Abstract
Genomic resources developed for domesticated species provide powerful tools for studying the evolutionary history of their wild relatives. Here we use 61K single-nucleotide polymorphisms (SNPs) evenly spaced throughout the canine nuclear genome to analyse evolutionary relationships among the three largest European populations of grey wolves in comparison with other populations worldwide, and investigate genome-wide effects of demographic bottlenecks and signatures of selection. European wolves have a discontinuous range, with large and connected populations in Eastern Europe and relatively smaller, isolated populations in Italy and the Iberian Peninsula. Our results suggest a continuous decline in wolf numbers in Europe since the Late Pleistocene, and long-term isolation and bottlenecks in the Italian and Iberian populations following their divergence from the Eastern European population. The Italian and Iberian populations have low genetic variability and high linkage disequilibrium, but relatively few autozygous segments across the genome. This last characteristic clearly distinguishes them from populations that underwent recent drastic demographic declines or founder events, and implies long-term bottlenecks in these two populations. Although genetic drift due to spatial isolation and bottlenecks seems to be a major evolutionary force diversifying the European populations, we detected 35 loci that are putatively under diversifying selection. Two of these loci flank the canine platelet-derived growth factor gene, which affects bone growth and may influence differences in body size between wolf populations. This study demonstrates the power of population genomics for identifying genetic signals of demographic bottlenecks and detecting signatures of directional selection in bottlenecked populations, despite their low background variability.
Keywords: bottleneck, effective population size, genetic differentiation, grey wolf, linkage disequilibrium, selection
Introduction
Studies on evolutionary processes in natural populations have been greatly enabled by technological advances related to whole genome sequence data from a variety of domesticated species (Allendorf et al., 2010). Access to large number of loci, often with annotated positions within the genome of the investigated species, permits researchers to overcome analytical limitations associated with the analysis of a small number of genetic markers. Examples include reconstruction of admixture patterns among closely related species (vonHoldt et al., 2011; Miller et al., 2012), identification of the genetic basis of parallel adaptations (Hohenlohe et al., 2010; Zulliger et al., 2013) and investigation of demographic effects of past climate change (Miller et al., 2012; Zhao et al., 2013). Here, we use a population genomic approach to study the genetic effects of demographic bottlenecks in European grey wolf populations.
Demographic bottlenecks have been extensively explored using classical population genetic methods, typically based on a small number of neutral microsatellite loci (as reviewed in Peery et al., 2012) or MHC loci, presumably under balancing selection (for example, Oliver and Piertney 2012). Given the limitations of using limited numbers of genetic markers (Peery et al., 2012), genome-wide studies based on data from natural populations that underwent population declines are needed. Considerable attention has been paid to population bottlenecks associated with domestication events and resulting problems with distinguishing true signals of selection from effects of drift (for example, Caicedo et al., 2007; Axelsson et al., 2013). However, in domestic species, a strong signal of artificial selection can be expected and predictions can be made regarding traits likely to be affected, while in wild species, the strength of selection and traits affected are less predictable.
Here, we assess genome-wide effects of population bottlenecks and identify signals of selection in European grey wolves (Canis lupus). The grey wolf is the direct ancestor of the domestic dog (Canis lupus familiaris), which is an important and emerging model for understanding the genetics of disease susceptibility and developmental biology. Therefore, genomic studies on the grey wolf benefit from the extensive genomic resources available for the domestic dog (for example, Lindblad-Toh et al., 2005; vonHoldt et al., 2010, 2011). Another advantage of focusing on the grey wolf is the extensive background knowledge regarding its ecology, recent demographic history and population genetics (reviewed in Musiani et al., 2010; Randi, 2011).
Genetic studies revealed a complex evolutionary history of the grey wolf, with no clear phylogeographic patterns worldwide (Vilà et al., 1999; Pilot et al., 2010), but with cryptic population genetic subdivisions related to environmental differences (for example, Geffen et al., 2004; Pilot et al., 2006; vonHoldt et al., 2011). Wolves had a continuous range in Europe throughout most of the Holocene, which was considerably reduced and fragmented in the last few centuries as a result of direct eradication and habitat loss (Boitani, 2003). Currently, wolves in Western Europe occur in isolated and partially protected populations in Italy (including the Apennine Peninsula and the western Italian Alps) and the Iberian Peninsula. In Eastern Europe, there are large and interconnected populations (Figure 1), most of which have experienced constant hunting pressures. Cryptic population structure has been observed in Eastern Europe (Pilot et al., 2006; Stronen et al., 2013), but this genetic differentiation is small compared to the differentiation between Eastern Europe and both Italian and Iberian populations. Therefore, herein, we use the term ‘Eastern European population' despite the lack of panmixia.
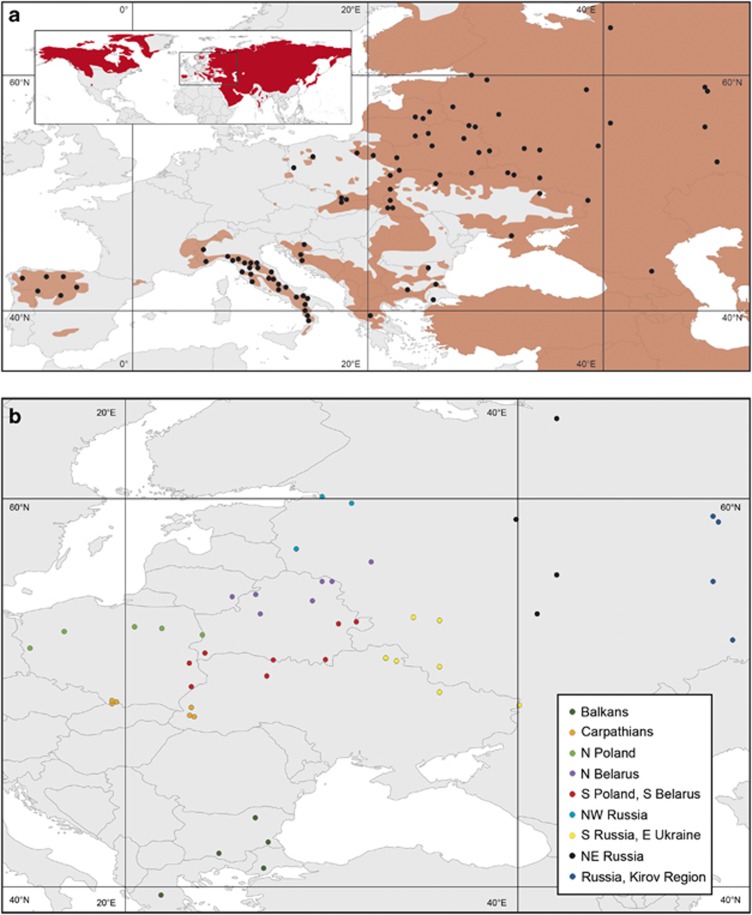
Figure 1.
(a) Map of sample distribution. The black circles represent sampling locations, which are exact, except for the Iberian Peninsula, where the exact sample locations were unknown. A sampling location may be shared by several individuals. The range of the grey wolf is marked in pale red on the main map and in red on the small map showing the worldwide distribution of this species based on the most recent data available from IUCN. Due to the recent expansion of wolf populations, some samples occur outside of the established range. (b) Subdivision into local populations in Eastern Europe based on the geographical proximity of the samples and data on their genetic differentiation from Pilot et al., (2006). Local populations with sample size of at least five individuals were used in the LD decay and ROH analyses.
Patterns of mtDNA variability suggest that Eastern Europe and the Iberian Peninsula were linked by gene flow before the extinction of intermediate populations, a conclusion supported by the presence of a shared haplotype between the Eastern European and the Iberian population (Pilot et al., 2010). By comparison, long-term isolation has been suggested for the Italian wolf population (Lucchini et al., 2004), which has a unique mtDNA haplotype not found elsewhere.
The three main European wolf populations have distinct demographic histories. The Iberian Peninsula currently contains the largest wolf population in Western Europe, numbering over 2000 individuals (Sastre et al., 2011). This population has been isolated at least since the extinction of the wolf from France at the end of the nineteenth century, and suffered a recent demographic bottleneck in the 1970s, when the population was reduced to about 700 individuals (Sastre et al., 2011). Since that time, the population has expanded in range and size. The current population has a small effective population size (about 50) and shows signs of the past genetic bottleneck (Sastre et al., 2011).
The Italian wolf population also experienced a severe demographic bottleneck in 1970s, when it was reduced to about 100 individuals, the effects of which are detectable at the genetic level (Randi, 2011). However, the history of this population may be more complex than a single recent bottleneck. Lucchini et al. (2004) used a Bayesian coalescent analysis to show that Italian wolves underwent a 100- to 1000-fold population contraction during the last 2000–10 000 years, which may be more important in defining their current genetic profiles. As a result of recent legal protection and abundance of prey, the Italian wolf has recovered to a range that includes the entire Apennines and the western Italian Alps, and is expanding to the Swiss and French Alps (Randi, 2011), eastern Italian Alps (Fabbri et al., in press) and even into Spain (Sastre, 2011).
The wolf distribution in Eastern Europe is relatively continuous, and is connected with Asian populations (Boitani, 2003; Figure 1). To the best of our knowledge, there is no account of any strong bottleneck that would affect this population, although there is some evidence for a large-scale population decline in the former Soviet Union and the neighbouring European countries in the 1970s (Boitani, 2003; Sastre et al., 2011). However, the Eastern European population has experienced strong hunting pressure for many generations, and the hunting continues in most of its range to this day. As a result of hunting pressures on both the wolves and their prey, the Eastern European wolves have suffered multiple local demographic fluctuations (for example, Spiridinov and Spassov, 1985; Jędrzejewska et al., 1996; Ozolins and Andersone, 2001; Sidorovich et al., 2003; Gomercic et al., 2010).
Most of the genetic studies on European grey wolves are based on a small number of markers (nuclear and mitochondrial), with few comparative studies across all the three populations (reviewed in Randi, 2011). The availability of validated tools for genome-wide analysis of single-nucleotide polymorphisms (SNPs) in the domestic dog opened new perspectives for population genetic studies of wild canids (Lindblad-Toh et al., 2005). The utility of this approach has been demonstrated by vonHoldt et al. (2011), who applied Affymetrix Canine SNP Genome Mapping Array to study genome-wide variability in wild wolf-like canids worldwide, with a focus on North America. That study addressed long-standing questions about diversification and admixture in wolf-like canids, including the systematic status of enigmatic taxa such as the red wolf and Great Lakes wolf (vonHoldt et al., 2011). Here we analyse genome-wide SNP variability in European grey wolves to test the following hypotheses: (1) the three European populations should show high levels of genetic differentiation, with the Italian population being particularly distinct, reflecting its supposed ancient divergence and long-term isolation (Lucchini et al., 2004; Pilot et al., 2010); (2) the Italian and Iberian populations should show evidence for strong genetic bottlenecks (Lucchini et al., 2004; Sastre et al., 2011); (3) a decline in effective size throughout the last few centuries should be observed in each population as a result of a direct extermination by humans and habitat loss (for example, Randi, 2011); and (4) the three European populations should show a signal of diversifying selection, reflecting their local adaptation to different types of habitat and available prey (for example, Geffen et al., 2004; Pilot et al., 2006; vonHoldt et al., 2011).
Materials and methods
Data set
This study utilized data derived from the CanMap project (Boyko et al., 2010; vonHoldt et al., 2010) that provided genome-wide SNP data from 912 domestic dogs and 337 wild canids, based on genotyping with an Affymetrix Canine SNP Genome Mapping Array (coordinates based on the CanFam2 assembly). Samples were genotyped at 60 584 high-quality autosomal SNPs (referred to as 61K) and 851 X chromosome SNP loci (Boyko et al., 2010; vonHoldt et al., 2010). Here, we used a subset of the CanMap SNP data set that consisted of 103 grey wolves: 54 from Eastern Europe, 19 from Italy, 6 from the Iberian Peninsula, 7 from Asia and 17 from North America, plus 5 coyotes that served as an outgroup (see Supplementary Table 1).
For linkage disequilibrium and autozygosity analyses (see below), we introduced subdivision by defining small groups of spatially proximate samples within Eastern Europe (Figure 1b). These groups were delimited based on both geographical proximity of sampling locations and results of an earlier study showing genetic structure within Eastern Europe (Pilot et al., 2006), and therefore in some cases, geographically proximate samples are assigned to different groups to reflect population differentiation found previously.
The initial set of 61K loci was pruned using PLINK (Purcell et al., 2007) for loci that were invariant among the sample set, or had very low minor allele frequency (<0.01), resulting in 53 793 SNPs. For many applications, using a data set pruned for loci in strong linkage disequilibrium (LD) is advised (for example, Alexander et al., 2009). Therefore, we further pruned the data set for SNPs with an r2<0.5 within 50 SNP sliding windows, shifted and recalculated every 10 SNPs. This data set consisted of 33 958 SNPs (referred to as 34K data set).
Screening the data set for related individuals
We screened the initial larger data set for the presence of close relatives by calculating pairwise identity-by-state estimates in PLINK. This approach alone was insufficient to identify all close relatives in the highly isolated and bottlenecked wolf populations from Italy and the Iberian Peninsula, as all pairs of individuals had identity-by-state values >0.8, which in an outbred population is the empirical threshold for close relatives (vonHoldt et al., 2011). Therefore, for the Italian and Iberian populations, we identified close relatives using maximum-likelihood approaches as implemented in CERVUS 3.0 (Marshall et al., 1998) and KINGROUP 2 (Konovalov et al., 2004). CERVUS was used for parentage analysis, and KINGROUP was used to identify individuals related at the full-sibling and half-sibling level.
For CERVUS analysis, we selected loci with no missing data and with allele frequencies between 0.45 and 0.55. There were 827 SNPs that met those conditions in the Italian population and 1442 in the Iberian population. For KINGROUP analysis, we randomly selected 100 SNPs from this set (which was the maximum number of loci accepted).
Using KINGROUP for the Italian population (initial N=23), we identified one pair of full-siblings and three pairs of half-siblings. Only one individual from each pair was retained in the data set. Among the Iberian wolves (initial N=10), KINGROUP identified two pairs and one trio of full-siblings. CERVUS identified two parent-offspring pairs and one parent-offspring trio, consistent with three out of four full-sibling groups identified by KINGROUP, and only one individual from each pair or trio was retained in the data set. The sample sizes after removing the closely related individuals were 19 for Italy, 6 for the Iberian Peninsula and 54 for Eastern Europe – this data set was used in all the subsequent analyses.
Population structure analysis
Analysis of genetic differentiation in European wolves
We analysed the population genetic structure for the entire data set consisting of European, Asian and North American grey wolves, with coyotes as an outgroup. Genetic structure analyses were performed using the 34K data set. We used the Bayesian inference of genetic structure with no prior population information as implemented in STRUCTURE (Pritchard et al., 2000) and ADMIXTURE (Alexander et al., 2009). We used the two programs to check for consistency of the inferred structure.
STRUCTURE was run for K (the number of groups) from 1 to 10, with 100 000 MCMC iterations preceded by 20 000 burn-in iterations, and with three replicates for each K value. We used the admixture model and correlated allele frequencies. For each K, we checked whether the run parameters (likelihood, posterior probability of data and alpha) reach convergence within the burn-in period. Selection of optimal K on the basis of STRUCTURE output was performed with the support of STRUCTURE HARVESTER software (Earl and vonholdt, 2012). We chose the optimal K value based on likelihood values, the Evanno et al. (2005) ΔK method and maximum biological information.
ADMIXTURE analysis was run for K from 2 to 10 using the default termination criterion, which stops iterations when the log-likelihood increases by less than ɛ=10−4 between iterations. The value of K for which the model was optimally predictive was identified using a cross-validation method in which runs are performed holding out 10% of the genotypes at random, with 10 repetitions (Alexander et al., 2009). The optimal K was selected as the value that exhibited the lowest cross-validation error compared to other K values. We also used ADMIXTURE to carry out a separate analysis for Eastern European wolves only. We performed this additional analysis because earlier studies suggested population structuring in this region (Pilot et al., 2006; Stronen et al., 2013), which could have remained undetected in the context of strongly differentiated wolf populations from other parts of the world.
Moreover, we performed a principal components analysis (PCA) using the package SMARTPCA from EIGENSOFT (Patterson et al., 2006) to visualize the dominant components of variability within the data set. This analysis was performed for: (1) the entire sample set; (2) European wolves; and (3) Eastern European wolves. EIGENSOFT was also used to assess pair-wise FST and average divergence between and within populations (for details, see Supplementary Material).
Analysis of genetic structure using X chromosome data
The X chromosome data, which included 851 SNPs, were analysed for 37 females from the three European populations. We excluded SNPs from the pseudoautosomal region (first 6 Mb of the X chromosome). Outside the pseudoautosomal region, we removed additional four loci that were heterozygous in six males (which suggested genotyping errors). At each of the remaining 508 SNPs, no more than two males genotyped were heterozygotes. These were most likely genotyping errors and we treated them as missing data. After this adjustment, we obtained X chromosome haplotypes for males, which were used as a reference to improve the phasing of the corresponding female genotypes, which was carried out using FastPhase (Scheet and Stephens, 2006). The inferred female haplotypes were used to construct a neighbour-joining tree in MEGA 5 (Tamura et al., 2011), using genetic distances calculated as the proportion of the number of different bases to the total number of SNP sites. This procedure was also carried out for 50 pure-breed domestic dogs available from the CanMap project (vonHoldt et al., 2010). We then selected three dog females to be included as an outgroup in the neighbour-joining tree. We also analysed population structure using ADMIXTURE (with the same parameter settings as described for the autosomal data) for the LD-pruned X chromosome data set consisting of 249 SNPs.
Heterozygosity, linkage disequilibrium and autozygosity analysis
We calculated observed and expected heterozygosity for the Iberian and Italian wolf populations, and local populations from Eastern Europe (see Figure 1b), on the basis of the 61K SNP data set. Because estimates of heterozygosity and other parameters (see below) are dependent on sample sizes, we included only the local populations with at least five individuals sampled, and selected a random subset of six individuals from each of the populations with more than six individuals. For these groups, we estimated LD between all pairs of autosomal SNPs with minor allele frequency >0.15 by calculating genome-wide pairwise genotypic association coefficient (r2), based on the 61K SNP data set. We estimated LD decay as the physical distance at which r2 coefficient decays below a threshold of 0.5.
In addition, we identified runs of homozygosity (ROHs) >100 kb spanning at least 25 SNPs in individuals from each population. Long ROHs (>1 Mb) are indicative of autozygosity (that is, homozygosity by descent) and are a product of recent demographic events such as inbreeding or admixture, whereas ROHs across shorter chromosome fragments (<1 Mb) are indicative of more ancient population processes (Boyko et al., 2010). As our goal was to find ROHs that represent autozygosity rather than simply occur by chance, this analysis was performed using the SNPs pruned for local LD (r2<0.5). In this case, the pruning was performed for each local population separately. All the above analyses were performed in PLINK (Purcell et al., 2007).
Estimation of past demographic changes in European wolf populations
Effective population sizes (NE) were estimated from the equation E(r2)=1/(1+4NE c)+1/n, where r2 is a squared correlation in genotype frequencies between autosomal SNPs (representing the extent of LD), c is the genetic distance between loci in Morgans and 1/n is the adjustment for small sample size (Tenesa et al., 2007). We assumed that 100 Mb=1 Morgan (as for example in Kijas et al., 2012). We estimated average values of r2 in 20 distance classes between 2.5 kb and 1 Mb (corresponding to 0.0025–1 cM). We used the same distance classes as in the LD decay analysis (see Figure 5a), but the smallest distance class was not used here because r2 estimates at small distances may be highly biased (Frisse et al., 2001; Gattepaille et al., 2013). Average r2 value for a particular genetic distance (c) provides a NE estimate t generations ago, where t≈1/(2c) (Hayes et al., 2003). Therefore, the distance classes considered here translate into demographic changes from 50 to 20 000 generations ago, which corresponds to 150–60 000 years ago, assuming a generation time of 3 years (Mech and Seal, 1987). The linear dependence between the recombination distance and time is approximate and holds best when population size is changing linearly (Hayes et al., 2003), which is not the case here (see RESULTS). Therefore, the timing of the demographic changes being inferred here is approximate.
Temporal NE changes were reconstructed for Eastern European wolves (pooled), Iberian and Italian wolves. As the correction for the small sample size was applied, we did not use equal sample sizes, but included all available individuals. However, we compared the results for the Italian population based on 19 and 6 individuals and found them to be similar (see RESULTS). We also estimated the demographic changes for local populations in Eastern Europe (the same as in the LD decay analysis) to compare them with the global estimate for the entire Eastern European population. In addition, the NE estimates were also obtained for the North American wolves. We expected them to have lower NE estimates than Eastern European wolves over time, because of a bottleneck (or, precisely, founder effect) during the colonization of North America from Eurasia (Nowak, 2003).
Estimation of divergence times between the European wolf populations
We used a method of Gautier and Vitalis (2013) implemented in the program KIM TREE, which estimates divergence times on a diffusion time scale (that is, forward in time), conditionally on a population history that is represented as a tree. The most likely tree topology is identified using the deviance information criterion (Spiegelhalter et al., 2002). The branch lengths are estimated as τ≈T/(2NE), where τ is the length of the branch leading to a particular population, NE is the effective size of this population, and T is time (in generations). We used this program to establish the order of splitting events between the three European populations, and the relative temporal distances between them. We also made an attempt to estimate divergence times in generation units as 2NEτ, and then in years assuming a 3-year generation time. However, there was a considerable uncertainty connected with these estimates (see Supplementary Material).
Identification of candidate loci under selection
We used the program BAYESCAN (Foll and Gaggiotti, 2008) to identify candidate loci under natural selection in European wolves. This analysis was performed for the three European populations (Eastern European, Iberian and Italian) using the entire 61K SNP set, but excluding loci that were monomorphic in European wolves (which gave 55 023 SNPs). BAYESCAN applies a Bayesian model developed by Beaumont and Balding (2004). It assumes an island model, where the difference in allele frequencies at each locus between each population and a common gene pool for all the populations is presented as a population-specific FST. Selection is introduced by decomposing FST coefficients for each locus into a population-specific component (β) shared by all loci, and a locus-specific component (α) shared by all populations considered (Foll and Gaggiotti, 2008). For example, three populations with a moderate level of genome-wide differentiation (for example, average FST=0.1), but fixed for three different alleles at a particular locus (locus-specific FST=1) would have a high, positive value of α coefficient for this particular locus. Departure from neutrality is assumed for these loci for which the α component is necessary to explain the observed pattern of diversity at a given locus. This corresponds to α being significantly different from 0, with positive values suggesting diversifying selection, and negative values – balancing or purifying selection (Foll and Gaggiotti, 2008). A threshold value to detect selection was set using a maximum false discovery rate (the expected proportion of false positives) at 0.05. This approach has been assessed as conservative in comparison with other methods of detecting selection (for example, Zhao et al., 2013), but because of the nature of our data (bottlenecked populations) we did not use the relaxed false discovery rate threshold of 0.1 applied elsewhere (for example, Zhao et al., 2013). BAYESCAN accounts for the uncertainty of allele frequency estimates associated with small sample sizes, and therefore it can be applied for very small samples without bias, but with the risk of low power (Foll and Gaggiotti, 2008). Therefore, our analysis has a low risk of detecting false positives, but it is likely that a number loci being under selection will remain undetected. For the SNPs identified as the candidate loci, we performed a search in UCSC Genome Browser for the closest protein-coding genes in CanFam2 dog genome assembly (SNP coordinates were based on this assembly), and also searched for homologous genes identified in humans and other mammals using this browser. Population differentiation at loci putatively under selection was assessed using the PCA implemented in the EIGENSOFT software.
Results
Genetic differentiation among European wolf populations in relation to other Holarctic populations
Population genetic structure at genome-wide loci set
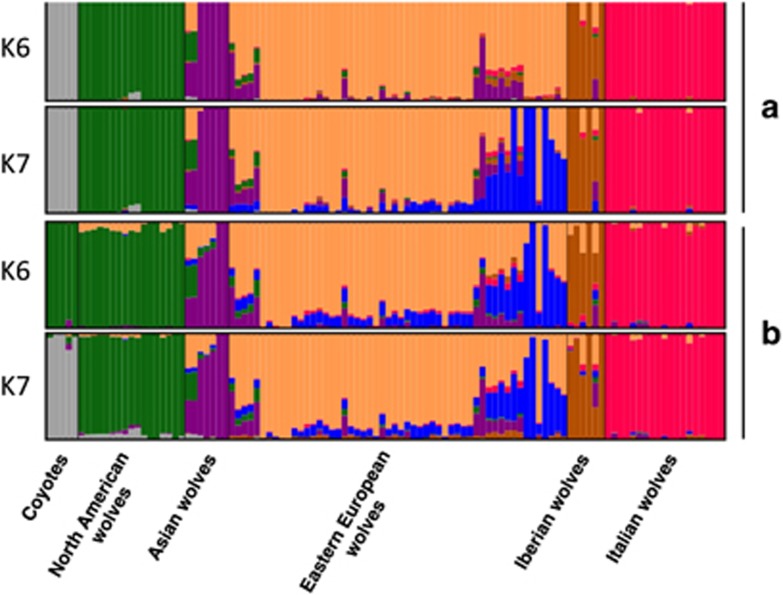
Both STRUCTURE and ADMIXTURE identified Italian wolves as the most distinct population at K=2, with North American canids (grey wolves and coyotes) identified as the third distinct group at K=3 (Supplementary Figure S1). The coyotes were not separated from wolves at K=2 because of the large differences in the sample sizes for these two groups (see DISCUSSION). For larger values of K, the subsequent groups emerged in different order depending on the program used. ADMIXTURE identified coyotes as a distinct group at K=4, and STRUCTURE at K=7. ADMIXTURE identified K=6 as the most informative genetic subdivision, with the clusters corresponding to phylogenetic and geographic subdivision of the samples: Italian, Iberian, Eastern European, Asian and North American wolves, and coyotes. STRUCTURE identified K=7 as the most informative genetic subdivision, both based on the maximum likelihood and the Evanno et al., (2005) method. The clusters identified were the same as in ADMIXTURE for K=6, but with one additional cluster that was represented in most Eastern European individuals as a secondary genetic component.
This cluster constituted the main component of the genetic variability for only three individuals from the Carpathian Mountains, with nine other individuals from the Carpathian Mountains and the Balkans showing levels of admixture with this cluster between 0.27 and 0.48. The same cluster was identified in ADMIXTURE at K=7. The differences in assignment probabilities to these clusters may suggest further differentiation between the Carpathian Mountains and the Balkans. Some Eastern European individuals, in particular those from the easternmost sampling area, that is, the Kirov region in Russia (see Figure 1b) showed mixed ancestry with Asian wolves (Figure 2). Although these results suggest some level of differentiation within Eastern Europe, the separate analysis including only Eastern European wolves detected no population structure (see Supplementary Material), which may be a result of uneven sample distribution and small sample sizes (see DISCUSSION).
Figure 2.
Results of (a) ADMIXTURE and (b) STRUCTURE clustering analysis of European wolf populations in comparison with other wolf populations and the coyotes, for K=6 and K=7. The analysis was performed for the LD-pruned 34K SNP set. Within Eastern Europe, the samples are sorted according to their geographical locations, from the Kirov region in Russia on the left to the Balkans and Carpathians on the right.
Principal component analysis
In the analysis including grey wolves from Europe and other continents as well as coyotes, the first axis (PC1; 8.8% variation) discriminated Italian wolves from other populations (Figure 3a). From positive to negative values, the second axis (PC2, 5.7% variation) separated coyotes, North American grey wolves, Italian wolves, Asian wolves and other European wolves. A similar trend of decreasing values on PC2 was also observed within Eastern European wolves, with individuals from regions geographically more proximate to Asia (from easternmost sampling locations in Russia and Ukraine) placed closer to Asian wolves in the PCA plot. Iberian wolves clustered with Eastern European wolves, but with some separation. The level of differentiation between Eastern European and Italian wolves (FST=0.195) was higher than that between Eastern European and North American wolves (FST=0.114; Figure 3a).
Figure 3.
Principal component analysis illustrating the extent of genetic diversification at the genome-wide 34K SNP set (a–c), and at loci putatively under diversifying selection in European wolves (d, e) among the following populations: (a) and (d) European, Asian and North American grey wolves, and coyotes; (b) and (e) European grey wolves; and (c) Eastern European grey wolves.
The analysis including only European wolves revealed that PC1 (8.3% variation) separated Italian wolves from Eastern European and Iberian wolves, while PC2 (2.9% variation) separated Iberian wolves from the other populations. Within Eastern European wolves, individuals from the Balkans (Bulgaria, Croatia and Greece) and northeastern Europe (Belarus, Latvia, Poland, Russia, Slovakia and Ukraine) formed two distinct subclusters (Figure 3b).
In the analysis including only Eastern European wolves, PC1 (2.1% variation) separated wolves from the Carpathians, the Balkans and northeastern Europe. PC2 (1.8% variation) separated different groups from northeastern Europe, but they were not geographically clustered.
Genetic differentiation among populations
As expected, the highest pair-wise FST values were observed between the coyotes and the grey wolves (Table 1A). Among wolves, the highest FST value (0.293) was observed between Italian and Iberian populations, whereas the lowest FST value (0.059) was observed between Eastern European and Asian populations. The Eastern European population was more divergent from the Italian and Iberian populations than from the Asian and North American populations (Table 1A).
Table 1. Genetic differentiation among grey wolf populations and coyotes calculated in EIGENSOFT based on (A) the 34K data set, and (B) the loci putatively under differential selection in European populations.
|
Grey wolves |
||||||
|---|---|---|---|---|---|---|
| Italy | Iberian Peninsula | Eastern Europe | Asia | North America | Coyotes | |
| (A) | ||||||
| Italy | 0.609 | 1.165 | 1.155 | 1.526 | 1.337 | 1.539 |
| Iberian Peninsula | 0.293 | 0.871 | 1.177 | 1.534 | 1.356 | 1.564 |
| Eastern Europe | 0.195 | 0.128 | 1.098 | 1.477 | 1.311 | 1.513 |
| Asia | 0.229 | 0.167 | 0.059 | 1.623 | 1.584 | 1.745 |
| North America | 0.284 | 0.221 | 0.114 | 0.112 | 1.095 | 1.505 |
| Coyotes | 0.467 | 0.404 | 0.296 | 0.275 | 0.305 | 0.706 |
| (B) | ||||||
| Italy | 1.274 | 3.509 | 4.856 | 4.484 | 4.364 | 4.288 |
| Iberian Peninsula | 0.925 | 0.923 | 2.761 | 2.732 | 2.490 | 2.376 |
| Eastern Europe | 0.848 | 0.758 | 0.938 | 1.233 | 1.136 | 1.559 |
| Asia | 0.796 | 0.694 | 0.086 | 1.290 | 1.279 | 1.605 |
| North America | 0.821 | 0.711 | 0.170 | 0.110 | 1.017 | 1.524 |
| Coyotes | 0.867 | 0.751 | 0.422 | 0.359 | 0.444 | 0.559 |
Above the diagonal: average divergence between populations; on the diagonal: average divergence within populations; below the diagonal: pair-wise FST between populations (in italics). All the pair-wise differences were significant (analysis of variance, P<0.05).
Average divergence values between populations did not follow the same pattern as FST, which was due to the lack of correction for intra-population divergence. Intra-population divergence was low in the Italian and Iberian populations (0.609 and 0.871, respectively), high in the Asian population (1.623), and had intermediate values in Eastern European and North American populations (Table 1A). Genetic differences between each pair of populations were significant (analysis of variance, P<0.0001).
Population structure based on X chromosome data
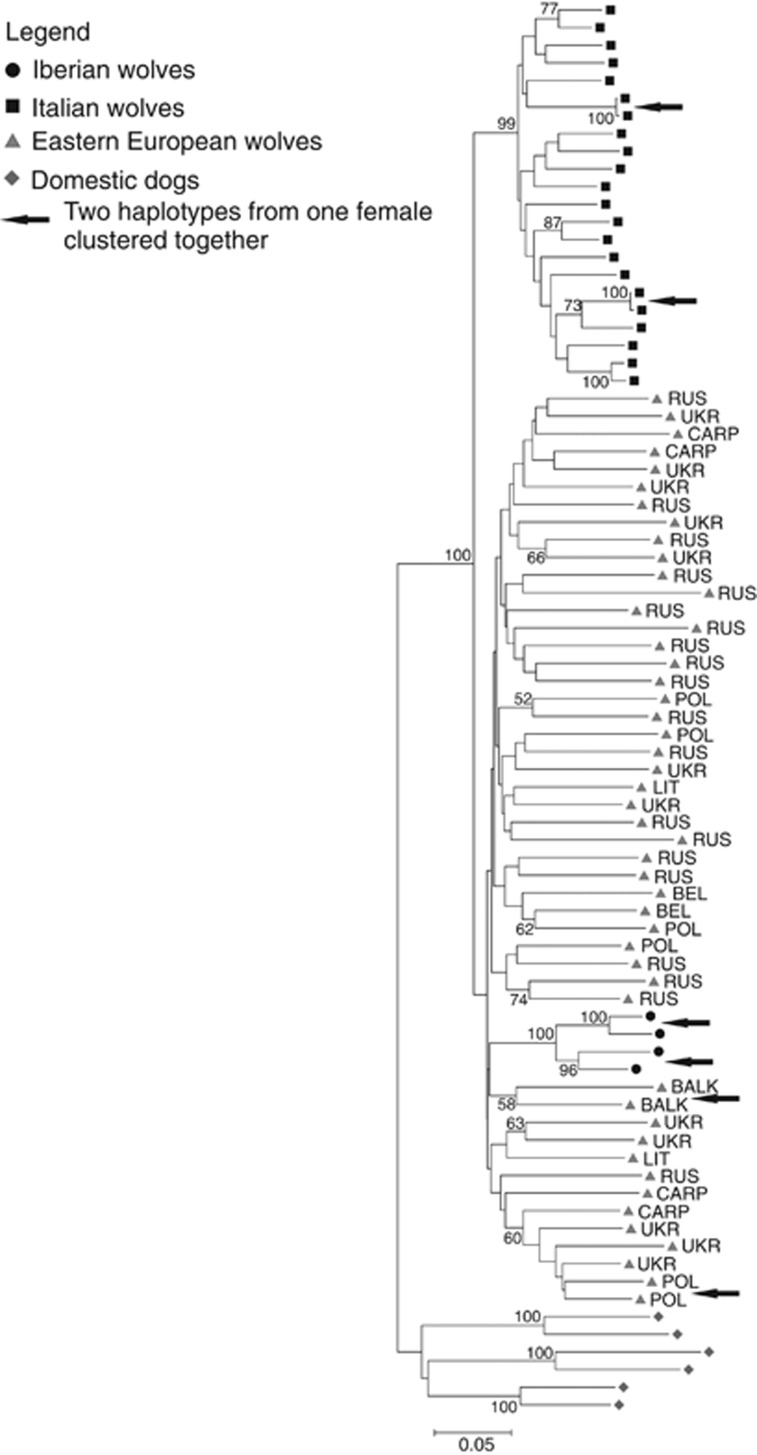
The neighbour-joining tree of female X chromosome haplotypes showed that Italian and Eastern European wolves were grouped in two distinct clades, but only the clade of Italian wolves was supported by the bootstrap analysis (Figure 4). Iberian haplotypes were clustered with Eastern European wolves, forming a distinct subclade with 100% support. There was no clear geographical structure among Eastern European wolves (Figure 4). In the majority of cases, the two X chromosome haplotypes of individual female wolves were not placed next to each other in the tree. The exceptions where two haplotypes of the same individuals were more similar to each other than to any other haplotype included two (100%) Iberian wolves, two (18%) Italian wolves and two (8%) Eastern European wolves.
Figure 4.
Evolutionary relationships among X chromosome haplotypes of females inferred using the neighbour-joining method. The distances were computed using the p-distance measure. Bootstrap support is shown if higher than 50% of 1000 replicates.
ADMIXTURE analysis for female X chromosome data distinguished Italian wolves from Eastern European wolves at K=2, with the two Iberian individuals grouped with Eastern European wolves. At K=3 (which was indicated as the most likely genetic structure), two clusters were identified for Eastern European wolves: one comprised of Carpathian and Balkan individuals, and second of the individuals from northeastern Europe. These clusters were consistent with those detected using the autosomal loci set.
Heterozygosity, linkage disequilibrium and autozygosity in local populations of European wolves
All local wolf populations from Eastern Europe had comparable levels of heterozygosity (HO=0.21–0.24, HE=0.22–0.26; Table 2), whereas populations from southwestern Europe exhibited lower heterozygosity (Iberian Peninsula: HO and HE=0.17; Italy: HO and HE=0.16). Eastern European wolves had low to moderate levels of LD (LD decayed below r2=0.5 between 2.5 and 10 kb), as expected for populations that have not experienced severe bottlenecks. The southernmost population from the Balkans had the highest LD levels within Eastern European populations (Figure 5a). In contrast to these populations, the Iberian population had high LD levels (257 kb), consistent with bottlenecks and subsequent inbreeding. In the Italian population, LD did not decay below 0.5 for the entire range of distances considered (up to 1 Mb), suggesting more severe and/or a longer bottleneck as compared to the Iberian population (Table 2, Figure 5a).
Table 2. Heterozygosity, linkage disequilibrium and autozygosity in local wolf populations from Europe.
| Local population | HO (s.e.) | HE (s.e.) | Distance (r2<0.5) (kb) | Average no. of homozygous segments per individual (s.e.) | Average length of homozygous segments (Kb) per segment per individual (s.e.) |
|---|---|---|---|---|---|
| S Poland and S Belarus | 0.235 (0.0010) | 0.232 (0.0008) | 5.00 | 2.7 (0.6) | 3634 (432) |
| N Poland | 0.234 (0.0010) | 0.220 (0.0008) | 10.00 | 2.2 (0.3) | 2950 (631) |
| N Belarus | 0.233 (0.0010) | 0.263 (0.0010) | 3.75 | 5.0 (1.1) | 4378 (1105) |
| NE Russia | 0.232 (0.0010) | 0.219 (0.0008) | 3.75 | 3.3 (0.8) | 3696 (1107) |
| Kirov region, Russia | 0.230 (0.0010) | 0.233 (0.0008) | 7.50 | 2.4 (0.9) | 3080 (773) |
| S Russia and E Ukraine | 0.228 (0.0009) | 0.233 (0.0008) | 2.50 | 4.7 (1.1) | 3502 (844) |
| Balkans | 0.217 (0.0010) | 0.223 (0.0008) | 10.00 | 3.2 (1.5) | 1771 (631) |
| Carpathians | 0.214 (0.0010) | 0.257 (0.0010) | 7.50 | 6.5 (0.7) | 5142 (399) |
| Iberian Peninsula | 0.173 (0.0010) | 0.169 (0.0008) | 275.00 | 1.5 (0.8) | 1902 (928) |
| Italy | 0.161 (0.0010) | 0.155 (0.0010) | >1000.00 | 1.6 (0.7) | 2449 (937) |
The extent of linkage disequilibrium is measured as the average distance between loci at which r2 falls below 0.5. Autozygosity is measured as average number of homozygous segments per individual and their average length.
Figure 5.
(a) Extent of linkage disequilibrium in European wolf populations. Average genotypic association coefficient r2 is presented as a function of inter-SNP distance for each local wolf population. (b) Frequency distribution of runs of homozygosity in European wolf populations. (c) Temporal changes of NE in European wolves, with North American wolves presented for a comparison.
Despite high LD levels in the Iberian and Italian populations, they had fewer fragments of ROH >1 Mb than Eastern European populations (Table 2, Figure 5b). In contrast, some Eastern European populations, such as the Carpathians, Northern Belarus, and Southern Russia/Eastern Ukraine, had an elevated number of ROH fragments of size 1–5 Mb.
Past demographic changes in European wolf populations
LD-based estimates suggest that effective population sizes of both European and North American wolves declined over the entire period considered (Figure 5c and Supplementary Figure S2). NE estimates for the Italian and Iberian populations were considerably lower as compared with the Eastern European population in each time interval (Figure 5c and Supplementary Figure S2). The most recent effective population sizes (at about 150 years ago) were estimated at 1366 for Eastern Europe, 71 for Italy and 59 for the Iberian Peninsula. The most ancient estimates (at about 60 000 years ago) were: ∼20 000 for Eastern Europe, ∼4500 for Italy and ∼10 000 for the Iberian Peninsula (Supplementary Table S2). Prior to the divergence of the European populations (which most likely occurred within the timeframe considered), their NE should be the same, which is not observed. This may be interpreted as an evidence for long-term bottlenecks in the Italian and Iberian populations (see DISCUSSION) or ancient population structure. NE estimates for the North American wolves (most recent: 358, most ancient: ∼18 000) do not converge on those of Eastern European wolves either, which may reflect a more complex demographic history of North America, including multiple founder effects and bottlenecks associated with glaciation events (Nowak, 2003). Most local groups of Eastern European wolves do not converge to the effective size of the total Eastern European population (Figure 5c), which may result from population structure (Pilot et al., 2006) and/or local bottlenecks.
Divergence times between the European wolf populations
The most likely tree topology inferred using the Kim Tree program suggests that the Iberian population diverged first from the common ancestor of all populations considered, which was followed by the split between the Italian and Eastern European populations (Supplementary Figure S3A). However, small differences in deviance information criterion values (92–227) between the alternative topologies and a very short internal branch (Supplementary Figure S3A) suggest that the splits between these three populations occurred within a short time period, and the topology is close to star-shaped.
We added North American grey wolves to the most-likely tree topology of the three European populations, assuming the reciprocal monophyly between European and North American wolves (as shown in vonHoldt et al., 2011). Using the time of flooding of the Bering Land Bridge (11 000 yBP, Keigwin et al., 2006), which separated Eurasian and North American wolves as a calibration point, we obtained the conservative estimates of divergence of European populations from their most recent common ancestor at 3200–5600 years ago (SD 33–123 years) (Supplementary Table S3, Supplementary Figure S3B). These estimates have considerable uncertainty resulting from a number of assumptions (see Supplementary Material for details), and therefore should be treated with caution.
Identification of candidate loci under selection
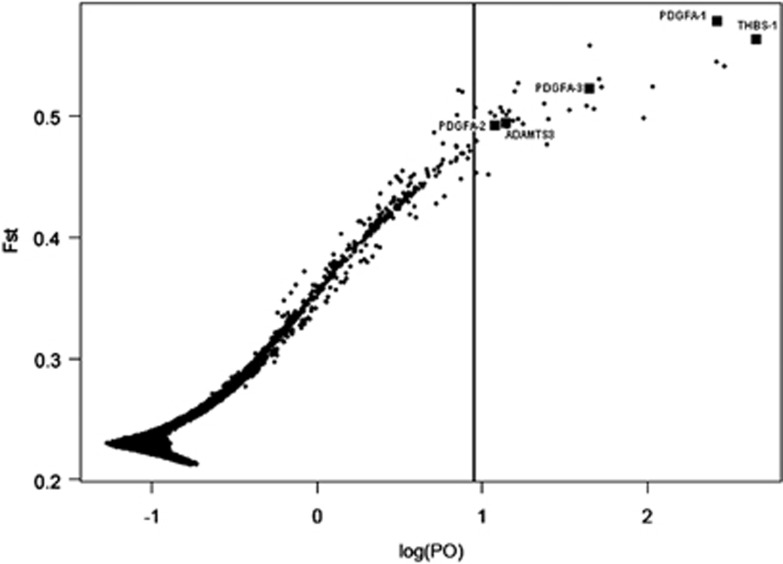
Using a 5% false discovery rate threshold, we identified 35 outlier SNPs (Figure 6, Supplementary Table S4). This threshold corresponded to posterior odds (PO) of 8.94 and false non-discovery rate (the expected proportion of false negatives) of 0.094, and P=0.90, respectively. Thirty-one of these outliers fitted within a threshold of PO<10. Each of the 35 outliers had positive α values between 1.27 and 2.03, suggestive of diversifying selection. FST coefficient averaged over populations ranged from 0.45 to 0.58 compared to the average value of 0.21 among the genome-wide 34K loci (Supplementary Figure S4). None of these 35 outlier loci showed evidence for directional selection within Eastern European wolves (see Supplementary Material).
Figure 6.
Signatures of selection in the Iberian, Italian and Eastern European wolf populations inferred using the program BAYESCAN. The vertical axis indicates mean FST values between each of the three populations, and the horizontal axis indicates the logarithm of posterior odds (log(PO)). The vertical line indicates the log(PO) value corresponding to the false discovery rate threshold of 0.05. Loci on the right of this line are putatively under selection.
A search of the CanFam2 dog genome assembly in the UCSC Genome Browser for the closest protein-coding genes indicated that two outlier SNPs from chromosome 6 are flanking the coding region of platelet-derived growth factor, alpha polypeptide (PDGFA). The first SNP (further referred to as locus PDGFA-1) was 4.6 kb downstream from the chromosomal fragment marked as a coding region, and the second SNP (PDGFA-2) was 30.7 kb upstream. Locus PDGFA-1 was among five loci with PO>100 (corresponding to P>0.99) and had the highest α value of all loci (α=2.03) and the highest level of differentiation (FST=0.58). In addition, one more putatively selected locus (PDGFA-3) was 425 kb upstream from this chromosomal fragment.
For the remaining 32 SNPs identified as putative loci under selection, we found adjacent regions analogous to genes described in the human genome and other mammalian genomes (Supplementary Table S4), which have not been annotated for the dog yet. One of these loci, which had the highest PO value and second highest FST of all the loci putatively under selection, was placed within a sequence analogous to thrombospondin type 1 gene (THBS1), which was annotated in humans, mice and rats. Another locus was placed within a sequence analogous to metallopeptidase with thrombospondin type 1 motif (ADAMTS3), which was annotated in humans, mice, rats and cows. Functions of thrombospondin type 1 include angiogenesis, apoptosis, and activation of transforming growth factor beta.
Population differentiation at loci putatively under selection
The PCA plot representing genetic differentiation among worldwide grey wolf populations and coyotes at loci putatively under selection showed different patterns as compared with that obtained for the 34K data set. While separation of Italian wolves from other wolves and coyotes at PC1 was consistent with the 34K data set, at PC2, Iberian wolves were the most distinct population, and they were more similar to coyotes than to Eastern European wolves (Figure 3d). There was no clear distinction between Eastern European, Asian and North American wolves. At PC1, PDGFA-3 and PDGFA-1 were the first and the third of loci showing the highest level of differentiation among populations. On PC3, which distinguished the coyotes from the grey wolves, PDGFA-2 showed the second highest level of differentiation among populations, after another locus from the same chromosome, but more distant from PDGFA gene.
The PCA plot representing the differentiation among the three European populations at loci putatively under selection using the PCA method showed a similar pattern as compared with the differentiation at 34K loci (Figure 3e), but as expected, with much stronger differentiation. For example, PC1 (distinguishing Italian and Eastern European wolves) explained 42.9% of genetic variation versus 8.3%. Similarly, PC2 (distinguishing Iberian wolves from the two other populations) explained 5.7% of genetic variation versus 2.9%. Differentiation between northeastern and southeastern Europe observed for the 34K data set was not observed here. At PC1, two loci showing the highest level of differentiation among populations were PDGFA-1 and PDGFA-3. At PC3, which distinguished the Italian wolves from other European wolves, PDGFA-2 showed the highest level of differentiation among populations.
As expected in case of diversifying selection, pair-wise FST values between the European populations were highly elevated (0.758–0.925), with the highest level of differentiation between the Italian and Iberian populations. A less obvious effect was the substantial elevation of FST values between the coyotes and each of the grey wolf populations (0.359–0.867; Table 1B).
Discussion
Genetic differentiation among European wolf populations in relation to other Holarctic populations
We detected six genetically distinct groups within the analysed data set, which were consistent with species-level and geographic subdivision of the samples. Specifically, Italian, Iberian, Eastern European, Asian and North American wolves, and coyotes formed distinct clusters. The population structure based on X chromosome haplotypes confirmed the high level of genetic differentiation among the three main European populations.
The genetic distinctiveness of the Italian, Iberian and Eastern European populations was expected given their geographic isolation and likely near complete lack of gene flow for at least the last 100 years (Lucchini et al., 2004), except for the last decade of wolf population expansion in Western Europe (Sastre, 2011; Fabbri et al., in press). These three populations spatially correspond to different glacial refugia: the Apennine and Iberian refugia for the two southwestern populations, and the Balkan refugium for the southeastern (Balkan) population (with northeastern European population possibly having a different or mixed origin-see Pilot et al., 2010). It has been unclear, though, whether the distinctiveness of these populations results from their long-term isolation or recent geographical separation resulting from extinction of the wolf in central-western Europe. The wolf range in Europe during the Last Glacial Maximum was not reduced to the southern refugia (see Sommer and Benecke, 2005), so the effect of Pleistocene glaciations on population structuring in this species may be overestimated. Our estimates support the ancient divergence of the three European populations (5600–3200 years ago), but this date is considerably later than the Last Glacial Maximum (∼20 000 years ago). Although this estimate has considerable uncertainty, when combined with other evidence it implies a new hypothesis concerning the events leading to the divergence of these populations (see below).
All the methods of population structure analysis indicate that the Italian population is the most genetically distinct of the three European populations considered here. This is consistent with an inference based on mtDNA data from modern and ancient European wolves, suggesting historic gene flow between Eastern Europe and the Iberian Peninsula through intermediate populations, and longer-term isolation of wolves in the Apennine Peninsula (Pilot et al., 2010). An analysis based on microsatellite loci also suggested the isolation of the Italian wolf population for thousands of generations (Lucchini et al., 2004).
In contrast, the genealogy of the European populations inferred using the Kim Tree method suggests that the Iberian population diverged first from the ancestral European population, which was followed by the divergence between the Italian and Eastern European populations. However, the support for this tree topology over the alternative topologies is weak, and the internal branch is short, suggesting that the splits between these three populations occurred within a short period, and the topology is close to star-shaped.
The effect of sampling on the analysis of population structure
PCA suggested some level of differentiation within the Eastern European wolves, as the Carpathian, Balkan and northeastern populations formed distinct sub-clusters. However, this separation was not well supported by Bayesian clustering methods. These methods detected two genetic clusters within Eastern Europe, one prevailing in northeastern Europe and another in the Carpathians and the Balkans, but with high level of admixture. Lack of clear, geographically clustered genetic subdivision within the Eastern European wolves contrasted with an earlier study that showed cryptic population structure in this region based on 14 microsatellite loci and mtDNA variability (Pilot et al., 2006), which was subsequently confirmed based on an independent sample set collected from a smaller area (Czarnomska et al., 2013). The discrepancy is likely due to much lower sample coverage, as 54 Eastern European wolves were analysed here versus 643 wolves in Pilot et al. (2006). In this case, the result based on a small number of loci, but large sample size is more reliable, which demonstrates the importance of the sample size in population structure studies, regardless of the number of loci.
The effect of sample size was also evident in the analysis of coyote data. Although their distinctiveness from grey wolf populations was clearly reflected in pair-wise FST values, it was less clear based on PCA and population structure plots. As grey wolves predominated in the sample and only five coyotes were tested, subdivisions within grey wolves dominated the results. By comparison, a study that analysed the same SNP data with a more balanced number of grey wolves and coyotes (vonHoldt et al., 2011) identified a clear distinction between these species, consistent with past phylogenetic studies (for example, Vilà et al., 1999; Lindblad-Toh et al., 2005). This is consistent with the simulation study showing that variation in sample size may affect the population clustering inferred in STRUCTURE (Kalinowski, 2011).
Although small sample sizes may affect the reliability of genetic structure analysis, the availability of a large number of loci with uniform genome-wide distribution enables other analyses that are largely independent of the sample sizes. Genome-wide data proved to be very effective in reconstructing past demographic changes and detecting signatures of selection based on small sample sizes (for example, Jones et al., 2012; Keller et al., 2013), which may be reduced even to single individuals when high-coverage genome sequences are available (for example, Miller et al., 2012; Zhao et al., 2013; Freedman et al., in press).
Genetic diversity and linkage disequilibrium in European wolf populations: detecting genome-wide signatures of population bottlenecks
Eastern European wolves had levels of heterozygosity comparable with large grey wolf populations from Canada and northwestern United States, which have a history of constant or recently expanding population size (vonHoldt et al., 2011). Italian and Iberian wolves had decreased heterozygosity and higher LD levels as compared with Eastern European wolves, which is consistent with earlier studies that reported signatures of bottlenecks in these populations based on microsatellite loci analysis (Lucchini et al., 2004; Sastre et al., 2011). Despite high LD levels, both the Italian and Iberian population had fewer ROHs over 1 Mb in length as compared to Eastern European wolves, suggesting that the high LD levels are likely due to ancient bottlenecks rather than recent inbreeding. Consistent with this result, the levels of observed and expected heterozygosity were comparable in both the Italian and Iberian population, while in the recently bottlenecked Mexican wolf population observed heterozygosity was much lower than expected (0.12 versus 0.18), implying recent inbreeding (vonHoldt et al., 2011).
LD levels in Italian and Iberian populations were also lower as compared to Mexican wolves and a small, isolated, recently founded population from the Isle Royale National Park (vonHoldt et al., 2011). These two North American wolf populations also had the highest fraction of autozygous segments across all chromosomal fragment sizes of all populations of North-American wolf-like canids (vonHoldt et al., 2011). This contrasts with the Italian and Iberian wolves, for which autozygosity levels are low compared with Eastern European populations. The analysis of genome-wide variability thus shows a clear distinction between populations that are inbred due to recent drastic demographic declines or founder events such as the Mexican and Isle Royale wolves, respectively, as compared with populations that have reduced levels of genetic variability due to long-term isolation and low population sizes lasting for a large number of generations, such as the Italian and Iberian wolves. Populations with these two different types of demographic history have been designated as ‘bottlenecked'. Here, we show that there is a clear difference in the genomic signature of their demographic histories. This result has important implications for studies where a genetic analysis is the only source of information on demographic history.
Analysis of phylogenetic relationships among female X chromosome haplotypes showed that all the female wolves from the Iberian Peninsula and two of the 11 female wolves from Italy had haplotypes that were more related to each other than to any other haplotype. This suggests that these populations have an increased probability of forming mating pairs between individuals sharing a recent common ancestry (even if not directly related). This is expected for populations that have experienced isolation and long-term bottlenecks. In contrast, in Eastern Europe, only two out of 24 individuals carried X chromosome haplotypes showing close phylogenetic similarity, while in other cases, haplotypes from distant locations were phylogenetically related. This result is consistent with substantial gene flow between different parts of Eastern Europe, which may counterbalance the effects of recent local inbreeding (see below).
The Italian population had lower variability as compared with the Iberian population (although more individuals were analysed), consistent with earlier studies based on mtDNA and microsatellite loci (Vilà et al., 1999; Pilot et al., 2010; Sastre et al., 2011). Moreover, the Italian population had higher LD levels as compared with the Iberian population, an indication of longer and/or more severe bottleneck events in Italian wolves. This finding is consistent with the conclusion based on population structure analyses, and with an earlier study suggesting long-term isolation of the Italian population based on microsatellite data (Lucchini et al., 2004). In contrast, mtDNA haplotype sharing between Iberian and Eastern European wolves suggested more recent gene flow between Iberian and Eastern European wolves, most likely through now-extinct intermediary populations (Pilot et al., 2010). The present study showed high pair-wise population divergence estimates between the Eastern European population and both Italian and Iberian populations, and the divergence between the Italian and Iberian populations is the highest of all pairs of the wolf populations studied. This inconsistency between genetic and geographical distance may be a result of strong genetic drift during population bottlenecks in the Iberian and Apennine Peninsulas. In contrast with the Italian and Iberian populations, wolves from some Eastern European regions had elevated levels of ROH, suggesting recent inbreeding. This was likely connected with the disruption of pack structure due to strong hunting pressure (for example, see Jędrzejewski et al., 2005). In one of the regions with elevated ROH levels, Northern Belarus, strong hunting pressure has been well documented (Sidorovich et al., 2003).
Past demographic changes in European wolf populations
Effective population sizes of European and North American wolves inferred from the LD patterns declined over the entire period considered (60 000 to 150 years ago). This is consistent with the growing evidence from ancient DNA studies showing that large mammal species experienced a considerable loss of genetic diversity since the late Pleistocene (reviewed in Hofreiter and Barnes 2010). In particular, the loss of mtDNA haplotypes has been documented in North American (Leonard et al., 2007) and European grey wolves (Pilot et al., 2010), and this was correlated with the loss of morphological and ecological diversity (Leonard et al., 2007; Germonpré et al., 2009).
While a general trend of NE decline in time is consistent with the expectation, we also expected a signal of population growth after the Last Glacial Maximum, reflecting the spatial expansion to the areas previously covered by the retreating ice sheet. The spatial expansion has been documented based on the sub-fossil record (Sommer and Benecke 2005), but it is possible that it was not accompanied by a substantial demographic expansion, for example, due to declines of large herbivore prey (see Hofreiter and Barnes 2010) and exponential growth of the human population (see for example, McEvoy et al., 2011). The demographic reconstruction based on high-coverage genome sequences shows a continuous decline of wolf populations in Europe, Middle East and East Asia since ∼20 000 years ago until the present (Freedman et al., in press). This is consistent with our result, but also shows that our upper time limit of 60 000 years for the decline may be overestimated due to an imprecision of time estimates based on recombination distance.
NE estimates in the most recent time period considered (∼150 years ago) show a good correspondence with estimates for the contemporary (21st century) populations. Sastre et al. (2011) reports NE ∼50 (43–54) for the contemporary Iberian population, which corresponds well with our NE estimate of 59 individuals about 150 years ago. The contemporary NE estimate for northeastern part of European Russia (138–312.5; Sastre et al., 2011) is also consistent with our estimates for three local populations from this region (159 in NE Russia, 224 in S Russia/E Ukraine and 239 in N Belarus). Importantly, the contemporary NE estimates result in NE to census size ratio of about 0.11 in Russia and 0.025 in the Iberian Peninsula, suggesting a severe bottleneck and/or an overestimation of the current census size in the Iberian population (Sastre et al., 2011).
Prior to the divergence of the European populations (which took place within the considered timeframe – see below), their NE estimates should converge, which is not observed. In the analogous analysis carried out for humans, NE estimates for non-African populations are lower than those of African populations instead of converging to the same values prior to the divergence time (McEvoy et al., 2011). This pattern was interpreted as a signature of the ‘out of Africa' bottleneck (McEvoy et al., 2011). A drastic reduction of population size inflates r2 estimates even for the small distance classes (representing distant time periods), leading to an underestimation of NE before the bottleneck (McEvoy et al., 2011). Therefore, the patterns observed in the Italian and Iberian populations may be interpreted as an evidence for bottlenecks, with the more severe bottleneck in the Italian population as compared with the Iberian population.
The timing of these bottlenecks cannot be inferred from the LD patterns. Continuous population decline observed for each population suggests that there was no recovery phase that would have marked the end of the bottleneck period. However, the timing of a strong bottleneck is expected to coincide with coalescence of lineages involved in this bottleneck, resulting in a genealogy with short internal branches close to the root (Gattepaille et al., 2013). The genealogy reconstructed for the European wolves has this topology, so it may be expected that the time of their divergence corresponds with a bottleneck period, or with an onset of a long-term bottleneck. This time was estimated at 5600–3200 years ago, which corresponds to the late Neolithic in Europe.
There is a considerable uncertainty associated with this estimate, resulting from a number of assumptions made. For example, we made an unrealistic assumption that there was no or little gene flow between the populations after the split, and therefore the divergence times are likely to be underestimated (see Gautier and Vitalis, 2013). However, in consistence with other evidence from this and earlier studies (for example, Lucchini et al., 2004), this estimate shows that the population bottlenecks in Italian and Iberian wolves were ancient rather than recent. Possibly, they could have resulted from the Neolithic expansion of the human population (for example, Bocquet-Appel, 2011) leading to increased hunting pressure and competition for resources (large game species) with humans, as well as habitat loss due to agricultural expansion. Human population growth and habitat loss have continued until present, preventing the recovery of wolf populations from past bottlenecks, which may explain the observed pattern of continuous decline. Contemporary expansion of the wolf populations in Europe (for example, Boitani, 2003; Randi, 2011), largely resulting from their release from hunting pressure, is too recent to be detected from LD patterns.
Signatures of diversifying selection among European populations
In populations that have experienced recent bottlenecks, large numbers of loci may display low levels of heterozygosity as a result of genetic drift, and therefore directional selection may be difficult to detect (for example, Axelsson et al., 2013). To account for this problem, we considered outliers in the empirical distribution as candidate targets of selection, and established a conservative outlier threshold. In addition, we compared variation at putatively selected loci in the populations for which selection test has been performed to that in non-tested populations, expecting that signatures of selection will be consistent across multiple populations or across closely related species, as has been shown in other studies (for example, Hohenlohe et al., 2010; Zulliger et al., 2013).
We identified 35 putative loci under diversifying selection among 55K SNPs tested. These estimates are conservative and associated with a nearly 10% false non-discovery rate. For most of the outlier SNPs, appropriately annotated genome data was unavailable and as a result, associations with particular genes are uncertain. However, three outlier SNPs were flanking the coding region of the canine platelet-derived growth factor, alpha polypeptide (PDGFA) gene. The presence of these three loci with the strong signature of selection near this gene (one of which had the highest FST from all the loci analysed; Figure 6) makes it a strong candidate gene under diversifying selection among wolf populations. This gene takes part in numerous developmental processes (Alvarez et al., 2006). Importantly, it interacts with insulin-like growth factor-1(IGF1) in the development of bone and cartilage tissues, which was described in humans (for example, Schmidt et al., 2006; Bassem and Lars, 2011) and dogs (Stefani et al., 2000). Sutter et al. (2007) found that a single allele of the IGF1 gene determines small size in dogs and this gene shows a signature of intense artificial selection. The small size allele was absent from a large worldwide sample of grey wolves (Gray et al., 2010), and we found no signature of selection on IGF1 in wolves. Consequently, rather than IGF1, PDGFA may be a major gene influencing body size differences observed in European grey wolves (see below). However, it should be noted that differences in body size between wolf populations across Europe are small as compared with differences between dog breeds.
In addition, a SNP that had the highest PO value was placed within a sequence analogous to human thrombospondin type 1 gene, and another SNP was located within a sequence analogous to human ADAMTS3 gene with thrombospondin type 1 motif. Thrombospondin type 1 takes part in a number of developmental processes, including activation of TGFβ, another growth factor produced by platelets and involved in bone development (for example, Reddi and Cunningham, 1990). Thus, diversifying selection on the European wolf populations appears to involve two different growth factors that possibly may be associated with differentiation of body size and shape.
The Italian and the Iberian wolf have been recognized as separate subspecies Canis lupus italicus (Altobello, 1921) and Canis lupus signatus (Cabrera, 1907) on the basis of morphological differences including overall body size, coat coloration and cranial measurements (Cabrera, 1907; Altobello, 1921; Vilà, 1993; Nowak and Federoff, 2002). Although body size differences across Europe are not large (Vilà, 1993) and may be due to phenotypic plasticity or genetic drift resulting from long-term isolation, it is also possible that they reflect local adaptation. Smaller body size in grey wolves may have a selective advantage in habitats with smaller prey (MacNulty et al., 2009), and the three European populations occupy distinct habitats that differ in species composition and the relative abundance of ungulate prey. Smaller species like the roe deer (Capreolus capreolus) and the wild boar (Sus scrofa) are common in the wolf diet in the Iberian Peninsula and Italy (for example, Barja, 2009; Mattioli et al., 2011), whereas larger prey such as the red deer (Cervus elaphus) and moose (Alces alces) are more frequent in the wolf diet of northeastern Europe (Jędrzejewski et al., 2010).
Importantly, although selection was inferred using the European data set only, the patterns of population differentiation at putatively selected loci among worldwide grey wolves and coyotes were substantially different when compared with that obtained for the 34K data set. Particularly striking is the position of the coyotes on the PCA plot (Figure 3d), showing reduced relative distance between this species and Iberian and Italian wolves as compared with the 34K data set. Coyotes are smaller than North American grey wolves and feed on smaller prey species, and their natural geographic range was south of the grey wolf range (Gompper, 2002). Therefore, parallel patterns of diversifying selection may exist among European grey wolves and North American large canids. The contrasting pattern between the putatively selected loci and genome-wide loci may reflect parallel adaptation involving the same genes. Further study is required to assess the role of these candidate genes in the adaptive diversification of wolf-like canids, which could involve DNA and protein sequence characterization in multiple populations, analysis of gene expression, quantitative analysis of relevant phenotypic traits and possibly functional in vitro studies.
Conclusions
Our analysis of genome-wide variability provided new insights into the evolutionary history of the grey wolf in Europe, revealing continuous population declines since the Late Pleistocene as well as long-term isolation and demographic bottlenecks in southwestern Europe. Eastern European wolves show more genetic similarity to Asian wolves than to Italian and Iberian wolves, and Italian wolves are particularly distinct from other wolf populations. This pattern results from strong genetic drift and does not reflect phylogenetic relationships among lineages. The Italian and Iberian populations show the genomic signature of long-term bottlenecks, which is clearly different from recent drastic population declines or founder events such as in the Mexican and Isle Royale wolves. The fact that these demographic histories can be distinguished based on genomic data may be important in cases where genetic variability is the only source of information.
We detected 35 loci putatively under diversifying selection between the three main European populations. Two of these loci were within 31 kb from the canine PDGF gene, which may influence differences in body size between wolves from eastern and southwestern Europe. The contrasting pattern of genetic differentiation among the populations of grey wolves and the coyotes at the putatively selected versus genome-wide loci may reflect parallel adaptation involving the same genes, a possibility that should be explored by resequencing studies of both species.
Data archiving
The genotyping data from the CanMap project are available at http://genome-mirror.bscb.cornell.edu/cgi-bin/hgGateway (see ‘SNPs' track under the Variations and Repeats heading).
Acknowledgments
We thank M Shkvyrya, I Dikiy, E Tsingarska, S Nowak and M Apollonio for providing samples from European wolves used in this project, A Moura for help with preparing the figures, and M Gautier for sharing the R script to plot the outputs from the Kim Tree program. We are grateful to Giorgio Bertorelle, Michael Bruford and four anonymous reviewers for their constructive comments on the manuscript. This project was supported by grants from the Foundation for Polish Science (MP), the Polish Committee for Scientific Research (MP and WJ), the US National Science Foundation (RKW), the Intramural Program of the National Human Genome Research Institute (EAO), the Italian Ministry of Environment and the Italian Institute for Environmental Protection and Research (ER and CG).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Allendorf FW, Hohenlohe PA, Luikart G. Genomics and the future of conservation genetics. Nat Rev Genet. 2010;11:697–709. doi: 10.1038/nrg2844. [DOI] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altobello G.1921[Fauna of Abruzzo and Molise] Mammiferi 438–45.[In Italian]. [Google Scholar]
- Alvarez RH, Kantarjian HM, Cortes JE. Biology of platelet-derived growth factor and its involvement in disease. Mayo Clin Proc. 2006;81:1241–1257. doi: 10.4065/81.9.1241. [DOI] [PubMed] [Google Scholar]
- Axelsson E, Ratnakuma A, Arendt ML, Maqbool K, Webster MT, Perloski M, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- Barja I. Prey and prey-age preference by the Iberian wolf Canis lupus signatus in a multiple-prey ecosystem. Wildlife Biol. 2009;15:147–154. [Google Scholar]
- Bassem MD, Lars S. Regulation of human adipose-derived stromal cell osteogenic differentiation by insulin-like growth factor-1 and platelet-derived growth factor-alpha. Plast Reconstr Surg. 2011;127:1022–1023. doi: 10.1097/PRS.0b013e318200ad65. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- Bocquet-Appel JP. When the world's population took off: the springboard of the Neolithic Demographic Transition. Science. 2011;333:560–561. doi: 10.1126/science.1208880. [DOI] [PubMed] [Google Scholar]
- Boitani L.2003Wolf conservation and recoveryIn Mech LD, Boitani L (eds).Wolves: Behavior, Ecology, and Conservation The University of Chicago Press: Chicago; 317–340. [Google Scholar]
- Boyko AR, Quignon P, Li L, Schoenebeck J, Degenhardt JD, Lohmueller KE, et al. A simple genetic architecture underlies quantitative traits in dogs. PLoS Biol. 2010;8:e1000451. doi: 10.1371/journal.pbio.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera A. Los lobos de Espana. Bol R Soc Esp Hist Nat. 1907;7:193–198. [Google Scholar]
- Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Feled-Alon A, York TL, et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3:e163. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnomska SD, Jędrzejewska B, Borowik T, Niedzialkowska M, Stronen AV, Nowak S, et al. Concordant mitochondrial and microsatellite DNA structuring between Polish lowland and Carpathian Mountain wolves. Cons Genet. 2013;14:573–588. [Google Scholar]
- Earl DA, vonholdt BM. Structure Harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Fabbri E, Caniglia R, Kusak J, Galov A, Gomerčić T, Arbanasić H, et al. Genetic structure of expanding wolf (Canis lupus) populations in Italy and Croatia, and the early steps of the recolonization of the Eastern Alps Mamm Biol(in press).
- Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman AH, Schweizer RM, Gronau I, Han E, Ortega – Del Vecchyo D, Silva PM, et al. Genome sequencing highlights the dynamic early history of dogs PLoS Genet(in press). [DOI] [PMC free article] [PubMed]
- Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Di Rienzo A. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet. 2001;69:831–843. doi: 10.1086/323612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattepaille LM, Jakobsson M, Blum MG. Inferring population size changes with sequence and SNP data: lessons from human bottlenecks. Heredity. 2013;110:409–419. doi: 10.1038/hdy.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M, Vitalis R. Inferring population histories using genome-wide allele frequency data. Mol Biol Evol. 2013;30:654–668. doi: 10.1093/molbev/mss257. [DOI] [PubMed] [Google Scholar]
- Geffen E, Anderson MJ, Wayne RK. Climate and habitat barriers to dispersal in the highly mobile grey wolf. Mol Ecol. 2004;13:2481–2490. doi: 10.1111/j.1365-294X.2004.02244.x. [DOI] [PubMed] [Google Scholar]
- Germonpré M, Sablin MV, Stevens RE, Hedges REM, Hofreiter M, Stiller M, et al. Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: osteometry, ancient DNA and stable isotopes. J Arch Science. 2009;36:473–490. [Google Scholar]
- Gomercic T, Sindicic M, Galov A, Arbanasic H, Kusak J, Kocijan I, et al. High genetic variability of the grey wolf (Canis lupus L.) population from Croatia as revealed by mitochondrial DNA control region sequences. Zool Stud. 2010;49:816–823. [Google Scholar]
- Gompper ME. Top carnivores in the suburbs? Ecological and conservation issues raised by colonization of north eastern North America by coyotes. Bioscience. 2002;52:185–190. [Google Scholar]
- Gray MM, Sutter NB, Ostrander EA, Wayne RK. The IGF1 small dog haplotype is derived from Middle Eastern grey wolves. BMC Biol. 2010;8:1–13. doi: 10.1186/1741-7007-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes BJ, Visscher PM, McPartlan HC, Goddard ME. Novel multilocus measure of linkage disequilibrium to estimate past effective population size. Genome Res. 2003;13:635–643. doi: 10.1101/gr.387103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M, Barnes I. Diversity lost: are all Holarctic large mammal species just relict populations. BMC Biol. 2010;8:46. doi: 10.1186/1741-7007-8-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jędrzejewska B, Jędrzejewski W, Bunevich AN, Minkowski L, Okarma H. Population dynamics of wolves Canis lupus in Bialowieza primeval forest (Poland and Belarus) in relation to hunting by humans, 1847–1993. Mammal Rev. 1996;26:103–126. [Google Scholar]
- Jędrzejewski W, Branicki W, Veit C, Meðugorac I, Pilot M, Bunevich AN, et al. Genetic diversity and relatedness within packs in an intensely hunted population of wolves Canis lupus. Acta Theriol. 2005;50:3–22. [Google Scholar]
- Jędrzejewski W, Jędrzejewska B, Andersone-Lilley Z, Balciauskas L, Mannil P, Ozolins J, et al. 2010Synthesizing wolf ecology and management in Eastern Europe: Similarities and contrasts with North AmericaIn: Musiani M, Boitani L, Paquet PC (eds).The World of Wolves. New perspectives on Ecology, Behavior and Management University of Calgary Press: Calgary; 207–233. [Google Scholar]
- Jones FC, Chan YF, Schmutz J, Grimwood J, Brady S, Southwick A, et al. A genome-wide SNP genotyping array reveals patterns of global and repeated species-pair divergence in sticklebacks. Curr Biol. 2012;22:83–90. doi: 10.1016/j.cub.2011.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST. The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity. 2011;106:625–632. doi: 10.1038/hdy.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keigwin LD, Donnelly JP, Cook MS, Neal W., Driscoll NW, Brigham-Grette J. Rapid sea-level rise and Holocene climate in the Chukchi Sea. Geology. 2006;34:861–864. [Google Scholar]
- Keller I, Wagner CE, Greuter L, Mwaiko S, Selz OM, Sivasundar A, et al. Population genomic signatures of divergent adaptation, gene flop and hybrid speciation in the rapid radiation of Lake Victoria cichlid fishes. Mol Ecol. 2013;22:2848–2863. doi: 10.1111/mec.12083. [DOI] [PubMed] [Google Scholar]
- Kijas JW, Lenstra JA, Hayes B, Boitard S, Porto Neto LR, San Cristobal M, et al. Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 2012;10:e1001258. doi: 10.1371/journal.pbio.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalov DA, Manning C, Henshaw MT. KINGROUP: a program for pedigree relationship reconstruction and kin group assignments using genetic markers. Mol Ecol Notes. 2004;4:779–782. [Google Scholar]
- Leonard JA, Vilà C, Fox-Dobbs K, Koch PL, Wayne RK, Van Valkenburgh B. Megafaunal extinctions and the disappearance of a specialized wolf ecomorph. Curr Biol. 2007;17:1146–1150. doi: 10.1016/j.cub.2007.05.072. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lucchini V, Galov A, Randi E. Evidence of genetic distinction and long-term population decline in wolves (Canis lupus) in the Italian Apennines. Mol Ecol. 2004;13:523–536. doi: 10.1046/j.1365-294x.2004.02077.x. [DOI] [PubMed] [Google Scholar]
- MacNulty DR, Smith DW, Mech LD, Eberly LE. Body size and predatory performance in wolves: is bigger better. J Anim Ecol. 2009;78:532–539. doi: 10.1111/j.1365-2656.2008.01517.x. [DOI] [PubMed] [Google Scholar]
- Marshall TC, Slate J, Kruuk LEB, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- Mattioli L, Capitani C, Gazzola A, Scandura M, Apollonio M. Prey selection and dietary response by wolves in a high-density multi-species ungulate community. Eur J Wildlife Res. 2011;57:909–922. [Google Scholar]
- McEvoy BP, Powell JE, Goddard ME, Visscher PM. Human population dispersal ‘Out of Africa' estimated from linkage disequilibrium and allele frequencies of SNPs. Genome Res. 2011;21:821–829. doi: 10.1101/gr.119636.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mech LD, Seal US. Premature reproductive activity in wild wolves. J Mammal. 1987;68:871–873. [Google Scholar]
- Miller W, Schuster SC, Welch AJ, Ratan A, Bedoya-Reina OC, Zhao F, et al. Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. PNAS. 2012;109:E2382–E2390. doi: 10.1073/pnas.1210506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiani M, Boitani L, Paquet P. The World of Wolves: New Perspectives on Ecology, Behaviour and Management. University of Calgary Press: Calgary; 2010. [Google Scholar]
- Nowak RM.2003Wolf evolution and taxonomyIn Mech LD, Boitani L (eds).Wolves: Behavior, Ecology, and Conservation The University of Chicago Press: Chicago; 239–258. [Google Scholar]
- Nowak RM, Federoff NE. The systematic status of the Italian wolf Canis lupus. Acta Theriol. 2002;47:333–338. [Google Scholar]
- Oliver MK, Piertney SB. Selection maintains MHC diversity through a natural population bottleneck. Mol Biol Evol. 2012;29:1713–1720. doi: 10.1093/molbev/mss063. [DOI] [PubMed] [Google Scholar]
- Ozolins J, Andersone Z.2001Status of large carnivore conservation in the Baltic States. Action plan for the conservation of wolf (Canis lupus) in Latvia European Commission: Strasbourg; T-PVS (2001) 73: add. 2, pp1–32. [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peery MZ, Kirby R, Reid BN, Stoelting R, Doucet-Bëer E, Robinson S, et al. Reliability of genetic bottleneck tests for detecting recent population declines. Mol Ecol. 2012;21:3403–3418. doi: 10.1111/j.1365-294X.2012.05635.x. [DOI] [PubMed] [Google Scholar]
- Pilot M, Branicki W, Jędrzejewski W, Goszczyński J, Jędrzejewska B, Dykyy I, et al. Phylogeographic history of grey wolves in Europe. BMC Evol Biol. 2010;21:10–104. doi: 10.1186/1471-2148-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot M, Jędrzejewski W, Branicki W, Sidorovich VE, Jędrzejewska B, Stachura K, et al. Ecological factors influence population genetic structure of European grey wolves. Mol Ecol. 2006;15:4533–4553. doi: 10.1111/j.1365-294X.2006.03110.x. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randi E. Genetics and conservation of wolves Canis lupus in Europe. Mammal Rev. 2011;41:99–111. [Google Scholar]
- Reddi AH, Cunningham NS. Bone induction by osteogenin and bone morphogenetic proteins. Biomaterials. 1990;11:33–34. [PubMed] [Google Scholar]
- Sastre N.2011Genética de la conservación: el lobo gris (Canis lupus)PhD thesisAutonomous, University of Barcelona: Spain [Google Scholar]
- Sastre N, Vila C, Salinas M, Bologov VV, Urios V, Sanchez A, et al. Signatures of demographic bottlenecks in European wolf populations. Conserv Genet. 2011;12:701–712. [Google Scholar]
- Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MB, Chen EH, Lynch SE. A review of the effects of insulin-like growth factor and platelet derived growth factor on in vivo cartilage healing and repair. Osteoarthr Cartilage. 2006;14:403–412. doi: 10.1016/j.joca.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Sidorovich VE, Tikhomirova LL, Jędrzejewska B. Wolf Canis lupus numbers, diet and damage to livestock in relation to hunting and ungulate abundance in northeastern Belarus during 1990–2000. Wildlife Biol. 2003;9:103–111. [Google Scholar]
- Sommer R, Benecke N. Late-Pleistocene and early Holocene history of canid fauna of Europe (Canidae) Mamm Biol. 2005;70:227–241. [Google Scholar]
- Spiegelhalter DJ, Best NG, Carlin BP, Linde AVD. Bayesian measures of model complexity and fit. J Roy Stat Soc B. 2002;64:583–639. [Google Scholar]
- Spiridonov G, Spassov N.1985Wolf—Canis lupus L., 1758In: Botev Peshev (ed) Red Data Book of Bulgaria Bulgarian Academy of Science: Sofia; 132 [Google Scholar]
- Stefani CM, Machado MA, Sallum EA, Sallum AW, Toledo S, Nociti FH., Jr Platelet-derived growth factor/insulin-like growth factor-1 combination and bone regeneration around implants placed into extraction sockets: a histometric study in dogs. Implant Dent. 2000;9:126–131. doi: 10.1097/00008505-200009020-00004. [DOI] [PubMed] [Google Scholar]
- Stronen AV, Jędrzejewska B, Pertoldi C, Demontis D, Randi E, Niedziałkowska M, et al. North-south differentiation and a region of high diversity in European wolves (Canis lupus) PLoS ONE. 2013;8:e76454. doi: 10.1371/journal.pone.0076454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using likelihood, distance, and parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenesa A, Navarro P, Hayes BJ, Duffy DL, Clarke GM, Goddard ME, et al. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 2007;17:520–526. doi: 10.1101/gr.6023607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilà C.1993Aspectos morfologicos y ecológicos del lobo iberico Canis lupus LPhD thesisUniversidad de Barcelona: Spain [Google Scholar]
- Vilà C, Amorim I. R, Leonard J. A, Posada D, Castroviejo J, Petrucci-Fonseca F, et al. Mitochondrial DNA phylogeography and population history of the grey wolf Canis lupus. Mol Ecol. 1999;8:2089–2103. doi: 10.1046/j.1365-294x.1999.00825.x. [DOI] [PubMed] [Google Scholar]
- vonHoldt B, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt BM, Pollinger JP, Earl DA, Knowles JC, Boyko AR, Parker H, et al. A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids. Genome Res. 2011;21:1294–1305. doi: 10.1101/gr.116301.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Pingping Z, Dong S, Zhan X, Wu Q, Guo X, et al. Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat Genet. 2013;45:67–71. doi: 10.1038/ng.2494. [DOI] [PubMed] [Google Scholar]
- Zulliger D, Schnyder E, Gugerli F. Are adaptive loci transferable across genomes of related species? Outlier and environmental association analyses in Alpine Brassicaceae species. Mol Ecol. 2013;22:1626–1639. doi: 10.1111/mec.12199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.