PubMed] [Google Scholar]
- Shankaran H, Wiley HS (2010) Oscillatory dynamics of the extracellular signal-regulated kinase pathway. Curr Opin Genet Dev 20: 650–655 [DOI] [PubMed] [Google Scholar]
- She Q-B, Solit DB, Ye Q, O’Reilly KE, Lobo J, Rosen N (2005) The BAD protein integrates survival signaling by EGFR/MAPK and PI3K/Akt kinase pathways in PTEN-deficient tumor cells. Cancer Cell 8: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelniec-Klotz I, Legewie S, Tchernitsa O, Witzel F, Klinger B, Sers C, Herzel H, Blüthgen N, Schäfer R (2012) Reverse engineering a hierarchical regulatory network downstream of oncogenic KRAS. Mol Syst Biol 8: 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm OE, Orton R, Grindlay J, Birtwistle M, Vyshemirsky V, Gilbert D, Calder M, Pitt A, Kholodenko B, Kolch W (2010) The mammalian MAPK/ERK pathway exhibits properties of a negative feedback amplifier. Sci Signal 3: ra90. [DOI] [PubMed] [Google Scholar]
- Tentner AR, Lee MJ, Ostheimer GJ, Samson LD, Lauffenburger DA, Yaffe MB (2012) Combined experimental and computational analysis of DNA damage signaling reveals context-dependent roles for Erk in apoptosis and G1/S arrest after genotoxic stress. Mol Syst Biol 8: 568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D (2009) Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer 9: 489–499 [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang M, Lazo JS, Carr BI (2002) Identification of epidermal growth factor receptor as a target of Cdc25A protein phosphatase. J Biol Chem 277: 19470–19475 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang B, Wang M, Carr BI (2005) Cdc25A and ERK interaction: EGFR-independent ERK activation by a protein phosphatase Cdc25A inhibitor, compound 5. J Cell Physiol 204: 437–444 [DOI] [PubMed] [Google Scholar]
- Ward AF, Braun BS, Shannon KM (2012) Targeting oncogenic Ras signaling in hematologic malignancies. Blood 120: 3397–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, Ribas A, Li J, Moffat J, Sutherlin DP, Koeppen H, Merchant M, Neve R, Settleman J (2012) Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487: 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi TM, Huang Y, Simon MI, Doyle J (2000) Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc Natl Acad Sci USA 97: 4649–4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y-K, Kim H-P, Han S-W, Hur H-S, Oh DY, Im S-A, Bang Y-J, Kim T-Y (2009) Combination of EGFR and MEK1/2 inhibitor shows synergistic effects by suppressing EGFR/HER3-dependent AKT activation in human gastric cancer cells. Mol Cancer Ther 8: 2526–2536 [DOI] [PubMed] [Google Scholar]
- Yoon Y-K, Kim H-P, Song S-H, Han S-W, Oh DY, Im S-A, Bang Y-J, Kim T-Y (2012) Down-regulation of mitogen-inducible gene 6, a negative regulator of EGFR, enhances resistance to MEK inhibition in KRAS mutant cancer cells. Cancer Lett 316: 77–84 [DOI] [PubMed] [Google Scholar]
- Yu CF, Liu Z-X, Cantley LG (2002) ERK negatively regulates the epidermal growth factor-mediated interaction of Gab1 and the phosphatidylinositol 3-kinase. J Biol Chem 277: 19382–19388 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures S1-S10, Supplementary methods 1 and 2
Model parameter