Abstract
Organic Lake is a shallow, marine-derived hypersaline lake in the Vestfold Hills, Antarctica that has the highest reported concentration of dimethylsulfide (DMS) in a natural body of water. To determine the composition and functional potential of the microbial community and learn about the unusual sulfur chemistry in Organic Lake, shotgun metagenomics was performed on size-fractionated samples collected along a depth profile. Eucaryal phytoflagellates were the main photosynthetic organisms. Bacteria were dominated by the globally distributed heterotrophic taxa Marinobacter, Roseovarius and Psychroflexus. The dominance of heterotrophic degradation, coupled with low fixation potential, indicates possible net carbon loss. However, abundant marker genes for aerobic anoxygenic phototrophy, sulfur oxidation, rhodopsins and CO oxidation were also linked to the dominant heterotrophic bacteria, and indicate the use of photo- and lithoheterotrophy as mechanisms for conserving organic carbon. Similarly, a high genetic potential for the recycling of nitrogen compounds likely functions to retain fixed nitrogen in the lake. Dimethylsulfoniopropionate (DMSP) lyase genes were abundant, indicating that DMSP is a significant carbon and energy source. Unlike marine environments, DMSP demethylases were less abundant, indicating that DMSP cleavage is the likely source of high DMS concentration. DMSP cleavage, carbon mixotrophy (photoheterotrophy and lithoheterotrophy) and nitrogen remineralization by dominant Organic Lake bacteria are potentially important adaptations to nutrient constraints. In particular, carbon mixotrophy relieves the extent of carbon oxidation for energy production, allowing more carbon to be used for biosynthetic processes. The study sheds light on how the microbial community has adapted to this unique Antarctic lake environment.
Keywords: metagenomics, Organic Lake, Antarctic microbial ecology, nutrient cycles, dimethylsulfide
Introduction
Molecular biology approaches have proven useful for describing the diversity and gene content of microorganisms in Antarctic lakes and for inferring the functional roles of the taxa present (Laybourn-Parry and Pearce, 2007; Wilkins et al., 2013b). However to date, only a few large-scale shotgun metagenome studies have been performed on the Antarctic continent and in the surrounding Southern Ocean (reviewed in Wilkins et al., 2013b). In the Vestfold Hills, metagenomics and metaproteomics have been used to study Ace Lake (Ng et al., 2010; Lauro et al., 2011) and Organic Lake (Yau et al., 2011).
Owing to the polar light cycle and low overall levels of photosynthetically active radiation, phototrophic growth and biomass production are restricted, being relatively high in summer and negligible in winter (Laybourn-Parry et al., 2005). In Ace Lake, phototrophic algae, particularly phytoflagellates, engage in mixotrophy, thereby supplementing their carbon and nutrient requirements to enable them to remain active during winter and poised to photosynthesize in summer (Laybourn-Parry et al., 2005). Resourcefulness has also been demonstrated by heterotrophic marine bacteria that exploit light energy, either directly through photoheterotrophic processes involving aerobic anoxygenic photosynthesis (AAnP), or indirectly through lithoheterotrophic processes utilizing inorganic compounds (for example, CO) formed from dissolved organic carbon reacting with light (Moran and Miller, 2007).
Organic Lake is located in the Vestfold Hills, on the eastern shore of Prydz Bay, East Antarctica (Supplementary Figure S1). It is shallow (6.8 m) and has variable surface water temperatures (−14 to +15 °C) while remaining sub-zero throughout most of its depth (Franzmann et al., 1987b; Gibson et al., 1991; Roberts et al., 1993; Gibson, 1999). The lake has a high organic load generated from autochthonous production, and input from penguins and terrestrial algae (for example, in soaks, shallow ephemeral streams and depressions), and nutrient turnover is expected to be slow due to the constraints imposed on microbial activity by the lake's hypersalinity (∼230 g l−1 maximum salinity) and low temperature (Franzmann et al., 1987b; Gibson et al., 1991; Roberts et al., 1993; Gibson, 1999). The salt and marine biota in the lake originate from seawater that was trapped in a basin ∼3000 years BP when the continental ice sheet receded and the land rose above sea level (Bird et al., 1991; Zwartz et al., 1998; Gibson, 1999). The bottom water of Organic Lake is unusual due to the absence of hydrogen sulfide and the high concentration of the gas dimethylsulfide (DMS), dimethylsulfoniopropionate (DMSP) and polysulfides (Deprez et al., 1986; Franzmann et al., 1987b; Gibson et al., 1991; Roberts and Burton 1993; Roberts et al., 1993). Concentrations of DMS as high as 5000 nM have been recorded in Organic Lake (Gibson et al., 1991), 100 times the maximum concentration recorded from seawater in the adjacent Prydz Bay and at least 1000 times that of the open Southern Ocean (Curran and Jones, 1998).
While it has been 40 years since DMS was identified as a carrier of sulfur from the oceans to the atmosphere (Lovelock and Maggs, 1972) and later proposed to have a regulatory effect on the global cloud cover (Charlson et al., 1987), the genes that encode for the enzymes involved in DMS production were only identified in the last 6 years (Todd et al., 2007). Rapid progress has been made in the short period since then, and the pathways and organisms involved in DMS transformations have been extensively reviewed (Johnston et al., 2008; Schäfer et al., 2010; Curson et al., 2011; Reisch et al., 2011; Moran et al., 2012). The main source of DMS in the marine environment is from the breakdown of DMSP. Eucaryal phytoplankton, in particular sea-ice diatoms, and dinoflagellates (for example, Prymnesiophytes such as Phaeocystis spp.) and haptophytes, produce DMSP, an organosulfur compound that is thought to function principally as an osmolyte (Yoch, 2002; Stefels et al., 2007; Hatton et al., 2012). DMSP is released due to cell lysis, grazing or leakage and follows two known fates: DMSP cleavage by DMSP lyases (DddD, -L, -P, -Q, -W and -Y) or demethylation by DMSP demethylase (DmdA). Both pathways are associated with diverse microorganisms that can utilize DMSP as a sole carbon and energy source. However, it is only the cleavage pathway that releases volatile DMS that can lead to sulfur loss through ventilation to the atmosphere.
The very high levels of DMS in Organic Lake make it an ideal system for identifying the microorganisms and the processes involved in DMS accumulation. The previous metagenome study of Organic Lake examined viruses from the 0.1-μm fraction of surface water that was collected from Organic Lake in December 2006, and November and December 2008 (Yau et al., 2011). In the present study, we focused on the cellular population rather than viruses in these samples. Our study determined the composition and functional potential of Organic Lake microbiota and, in conjunction with historic and contemporary physicochemical data, generated an integrative understanding of the whole-lake ecosystem.
Materials and methods
Metagenomic analyses (2.4 Gbp) were performed on biomass captured by sequential filtration through a 20-μm pre-filter onto 3.0, 0.8 and 0.1 μm filters based on the design of the Global Ocean Sampling expedition (Rusch et al., 2007). Water was sampled (including for epifluorescence microscopy, and physical and chemical measurements) from a depth profile (1.7, 4.2, 5.7, 6.5 and 6.7 m) taken in November 2008 (Supplementary Figures S1–S3 and Supplementary Tables S1–S3), and cellular diversity and functional potential were determined based on approaches developed for studying Antarctic aquatic microbial communities (Ng et al., 2010; Lauro et al., 2011; Yau et al., 2011; Brown et al., 2012; Wilkins et al., 2013a; Williams et al., 2013). Full details of Materials and methods are provided in Supporting Information.
Results and discussion
Abiotic properties and water column structure
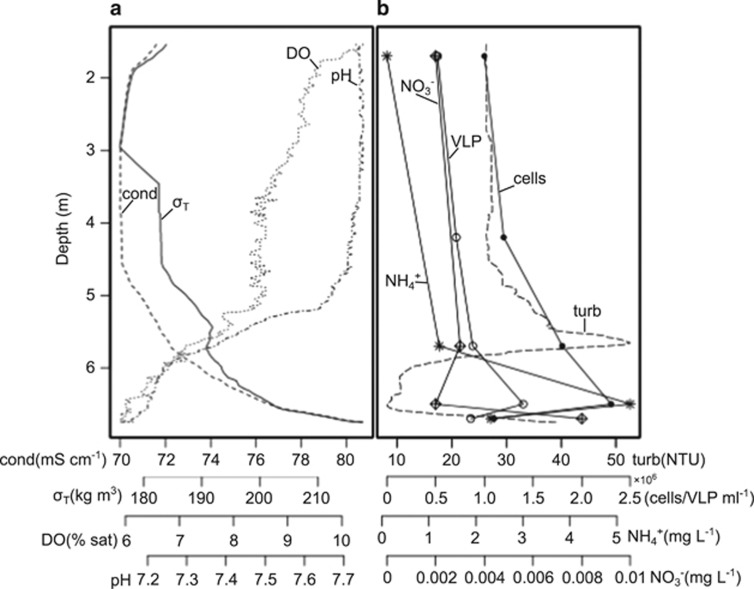
In situ physicochemical profiles (Supplementary Figure S2) were measured at the deepest point in the lake (Supplementary Figure S3), and samples were collected from the upper mixed (1.7, 4.2 and 5.7 m) and suboxic deep (6.5 and 6.7 m) zones (Figure 1a). All nutrients, except for nitrate and nitrite, reached maximum concentrations at 6.5 m (Table 1), suggesting a layer of high biological activity above the lake bottom. Consistent with this, cell and virus-like particle counts were highest at 6.5 m. However, turbidity was lowest at this depth, demonstrating that turbidity was not principally determined by cell density (Figure 1b). Microscopy images did not show a shift in cell morphology that could account for the large drop in turbidity (Supplementary Figure S4), which suggests that particulate matter primarily contributed to turbidity readings. The low turbidity and peak in cell counts and nutrients at the oxycline at 6.5 m may be caused by an active microbial community degrading particulate matter. This inference is supported by the report of high concentrations of dissolved organic acids and free amino acids in the deep zone (Gibson et al., 1994), as these nutrients are indicative of the breakdown of high molecular weight carbohydrates, lipids and proteins. Furthermore, the C:N and C:P ratios throughout the lake were high compared with the Redfield ratio (Redfield et al., 1963) except at 6.5 m, indicating that this was the only depth where dissolved nitrogen and phosphorus were not relatively limited (Table 1). Principal component analysis of physicochemical parameters showed all samples, except the 6.5 m sample, separated with depth along the principal component 1 axis (Supplementary Figure S5). Accordingly, turbidity, total sulfur and cell density were the strongest explanatory variables for the separation of the 6.5 m sample from the other deep sample, indicating that increased activity at 6.5 m was related to the breakdown of particulate matter and sulfur chemistry.
Figure 1.
Vertical structure of Organic Lake. (a) Parameters that varied unimodally with depth showed two zones: an aerobic mixed zone above 5.7 m and a denser suboxic deep zone below. (b) Additional factors that revealed stratification within the deep zone. The peak in concentration at 6.5 m for ammonia was also observed for all other nutrients assayed except nitrate and nitrite (see Table 1). σT=(1000−density); cond, conductivity; DO, dissolved oxygen; turb, turbidity.
Table 1. Physicochemical properties, cell counts and VLP counts of Organic Lake samples.
|
Sample depths (m) |
|||||
|---|---|---|---|---|---|
| 1.7 | 4.2 | 5.7 | 6.5 | 6.7 | |
| Ammonia (mg l−1) | 0.108 | ND | 1.22 | 5.29 | 2.32 |
| Nitrate (mg l−1) | <0.002 | ND | 0.003 | <0.002 | 0.008 |
| Nitrite (mg l−1) | <0.002 | ND | <0.002 | 0.010 | 0.010 |
| SRP (mg l−1) | 0.08 | ND | 0.10 | 0.20 | 0.18 |
| TOC (mg l−1) | 88 | 87 | 110 | 170 | 130 |
| DOC (mg l−1) | 69 | ND | 97 | 150 | 120 |
| TN (mg l−1) | 7.70 | 7.50 | 11 | 24 | 13 |
| TDN (mg l−1) | 0.112 | ND | 1.225 | 5.302 | 2.338 |
| TP (mg l−1) | 1.5 | 1.4 | 3.0 | 7.6 | 3.7 |
| TDP (mg l−1) | 0.509 | ND | 0.805 | 4.5 | 2 |
| TS (mg l−1) | 1010 | 974 | 1020 | 1410 | 950 |
| TDS (mg l−1) | 996 | ND | 1250 | 1290 | 995 |
| Particulate C:N:P (molar ratios) | 49:7:1 | ND | 15:2:1 | 17:3:1 | 15:1:1 |
| Dissolved C:N:P (molar ratios) | 350:20:1 | ND | 311:26:1 | 86:10:1 | 155:13:1 |
| Practical salinity | 166 | 166 | 172 | 178 | 186 |
| Temperature (°C) | −13 | −13.5 | −13 | −12.5 | −12 |
| Cells ml−1 | 1.0±0.4 × 106 | 1.2±0.3 × 106 | 1.8±0.5 × 106 | 2.3±0.8 × 106 | 1.1±0.4 × 106 |
| VLP ml−1 | 5.2±2.1 × 105 | 7.1±1.3 × 105 | 8.8±3.4 × 105 | 14.0±3.0 × 105 | 8.6±3.3 × 105 |
Abbreviations: DOC, dissolved organic carbon; ND, data not determined; SRP, soluble reactive phosphate; TDN, total dissolved nitrogen; TDP, total dissolved phosphorus; TDS, total dissolved sulfur; TN, total nitrogen; TOC, total organic carbon; TP, total phosphorus; TS, total sulfur; VLP, virus-like particles. One s.d. shown for cell and VLP counts.
Variation of microbial composition according to size and depth
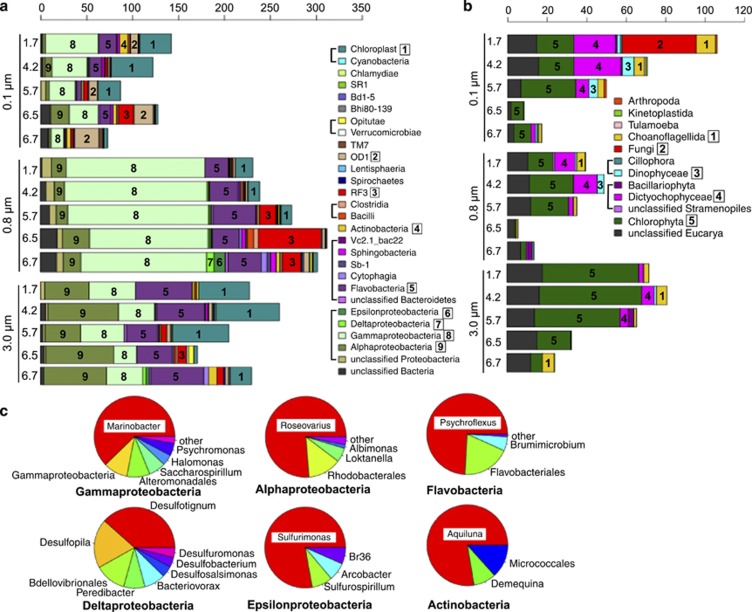
SSU rRNA genes (3959 reads) that were retrieved from the metagenome data grouped into 983 operational taxonomic units (OTUs). OTUs for Bacteria comprised 76.2% and, Eucarya 16.3%, whereas 7.5% of SSU rRNA gene sequences could not be classified (Figure 2). Only two reads were assigned to a deep-sea hydrothermal clade of Halobacteriales (Supplementary Table S4), indicating that Archaea were rare in Organic Lake.
Figure 2.
Diversity of Bacteria and Eucarya in Organic Lake. (a) Bacteria and (b) Eucarya from each size fraction (0.1, 0.8 and 3.0 μm) at each sample depth (1.7, 4.2, 5.7, 6.5 and 6.7 m) aggregated according to class. The x-axis shows counts of SSU rRNA gene sequences normalized to average reads acquired per sample filter. Taxa that belong to the same higher rank are shown grouped with a square bracket in the legend. Abundant taxa are labeled in the plot with a number that corresponds to the numbered boxes in the legend. (c) Composition of abundant bacterial classes across all size fractions combined. See Supplementary Table S4 for lower taxonomic rank assignments.
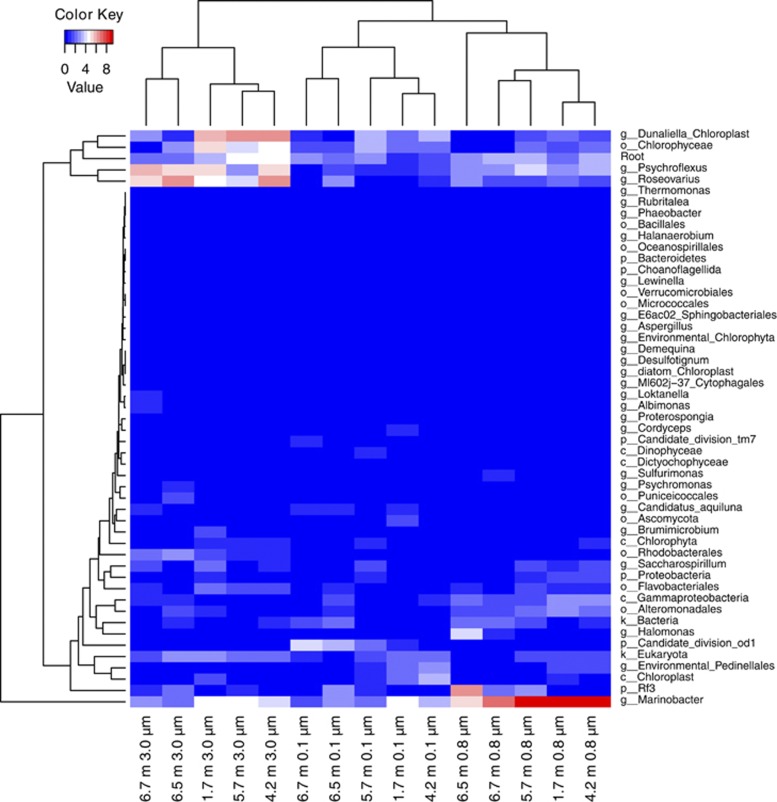
Community composition varied with size fraction and depth. This was supported by seriation analysis that showed samples clustered according to size fraction, and those clusters further separated into upper mixed and deep-zone groups (Figure 3). A significant difference in genus-level composition between the upper mixed and deep zones was supported by the analysis of similarity test (ρ: 0.53, significance: 0.1%). Taxa were differentially distributed consistent with vertical, physical and chemical stratification, indicating that ecological functions were partitioned in the lake.
Figure 3.
Heatmap and biclustering plot of the SSU rRNA gene composition in Organic Lake. Samples are shown according to size fractionate (3.0, 0.8 and 0.1 μm) and depth (1.7, 4.2, 5.7, 6.5 and 6.7 m). SSU rRNA genes were classified to the lowest taxonomic rank that gave bootstrap confidence>85% until the rank of genus. SSU rRNA gene counts were normalized and square root transformed. Taxa that comprised <2% of the sample were not included.
20–3.0-μm fraction community composition
The upper mixed zone samples had a relatively high OTU abundance of Dunaliella chloroplasts and chlorophyte algae, consistent with the large, active photosynthetic organisms concentrated near surface light. They are likely the main source of primary production in Organic Lake and have previously been reported to be the dominant algae (Franzmann et al., 1987b). The SSU rRNA gene sequences for these algae at the bottom of the lake are likely to be due to sedimentation of dead cells or resting cysts.
Psychroflexus OTUs were over-represented in the surface and 6.7 m samples. Consistent with enrichment on the 3.0 μm filters, Psychroflexus (formerly Flavobacterium) gondwanensis (Bowman et al., 1998) isolated from Organic Lake (Franzmann et al., 1987b) had cells 1.5–11.5 μm in length (Dobson et al., 1991). Flavobacteria associate with phytoplankton blooms in the Southern Ocean (Abell and Bowman 2005a, 2005b; Williams et al., 2013) and have specialized abilities to degrade polymeric substances from algal exudates and detritus (reviewed in Kirchman, 2002; Williams et al., 2013). It is likely that Organic Lake Psychroflexus fills a similar ecological role. In support of this, Psychroflexus OTUs cluster with Dunaliella chloroplasts in the seriation analysis (Figure 3) and P. gondwanensis abundance in Organic Lake has been correlated with average hours of sunshine per day, indicating population dynamics that are related to summer algal blooms (James et al., 1994). The Psychroflexus OTUs in the deep zone are most likely due to sedimentation, as P. gondwanensis is nonmotile and strictly aerobic (Dobson et al., 1991; Dobson et al., 1993).
Roseovarius OTUs were enriched at 4.2 and 6.5 m, suggesting that different ecotypes may be present in the upper mixed zone compared with the deep zone. Roseovarius tolerans, an isolate from Ekho Lake in the Vestfold Hills, Antarctica, has a cell size (1.1–2.2 μm; Labrenz et al., 1999) that would be expected to be captured on the 0.8-μm filter. The Roseovarius captured on the 3-μm filter may, therefore, be a different species, or a strain similar to R. tolerans from Ekho Lake that exhibits different growth characteristics (that is, larger cell size or forms aggregates). A strain of R. tolerans from Ekho Lake is capable of microaerobic growth (Labrenz et al., 1999). Over-representation at 6.5 m may, therefore, be indicative of growth at that depth rather than sedimentation because sinking cells would be more abundant close to the lake bottom at 6.7 m. Roseovarius OTUs cluster with Dunaliella chloroplast and Psychroflexus OTUs in the seriation analysis (Figure 3), suggesting that Organic Lake Roseovarius may be utilizing compounds released from algal-derived particulate matter, or made available by processing of complex organic matter by Psychroflexus. Roseovarius is a member of the Roseobacter clade, which is inferred to have an opportunistic ecology frequently associated with nutrient-replete plankton aggregates, including by-products of flavobacterial exoenzymatic attack (Moran et al., 2007; Teeling et al., 2012). Additionally, the diverse metabolic capabilities of the Roseobacter clade include DMSP degradation, AAnP and CO oxidation (reviewed in Wagner-Döbler and Biebl, 2006). All of these capabilities should facilitate growth in both the upper mixed and deep zones of Organic Lake (see Carbon resourcefulness in dominant heterotrophic bacteria below).
3–0.8-μm size fraction community composition
On the 0.8-μm filter, OTUs for Marinobacter dominated at all depths except 6.5 m. Their capture on this size fraction is consistent with the cell size of isolates (1.2–3 μm) (Gauthier et al., 1992). The genus is metabolically versatile, which likely permits it to occupy the entire water column. Marinobacter is heterotrophic and the genus includes hydrocarbon-degrading strains (e.g., Gauthier et al., 1992; Huu et al., 1999), although deep-sea metal-oxidizing autotrophs have also been reported (Edwards et al., 2003). Marinobacter isolates from Antarctic lakes are capable of anaerobic respiration using dimethylsulfoxide (DMSO) (Matsuzaki et al., 2006) or nitrate (Ward and Priscu, 1997). Analysis of functional potential linked to Marinobacter revealed additional metabolic capabilities potentially related to its dominance in Organic Lake (see Carbon resourcefulness in dominant heterotrophic bacteria and Molecular basis for unusual sulfur chemistry below).
OTUs for RF3 and Halomonas were over-represented at 6.5 m, and RF3 sequences were more abundant (Figures 2 and 3). Their relative abundance in the deep zone indicates a role in microaerobic processes. The majority of RF3 sequences retrieved to date are from anaerobic environments including mammalian gut (Tajima et al., 1999; Ley et al., 2006; Samsudin et al., 2011), sediment (Yanagibayashi et al., 1999; Röske et al., 2012), municipal waste leachate (Huang et al., 2005), anaerobic sludge (Chouari et al., 2005; Goberna et al., 2009; Rivière et al., 2009; Tang et al., 2011), a subsurface oil well head (Yamane et al., 2011), and the anaerobic zone of saline lakes (Bowman et al., 2000; Humayoun et al., 2003; Schmidtova et al., 2009). However, some members have been found in surface waters (Demergasso et al., 2008; Xing et al., 2009; Yilmaz et al., 2012) suggesting that not all members are strict anaerobes. Several Halomonas isolates have been sourced from Organic Lake including two described species Halomonas subglaciescola and H. meridiana, both of which grow as rods with dimensions consistent with capture on this size fraction (Franzmann et al., 1987a; James et al., 1990). Despite these isolates being aerobic, Halomonas has been reported to be enriched at the oxycline in Organic Lake (James et al., 1994) indicating Halomonas in the lake plays an ecological role in the suboxic zone. This capacity may be linked to the ability of free amino acids and organic acids (which are abundant in the deep zone) to stimulate the growth of isolates (Franzmann et al., 1987a).
0.8–0.1-μm size fraction community composition
A large number of eucaryal sequences were evident in the 0.1-μm size fraction. The upper zone was over-represented by OTUs for Pedinellales (silicoflagellate algae) that co-varied with chloroplasts (Figures 2 and 3). Pedinellales have only been detected in Antarctic lakes from molecular studies (Unrein et al., 2005; Lauro et al., 2011), including Organic Lake (Yau et al., 2011), and light microscopy studies of Antarctic Peninsula freshwater lakes reported 5–8-μm diameter cells resembling Pseudopedinella (Unrein et al., 2005). It is possible that in Organic Lake, small (0.8–0.1 μm) free-living members or chloroplast-containing cyst forms (Thomsen, 1988) exist. However, without evidence to support this (for example, by microscopy), it seems more likely that the lake sustains a relatively small number of active photosynthetic cells and the sequences detected arise from cysts or degraded cellular material.
OTUs for Candidatus ‘Aquiluna', in the Luna-1 cluster of Actinobacteria, (Hahn et al., 2004; Hahn, 2009) were most abundant at 1.7 m. The genus has small cells (<1.2 μm; Hahn, 2009), accounting for their concentration on this size fraction. Although originally described in freshwater lakes, the same clade was detected in abundance in Ace Lake (Lauro et al., 2011) and surface Arctic seawater (Kang et al., 2012), demonstrating that they have ecological roles in polar saline systems. In Ace Lake surface waters, they were associated with utilization of labile carbon and nitrogen substrates (Lauro et al., 2011), and in Organic Lake surface waters, they probably perform similar functions. The presence of this clade in the deep zone implies a facultative anaerobic lifestyle or sedimented cells.
The bottom of the water column was distinguished by the presence of OTUs for candidate divisions OD1 and TM7. OD1 was more abundant, and its prevalence on this size fraction is consistent with similar findings for size fractionation of ground water (Miyoshi et al., 2005). OD1 is consistently associated with reduced, sulfur-rich, anoxic environments (Harris et al., 2004; Elshahed et al., 2005). OD1 from Zodletone Spring, Oklahoma, was reported to possess enzymes related to those from anaerobic microorganisms (Elshahed et al., 2005). Genomic analyses identified OTUs for OD1 in the anoxic zone of Ace Lake (Lauro et al., 2011). The distribution of OD1 in Organic Lake is consistent with an anaerobic metabolism and potential involvement in sulfur chemistry.
Organic Lake functional potential
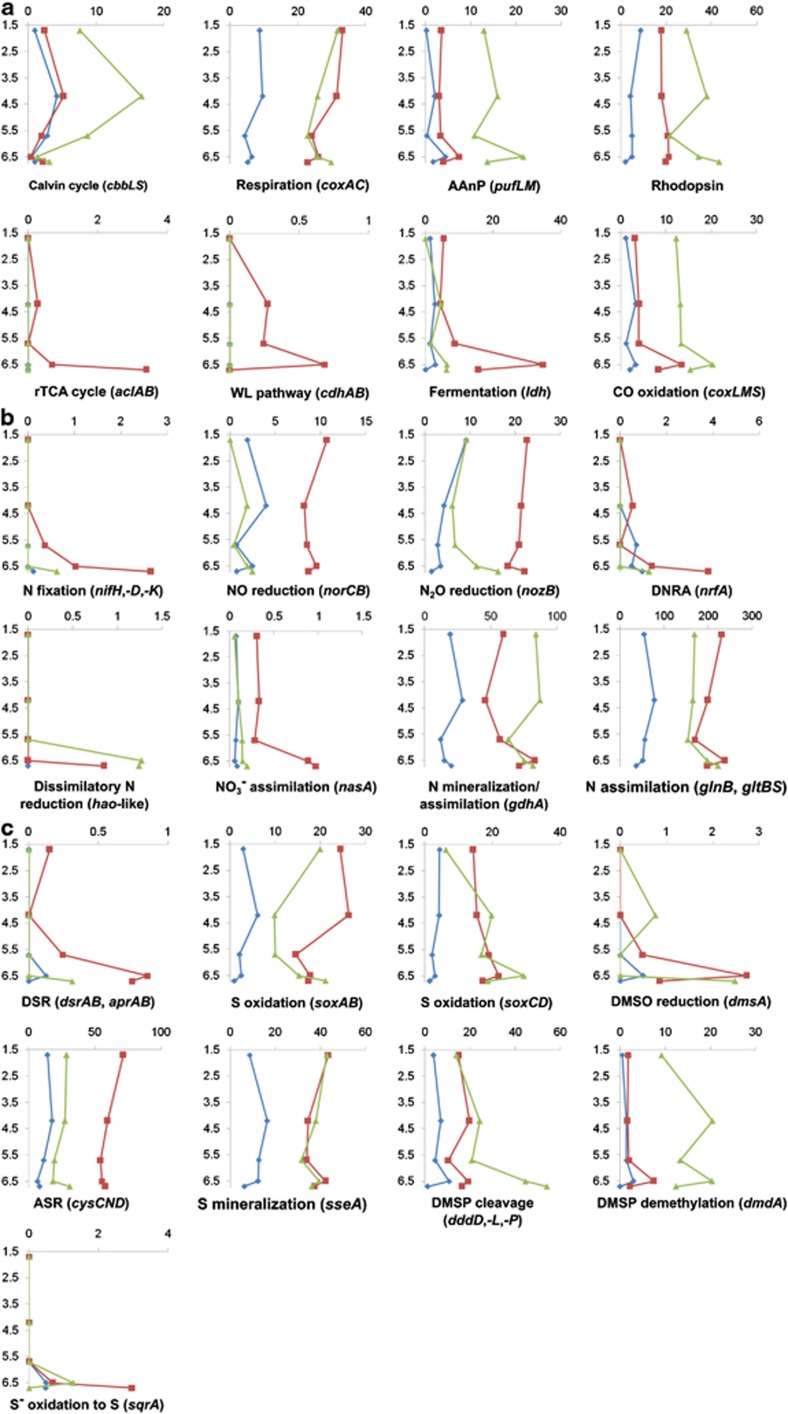
To determine the potential for functional processes in Organic Lake, gene markers for carbon, nitrogen and sulfur conversions (Figure 4) were retrieved from metagenomic reads. BEST analysis, which examines at all the abiotic variables in combination and finds a subset sufficient to ‘best' explain the biotic structure, showed that variation in the population structure was significantly correlated (ρ: 0.519, significance: 0.3%) with the abiotic parameters, dissolved oxygen, temperature, total sulfur and total nitrogen. The dissolved oxygen gradient has an obvious effect of separating aerobic from anaerobic taxa, and allows oxygen-sensitive nitrogen and sulfur processes to occur in the deep zone.
Figure 4.
Carbon, nitrogen and sulfur cycles in Organic Lake. Vertical profiles of genetic potential for (a) carbon (b) nitrogen and (c) sulfur conversions for each size fraction. The y-axis shows sample depths (m) and the x-axis shows counts of marker genes normalized to 100 Mbp of DNA sequence. The 0.1, 0.8 and 3.0 μm size fractions are shown as blue, red and green, respectively. Counts for marker genes for the same pathway or enzyme complex were averaged and those from different pathways were summed. For marker gene descriptions see Supplementary Tables S2 and S3. AAnP, aerobic anoxygenic photosynthesis; rTCA, reverse TCA; WL, Wood-Ljungdahl; DNRA, dissimilatory nitrate reduction to ammonia; DSR, dissimilatory sulfate reduction; DMSO, dimethylsulfoxide; ASR, assimilatory sulfate reduction; DMSP, dimethylsulfoniopropionate.
Carbon resourcefulness in dominant heterotrophic bacteria
In both the upper mixed and deep zones, potential for carbon fixation was much lower than for degradative processes, indicating the potential for net carbon loss (Figure 4a). Potential for carbon fixation via the oxygen-tolerant Calvin cycle (Figure 4a) was originally assessed by presence of the marker genes ribulose-bisphosphate carboxylase oxygenase (RuBisCO) and phosphoribulokinase (prkB) (Hügler and Sievert, 2011). The majority of RuBisCO homologs were related to Viridiplantae (Table 2) supporting the ecological role of green algae as the principle photosynthetic organisms. RuBisCO was only associated with a small proportion of Gammaproteobacteria (Table 2), principally from sulfur-oxidizing Thiomicrospira, indicating some Gammaproteobacteria are autotrophs. However, the majority of prkB matched to Gammaproteobacteria (Table 2), predominantly Marinobacter. Although deep-sea, iron-oxidizing autotrophic members of Marinobacter have been isolated (Edwards et al., 2003), all genomes reported for Marinobacter have prkB, but lack RuBisCO genes. Across Marinobacter genomes, the prkB homolog is consistently adjacent to a gene for a putative phosphodiesterase, suggesting that the enzymes expressed by these genes may be involved in pentose phosphate metabolism unrelated to carbon fixation. Albeit exceptional, this decoupling of prkB from RuBisCO involved in carbon fixation (forms I and II), also observed in Ammonifex (Hügler and Sievert, 2011), undermines the utility of prkB as a marker gene for the Calvin cycle within certain groups. Thus, there is no evidence for autotrophy in Organic Lake mediated by Marinobacter.
Table 2. Contribution of different taxonomic groups to counts of marker genes involved in carbon, nitrogen and sulfur conversions.
| Taxon | Calvin cycle | prkB | Respiration | Fermentation | rTCA | WL | CO oxidation | AAnP | N fixation | NO reduction | N2O reduction | DNRA | hao | N mineralization | NO3−assimilation | N assimilation | DSR | S oxidation | S asssimilation | S mineralization | DMSO reduction |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acidobacteria | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.07 | 0 | 0.08 | 0 | 0 | 0.03 | 1.03 | 0 |
| Actinobacteria | 0 | 0 | 0.64 | 0.23 | 0 | 0 | 0.08 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0.32 | 0 | 5.41 | 0 | 0 | 0.15 | 0 | 0 |
| Alphaproteobacteria | 0.05 | 0 | 4.84 | 0 | 0 | 0 | 6.74 | 6.98 | 0.01 | 0.12 | 0 | 0 | 0 | 6.39 | 5.49 | 49.4 | 0 | 2.05 | 0.85 | 11.7 | 0 |
| Aquificae | 0 | 0 | 0.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.26 | 0 | 0 | 0 | 0 | 0.06 | 0 | 0 | 0 | 0 | 0 |
| Bacteroidetes | 0 | 0 | 3.42 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.00 | 0.27 | 0 | 3.90 | 0.03 | 15.5 | 0 | 0 | 5.06 | 0.20 | 0 |
| Betaproteobacteria | 0.04 | 0.06 | 0.07 | 0.09 | 0 | 0 | 0.22 | 0 | 0 | 0.03 | 0 | 0 | 0 | 0.06 | 0.41 | 19.2 | 0 | 1.09 | 2.07 | 0.69 | 0 |
| Chlorobi | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0.2 | 0 | 0.31 | 0 | 0 | 0.03 | 0.15 | 0 |
| Chloroflexi | 0 | 0 | 0.02 | 0 | 0 | 0 | 0.07 | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0.03 | 0 | 0 | 0 | 0.30 | 0 |
| Chrysiogenetes | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0.06 | 0 | 0 | 0.01 | 0 | 0 |
| Cyanobacteria | 0.09 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.29 | 0 | 0.10 | 0 | 0 | 0.13 | 0.05 | 0 |
| Deferribacteres | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Deinococcus–Thermus | 0.01 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0.11 | 0 | 0 | 0 | 0.09 | 0 |
| Deltaproteobacteria | 0 | 0 | 0.09 | 0 | 0 | 0.06 | 0.21 | 0 | 0.04 | 0.01 | 0 | 0.07 | 0.22 | 0.23 | 0 | 0.58 | 0.19 | 0 | 0.56 | 0.20 | 0.03 |
| Epsilonproteobacteria | 0 | 0 | 0 | 0 | 0.28 | 0 | 0 | 0 | 0.32 | 0 | 0 | 0 | 0 | 0.05 | 0 | 1.49 | 0 | 0.03 | 0.13 | 0 | 0 |
| Firmicutes | 0.01 | 0 | 0.01 | 4.90 | 0 | 0.02 | 0.15 | 0 | 0.03 | 0 | 0 | 0.03 | 0 | 0.70 | 0 | 3.16 | 0 | 0 | 0.22 | 0.09 | 0.36 |
| Fornicata | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Fusobacteria | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.04 | 0 | 0 | 0.03 | 0.13 | 0 |
| Gammaproteobacteria | 0.05 | 12.1 | 9.86 | 1.03 | 0 | 0 | 0.06 | 0.04 | 0 | 3.91 | 8.28 | 0 | 0 | 4.75 | 14.1 | 50.6 | 0 | 2.64 | 22.4 | 14.0 | 0 |
| Nitrospirae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Planctomycetes | 0 | 0 | 0.02 | 0.08 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0.16 | 0 | 0 | 0 | 0.26 | 0 | 0 | 0.03 | 0 | 0 |
| Spirochaetes | 0 | 0 | 0 | 0.03 | 0 | 0 | 0.16 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0.11 | 0 | 0.15 | 0 | 0 | 0 | 0.03 | 0.12 |
| Thermobaculum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.10 | 0 | 0 | 0 | 0 | 0 | 0.08 | 0 |
| Thermotogae | 0.01 | 0 | 0 | 0 | 0 | 0 | 0.17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0 |
| Verrucomicrobia | 0 | 0 | 0.13 | 0.05 | 0 | 0 | 0 | 0 | 0 | 0 | 0.17 | 0.03 | 0 | 0.25 | 0 | 0.82 | 0 | 0 | 0.18 | 0 | 0 |
| Crenarchaeota | 0 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0.02 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 |
| Euryarchaeota | 0.04 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.12 | 0.09 | 0.10 | 0 | 0 | 0.02 | 0.12 | 0 |
| Alveolata | 0 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.18 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0 |
| Euglenozoa | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 |
| Opistokonta | 0 | 0 | 0.16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.15 | 0 | 0.13 | 0 | 0 | 0.11 | 0.03 | 0 |
| Rhodophyta | 0.16 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 |
| Stramenopiles | 0.34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0.15 | 0 | 0 | 0 | 0 | 0 |
| Viridiplantae | 3.10 | 0.06 | 1.10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0.35 | 0 | 0 | 0.03 | 0.15 | 0 |
Abbreviations: AAnP, aerobic anoxygenic photosynthesis; CO, carbon monoxide; DMSO, dimethylsulfoxide; DNRA, dissimilatory nitrate reduction to ammonia; DSR, dissimilatory sulfate reduction; N, nitrogen; prkB, phosphoribulokinase; rTCA, reverse tricarboxylic acid; S, sulfur; WL, Wood-Ljungdahl
The values shown for each taxon are the average number of matches from all samples to marker genes for a process expressed per 100 Mbp of metagenomic sequence. Counts from the taxonomic group with the greatest contribution to each process is shown in bold. Genes used as markers for each process are the same as described in Figure 4.
Evidence for carbon fixation via the reverse tricarboxylic acid (rTCA) cycle was also indicated (Figure 4a), with genes for ATP citrate lyase (aclAB) linked to sulfur-oxidizing Epsilonproteobacteria (Table 2). In general, the rTCA cycle is restricted to anaerobic and microaerobic bacteria (Hügler and Sievert, 2011), which is consistent with the detection of Epsilonproteobacteria in the lake bottom where oxygen is lowest, and the microaerophilic/anaerobic metabolisms characteristic of the group (Campbell et al., 2006). Anaerobic carbon fixation was represented by potential for the Wood-Ljungdahl (or reductive acetyl-CoA) pathway (Figure 4a). Wood-Ljungdahl-mediated carbon fixation, for which CO dehydrogenase/acetyl-CoA synthase is the key enzyme, was linked to Firmicutes and Deltaproteobacteria that are known to grow autotrophically using this pathway (Hügler and Sievert, 2011).
Potential for carbon loss by respiration was indicated by an abundance of cytochrome c oxidase genes (coxAC) throughout the water column. In the deep zone, potential for fermentation was greatest at 6.5 m (Figure 4a) and likely the main biological activity that was occurring at that depth. Fermentation was indicated by the marker gene lactate dehydrogenase (ldh). These genes were linked to Firmicutes (Table 2), which was only present at 6.5 m and represented by the classes Clostridia and Bacilli (Figure 2a). As the related candidate division RF3 (Tajima et al., 1999) also has relatively high abundance in this zone (Figure 2a) (see 0.8–3.0-μm size fraction community composition above), there is circumstantial evidence that RF3 possesses a fermentative metabolism and may, therefore, have an important ecological role in Organic Lake by degrading high molecular weight compounds to organic acids that other organisms can utilize. Assimilation of fermentation products appears to have a greater role in Organic Lake rather than complete anaerobic oxidation involving methanogens or sulfate-reducing bacteria; the former were absent and the latter were present in low abundance (Figures 2a and c).
Alphaproteobacteria, predominantly Roseovarius (Figure 2c), were implicated in CO oxidation (Table 2), which is used to generate energy for lithoheterotrophic growth (Moran and Miller, 2007), although CO oxidation may also be involved in anaplerotic C fixation (Moran et al., 2007). The CO oxidation capacity was at a maximum at 6.5 m (Figure 4a), and therefore, associated with the putative deep-zone Roseovarius ecotype of Organic Lake. CO oxidation can function as a strategy to limit oxidation of organic carbon for energy so that a greater proportion can be directed towards biosynthesis (Moran and Miller, 2007).
Photosynthesis reaction center genes pufLM, involved in photoheterotrophy via AAnP, were abundant in Organic Lake (Figure 4a; Table 2). These were linked to the Roseobacter clade of Alphaproteobacteria (Table 2), major contributors to AAnP in ocean surface waters (Béjà et al., 2000; Béjà et al., 2002; Moran et al., 2007). This is consistent with the known metabolic potential of bacteriochlorophyll-a-producing R. tolerans from Ekho Lake (Labrenz et al., 1999). Photoheterotrophy can also be rhodopsin-dependent, with proteorhodopsins of marine Flavobacteria and Vibrio previously linked to light-dependent energy generation to supplement heterotrophic growth, particularly during carbon limitation (Gómez-Consarnau et al., 2007, 2010). However, the function(s) of rhodopsins are diverse, and PRs are also hypothesized to be involved in light or depth sensing (Fuhrman et al., 2008).
Rhodopsin genes were abundant in Organic Lake (Figure 4a), and were associated with all the dominant Organic Lake aerobic heterotrophic lineages (Supplementary Figure S6). Phylogenetic analysis revealed six well-supported Organic Lake rhodopsin groups (Supplementary Figure S6). All groups had an L or M residue at position 105 (versus the SAR86 proteorhodopsin), denoting tuning to surface green light (Man et al., 2003; Gómez-Consarnau et al., 2007), and is characteristic of oceanic coastal samples (Rusch et al., 2007). Four of the groups clustered with homologs of genera detected in the lake, namely Marinobacter, Psychroflexus, Octadecabacter and ‘Ca. Aquiluna' (Supplementary Figure S6 and Supplementary Table S4). Another group (SAL-R group) originates from the sphingobacterium Salinibacter ruber, which produces xanthorhodopsin (Balashov et al., 2005); it is therefore likely that Organic Lake Sphingobacteria (Supplementary Table S4) were the origin of this rhodopsin group. The most abundant group (OL-R1; Supplementary Figure S6) had no close homologs from GENBANK, but it was abundant on the 3.0-μm fraction and has a distribution, suggesting that it originates from Organic Lake members of the Roseobacter clade (Figure 4a). All ORFs adjacent to OL-R1 rhodopsin-containing scaffolds were related to Octadecabacter, further supporting their Roseobacter clade provenance (Supplementary Figure S7). Genes downstream of OL-R1 were involved in carotenoid synthesis, indicating that OL-R1 is a xanthorhodopsin occurring as a retinal protein or in a carotenoid complex (Balashov et al., 2005).
Photoheterotrophic potential of Organic Lake was compared with other aquatic environments including nearby Ace Lake, Southern Ocean and Global Ocean Sampling expedition samples. The Organic Lake 0.1-μm fraction had the lowest rhodopsin counts and percentage of cells containing rhodopsin genes (estimated from the ratio of rhodopsin genes to the single copy of recA gene) of all size-matched samples surveyed (Table 3). Non-marine Global Ocean Sampling samples from the 0.1-μm fraction have been noted to have lower rhodopsin abundance (Sharma et al., 2008), which was similarly evident from our analysis (Table 3). In contrast, the 3.0-μm Organic Lake size fractions had higher rhodopsin counts than Ace Lake and comparable counts to the Southern Ocean samples, although the percentage of cells containing rhodopsin genes was still lower than that of the Southern Ocean. The paucity of rhodopsins in the Organic Lake 0.1-μm fraction is likely due to the lack of SAR11 clade, which is expected to be the main source of rhodopsin genes in Ace Lake and marine samples. This indicates that although Organic Lake has an overall lower frequency of rhodopsin genes compared with sites for which size fraction-matched metagenomes are available, the rhodopsins associated with larger or particle-associated cells are as abundant as in the marine environment.
Table 3. Counts of genes involved in DMSP catabolism and photoheterotrophy.
| Site | size (μm) | dddD (%) | dddL (%) | dddP (%) | dmdA (%) | Rhodopsin (%) | pufLM (%)a | recA |
|---|---|---|---|---|---|---|---|---|
| Organic lake 1.7 m (GS374) | 0.1 | 2 (9) | 4 (19) | 0 | 0* (2) | 1 (5) | 0* (1) | 21 |
| 0.8 | 10 (36) | 10 (39) | 1 (2) | 2 (7) | 5 (20) | 4 (14) | 26 | |
| 3.0 | 11 (50) | 5 (21) | 2 (7) | 9 (43) | 12 (57) | 13 (61) | 21 | |
| Organic lake 4.2 m (GS375) | 0.1 | 5 (34) | 5 (34) | 0 | 1 (10) | 1 (10) | 2 (16) | 14 |
| 0.8 | 15 (54) | 9 (31) | 0 | 2 (6) | 7 (23) | 3 (11) | 28 | |
| 3.0 | 23 (75) | 2 (8) | 1 (2.5) | 20 (68) | 14 (45) | 16 (53) | 30 | |
| Organic lake 5.7 m (GS376) | 0.1 | 4 (43) | 1 (7) | 0 | 1 (14) | 2 (21) | 0* (4) | 10 |
| 0.8 | 6 (20) | 9 (32) | 0 | 2 (7) | 6 (22) | 3 (12) | 29 | |
| 3.0 | 19 (68) | 3 (12) | 0 | 13 (47) | 6 (21) | 11 (38) | 28 | |
| Organic lake 6.5 m (GS377) | 0.1 | 10 (51) | 0* (2) | 0 | 3 (15) | 1 (7) | 4 (22) | 20 |
| 0.8 | 14 (38) | 9 (23) | 1 (2) | 7 (20) | 6 (16) | 7 (20) | 28 | |
| 3.0 | 42 (106) | 5 (13) | 0 | 20 (52) | 6 (16) | 22 (55) | 29 | |
| Organic lake 6.7 m (GS378) | 0.1 | 1 (7) | 0* (4) | 0 | 0 | 1 (7) | 2 (13) | 13 |
| 0.8 | 12 (26) | 8 (17) | 0 | 2 (5) | 8 (16) | 4 (9) | 47 | |
| 3.0 | 50 (174) | 5 (17) | 4 (13) | 12 (43) | 12 (43) | 14 (48) | 29 | |
| Ace Lake mixolimnion | 0.1 | 0* (2) | 0 | 1 (2) | 15 (56) | 15 (53) | 0* (1) | 28 |
| 0.8 | 2 (3) | 1 (2) | 0 | 2 (4) | 12 (27) | 3 (12) | 45 | |
| 3.0 | 0 | 0 | 0* (4) | 0 | 5 (42) | 0 | 11 | |
| Newcomb Bay (GS235) | 0.1 | 6 (14) | 0 | 3 (7) | 50 (111) | 89 (196) | 0 | 45 |
| 0.8 | 5 (12) | 0 | 0 | 18 (41) | 55 (123) | 0 | 45 | |
| 3.0 | 0 | 0 | 0 | 2 (17) | 4 (33) | 0 | 11 | |
| Southern Ocean SZ | 0.1 | 2 (3) | 0 | 6 (9) | 71 (101) | 98 (139) | 0 | 70 |
| 0.8 | 3 (6) | 0* (0*) | 5 (12) | 32 (81) | 43 (108) | 0 | 39 | |
| 3.0 | 0* (7) | 0 | 0* (4) | 4 (66) | 5 (84) | 0 | 6 | |
| Southern Ocean NZ | 0.1 | 0* (1) | 0 | 5 (7) | 124 (159) | 111 (142) | 1 (1) | 78 |
| 0.8 | 0* (2) | 0 | 9 (30) | 28 (84) | 35 (107) | 2 (7) | 33 | |
| 3.0 | 0* (3) | 0 | 1 (9) | 7 (54) | 11 (89) | 0* (4) | 12 | |
| GOS coastal | 0.1 | 0* (0*) | 0 | 5 (6) | 44 (52) | 74 (87) | 5 (6) | 85 |
| GOS open ocean | 0.1 | 0 | 0 | 7 (8) | 45 (50) | 66 (74) | 5 (5) | 90 |
| GOS estuary | 0.1 | 0 | 0 | 1 (1) | 29 (36) | 61 (77) | 2 (3) | 80 |
| GOS embayment (GS005) | 0.1 | 4 (8) | 0 | 6 (12) | 28 (54) | 58 (112) | 3 (6) | 52 |
| GOS Lake Gatun (GS020) | 0.1 | 0 | 0 | 0 | 4 (4) | 48 (53) | 2 (2) | 90 |
| GOS fringing reef (GS025) | 0.8 | 0 | 0 | 0 | 0 | 7 (39) | 0 | 18 |
| GOS warm seep (GS030) | 0.1 | 0 | 0 | 7 (6) | 75 (63) | 83 (69) | 6 (5) | 120 |
| GOS Upwelling, Fernandina (GS031) | 0.1 | 0 | 0 | 4 (4) | 81 (77) | 81 (76) | 4 (4) | 106 |
| GOS mangrove (GS032) | 0.1 | 0 | 0 | 2 (3) | 24 (34) | 25 (36) | 1 (1) | 71 |
| GOS Punta Cormorant Lagoon (GS033) | 0.1 | 0 | 11 (15) | 14 (21) | 4 (6) | 31 (43) | 15 (21) | 72 |
| GOS Rangirora Atoll (GS051) | 0.1 | 0 | 0 | 11 (15) | 38 (49) | 73 (94) | 3 (4) | 77 |
Abbreviations: DMSP, dimethylsulfoniopropionate; GOS, Global Ocean sampling; NZ, Northern zone; SZ, Southern zone.
Counts shown for Organic Lake, Ace Lake mixolimnion, Southern Ocean and GOS metagenomes per 100 Mbp of metagenomic sequence. Percentages shown in parentheses are estimates of cells containing that marker gene, which is the percentage of the marker gene to the single-copy gene recA. The sample ID for each site is shown in parentheses after the site description. Values marked with an asterisk are >0 but <0.5. Counts for the following sites are averages of several samples: Ace Lake mixolimnion (GS232, GS231); Southern Ocean SZ (GS349, GS351–GS353, GS356–GS360); Southern Ocean NZ (GS363, GS346, GS364, GS366–GS368); GOS coastal (GS002–GS004, GS007–GS010, GS012–GS016, GS019, GS021, GS027–GS029, GS034–GS036); GOS open ocean (GS017, GS018, GS022, GS023, GS026, GS037, GS047); GOS estuary (GS006, GS011, GS012). Values shown in bold are the highest for that marker gene.
Counts of pufLM genes in the Organic Lake 0.1-μm size fraction were similar to Global Ocean Sampling samples, except for Punta Cormorant hypersaline lagoon that had the highest pufLM counts and percentage of cells containing AAnP genes (Table 3). However, the highest overall counts of pufLM were from the 3.0-μm size fraction of Organic Lake, likely due to the high proportion of members of the Roseobacter clade. Notably, pufLM genes were not detected in high abundance in Ace Lake or the Southern Ocean samples, indicating that AAnP is a unique adaptation in Organic Lake among these polar environments. The similarly high abundance of pufLM genes in Punta Cormorant hypersaline lagoon indicates that AAnP may be advantageous in environments with salinity above marine levels.
The contribution of light-driven energy generation processes to the carbon budget is difficult to infer from genetic potential alone. For example, the relative abundance of AAnP and proteorhodopsin genes in Arctic bacteria has been reported to be the same in winter and summer (Cottrell and Kirchman, 2009). Furthermore, regulation of pigment synthesis is complex; for example, bacteriochlorophyll a expression in R. tolerans occurs in the dark but is inhibited by continuous dim light (Labrenz et al., 1999). However, it is possible that the apparent negative balance in carbon conversion potential could be ameliorated by photoheterotrophy performed by bacterial groups that are abundant in Organic Lake. In particular, the Organic Lake Psychroflexus could have a particular role, as it has a proteorhodopsin related to Dokdonia, which was shown to function under carbon limitation (Gómez-Consarnau et al., 2007). Furthermore, the detection of higher AAnP potential in Organic Lake than other aquatic environments, and the presence of taxa known to be capable of AAnP, suggests that AAnP may have a relatively important role in the carbon budget of Organic Lake.
Regenerated nitrogen is predominant in the nitrogen cycle
Nitrogen cycling potential throughout the lake was dominated by assimilation and mineralization/assimilation pathways (Figure 4b). Glutamate dehydrogenase (GDH) genes (gdhA) were abundant (Figure 4b), and linked predominantly to Alpha- and Gammaproteobacteria, and to a lesser extent Bacteroidetes (Table 2). However, the functional significance of the readily reversible GDH depends on its origin; Bacteroidetes are likely to use GDH in the oxidative direction for glutamate catabolism (Takahashi et al., 2000; Williams et al., 2013), whereas the use of GDH in the oxidative or reductive directions by Proteobacteria is likely to depend upon the available source of reduced nitrogen (ammonia versus amino acids). Glutamine synthetase (glnB) and glutamate synthase genes (gltBS) were predominantly linked to Alpha- and Gammaproteobacteria (Table 2), indicating the potential for high-affinity ammonia assimilation by these groups in Organic Lake. The high ammonia concentration in the deep zone (Figure 1b; Table 1) would result from a higher rate of mineralization (ammonification) than assimilation. This is consistent with abundant OTUs for Psychroflexus (Bacteroidetes) in this zone, and due to either turnover of organic matter or lysis of Bacteroidetes cells after sedimentation in anoxic water. In addition, the gene for ammonia-generating nitrite reductase (nrfA) was linked to Bacteroidetes and Planctomycetes (Table 2), indicating that ammonia may also be produced by these putative aerobic heterotrophs. Overall, the data suggest that ammonia is actively assimilated in the aerobic upper mixed zone, but is permitted to accumulate in the anaerobic deep zone.
Potential for nitrogen conversions typically found in other aquatic environments was greatly reduced in Organic Lake. There was a very low potential for nitrogen fixation that was confined to the deep zone (Figure 4b) and principally linked to anaerobic Epsilonproteobacteria (Table 2). This diazotrophic potential may not be realized by Epsilonproteobacteria, given the high ammonia concentration present in the deep zone. No ammonia monooxygenase genes (amoA) were detected. The potential for ammonia oxidation was only represented by hydroxylamine/hydrazine oxidase-like (hao) genes, which were in low abundance and linked to Deltaproteobacteria (Table 2). hao genes are present in non-ammonia-oxidizing bacteria (Bergmann et al., 2005), and those from Organic Lake belong to a family of multiheme cytochrome c genes present in sulfate-reducing Deltaproteobacteria that have no proven role in ammonia oxidation. In the genomes of sulfate-reducing Deltaproteobacteria, the hao gene is invariably situated adjacent to a gene for a NapC/NirT protein, which suggests a role in dissimilatory nitrate reduction. Collectively, these data indicate an inability for nitrification to occur in the upper mixed zone and no potential for ammonia loss in the deep zone.
Denitrification genes (norCB and nozB) and genes for nitrate assimilation (nasA) were present throughout the water column (Figure 4b), and were linked primarily to Gammaproteobacteria (Table 2). One hypothesis is that low nitrate and nitrite in the deep zone (Figure 1b Table 1) is the result of oxidized nitrogen being depleted by dissimilatory or assimilitory reduction by heterotrophic Gammaproteobacteria. Denitrification genes are phylogenetically widespread and usually induced by low oxygen or oxidized nitrogen species (Kraft et al., 2011), and thus expected to be active in the deep zone or oxycline. However, denitrification may be inhibited even if conditions appear appropriate. For example, in Lake Bonney, Antarctica, denitrification occurs in the west lobe, but not in the east lobe of the lake despite the presence of anoxia, nitrate and denitrifying Marinobacter species (Ward and Priscu, 1997; Ward et al., 2005). Moreover, in the absence of nitrification, denitrification and nitrate assimilation would be limited by the lack of potential to reform oxidized nitrogen. While exogenous input may augment the carbon and nitrogen budget (see Conclusion below), the preponderance of assimilation/mineralization pathways geared towards reduced nitrogen appears to reflect a ‘short circuit' of the typical nitrogen cycle that would conserve nitrogen in a largely closed system. Therefore, the predominant nitrogen source is the regenerated fixed nitrogen. Similar findings were also made for Ace Lake, although in this system the presence of a dense layer of green sulfur bacteria with the potential to fix nitrogen augments the nitrogen cycle (Lauro et al., 2011).
Molecular basis for unusual sulfur chemistry
Several meromictic hypersaline lakes in the Vestfold Hills, including Organic Lake, with practical salinity greater than ∼150 are characterized by an absence of hydrogen sulfide and photoautotrophic sulfur bacteria (Burke and Burton, 1988). Although sulfate is present (Franzmann et al., 1987b), geochemical conditions of these lakes are not conducive to dissimilatory sulfur cycling between sulfur-oxidizing and sulfate-reducing bacteria typical of other stratified systems such as Ace Lake (Ng et al., 2010; Lauro et al., 2011). Consistent with this, potential for dissimilatory sulfate reduction represented by dissimilatory sulfite reductase (dsrAB) and adenylylsulfate reductase (aprAB) linked to sulfate-reducing Deltaproteobacteria (Table 2) was low in Organic Lake. Sulfate reduction potential was confined to the 6.7 m sample (Figure 4c) where oxygen concentration was lowest and Deltaproteobacteria were present (Figure 2a).
Capacity for oxidation of reduced sulfur compounds, represented by the sulfur oxidation multienzyme genes (soxAB), was present throughout the water column (Figure 4c) and linked primarily to Alpha- and Gammaproteobacteria (Table 2). Sulfur-oxidizing Gammaproteobacteria and Alphaproteobacteria are known to oxidize sulfur compounds, such as thiosulfate, aerobically. Although a small proportion of Gammaproteobacteria had the capacity for autotrophy (see Carbon resourcefulness in dominant heterotrophic bacteria), the majority of sulfur oxidizers were likely chemolithoheterotrophs as they were related to heterotrophic Marinobacter and Roseobacter clade. The sulfur dehydrogenase genes soxCD linked to Alpha- and Gammaproteobacteria were similarly present throughout the water column. soxCD are accessory components of the Sox enzyme system without which complete oxidation of thiosulfate cannot occur (Friedrich et al., 2005). Thus, the presence of soxCD indicates that complete oxidization likely occurs, although the different distribution of soxAB and soxCD in the water column (Figure 4c) suggests that a proportion of the community may lack soxCD and deposit sulfur. Sulfur-oxidizing Epsilonproteobacteria possessing soxAB genes (Table 2) were present only in the deep zone of Organic Lake (Figures 2a and c) and were related to autotrophic deep-sea sulfur-oxidizers, some members of which are capable of anaerobic sulfur oxidation using nitrate (Yamamoto and Takai, 2011). It is unlikely that appreciable sulfur oxidation occurs in the deep zone as the known terminal electron acceptors, oxygen and nitrate are depleted and the abundance of sulfur-oxidizing Epsilonproteobacteria is low (Figure 2a). Epsilonproteobacteria were also linked to a capacity for oxidation of sulfide to elemental sulfur by utilizing sulfide:quinone oxidoreductase (sqrA) (Figure 4a, Table 2). In this pathway, sulfur is released as polysulfides, which is a potential biological source of the abundant polysulfides that have been detected in the lake (Roberts et al., 1993).
It is likely that the limited anaerobic dissimilatory sulfur cycle contributes to the accumulation of DMS in Organic Lake in the deep zone. In the upper mixed zone, DMS could potentially be oxidized as a carbon and energy source or utilized as an electron donor by sulfur-oxidizing bacteria (Schäfer et al., 2010). In anoxic zones, methanogenic Archaea or sulfate-reducing bacteria are the main organisms known to break down DMS (Schäfer et al., 2010). As the genes that encode enzymes involved in DMS catabolism have not been identified, potential for DMS breakdown in the deep zone was assessed from the taxonomic composition. Methanogens and genes involved in methanogenesis were not detected, nor has methane been detected (Gibson et al., 1994), leaving sulfate reduction the most likely route of DMS catabolism. The low dissimilatory sulfate reduction potential in the deep zone coupled with the relatively stagnant water would likely minimize DMS oxidation and loss by ventilation. DMS would therefore be expected to accumulate in the deep zone if production rates were higher than breakdown.
To determine the source of high DMS in the bottom water of Organic Lake, the genes involved in DMS formation were surveyed (Supplementary Table S3). Genes for DMSP lyases dddD, dddL and dddP were detected in Organic Lake at levels comparable to other dominant processes such as respiration and fermentation (Figure 4c), indicating that DMSP is an important carbon and energy source in Organic Lake. The other known DMSP lyases, dddQ, dddW and dddY (Todd et al., 2012), were not detected. dddD was the most abundant of the Organic Lake DMSP lyases (Table 3) and comprised two main types: MAR-dddD and OL-dddD (Supplementary Figure S8). Neither of these types clustered with the non-functional Dinoroseobacter shibae DFL 12 and Ruegeria pomeroyi DSS-3 dddD homologs (Todd et al., 2011) or carnitine coenzyme A transferase outgroups, thereby providing support for their proposed role as functional DMSP lyases. The MAR-dddD type includes the Marinobacter sp. ELB17 dddD homolog, and MAR-dddD sequences were most abundant on the 0.8-μm fraction where Marinobacter OTUs were also abundant, indicating that MAR-dddD derives from Organic Lake Marinobacter (Supplementary Figure S8). OL-dddD had ∼83% identity to the dddD of Halomonas sp. HTNK1, but was designated as a separate type due to poor support for the Organic Lake dddD and Halomonas sp. HTNK1 dddD cluster (Supplementary Figure S8). Although Halomonas OTUs were detected in Organic Lake (Figure 3, Supplementary Table S4), they were only detected on the 0.1 and 0.8 μm size fractions, which does not correlate with the distribution of the OL-dddD genes (data not shown). The abundance of OL-dddD on the 3.0-μm fraction suggests that it originates from Alphaproteobacteria. OL-dddD containing contigs carried genes of mixed Alpha- and Gammaproteobacterial origin supporting its provenance from one of these classes and consistent with the ‘pick ‘n' mix' arrangement of genes found beside sequenced dddD regions (Johnston et al., 2008). Adjacent to OL-dddD was dddT (Supplementary Figure S9), a betaine/choline/carnitine transporter family protein that likely functions in DMSP import, demonstrating that OL-dddD forms an operon-like structure, similar to Halomonas sp. HTNK1 (Todd et al. 2010). DMSP lyase DddD converts DMSP to 3-OH-propionate and DMS. A dddA gene was also located next to dddD (Supplementary Figure S9); this encodes an alcohol dehydrogenase (DddA; originally annotated as ‘choline dehydrogenase') for the conversion of 3-OH-propionate to malonate semialdehyde in the second step of DMSP catabolism (Todd et al., 2010).
Two dddL groups were detected in Organic Lake: SUL-dddL and MAR-dddL (Supplementary Figure S10). The former includes the Sulfitobacter sp. EE-36 dddL and the latter the Marinobacter manganoxydans MnI7-9 homolog, indicating that they originate from Roseobacter clade and Gammaproteobacteria, respectively. Sulfitobacter sp. EE-36 has demonstrated DMSP lyase activity and the dddL gene alone is sufficient for DMS generation (Curson et al., 2008). These data indicate that the Organic Lake members of the SUL-dddL group perform the same functional role. The MAR-dddL clade appears to be an uncharacterized branch of the dddL family. dddP was detected as the least abundant of the DMSP lyases (Table 3). Phylogenetic analyses showed that Organic Lake dddP likely originate from Roseovarius (Supplementary Figure S11). The Organic Lake sequences formed a clade with the functionally verified Roseovarius nubinhibens ISM dddP (Todd et al., 2009).
A single type of DMSP demethylase, dmdA, was identified. It clustered with Roseobacter clade dmdA (Supplementary Figure S12), corresponding to the marine clade A (Howard et al., 2008), and includes the functionally verified R. pomeroyi DSS-3 homolog. These data indicate that the Organic Lake sequences correspond to true DMSP demethylases and not the related glycine cleavage T proteins or other aminomethyltransferases (Howard et al., 2008).
DMSP cleavage appears to be a significant source of DMS in Organic Lake. DMSP likely originates from Bacillariophyta or Dinoflagellida as Organic Lake Dunaliella have been reported not to produce DMSP in culture (Franzmann et al., 1987b). Based on the abundance of marker genes, DMSP cleavage is predicted to occur at highest levels in the deep zone (Figure 4c) where the DMS concentration has been measured to be highest (Deprez et al., 1986; Franzmann et al., 1987b; Gibson et al., 1991; Roberts and Burton, 1993; Roberts et al., 1993). Consistent with DMS levels, the concentration of DMSP (Gibson et al., 1991) and DMSO (J Gibson, unpublished results) in the bottom zone of Organic Lake has also been found to be high, although DMSP levels reported in a separate study were not as high (Roberts et al., 1993), possibly reflecting technical difficulties measuring methylated sulfur compounds. In these studies (Gibson et al., 1991; Roberts et al., 1993), chlorophyll a levels were not measured.
DMS can also be produced in anoxic environments from the reduction of DMSO, degradation of sulfur-containing amino acids and sulfide methylation (Schäfer et al., 2010). Our data indicate that some DMSO reduction linked to Firmicutes could occur, but is not likely a major pathway because DMSO reductase genes were not abundant (Table 2, Figure 4c). The potential for the other DMS-yielding processes could not be determined because the enzymes involved in these pathways have not been established. When cultivated, Halomonas isolates from Organic Lake produced DMS from cysteine (Franzmann et al., 1987b), providing some evidence that DMS production from anaerobic degradation of amino acids can occur. Abiotic pathways for anaerobic production of DMS have also been proposed (Roberts et al., 1993).
The potential for DMSP cleavage was more than twice that of DMSP demethylation (Figure 4c). This is unusual compared with the marine environment or Ace Lake where DMSP demethylation potential is much higher than cleavage (Table 3). Previous estimates have similarly shown marine environments to have demethylation potential up to two orders of magnitude higher than cleavage (Howard et al., 2008; Todd et al., 2009; Reisch et al., 2011; Todd et al., 2011). The frequency of DMSP lyase genes dddD and dddL in Organic Lake exceeded those of all other environments examined, except Punta Cormorant hypersaline lagoon, where dddL abundance was comparable (Table 3). This suggests the selection in Organic Lake for DMSP cleavage due to functional advantage and/or selection for taxa that carry DMSP lyase genes. There is evidence that high DMSP cleavage potential is adaptive in hypersaline systems, as a high proportion of ddd genes were similarly detected in Punta Cormorant hypersaline lagoon and saltern ponds (Raina et al., 2010). The accumulated DMS in Organic Lake suggests that conditions in Organic Lake favor the relatively inefficient lysis pathway, where both sulfur and carbon are lost to the organism performing the DMSP lysis, over the more ‘thrifty' demethylation pathway. This is particularly pertinent to the Roseobacter lineages that can also perform either process.
Conclusion
Through the use of shotgun metagenomics and size partitioning of samples, we discovered that the Organic Lake system is dominated by heterotrophic bacteria related to Psychroflexus, Marinobacter and Roseovarius, with primary production provided largely by chlorophyte algae related to Dunaliella. Genetic potential for oxidation of fixed carbon by heterotrophic bacteria occurs greatly in excess of the potential for carbon fixation, suggesting possible net carbon loss. However, by linking key metabolic processes to the dominant heterotrophic lineages we uncovered processes that were unusually abundant in Organic Lake that may serve to maximize exploitation of limited resources and minimize loss. Recalcitrant polymeric algal material and particulate matter are likely remineralized by Psychroflexus in the upper mixed zone and by Firmicutes in the deep zone to provide labile substrates for use by other heterotrophic bacteria. The generalist Marinobacter and Roseovarius lineages were associated with abundant genes involved in rhodopsin-mediated and AAnP photoheterotrophy, the latter of which was more abundant in Organic Lake than any other system surveyed in an analogous manner. Potential for chemolithoheterotrophy, sulfur oxidation and CO oxidation was also high, and along with photoheterotrophy, may provide a supplementary energy source if organic carbon becomes limiting.
In addition to being able to describe the functional capacities and potential importance of poorly understood microbial processes occurring in the lake (for example, photoheterotrophy by Alphaproteobacteria), we were able to answer targeted questions about the biology of the unusual lake sulfur chemistry. The low potential for dissimilatory sulfur cycling (both sulfur oxidation and DSR) and relatively stable waters of the deep zone, combined with the generation of DMS from DMSP, facilitate the accumulation of a high level of DMS in the lake. It appears Marinobacter and Roseovarius have a key role in DMS formation by cleaving DMSP generated by upper mixed zone eucaryal algae. The remarkable abundance of DMSP lyase genes suggests that DMSP is a significant carbon source in Organic Lake, and the cleavage pathway provides a selective advantage under the unique constraints of the Organic Lake environment.
In view of the minimal capacity for biological fixation of carbon and nitrogen, and yet organic richness, including high levels of DMS in Organic Lake, we evaluated what input the lake may have received throughout its relatively brief ∼3000 year history. The volume of the lake is small (∼6 × 104 m3), and exogenous input may occur from guano deposited in a small penguin rookery nearby the lake, through giant petrel or skua grazing and defecation, and/or by decaying animal carcasses such as elephant seals, which can weigh on the order of 1 ton and are present near the lake. Precipitation in Antarctica tends to be high in nitrate and relatively high levels have been reported in snow, ice, inland drainage systems and isolated water bodies (Vincent and Howard-Williams, 1994; Webster-Brown et al., 2010). It is also possible that during isolation from the ocean, the base of the water column in the marine basin that formed the lake may have acted as a sump for organic material. Phytoplankton blooms and benthic mats (for example, fixation of nitrogen by cyanobacteria and oxidation of nitrogen compounds) tend to make coastal marine basins very productive, and organic matter that sediments out of the surface waters will become trapped in the denser, more saline bottom layers (Bird et al., 1991). Retention of captured organic matter in the lake may also have been facilitated by Organic Lake having become highly saline quickly (Bird et al., 1991). Studies in the future that experimentally determine exogenous input and historical lake dynamics (for example, stable isotope and biomarker analyses of lake sediment), the role of benthic communities (for example, in reoxidation of reduced nitrogen compounds), and metaproteogenomic analyses of interannual community composition and function, will provide improved knowledge of the unusual biogeochemistry of Organic Lake and better enable predictions to be made about how the lake may be affected by ecosystem changes.
Acknowledgments
This work was supported by the Australian Research Council and the Australian Antarctic Science program and undertaken with the assistance of resources provided at the NCI National Facility systems at the Australian National University through the National Computational Merit Allocation Scheme supported by the Australian Government. We acknowledge the assistance from the J Craig Venter Institute, and the Gordon and Betty Moore Foundation. We thank John Bowman for providing unpublished rhodopsin sequence data. We acknowledge those involved in the formal review process for their positive views and insightful comments. We acknowledge all financial support and any other personal connections.
The authors declare no conflict of interest
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abell GCJ, Bowman JP. Ecological and biogeographic relationships of class Flavobacteria in the Southern Ocean. FEMS Microbiol Ecol. 2005a;51:265–277. doi: 10.1016/j.femsec.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Abell GCJ, Bowman JP. Colonization and community dynamics of class Flavobacteria on diatom detritus in experimental mesocosms based on Southern Ocean seawater. FEMS Microbiol Ecol. 2005b;53:379–391. doi: 10.1016/j.femsec.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Balashov SP, Imasheva ES, Boichenko VA, Antón J, Wang JM, Lanyi JK. Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science. 2005;309:2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann DJ, Hooper AB, Klotz MG. Structure and sequence conservation of hao cluster genes of autotrophic ammonia-oxidizing bacteria: evident for their evolutionary history. Appl Environ Microbiol. 2005;71:5371–5382. doi: 10.1128/AEM.71.9.5371-5382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird MI, Chivas AR, Radnell CJ, Burton HR. Sedimentological and stable-isotope evolution of lakes in the Vestfold Hills, Antarctica. Palaeogeogr Palaeoclimatol Palaeoecol. 1991;84:109–130. [Google Scholar]
- Bowman JP, McCammon SA, Lewis T, Skerratt JH, Brown JL, Nichols DS, et al. Psychroflexus torquis gen. nov., sp. nov., a psychrophilic species from Antarctic sea ice, and reclassification of Flavobacterium gondwanense (Dobson et al. 1993) as Psychroflexus gondwanense gen. nov., comb. nov. Microbiology. 1998;144:1601–1609. doi: 10.1099/00221287-144-6-1601. [DOI] [PubMed] [Google Scholar]
- Bowman JP, McCammon SA, Rea SM, McMeekin TA. The microbial composition of three limnologically disparate hypersaline Antarctic lakes. FEMS Microbiol Lett. 2000;183:81–88. doi: 10.1111/j.1574-6968.2000.tb08937.x. [DOI] [PubMed] [Google Scholar]
- Brown MV, Lauro FM, DeMaere MZ, Muir L, Wilkins D, Thomas T, et al. Global biogeography of SAR11 marine bacteria. Mol Syst Biol. 2012;8:595. doi: 10.1038/msb.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke CM, Burton HR. Photosynthetic bacteria in meromictic lakes a stratified fjords of the Vestfold Hills, Antarctica. Hydrobiologia. 1988;165:13–23. [Google Scholar]
- Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Béjà O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T, et al. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature. 2002;6872:630–633. doi: 10.1038/415630a. [DOI] [PubMed] [Google Scholar]
- Campbell BJ, Engel AS, Porter ML, Takai K. The versatile ɛ-proteobacteria: key players in sulfidic habitats. Nat Rev Microbiol. 2006;4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- Charlson RJ, Lovelock JE, Andreae MO, Warren SG. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature. 1987;326:655–661. [Google Scholar]
- Chouari R, Le Paslier D, Daegelen P, Ginestet P, Weissenbach J, Sghir A. Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digester. Environ Microbiol. 2005;7:1104–1115. doi: 10.1111/j.1462-2920.2005.00795.x. [DOI] [PubMed] [Google Scholar]
- Cottrell MT, Kirchman DL. Photoheterotrophic microbes in the Arctic Ocean in summer and winter. Appl Environ Microbiol. 2009;75:4958–4966. doi: 10.1128/AEM.00117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran MAJ, Jones GB. Spatial distribution of dimethylsulfide and dimethylsulfonioproprionate in the Australasian sector of the Southern Ocean. J Geophys Res. 1998;103:16 677–16 689. [Google Scholar]
- Curson ARJ, Rogers R, Todd JD, Bearley CA, Johnston AWB. Molecular genetic analysis of a dimethysulfonioproprionate lyase that liberates the climate-changing gas dimethylsulfide in several marine α-proteobacteria and Rhodobacter sphaeroides. Environ Microbiol. 2008;10:757–767. doi: 10.1111/j.1462-2920.2007.01499.x. [DOI] [PubMed] [Google Scholar]
- Curson ARJ, Todd JD, Sullivan MJ, Johnston AWB. Catabolism of dimethylsulphonioproprionate: microorganisms, enzymes and genes. Nat Rev Microbiol. 2011;9:849–859. doi: 10.1038/nrmicro2653. [DOI] [PubMed] [Google Scholar]
- Demergasso C, Escudero L, Casamayor EO, Chong G, Balagué V, Pedrós-Alió C. Novelty and spatio-temporal heterogeneity in the bacterial diversity of hypersaline Lake Tebenquiche (Salar de Atacama) Extremophiles. 2008;12:491–504. doi: 10.1007/s00792-008-0153-y. [DOI] [PubMed] [Google Scholar]
- Deprez PP, Franzmann PD, Burton HR. Determination of reduced sulfur gases in Antarctic lakes and seawater by gas chromatography after solid adsorbent preconcentration. J Chromatogr. 1986;362:9–21. [Google Scholar]
- Dobson SJ, Colwell RR, McMeekin TA, Franzmann PD. Direct sequencing of the polymerase chain reaction-amplified 16S rRNA gene of Flavobacterium gondwanense sp. nov. and Flavobacterium salegens sp. nov., two new species from a hypersaline Antarctic lake. Int J Syst Bacteriol. 1993;43:77–83. doi: 10.1099/00207713-43-1-77. [DOI] [PubMed] [Google Scholar]
- Dobson SJ, James SR, Franzmann PD, McMeekin TA. A numerical taxonomic study of some pigmented bacteria isolated from Organic Lake, an antarctic hypersaline lake. Arch Microbiol. 1991;156:56–61. [Google Scholar]
- Edwards KJ, Rogers DR, Wirsen CO, McCollom TM. Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithoautotrophic α- and γ-Proteobacteria from the deep sea. Appl Environ Microbiol. 2003;69:2906–2913. doi: 10.1128/AEM.69.5.2906-2913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshahed MS, Najar FZ, Aycock M, Qu C, Roe BA, Krumholz LR. Metagenomic analysis of the microbial community at Zodletone Spring (Oklahoma): Insights into the genome of a member of the novel candidate division OD1. Appl Environ Microbiol. 2005;71:7598–7602. doi: 10.1128/AEM.71.11.7598-7602.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann PD, Burton HR, McMeekin TA. Halomonas subglaciescola, a new species of halotolerant bacteria isolated from Antarctica. Int J Syst Bacteriol. 1987a;37:27–34. [Google Scholar]
- Franzmann PD, Deprez PP, Burton HR, van den Hoff J. Limnology of Organic Lake, Antarctica, a meromictic lake that contains high concentrations of dimethyl sulfide. Aust J Mar Freshw Res. 1987b;38:409–417. [Google Scholar]
- Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J. Prokaryotic sulfur oxidation. Curr Opin Microbiol. 2005;8:253–259. doi: 10.1016/j.mib.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Schwalbach MS, Stingl U. Proteorhodopsins: an array of physiological roles. Nat Rev Microbiol. 2008;6:488–494. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- Gauthier MJ, Lafay B, Christen R, Fernandez L, Acquaviva M, Bonin P, et al. Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int J Syst Bacteriol. 1992;42:568–576. doi: 10.1099/00207713-42-4-568. [DOI] [PubMed] [Google Scholar]
- Gibson JAE. The meromictic lakes and stratified marine basins of the Vestfold Hills, East Antarctica. Antarct Sci. 1999;11:175–192. [Google Scholar]
- Gibson JAE, Garrick RC, Franzmann PD, Deprez PP, Burton H. Reduced sulfur gases in saline lakes of the Vestfold Hills, Antarctica. Palaeogeo Palaeoclimatol Palaeoecol. 1991;84:131–140. [Google Scholar]
- Gibson JAE, Qiang XL, Franzmann PD, Garrick RC, Burton HR. Volatile fatty and dissolved free amino acids in Organic Lake, Vestfold Hills, East Antarctica. Polar Biol. 1994;14:545–550. [Google Scholar]
- Goberna M, Insam H, Franke-Whittle IH. Effect of biowaste sludge maturation on the diversity of thermophilic bacteria and archaea in an anaerobic reactor. Appl Environ Microbiol. 2009;75:2566–2572. doi: 10.1128/AEM.02260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Consarnau L, Akram N, Lindell K, Pedersen A, Neutze R, Milton DL, et al. Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol. 2010;8:e1000358. doi: 10.1371/journal.pbio.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Consarnau L, González JM, Coll-Lladó M, Gourdon P, Pascher T, Neutze R, et al. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature. 2007;445:210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Stadler P, Wu QL, Pöckl M. The filtration–acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J Microbiol Methods. 2004;57:379–390. doi: 10.1016/j.mimet.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Hahn MW. Description of seven candidate species affiliated with the phylum Actinobacteria, representing planktonic freshwater bacteria. Int J Syst Evol Microbiol. 2009;59:112–117. doi: 10.1099/ijs.0.001743-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JK, Kelley ST, Pace NR. New perspective on uncultured bacterial phylogenetic division OP11. Appl Environ Microbiol. 2004;70:845–849. doi: 10.1128/AEM.70.2.845-849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton AD, Shenoy DM, Hart MC, Mogg A, Green DH. Metabolism of DMSP, DMS and DMSO by the cultivable bacterial community associated with the DMSP-producing dinoflagellate Scrippsiella trochoidea. Biogeochem. 2012;110:131–146. [Google Scholar]
- Howard EC, Sun S, Biers EJ, Moran MA. Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ Microbiol. 2008;10:2397–2410. doi: 10.1111/j.1462-2920.2008.01665.x. [DOI] [PubMed] [Google Scholar]
- Huang L, Zhu S, Zhou H, Qu L. Molecular phylogenetic diversity of bacteria associated with the leachate of a closed municipal solid waste landfill. FEMS Microbiol Lett. 2005;242:297–303. doi: 10.1016/j.femsle.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Humayoun SB, Bano N, Hollibaugh JT. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl Environ Microbiol. 2003;69:1030–1042. doi: 10.1128/AEM.69.2.1030-1042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huu NB, Denner EB, Ha DT, Wanner G, Stan-Lotter H. Marinobacter aquaeolei sp. nov., a halophilic bacterium isolated from a Vietnamese oil-producing well. Int J Syst Bacteriol. 1999;49:367–375. doi: 10.1099/00207713-49-2-367. [DOI] [PubMed] [Google Scholar]
- Hügler M, Sievert SM. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Annu Rev Mar Sci. 2011;3:261–289. doi: 10.1146/annurev-marine-120709-142712. [DOI] [PubMed] [Google Scholar]
- James SR, Burton HR, McMeekin TA, Mancuso CA. Seasonal abundance of Halomonas meridiana, Halomonas subglaciescola, Flavobacterium gondwanense and Flavobacterium salegens in four Antarctic Lakes. Antarct Sci. 1994;6:325–332. [Google Scholar]
- James SR, Dobson SJ, Franzmann PD, McMeekin TA. Halomonas meridiana, a new species of extremely halotolerant bacteria from Antarctic saline lakes. System Appl Microbiol. 1990;13:270–278. [Google Scholar]
- Johnston AWB, Todd JD, Sun L, Nikolaidou-Katsaridou MN, Curson ARJ, Rogers R. Molecular diversity of bacterial production of the climate changing gas, dimethyl sulphide, a molecule that impinges on local and global symbioses. J Exp Bot. 2008;59:1059–1067. doi: 10.1093/jxb/erm264. [DOI] [PubMed] [Google Scholar]
- Kang I, Lee K, Yang S-J, Choi A, Kang D, Lee YK, et al. Genome sequence of ‘Candidatus Aquiluna' sp. strain IMCC13023, a marine member of the Actinobacteria isolated from an Arctic Fjord. J Bacteriol. 2012;194:3550–3551. doi: 10.1128/JB.00586-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman DL. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol. 2002;39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- Kraft B, Stous M, Tegetmeyer HE. Microbial nitrate respiration—genes, enzymes and environmental distribution. J Biotechnol. 2011;155:104–117. doi: 10.1016/j.jbiotec.2010.12.025. [DOI] [PubMed] [Google Scholar]
- Labrenz M, Collins MD, Lawson PA, Tindall BJ, Schumann P, Hirsch P. Roseovarius tolerans gen. nov., sp. nov., a budding bacterium with variable bacteriochlorophyll a production from hypersaline Ekho Lake. Int J Syst Bacteriol. 1999;49:137–147. doi: 10.1099/00207713-49-1-137. [DOI] [PubMed] [Google Scholar]
- Lauro FM, DeMaere MZ, Yau S, Brown MV, Ng C, Wilkins D, et al. An integrative study of a meromictic lake ecosystem in Antarctica. ISME J. 2011;5:879–895. doi: 10.1038/ismej.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn-Parry J, Marshall WA, Marchant HJ. Flagellate nutritional versatility as a key to survival in two contrasting Antarctic saline lakes. Freshw Biol. 2005;50:830–838. [Google Scholar]
- Laybourn-Parry J, Pearce D. The biodiversity and ecology of Antarctic lakes: models for evolution. Phil Trans R Soc B. 2007;364:2273–2289. doi: 10.1098/rstb.2006.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Lovelock JE, Maggs RJ. Atmospheric dimethyl sulfide and the natural sulphur cycle. Nature. 1972;237:452–453. [Google Scholar]
- Man D, Wang W, Sabehi G, Aravind L, Post AF, Massana R, et al. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 2003;22:1725–1731. doi: 10.1093/emboj/cdg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Kubota K, Satoh T, Kunugi M, Ban S, Imura S. Dimethyl sulfoxide-respiring bacteria in Suribati Ike, a hypersaline lake, in Antarctica and the marine environment. Polar Biosci. 2006;20:73–87. [Google Scholar]
- Miyoshi T, Iwatuski T, Naguma T. Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2 μm-pore-size filters. Appl Environ Microbiol. 2005;71:1084–1088. doi: 10.1128/AEM.71.2.1084-1088.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA, Belas R, Schell MA, González JM, Sun F, Binder BJ, et al. Ecological genomics of marine Roseobacters. Appl Environ Microbiol. 2007;73:4559–4569. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA, Miller WL. Resourceful heterotrophs make the most of light in the coastal ocean. Nat Rev Microbiol. 2007;5:792–799. doi: 10.1038/nrmicro1746. [DOI] [PubMed] [Google Scholar]
- Moran MA, Reisch CR, Kiene RP, Whitman WB. Genomic insights into bacterial DMSP transformations. Ann Rev Marine Sci. 2012;4:523–542. doi: 10.1146/annurev-marine-120710-100827. [DOI] [PubMed] [Google Scholar]
- Ng C, DeMaere MZ, Williams TJ, Lauro FM, Raftery M, Gibson JAE, et al. Metaproteogenomic analysis of a dominant green sulfur bacterium from Ace Lake, Antarctica. ISME J. 2010;4:1002–1019. doi: 10.1038/ismej.2010.28. [DOI] [PubMed] [Google Scholar]
- Raina J-P, Dinsdale EA, Willis BL, Bourne DG. Do the organic sulfur compounds DMSP and DMS drive coral microbial associations. Trends Microbiol. 2010;3:101–108. doi: 10.1016/j.tim.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Redfield AC, Ketchum BH, Richards FA.1963The influence of organisms on the composition of seawaterIn: Hill MN, (ed).The Sea John Wiley and Sons: New York; 26–77. [Google Scholar]
- Reisch CR, Moran MA, Whitman WB. Bacterial catabolism of dimethylsulfonioproprionate (DMSP) Front Microbiol. 2011;2:1–12. doi: 10.3389/fmicb.2011.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, Weissenbach J, et al. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 2009;3:700–714. doi: 10.1038/ismej.2009.2. [DOI] [PubMed] [Google Scholar]
- Roberts NJ, Burton HR, Pitson GA. Volatile organic compounds from Organic Lake, an Antarctic hypersaline, meromictic lake. Polar Biol. 1993;13:361–366. [Google Scholar]
- Roberts NJ, Burton HR. Sampling volatile organics from a meromictic Antarctic lake. Polar Biol. 1993;13:359–361. [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:398–431. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röske K, Sachse R, Scheerer C, Röske I. Microbial diversity and composition of the sediment in the drinking water reservoir Saidenbach (Saxonia, Germany) Syst Appl Microbiol. 2012;35:35–44. doi: 10.1016/j.syapm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Samsudin AA, Evans PN, Wright AG, Al Jassim R. Molecular diversity of the foregut bacteria community in the dromedary camel (Camelus dromedariusi) Environ Microbiol. 2011;13:3024–3035. doi: 10.1111/j.1462-2920.2011.02579.x. [DOI] [PubMed] [Google Scholar]
- Schmidtova J, Hallam SJ, Baldwin SA. Phylogenetic diversity of transition and anoxic zone bacterial communities within a near-shore anoxic basin: Nitinat Lake. Environ Microbiol. 2009;11:3233–3251. doi: 10.1111/j.1462-2920.2009.02044.x. [DOI] [PubMed] [Google Scholar]
- Schäfer H, Myronova N, Boden R. Microbial degradation of dimethylsulphide and related C1-sulphur compounds: organisms and pathways controlling sulphur in the biosphere. J Exp Bot. 2010;61:315–334. doi: 10.1093/jxb/erp355. [DOI] [PubMed] [Google Scholar]
- Sharma AK, Zhaxybayeva O, Papke RT, Doolittle WF. Actinorhodopsins: proteorhodopsin-like gene sequences found predominantly in non-marine environments. Environ Microbiol. 2008;10:1039–1056. doi: 10.1111/j.1462-2920.2007.01525.x. [DOI] [PubMed] [Google Scholar]
- Stefels J, Steinke M, Turner S, Malin G, Belviso S. Environmental constraints on the production and removal of the climatically active gas dimethylsulphide (DMS) and implications for ecosystem modeling. Biogeochem. 2007;83:245–275. [Google Scholar]
- Tajima K, Aminov RI, Nagamine T, Ogata K, Nakamura M, Matsui H, et al. Rumen bacterial diversity as determined by sequence analysis of 16S rDNA. FEMS Microbiol Ecol. 1999;29:159–169. [Google Scholar]
- Takahashi N, Sato T, Yamada T. Metabolic pathways for cytotoxic end product formation from glutamate- and aspartate-containing peptides by Porphyromonas gingivalis. J Bacteriol. 2000;182:4704–4710. doi: 10.1128/jb.182.17.4704-4710.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Ji P, Hayashi J, Koike Y, Wu X, Kida K. Characteristic microbial community of a dry thermophilic methanogenic digester: its long-term stability and change with feeding. Appl Microbiol Biotechnol. 2011;91:1477–1461. doi: 10.1007/s00253-011-3479-9. [DOI] [PubMed] [Google Scholar]
- Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- Thomsen HA. Ultrastructural studies of the flagellate and cyst stages of Pseudopedinella tricostata (Pedinellales, Chrysophyceae) Brit Phycol J. 1988;23:1–16. [Google Scholar]
- Todd JD, Curson ARJ, Dupont CL, Nicholson P, Johnston AWB. The dddP gene, encoding a novel enzyme that converts dimethylsulfonioproprionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environ Microbiol. 2009;11:1376–1385. doi: 10.1111/j.1462-2920.2009.01864.x. [DOI] [PubMed] [Google Scholar]
- Todd JD, Curson ARJ, Kirkwood M, Sullivan MJ, Green RT, Johnston AWB. DddQ, a novel, cupin-containing, dimethylsulfonioproprionate lyase in marine roseobacters and in uncultured marine bacteria. Environ Microbiol. 2011;13:427–438. doi: 10.1111/j.1462-2920.2010.02348.x. [DOI] [PubMed] [Google Scholar]
- Todd JD, Curson ARJ, Nikolaidou-Kataraidou N, Brearley CA, Watmough NJ, Chan Y, et al. Molecular dissection of bacterial acrylate catabolism—unexpected links with dimethylsulfonioproprionate catabolism and dimethyl sulfide production. Environ Microbiol. 2010;12:327–343. doi: 10.1111/j.1462-2920.2009.02071.x. [DOI] [PubMed] [Google Scholar]
- Todd JD, Kirkwood M, Newton-Payne S, Johnston AWB. DddW, a third DMSP lyase in model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J. 2012;6:223–226. doi: 10.1038/ismej.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JD, Rogers R, Li YG, Wexler M, Bond PL, Sun L, et al. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science. 2007;315:666–669. doi: 10.1126/science.1135370. [DOI] [PubMed] [Google Scholar]
- Unrein F, Izaguirre I, Massana R, Balagué V, Gasol JM. Nanoplankton assemblages in maritime Antarctic lakes: characterisation and molecular fingerprinting comparison. Aquat Microb Ecol. 2005;40:269–282. [Google Scholar]
- Vincent WF, Howard-Williams C. Nitrate-rich inland waters of the Ross Ice Shelf region, Antarctica. Antarctic Sci. 1994;6:339–346. [Google Scholar]
- Wagner-Döbler I, Biebl H. Environmental biology of the marine Roseobacter lineage. Ann Rev Microbiol. 2006;60:255–280. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- Ward BB, Granger J, Maldonado MT, Casciotti KL, Harris S, Wells ML. Denitrification in the hypolimnion of permanently ice-covered Lake Bonney, Antarctica. Aquat Microb Ecol. 2005;38:295–307. [Google Scholar]
- Ward BB, Priscu JC. Detection and characterization of denitrifying bacteria from a permanently ice-covered Antarctic lake. Hydrobiologia. 1997;347:57–68. [Google Scholar]
- Webster-Brown J, Gall M, Gibson J, Wood S, Hawes I. The biogeochemistry of meltwater habitats in the Darwin Glacier region (80°S), Victoria Land, Antarctica. Antarct Sci. 2010;22:646–661. [Google Scholar]
- Wilkins D, Lauro FM, Williams TJ, DeMaere MZ, Brown MV, Hoffman JM, et al. Biogeographic partitioning of Southern Ocean microorganisms revealed by metagenomics. Environ Microbiol. 2013a;15:1318–1333. doi: 10.1111/1462-2920.12035. [DOI] [PubMed] [Google Scholar]
- Wilkins D, Yau S, Williams TJ, Allen MA, Brown MV, DeMaere MZ, et al. Key microbial drivers in Antarctic aquatic environments. FEMS Microbiol Rev. 2013b;37:303–335. doi: 10.1111/1574-6976.12007. [DOI] [PubMed] [Google Scholar]
- Williams TJ, Wilkins D, Long E, Evans F, DeMaere MZ, Raftery MJ, et al. The role of planktonic Flavobacteria is processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ Microbiol. 2013;15:1302–1317. doi: 10.1111/1462-2920.12017. [DOI] [PubMed] [Google Scholar]
- Xing P, Hahn MW, Wu QL. Low taxon richness of bacterioplankton in high-altitude lakes of the eastern Tibetan Plateau, with a predominance of Bacteroidetes and Synechoccocus spp. Appl Environ Microbiol. 2009;75:7017–7025. doi: 10.1128/AEM.01544-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Takai K. Sulfur metabolisms in epsilon- and gamma-Proteobacteria in deep-sea hydrothermal fields. Front Microbiol. 2011;2:1–8. doi: 10.3389/fmicb.2011.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Hattori Y, Ohtagaki H, Fujiwara K. Microbial diversity with dominance of 16S rRNA genes sequences with high GC contents at 74 and 98°C subsurface crude oil deposits in Japan. FEMS Microbiol Ecol. 2011;76:220–235. doi: 10.1111/j.1574-6941.2011.01044.x. [DOI] [PubMed] [Google Scholar]
- Yanagibayashi M, Nogi Y, Li L, Kato C. Changes in the microbial community in Japan Trench sediment from a depth of 6292 m during cultivation without decompression. FEMS Microbiol Lett. 1999;170:271–279. doi: 10.1111/j.1574-6968.1999.tb13384.x. [DOI] [PubMed] [Google Scholar]
- Yau S, Lauro FM, DeMaere MZ, Brown MV, Thomas T, Raftery MJ, et al. Virophage control of antarctic algal host-virus dynamics. Proc Natl Acad Sci USA. 2011;108:6163–6168. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P, Iversen MH, Hankeln W, Kottman R, Quast C, Glöckner FO. Ecological structuring of bacterial and archaeal taxa in surface ocean waters. FEMS Microbiol Ecol. 2012;81:373–385. doi: 10.1111/j.1574-6941.2012.01357.x. [DOI] [PubMed] [Google Scholar]
- Yoch DC. Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl Environ Microbiol. 2002;68:5804–5815. doi: 10.1128/AEM.68.12.5804-5815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwartz D, Bird M, Stone J, Lambeck K. Holocene sea-level change and ice-sheet history in the Vestfold Hills, East Antarctica. Earth Planet Sci Lett. 1998;155:131–145. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.