Abstract
Adipocyte differentiation and function have become areas of intense focus in the field of energy metabolism; however, understanding the role of specific genes in the establishment and maintenance of fat cell function can be challenging and complex. In this review, we offer practical guidelines for the study of adipocyte development and function. We discuss improved cellular and genetic systems for the study of adipose biology and highlight recent insights gained from these new approaches.
Keywords: energy metabolism, adipogenesis, obesity
Since the 1970s, fat cell differentiation of immortalized fibroblast cell lines has been an extensively studied model of cellular differentiation (1). The ability to study this process in a tissue culture dish has allowed cellular biologists and biochemists to explore general mechanisms of cell fate determination and cellular differentiation, and to isolate novel regulatory proteins and signaling cascades that initiate these processes. However, it is now clear that adipocytes represent critical regulators of nearly all aspects of energy balance. Adipose tissues exhibit high plasticity with a tremendous capacity for expansion and contraction in response to the energy demands of an organism. The alarming rise in obesity and obesity-linked metabolic disorders has directed the focus of energy metabolism researchers to the field of adipose development (2). Over the past several years an increasing number of mouse models exhibiting a lean or overweight phenotype have been described; these phenotypes have often been linked to primary defects in adipose biology. However, as our knowledge of energy metabolism and adipose biology expands, greater caution must be taken in interpreting studies of adipogenesis and adipocyte metabolism.
In this review, we offer a consolidated view of the wide-ranging approaches that can be taken to study adipose biology and discuss important considerations and pitfalls associated with each method. We divide the discussion into three sections; the first section describes model systems, ways to understand where and when molecules of interest may play a role in the adipose lineage, and recent insights gained from these approaches. The second and third sections provide guidelines for the study of adipocyte biology in vivo and in cellular systems, respectively. Along the way, we highlight improved genetic and cellular models that can aid researchers interested in studying the role of their favorite protein in the adipose tissue and discuss the insights gained from each approach.
IS MY FAVORITE GENE EXPRESSED IN THE ADIPOSE LINEAGE? WHERE DO I LOOK?
The establishment and maintenance of adipocytes in principle involves key regulatory steps (3). First, multipotent progenitor cells must commit to the adipocyte lineage, a process referred to as preadipose cell determination. Next, in response to appropriate spatial and temporal cues, committed preadipose cells undergo adipocyte differentiation, a morphological and biochemical transition into mature adipocytes. Finally, mechanisms must exist that function to maintain the differentiated state of the mature adipocyte (adipocyte maintenance). This includes factors critical for regulating the functional properties of adipocytes. Any gene suspected of playing a direct role in the adipocyte must obviously be expressed at some point in the life cycle of the fat cell or its progenitor. Thus, a logical first question that must be addressed is: “Where and when is my gene and protein of interest expressed and/or active in the adipose lineage?”
Expression in white and brown adipose tissue depots
Expression in white adipose tissue versus brown adipose tissue; subcutaneous versus visceral white adipose tissue.
Adipose tissues exist in mammals in two general forms, termed “white” and “brown”. White adipose tissue (WAT) serves as the principle site for safe energy storage. White adipocytes contain a single large lipid droplet and the enzymatic machinery to both synthesize and hydrolyze TGs. Brown adipose tissue (BAT) is specialized to dissipate chemical energy in the form of heat, and serves to protect mammals from hypothermia and defend against obesity (4). However, in more recent years, it has become more widely appreciated that the designation of an adipocyte as simply white or brown may in fact be too simplistic. For instance, white adipose depots located in different anatomical regions contain adipocytes that exhibit stable and intrinsic differences in gene expression and adipokine secretion (5–7). This implies that multiple types of white adipocytes exist in mammals and make different contributions to the maintenance of energy balance. In fact, numerous studies indicate that obese individuals accumulating fat in the visceral (abdominal) compartment are at much greater risk for developing type 2 diabetes and cardiovascular events than BMI-matched individuals whose adiposity is more prevalent in the subcutaneous regions (8). Furthermore, it is becoming increasingly clear that not all thermogenic adipocytes are created equally. It has long been appreciated that multi-locular uncoupling protein 1 (Ucp1)-expressing cells can be found within white adipose depots; however, lineage tracing and molecular analyses of these cells now reveal that these thermogenic cells within WAT (referred to as “beige” or “BRITE” cells) are in fact developmentally, molecularly, and perhaps functionally distinct from brown adipocytes located within the developmentally preformed BAT (“classical brown adipocytes”) (9–11).
Tremendous insight can be gained by analyzing the expression of genes of interest in these different adipose depots. Notably, many of the well-characterized determinants of thermogenic function in adipocytes, including Prdm16, Ppargc-1, Tle3, and Ebf2, are differentially expressed between white and brown adipocytes. Tle3, a recently described antagonist of thermogenic gene expression, is more abundant in white adipocytes than in brown adipocytes (12). Prdm16, Pgc-1, and Ebf2, all activators of brown adipocyte development and/or thermogenic gene expression, are relatively enriched in brown adipocytes (9, 13–16). Importantly, in rodents, many of these same genes will be expressed at intermediate levels in the subcutaneous inguinal WAT where beige fat cells are bountiful (10). Molecular determinants of WAT depot-specific adipocyte development and function are also coming in focus. Most recently, Cohen et al. (17) determined that Prdm16, whose expression is enriched in subcutaneous versus visceral adipose depots, suppresses a significant portion of the visceral adipocyte gene program and phenotype in subcutaneous (inguinal) WAT of mice. Kahn and colleagues discovered that the transcription factor, Shox2, is expressed more abundantly in subcutaneous fat than in visceral WAT depots in both mice and humans, and controls lipolysis preferentially in subcutaneous adipocytes (18). Moreover, Mueller and colleagues have identified Foxa3 as a visceral WAT-enriched transcription factor that regulates peri-gonadal, but not subcutaneous, adipogenesis in vivo (19).
Response to metabolic challenges.
The ability of adipose tissues to respond to rapidly changing environments is conferred by complex signaling cascades that are regulated at the transcriptional and posttranscriptional levels. For example, during fasting, adipocytes rapidly mobilize stored TG through the hydrolytic action of lipases, resulting in the release of NEFAs into circulation (20). Importantly, many of the factors that regulate lipolysis are also regulated at the level of gene expression. This includes hormone-sensitive lipase (HSL) itself, as well as transcription factors that control the expression of the lipolytic gene program (e.g., Tblr1, Tfe3, and Irf4) (21–23). Upon cold exposure, brown and beige adipocytes respond to adrenergic signals to induce thermogenic regulatory pathways. This involves acute regulatory events as well as alterations in gene expression programs. Proteins that coordinate these cellular adaptations, such as Pgc-1α, are often themselves regulated at the level of gene expression. Therefore, understanding how genes of interest are behaving under conditions of metabolic stress may shed insight into their functional role in the adipose tissue.
Expression programs within different adipose tissue depots.
It is critical to note that adipose tissue depots are heterogeneous. A majority of the mass of the tissue is attributable to mature adipocytes; however, the nonadipocyte compartment of adipose tissue (the “stromal vascular” component) consists of an abundant variety cell types, including endothelial cells, immune cells, fibroblasts, and adipose progenitor cells (APCs). The relative proportion of these diverse cell types can vary considerably between depots. A detailed understanding of the cell types that express genes of interest is essential to uncovering any potential function in the adipose lineage.
Methods for separating adipocytes from the stromal and vascular cells of the adipose tissue were pioneered in the 1960s (24). These methods, still in use today, involve digestion of adipose fragments with collagenase and physical separation of lipid-containing adipocytes from the stromal vascular fraction (SVF) by differential centrifugation. Hollenberg and Vost (25) made the seminal discovery that preadipocytes in adult animals exist within the adipose tissue SVF. They utilized tritiated thymidine to monitor DNA synthesis in the adipose tissue and found that radioactivity initially found exclusively in the SVF progressively appeared in the adipocyte fraction. Because adipocytes are postmitotic, this implied the existence of a replicating progenitor within the SVF. Today, it is still widely accepted that at least most preadipocytes reside within the adipose tissue depot, although other potential sources may exist (26–29). It is now common practice to examine the mRNA levels of candidate regulators in fractionated adipose tissues, documenting the relative expression pattern within the SVF and adipocyte compartment in both WAT and BAT.
Recent methodological advances in the adipose field now allow for highly adipogenic APCs within the SVF to be purified and separated from endothelial and hematopoietic lineage cells (26). One such method combines antibodies raised against combinations of known stem cell cell-surface proteins and fluorescent-activated cell sorting (FACS) to selectively purify APCs from freshly isolated adipose SVF. Elegant cell transplantation studies by Rodeheffer and colleagues determined that at least two distinct APC populations exist; a primitive population defined as CD31−;CD45−;CD29+;Sca1+;CD34+;CD24+ (CD24+ APCs) and a descending preadipocyte population defined as CD31−;CD45−;CD29+;Sca1+;CD34+;CD24− (CD24− APCs) (30, 31). Both the CD24+ and CD24− APC populations uniformly express platelet-derived growth factor receptor α (Pdgfrα). Pdgfrα is a cell surface tyrosine kinase receptor for the family of platelet-derived growth factors (Pdgfs). Pdgfrα is expressed broadly in the paraxial mesoderm during embryogenesis and is commonly found in fibroblastic/mesenchymal cells in various adult tissues (32). Importantly, lineage-tracing approaches reveal that adipocytes descend from cells that express Pdgfrα at some stage during their genesis (31); this includes adipocytes formed during development as well as new fat cells formed in adult animals (33–35). This is in line with historical observations that adipogenesis originates from fibroblast-like cells (36). Clonal analyses indicate that ∼70–80% of Pdgfrα+;CD31−;CD45− cells are capable of adipogenesis in vitro (33–35). Thus, the isolation of Pdgfrα+;CD31−;CD45− has become a convenient method to purify adipogenic cells. Other methods utilize transgenic reporter strains in which the expression of fluorescent proteins is driven by the locus/promoters of key regulators/markers of adipose lineage commitment. Most notably, Tang et al. (37) developed a lineage marking system that allows for purification and localization of stromal cells expressing, or once-expressing, Pparγ (PparγtTA;TRE-H2B-GFP reporter mice). In addition, we recently described the derivation of Zfp423GFP BAC transgenic mice in which GFP is expressed under the control of the Zfp423 locus (38). These reporter models have provided much needed molecular evidence to support a long-standing hypothesis that committed preadipocytes reside in the adipose tissue vasculature within the mural cell compartment (36, 39, 40). This is supported by a recent study by Kuang and colleagues demonstrating the expression of Fabp4 in PdgfRα+ adipose progenitors located in the adipose tissue vasculature (41). It is important to note that the precise relationship between all of the aforementioned APC populations remains unclear; however, these methods provide a starting point for defining the expression pattern and function of genes of interest in the early stages of adipogenesis.
Regulation during adipogenesis
Immortalized preadipose and mesenchymal cell lines.
Monitoring the expression and/or protein activity of potential adipocyte regulators during adipogenesis can provide insights into the detailed timing when candidates are functionally contributing to the adipogenic program. Numerous cellular models have been employed to study adipocyte differentiation. These include the multi-potent C3H 10T1/2 cell line, OP9 preadipocytes, and various 3T3-immortalized fibroblasts (42–44). Historically, the 3T3-L1 fibroblast cell line has been the standard model system to identify regulators of adipogenesis and fat cell function (such as lipogenesis, lipolysis, etc.). In the 1970s, Green and colleagues originally derived 3T3-L1 cells as a clonal subline of Swiss 3T3 mouse embryonic fibroblasts (45, 46). 3T3-L1 cells are viewed as being a “determined” cell line; the cells are morphologically indistinguishable from less- or nonadipogenic fibroblasts, but are highly competent to undergo adipogenesis in response to a hormonal/pharmacological cocktail consisting of dexamethasone, iso-butyl-methyl-xanthine, and insulin (generally referred to as “DMI medium”). Green and colleagues also derived 3T3-F442A cells (47). These cells were derived as a subclone of a nonadipogenic clonal line and spontaneously acquired its ability to undergo adipogenesis. 3T3-F442A cells are also highly committed. Unlike 3T3-L1 cells, they are able to differentiate simply by stimulating with insulin and, unlike 3T3-L1 cells, can form ectopic fat pads when implanted into the sternum of immunodeficient mice (48).
It is now standard practice to examine the expression of candidate adipogenesis factors during the time course of DMI-induced adipocyte differentiation in vitro. Often, the temporal pattern of expression can help predict the stage of differentiation that is influenced by the gene of interest. Genes that are upregulated in the early stages of differentiation are often, but not necessarily, positive regulators of adipogenesis. Genes whose expression is downregulated during differentiation are often negative regulators of adipose cell formation. Numerous transcriptional components that drive adipocyte differentiation have been identified through expression analysis of differentiated 3T3 cells; insightful reviews have been written on this matter (3, 49). Furthermore, differential protein expression or activity between preadipose and nonadipogenic fibroblast cell lines can indicate a role in preadipose cell determination. Indeed, preadipocyte commitment factors, such as Zfp423 and Tcf7l2, are enriched in preadipose fibroblast cell lines relative to nonadipogenic cells (50, 51).
The clonal fibroblast lines developed by Green and colleagues have been instrumental in elucidating many critical aspects of adipocyte biology (52). This includes the transcriptional regulatory network of fat cell differentiation (49), the dynamic alterations in the epigenomic landscape of differentiating fat cells (53–56), and adipokines controlling lipid and glucose metabolism (57–59). The function of nearly every critical factor of adipogenesis, including the master regulatory protein Pparγ, has been identified through analysis in 3T3 cells (60, 61). However, it is widely recognized that there are potential limitations to the use of immortalized cell lines. Most notably, 3T3-L1 cells are significantly aneuploid (52). 3T3-L1 preadipocytes are highly committed preadipose cells; however, the precise relationship between these cultured preadipocytes and adipose progenitors found in adult and fetal animals in vivo remains uncertain. 3T3 adipocytes in vitro can differ significantly from white adipocytes found in vivo (62). For instance, 3T3-L1 adipocytes express lower amounts of leptin than adipocytes found in vivo (63). Morphologically, 3T3-L1 preadipocytes differentiate into multilocular adipocytes, which are largely different from the unilocular mature white adipocytes in vivo. Importantly, 3T3-L1 adipocytes are white adipocyte-like; however, to what extent they resemble any particular depot-specific adipocyte found in vivo remains unclear. In vitro experiments based solely on evidence from a 3T3-L1 paradigm should therefore be interpreted with caution.
Adipose-derived stromal vascular cultures.
Studies from the 1970s first revealed that fibroblast-like cells from the cultures of the SVF [stromal vascular cultures (SVCs)] could be propagated and differentiated, in a similar way to 3T3-L1 fibroblasts, into mature adipocytes in vitro (64–66). Cells within SVCs, however, can also give rise to other osteoblasts, myoblasts, and chondrocytes (68). As recently described elsewhere, this has led to an expanded nomenclature to describe SVCs, which includes “adipose stem cells” or “mesenchymal stem cells” (68). Importantly, these in vitro SV-derived adipocytes molecularly resemble the adipocytes found in their depot of origin; BAT-derived SVCs differentiate into brown adipocytes and SVCs from WAT differentiate into white adipocytes (69, 70). Furthermore, WAT depot-specific characteristics are preserved in the in vitro-derived adipocytes. SVCs from human and mouse WAT give rise to adipocytes in culture that molecularly and functionally resemble adipocytes located in their tissue of origin; this suggests that at least some of the depot-specific properties of anatomically distinct white fat tissues are intrinsic, programmed at the level of the preadipocyte (70, 71).
Adipose SVCs offer distinct advantages over 3T3-L1 cells. First, they retain many of the properties of their depot of origin; to some extent, this allows for an analysis of depot-specific gene expression and function during adipogenesis. Recently, adipose SVCs from inguinal WAT have been instrumental in identifying and studying beige and white adipogenesis and function (10). Second, using primary cells circumvents the caveats associated with immortalized cells. However, these cells can easily become immortalized, if desired, through classic 3T3 passaging protocols. This has been particularly useful in the context of generating 3T3 immortalized clonal cell lines of beige fat progenitors (10). Furthermore, the primary cells can be transduced with retroviral or adenoviral vectors in an effort to create gain- or loss-of-function cellular models. It should be noted however, that adipose SVCs remain heterogeneous, even after extensive culturing. Methods to prospectively identify preadipose fibroblasts from these SVCs now exist. Preadipocytes from SVCs can be isolated on the basis of Zfp423, PdgfRα, or Fabp4 (aP2) expression when using corresponding reporter mouse strains (38, 41).
A potential caveat to the sole reliance on these cells is that the precise relationship between these cultured preadipocytes and native APCs found in adult animals in vivo remains uncertain. In fact, we have recently shown that the adipogenic capacity of depot-specific SVCs in vitro does not mirror the capacity of adipogenesis in vivo. SVCs from subcutaneous WAT differentiate more robustly in vitro than SVCs from visceral WAT (71, 72). However, in vivo, there is limited adipogenesis occurring in the inguinal WAT of male mice upon high-fat feeding; this depot expands predominantly through adipocyte hypertrophy (73). On the contrary, there is robust adipogenesis under numerous conditions in the epididymal fat of these same mice (73). It is possible that the in vitro cell culture conditions cannot fully mimic the stimulations these APCs are encountering in vivo under various physiological conditions. Also, it is unknown if the APCs at the embryogenic stage share the same characteristics with the APCs in adult or aged individuals, and if they respond to the same stimulations to undergo proliferation and differentiation. Further studies will be needed to better understand the precise relationship between the commonly used preadipocyte cultures and native APCs found in vivo.
Additional considerations
In most studies describing the function of genes/protein in adipose biology and energy metabolism, the experiments utilize rodents that are male; this reflects the widely accepted notion that male animals are more susceptible to metabolic disturbances than female animals and thus, phenotypes are more readily observed in these male models. It is important to note that there is a tremendous sexual dimorphism to the adipose phenotype in obesity. These differences have been recently highlighted elsewhere in numerous reviews (74, 75). The phenotypic response of the adipose tissue to high-fat feeding is regulated, in part, by the direct actions of estrogen signaling in the adipocyte itself (76). Likewise, age is another variable that may affect adipose function and expansion in obesity (77). Understanding the dependence of sex and age on the expression of genes of interest can shed light on the functional role of these factors.
It is also important to consider the impact of circadian rhythmicity on WAT and BAT gene expression. The circadian clock machinery is functional and regulated in adipocytes within all the major depots in mice (78, 79). A white adipocyte in vivo must adjust rates of de novo lipogenesis (DNL) and TG storage with the rate of hydrolysis of TG according to the circadian clock (80). Gerhart-Hines et al. (81) recently provided clear evidence that the thermogenic program of classical BAT is under circadian control. Thus, any interpretations of adipose gene expression data should take into account the precise time of day in which the tissues are harvested.
The paragraphs above highlight the importance of understanding the temporal and spatial expression pattern of candidate regulators of adipocyte biology. However, it is important to note that a gene or protein does not have to be actively regulated during adipogenesis or at the level of the mature adipocyte in order to exert a function in differentiation and/or maintenance of phenotype; it may simply have to be present. Moreover, understanding the activity, localization, secretion, or posttranslational modifications, rather than mere expression, of candidate enzymes, kinases, or secreted factors may be of equal importance. Ultimately, functional analyses in vitro and in vivo, as outlined below, are needed to fully understand the role of a specific protein in adipocyte development and function.
HOW DO I STUDY ADIPOCYTE DEVELOPMENT AND FUNCTION IN VIVO?
Tracking adipogenesis in vivo
Human and rodent studies suggest that most adipocytes are formed early in life and there is little adipocyte turnover in older animals (82–85). However, it is clearly evident that in response to high-fat feeding, when the demand for energy storage increases, there is an expansion in both adipocyte size (“adipocyte hypertrophy”) and adipocyte number (“adipocyte hyperplasia”) (82). Adipocytes are postmitotic; therefore, adipocyte hyperplasia results in the formation of new fat cells through adipogenesis. Understanding how adipocyte hypertrophy and adipocyte hyperplasia are regulated is of profound clinical significance; adipocyte hypertrophy is associated with WAT inflammation, fibrosis, and insulin resistance (86). WAT expansion through adipocyte hyperplasia, rather than adipocyte hypertrophy, correlates with improved insulin sensitivity (87, 88).
Historically, methods to monitor adipocyte hypertrophy and hyperplasia have relied upon: 1) quantification of cell size and number (82, 89), and 2) BrdU or radiolabeling “pulse-chase” experiments to quantify the incorporation of label into mature adipocytes (34, 90). Much has been learned using these approaches; however, there are a number of key assumptions that are made in interpreting these experiments. First, one assumes that cell size is a surrogate for the age of the cell; smaller fat cells corresponding to new fat cells, while large fat cells represent previously existing adipocytes undergoing hypertrophy. However, differences in fat cell size may simply reflect alterations in lipid handling. From a technical point of view, quantifying adipocyte number can be challenging given the heterogeneity of the adipose tissue. 3D reconstruction of histological sections may be needed for more accurate quantification. Second, when utilizing BrdU incorporation, one assumes that progenitors must proliferate before undergoing differentiation. This assumption is reasonable; however, recent data, described further below, suggests that a proliferative step may not be required for adipogenesis in some cases.
Lineage tracing approaches to track adipogenesis.
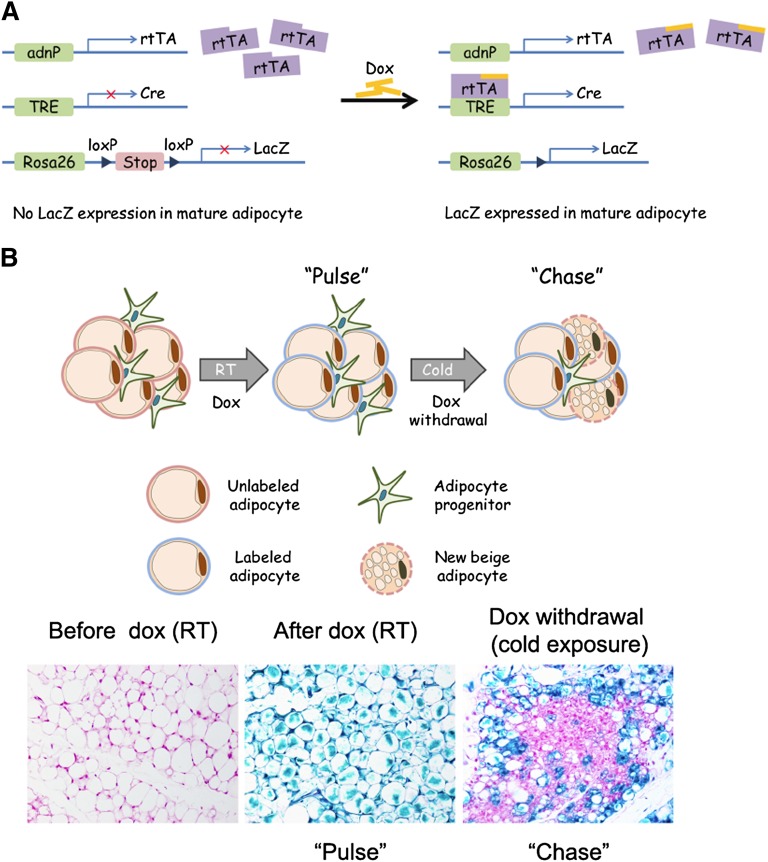
In recent years, inducible genetic lineage tracing systems have been used successfully to monitor de novo cell formation in vivo under different physiological or pathological conditions in adult organisms (91, 92). Recently, we developed a simple pulse-chase lineage tracing approach (referred to as the “AdipoChaser” mouse for convenience) to identify new adipocytes that are formed during high-fat feeding, or in response to cold exposure (73). AdipoChaser mice utilize a combination of three published transgenic lines (Fig. 1A): 1) transgenic mice expressing the “tet-on” transcription factor rtTA under the control of the adiponectin gene promoter (Adn-rtTA) (93); 2) a tet-responsive CRE (TRE-Cre) line that can be activated with rtTA in the presence of doxycycline (94); and 3) Rosa26 reporter mice expressing a reporter gene from the Rosa26 locus in a Cre dependent manner (Rosa26-loxP-STOP-loxP-reporter) (95). In the absence of doxycycline, there is no reporter expression in mature adipocytes. Upon treatment with doxycycline, rtTA activates the TRE promoter to induce Cre expression, and Cre protein will subsequently eliminate the floxed transcriptional stop cassette and permanently turn on reporter gene expression in every mature adipocyte present during doxycycline exposure (Fig. 1B, C). Importantly, within 12 h of removing doxycycline-containing food, Cre expression returns to baseline levels seen prior to doxycycline treatment (73). Thus, any new adipocyte generated after this stage will be LacZ negative. AdipoChaser mice were used to monitor adipocyte development in embryos and perinatal animals, as well as adipocyte hyperplasia occurring in obese adult mice. This led to the observation that most inguinal white adipocytes, like classic brown adipocytes, are formed during embryogenesis while the epididymal fat cells are formed only after birth. In adults, the inguinal WAT of male mice expands almost exclusively by adipocyte hypertrophy during high-fat diet feeding. In contrast, the epididymal WAT of these same mice exhibits both adipocyte hypertrophy and adipocyte hyperplasia, highlighting the depot-specific mechanisms of WAT expansion (73).
Fig. 1.
Tracking adipogenesis in vivo using the AdipoChaser mouse. A: The AdipoChaser mouse. AdipoChaser mice are derived from interbreeding three transgenic strains: 1) transgenic mice expressing the “tet-on” transcription factor rtTA under the control of the adiponectin gene promoter (Adn-rtTA); 2) a tet-responsive CRE (TRE-Cre) line that can be activated with rtTA in the presence of doxycycline (Dox); and 3) Rosa26 LacZ reporter mice expressing a reporter gene from the Rosa26 locus in a Cre-dependent manner (Rosa26-loxP-STOP-loxP-LacZ). In the absence of doxycycline, rtTA remains inactive and there is no Cre expression. Upon treatment with doxycycline, rtTA activates the TRE promoter to induce Cre expression, and Cre protein will subsequently eliminate the floxed transcriptional stop cassette and permanently turn on reporter gene expression in every mature adipocyte present during doxycycline exposure. B: Tracking cold-induced beige adipogenesis in inguinal WAT. Prior to doxycycline treatment, inguinal adipocytes are devoid of LacZ expression. Upon doxycycline treatment at room temperature, all adipocytes become LacZ positive (pulse labeling). After doxycycline withdrawal, mice were switched exposed to 4°C for 3 days; accumulated multilocular adipocytes are observed as LacZ negative, indicating that they were formed from cells originally unlabeled rather than preexisting mature adipocytes.
Cre-loxP-mediated lineage tracing approaches have also been used recently to address the origin and fate of cold-induced beige adipocytes in the inguinal WAT depot. The prevailing hypothesis in the field has been that UCP1+ adipocytes that arise within the WAT depot during cold exposure originate through a “transdifferentiation” of mature unilocular white adipocytes into thermogenic multilocular cells (96, 97). This hypothesis is supported by elegant morphological analyses, as well as numerous studies revealing the absence of BrdU incorporation in newly derived UCP1+ adipocytes (34). Unclear, however, was the fate of these UCP1+ adipocytes after mice returned to thermoneutral conditions. Our recent lineage tracing studies with the AdipoChaser model indicate that most cold-inducible UCP1+ cells emerging in the inguinal WAT actually derive from cells initially not expressing adiponectin (Fig. 1C); this indicates that the majority of inguinal beige adipocytes arise through adipogenesis of beige precursors (73). This is in line with recent studies indicating the presence of distinct beige precursor cells residing in the inguinal adipose SVF (10). Importantly, we found that beige adipogenesis occurred in a depot where BrdU incorporation in adipocytes was not found (34). This suggests that proliferation is not required before differentiation. Wolfrum and colleagues recently utilized tamoxifen-inducible Ucp1-CreERT2 mice to reveal that these beige adipocytes adopt a white adipocyte phenotype (unilocular) upon warm reacclimation (98). Going forward, the AdipoChaser model and Ucp1-CreERT2 mice represent genetic tools to study the development of both white and brown/beige adipocytes under different physiological conditions.
The AdipoChaser model can be used to provide clear evidence of adipocyte hyperplasia; however, the precise origin of the newly derived adipocytes cannot be determined with this model alone. As described further below, Cre lines driven by true preadipocyte-specific promoters do not exist; suitable markers have yet to be discovered. However, Granneman and colleagues have utilized tamoxifen-inducible Pdgfrα-CreERT2 mice to demonstrate that cold-induced beige adipocytes and high-fat diet-induced adipogenesis originate from Pdgfrα+ cells (34). Thus, combining the Pdgfrα-CreERT2 (99) with Rosa26 reporter lines offers a unique model to track adipogenesis in vivo.
Regulation of adipogenesis
Analysis of adipose development in germline “knockout” mouse models.
The analysis of adipose tissue development in global knockout mouse models is complex and relies on thorough understanding of the nutritional status of the animal. A common misconception is that aberrant adipogenesis per se drives the development of a lean or obese phenotype. In fact, obesity is fundamentally a disorder of energy balance; as energy intake exceeds energy expenditure, the demand for energy storage increases (100). Thus, adipose tissue expansion or contraction is a natural consequence of altered energy balance. A lean mouse is not necessarily a lipodystrophic mouse (see Fig. 2); low fat mass may simply reflect high levels of energy expenditure. In addition, unless the gene of interest is expressed exclusively in adipocytes (e.g., adiponectin or UCP1), the expression and function of the inactivated gene in other tissues needs to be understood in order to interpret any adipose phenotype. A number of studies, including our previous study of Zfp423 function, have analyzed embryonic adipose tissue formation in whole-body knockout models, because BAT and some subcutaneous WAT appear late in the gestation period (50). Studying adipose development during embryogenesis captures the early events of WAT and BAT formation and circumvents a number of potential metabolic confounders that may arise in adult animals; however, this approach alone still cannot determine if any observed defects in adipocyte formation represent a primary defect within the adipose lineage.
Fig. 2.
Lean or lipodystrophic? A commonly observed phenotype in animal models is a change in adiposity. In recent years, more and more mouse models exhibiting protection from diet-induced obesity have been described. Ultimately, the level of adiposity is dictated by the demand for energy storage. An engineered mouse with reduced adiposity may exhibit reduced fat mass simply because of reduced food intake or because of elevated energy expenditure. Such a mouse can be considered “lean” low adiposity is a normal consequence of a negative energy balance. A lean mouse (e.g., C57BL/6 mice on chow diet) exhibits greater glucose tolerance when compared with metabolically unhealthy obese animals (e.g., C57BL/6 mice on high-fat diet). A true adipose deficiency, or “lipodystrophy”, occurs when energy storage in adipose tissue cannot meet the demand for retention of fatty acids. This occurs when there is a primary defect in adipose tissue function or development (e.g., Fat-ATTAC, A-ZIP, or aP2-SREBP-1c transgenic animals; various Pparγ knockout models) (220). Lipodystrophic animals exhibit a metabolically unhealthy phenotype, similar (or worse than) that observed in unhealthy obese animals. This includes severe insulin resistance, glucose intolerance, elevated TGs, and ectopic lipid deposition and ceramide accumulation in the liver, pancreas, and/or skeletal muscle. The residual adipose tissue, though lacking adipocytes, actually exhibits features of pathological adipose tissue seen in unhealthy obese animals. This includes increased fibrosis and inflammation. Thus, it is not necessarily obesity per se that leads to metabolic disease. In fact, mice can be engineered (e.g., aP2-Mitoneet, aP2-Glut4, aP2-adiponectin transgenic animals) (87, 88, 221) to exhibit a “healthy-obese” phenotype. This obese phenotype includes preferential expansion of subcutaneous WAT, and insulin sensitivity, glucose tolerance, and adiponectin levels seen in normal BMI individuals. Importantly, the adipose tissue appears histologically similar to WAT of lean individuals; adipose expansion occurs through adipocyte hyperplasia and limited fibrosis occurs. Furthermore, the “healthy-obese” phenotype correlates with low ectopic lipid deposition. The image details the adipose and metabolic phenotypes commonly observed in rodent models and in humans. sWAT, subcutaneous adipose tissue; gWAT, gonadal adipose tissue.
Adipocyte-specific gene targeting.
The Cre-LoxP recombination system has been instrumental in elucidating gene function in specific organs or cell types (101). To date, most “adipocyte-specific” knockout models have been generated by utilizing strains in which Cre expression is driven by promoters of genes activated in the terminal stages of adipocyte differentiation (Table 1). The first and most commonly used Cre-driver has been the Fabp4-Cre mouse that was developed independently by a number of groups (102, 103). In these models, a Pparγ-dependent 5.4 kb enhancer located upstream of the Fabp4 transcriptional start-site drives the expression of Cre recombinase. The activity of this enhancer was previously characterized as being a differentiation-dependent regulatory element (104, 105). However, the precise timing of the activation of the Fabp4-driven transgenes has varied in different transgenic models; some Fabp4-Cre mice exhibit activity only in terminally differentiated adipocytes while others show Cre-mediated recombination at the level of the progenitor cell (37, 41). Fabp4-Cre mice have been used in numerous studies; however, very recent publications now highlight the serious pitfalls of this tool (106, 107). In particular, Fabp4-Cre-mediated recombination can occur in cells other than adipocytes, including macrophages, endothelial cells, neural cells, and cardiomyocytes (106, 108, 109). Furthermore, Fabp4-Cre-mediated recombination of floxed alleles in the germline can sometimes occur (106).
TABLE 1.
Available mouse strains for genetic manipulation of the adipose lineage
| Strain | Specificity | Expression in Adipose Lineage | Promoter |
| Fabp4-Cre (103), Fabp4-Cre (102), Fabp4-CreERT2 (123) | WAT and BAT. CNS, macrophages, endothelial cells, and cardiomyocytes (depending on strain) | Committed preadipocytes (some strains) and mature adipocytes | 5.4 kb |
| Adipo-Cre (110), Adipo-rtTA (73) | WAT, mosaic expression in BAT | Differentiating adipocytes, mature adipocytes | 5.4 kb |
| Adipo-Cre (23) | WAT and BAT | Differentiating adipocytes, mature adipocytes | BAC |
| Adipo-CreERT2 (124) | WAT, mosaic expression in BAT | Differentiating adipocytes, mature adipocytes | BAC |
| Resistin-Cre (107) | WAT and BAT. Some expression in CNS | Differentiating adipocytes, mature adipocytes | BAC, 33 kb fragment |
| Pparγ-tTA (37) | White and brown adipocytes, other tissues expressing Pparγ | Adipocyte progenitors and mature adipocytes | Knock-in endogenous allele |
| Ucp1-Cre (122), Ucp1-CreERT2 (98) | Beige and brown adipocytes | Mature adipocytes | BAC |
| Myf5-Cre (115) | Muscle lineage and brown adipocytes, mosaic expression in other cell types | Classical brown adipocyte precursors | Knock-in endogenous allele |
| Pax7-Cre (222), Pax7-CreERT2 (223) | Satellite cells and embryonic brown adipocyte precursors | Classical embryonic brown adipocyte precursors | Knock-in endogenous allele |
Alternative adipose-selective Cre lines are now becoming widely available and the Fabp4-Cre lines are likely to fall out of fashion. Most notably, models in which Cre expression is driven by the adiponectin gene promoter have been generated. Adpn-Cre, derived by Wang et al. (110), utilizes a 5.4 kb adiponectin promoter fragment that is sufficient to drive adiponectin transcription specifically in adipocytes (referred to as Adpn-Cre5.4). The line described by Eguchi et al. (23) is a BAC transgenic expressing Cre under the control of a very large portion of the adiponectin locus (referred to as Adpn-CreBAC). Adpn-Cre5.4 has recently been used successfully to delete ERα in an adipocyte-specific manner (76); however, in our experience, Cre expression is relatively weak and the Cre transgene may need to be bred to homozygosity for recombination of some floxed alleles. Thus, the most efficient and adipocyte-specific Cre line to date is the publically available Adpn-CreBAC mouse line developed by Eguchi et al. (23). This line, currently distributed through the Jackson Laboratory, has been used in a number of studies and has been demonstrated to be specific to adipocytes within WAT and BAT (12, 31, 112, 113).
An important consideration is that Cre expression is driven by promoters activated during the terminal differentiation of adipocytes in nearly all of the existing models (Table 1). Thus, the expression of floxed genes of interest will be lost only in actively differentiating or fully differentiated fat cells. An exception to this is the aforementioned Pparγ-tTA mouse (37). When combined with the TRE-Cre transgene and floxed alleles of interest, tTA is expressed in both adipose progenitors and mature adipocytes to drive Cre expression. This tool has recently been used successfully to examine the impact of activated WNT signaling on early events in adipose tissue formation (114). However, it should be noted that Pparγ expression is not limited to the adipose lineage; true preadipocyte-specific Cre lines currently do not exist.
BAT-selective gene targeting.
Lineage tracing studies using independent approaches discovered that the well-characterized brown adipocytes that arise during embryonic development in rodents (classical brown adipocytes) share a common lineage with skeletal muscle, rather than white adipocytes. The Myf5-Cre line (115) and Pax7-CreERT2 (tamoxifen-inducible Cre) (116) used to establish this finding can now be used to ablate floxed genes of interest early in development of BAT. These promoters are active in the earliest progenitors that give rise to skeletal muscle and brown adipocytes, but importantly, are not active in the mature brown adipocyte. The Myf5-Cre mouse has recently become a tool of choice to study classical brown adipogenesis; studies of chromatin regulators, PTEN, and BMP signaling in BAT development have recently employed this tool with great success (117–119). However, investigators should be reminded that Myf5, a muscle determination factor, is expressed throughout the skeletal muscle lineage, and activated quite early in the presomitic mesoderm of fetal rodents (120, 121). Therefore, any interpretation of phenotypes observed in models using the Myf5-Cre must consider the potential functions of the inactivated gene in the primitive and mature skeletal muscle. Cre lines that are specifically active in the classical brown adipose lineage currently do not exist; however, the UCP1-Cre line can be used to specifically target all mature thermogenic adipocytes, both brown and beige (122).
Regulation of adipocyte function in adult animals
Inducible adipocyte-specific gene targeting.
A significant limitation to the use of constitutively active Cre-expressing strains is the inability to separate gene function during development from gene function in adult animals. The precise response of adipose tissue to different metabolic challenges (e.g., high-fat feeding, cold exposure, etc.) in the adult will undoubtedly be influenced by defects manifested during development. To overcome this limitation, inducible adipocyte-selective Cre drivers have been described. Imai et al. (123) have utilized a tamoxifen-inducible Fabp4-CreERT2 strain to drive Pparγ deletion in adult adipocytes. However, this Cre strain, like other Fabp4-driven Cre lines described above, appears to target other nonadipose cell types and thus may ultimately be limited in use. Sassman, Offermanns, and Wettschureck (124) have described an alternative model in which CreERT2 is expressed from an adiponectin BAC, resulting in efficient inducible Cre-mediated recombination in WAT depots. Similarly, the Adn-rtTA transgenic allele, when combined with the TRE-Cre transgene and floxed alleles of interest, can yield doxycycline-inducible fat cell-specific knockout models (73). For inducible gene knockout in brown adipocytes, Rosenwald et al. (98) recently generated a BAC transgenic UCP1-CreERT2 line.
An important consideration in utilizing these inducible models is the potential metabolic effects of tamoxifen or doxycycline itself. Tamoxifen, an estrogen receptor antagonist, can influence estrogen receptor signaling throughout the mouse and has been observed to elicit glucose intolerance in male mice (125). Additional side effects of tamoxifen administration in mice, including gastric toxicity, have been reported (126). The importance of estrogen receptor signaling in the adipose tissue is becoming clearer (76, 127); therefore, greater caution must be taken with the use of tamoxifen. High doses of doxycycline may also elicit metabolic effects in mice. As an antibiotic, the potential side effect of doxycycline on metabolism is an alteration of gut microbiome; bacteria critical for normal energy balance may be replaced by doxycycline-resistant strains. However, it is evident that doxycycline can be removed from the system within 24 h (73), while tamoxifen is much harder to wash out and its prolonged effect lasts for weeks (128). A head-to-head comparison of the impact of tamoxifen and doxycycline on metabolism and adipocyte function is still needed; therefore, independent of the system used, it is good practice to study cohorts of mice in which experimental and control animals both receive the Cre-activating drug.
Does my mouse have an adipose phenotype?
A singular recipe or “standard protocol” for the analysis of metabolic phenotypes is difficult to prescribe; the precise approach to analyzing the phenotype of engineered mouse models depends on the nature and location of the genetic manipulation. In most cases, a thorough analysis of energy metabolism in rodents begins with measurements of energy intake (food intake) and energy expenditure. Excellent guidelines for conducting these assays have been described (129). Below, we highlight assays relevant to the study of adipose tissue in mouse models (Table 2).
TABLE 2.
Assays of adipose tissue function
| Function | Assays |
| Adipose tissue mass and morphology | Adipose tissue mass: NMR, MRI, or CT scan; endpoint tissue weight. |
| Adipocyte size and structure: HE and Oil Red O staining. | |
| Adipose tissue inflammation and macrophage infiltration | Immune cell profiling: FACS analysis or immunostaining of CD45+ (immune cells), CD3+CD4+ (T cells), CD3+CD8+ (T cells), B220+ (B cells), CD3−NKp46+ (NK cells), and CD11b+F4/80+ (macropahges). |
| Macrophage infiltration: immunohistochemical staining of F4/80. | |
| M1/M2 macrophage ratio: qPCR analysis of macrophage markers: CD206, CD301, and IL-10 for M1 macrophage; NOS2 and IFNγ for M2 macrophage. | |
| Alive or dead adipocytes: immunohistochemical staining of perilipin (living adipocytes), caspase 3 or TUNEL (apoptotic adipocytes). | |
| Adipose tissue hypoxia and fibrosis | Adipose tissue hypoxia: qPCR/Western blot analysis or immunostaining of Hif-α and ER stress markers; pimonidazole staining. |
| Adipose tissue fibrosis (increased ECM deposition): trichrome and picrosirius red staining and collagen VI expression. | |
| Adipose tissue glucose metabolism and insulin sensitivity | Glucose uptake: insulin-stimulated glucose uptake in primary adipocytes or in vivo. |
| Glucose clearance from the bloodstream: OGTT or IPGTT. | |
| Insulin sensitivity: ITT; hyperinsulinemic-euglycemic clamp; insulin induced phosphorylation and activation of insulin downstream targets (Akt, GSK3β, ERK1/2) in primary adipocytes or in adipose tissue. | |
| Adipose tissue lipolysis, TGs clearance and Lipogenesis | Lipolysis: basal, fasting and β3 agonist-induced induced NEFA or glycerol release in circulation; phosphorylation of HSL/PKA substrate in adipose tissue. |
| TGs clearance: whole-body TGs clearance upon oral TG gavage; 3H-triolein tracing lipid clearance in adipose tissue; LPL activity assay in adipose tissue. | |
| Lipogenesis: qPCR or Western-blot assay of enzymes involved in DNL pathways, such as FAS, ACL, and ACC; lipogenesis rate in primary adipocytes or in adipose tissue in vivo. | |
| Adipose tissue mitochondrial function | Mitochondrial gene expression profiling: qPCR analysis of mDIC. |
| Oxygen consumption rate: measured by the Seahorse analyzer system (mitochondrial respiration). | |
| Extracellular acidification rate: measured by the Seahorse analyzer system (mitochondrial glycolysis). | |
| Adipokines | Adipokine expression levels in adipose tissue: qPCR and Western blot analysis on adipose tissue extracts. |
| Adipokine concentrations in circulation: Western blot, ELISA, or RIA analysis on serum samples. |
CT, computed tomography; ER, endoplasmic reticulum; OGTT, oral glucose tolerance test; IPGTT, intraperitoneal glucose tolerance test; ITT, insulin tolerance test; GTT, glucose tolerance test; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
Adipose tissue mass and morphology.
As described above, alterations in WAT mass may reflect either a normal adaptation to changes in energy homeostasis or an actual defect in adipogenesis. The adipose tissue mass can be measured by simply weighing tissues; however, NMR (130), MRI (131), or CT scans (132) can provide insight into relative lean versus fat mass composition and body fat distribution. Importantly, and also described above, the size and number of adipocytes may give insight into how adipose tissue expands or handles lipid (adipocyte hyperplasia or hypertrophy). Functional adipose tissue deficiency can lead to accumulation of lipid in other tissues, such as pancreas, muscle, and liver (ectopic lipid deposition). Thus, histological analyses of these metabolically active tissues are critical for explaining an adipose phenotype at the whole-body level (see Fig. 2).
Adipose tissue inflammation and macrophage infiltration.
There is a widely accepted link between adipose tissue inflammation and systemic insulin resistance (133). During high-fat diet feeding, adipocytes and resident immune cells secrete an array of proinflammatory cytokines and chemoattractants that further enhances macrophage infiltration (133, 134). Accumulated adipose tissue macrophages (ATMs) secrete TNFα, IL1β, and other molecules that can directly impair adipocyte lipid and glucose metabolism (133). ATMs are composed of M1 (classically activated) and M2 (alternatively activated) macrophages (132); studies have shown that ATMs in normal insulin-sensitive lean mice have a larger M2 population, while increased adiposity leads to an accumulation of M1 population (135, 136). Therefore, besides the classic macrophage infiltration staining by F4/80 (137), it is useful to analyze the M1/M2 population in adipocytes by measuring the expression of selective markers (CD206, CD301, and IL-10 for M1 macrophages; NOS2 and IFNγ for M2 macrophages) (132). We have to remember though that the differentiation into M1 and M2 macrophages is a functional one and many macrophages do not strictly qualify to be placed into one of these distinct subclasses. Studies also show that numbers of CD8+ effector T cells are infiltrated in adipose tissue of obese mice, whereas CD4+ helper and regulatory T cells are diminished (138). Taken together, it is becoming more important to have a relatively full picture of the alteration of immune cell populations in adipose tissue; this can be achieved in part through FACS analysis or immunostaining of CD45+ (immune cells), CD3+CD4+ (T cells), CD3+CD8+ (T cells), B220+ (B cells), CD3−NKp46+ (NK cells), and CD11b+F4/80+ (macrophages) (139). Apoptotic adipocytes can be identified by immunohistochemistry; a deceased adipocyte appears negative for perilipin immunostaining, positive for caspase 3 or terminal deoxynucleotidyl transferase dUTP nick end labeling staining, and is surrounded by macrophages (F4/80 staining) (137, 140).
Adipose tissue hypoxia and fibrosis.
The ability of adipose tissue to expand depends, much like a tumor, on the ability of the vasculature to maintain oxygen supply. Over the past several years, it has become clear that a common feature of pathological adipose tissue expansion is the inability of the adipose tissue vasculature to keep pace with rapidly expanding adipocytes, leading ultimately to local hypoxia. This hypoxic response is characterized by increased hypoxia-inducible factor-1α (Hif-1α) expression (141, 142), unfolded protein response, and endoplasmic reticulum stress in adipocytes (143). Regions of hypoxia can be detected histologically by pimonidazole staining and are seen in most rodent models of diet-induced obesity (137, 144).
Functional studies of Hif-1α indicate that this protein, unlike its actions in tumor environments, fails to initiate a pro-angiogenic response. Instead, elevated Hif-1α drives the expression of proteins of the extracellular matrix (ECM). Adipocytes are embedded in a unique ECM; proteins within the ECM provide both mechanical support and participate in various signal pathways (145). However, excessive ECM production, defined as “fibrosis,” can reduce the plasticity of adipose tissue and can lead to adipocyte dysfunction (86). In recent years it has become clear that adipose tissue fibrosis is a hallmark of metabolically challenged adipocytes (146–148). Adipose tissue fibrosis can be visualized by trichrome and picrosirius red staining of histological sections (149). Molecularly, collagen VI is believed to be positively correlated with adipose tissue fibrosis and decreased collagen VI leads to improvement in adipose tissue energy homeostasis (146).
Glucose metabolism and insulin sensitivity.
Adipose tissue is a critical regulator of systemic glucose homeostasis and insulin sensitivity. Insulin-stimulated glucose uptake is a major function of adipocytes in response to insulin stimulation. In rodents, insulin-stimulated glucose uptake can be measured in primary adipocytes (150–152) or by in vivo glucose uptake studies (153, 154). Simple oral or intraperitoneal glucose tolerance tests can be utilized to assess the rate and magnitude of glucose clearance from the bloodstream (88), while circulating insulin levels during glucose tolerance tests reflect acute glucose-induced insulin secretion (155). Insulin tolerance tests can be used to measure systemic insulin sensitivity. Specific measurements of adipose tissue insulin sensitivity require more specialized approaches, such as the hyperinsulinemic-euglycemic clamp or the use of ex vivo and in vitro models as described below. To determine if an alteration in insulin sensitivity results from impaired insulin signaling in adipose tissue, phosphorylation/activation of Akt, GSK3β, and ERK can be analyzed by Western blotting of primary adipocytes treated by insulin (156, 157) or in vivo insulin injections into fasted animals (158–160). Circulating glucagon, another important hormone that has the opposite effect of insulin, is also worth measuring; blood samples should be collected with protease inhibitors (161).
Lipolysis, TG clearance, and lipogenesis.
The ability of adipocytes to store TG during periods of nutritional excess and release free fatty acids during fasting is achieved through coordinated cycles of lipogenesis and lipolysis (162). During normal cycles of fasting and feeding, the balance between lipogenesis and lipolysis is controlled by nutrients such as carbohydrates and polyunsaturated fatty acids, and by hormones such as insulin and catecholamines. Insulin resistance and dysfunctional adipose tissue in obesity is characterized by inappropriate regulation of these pathways (163, 164). In recent years, studies have shown that TG hydrolysis in WAT is not only controlled by HSL, but also by desnutrin/adipose TG lipase (ATGL) (165–169) and other lipases. Actually, although HSL has hydrolase activity for both TG and diacylglycerol, the enzyme may be more prone to hydrolysis of diacylglycerol (162, 170, 171). Desnutrin/ATGL is a TG-specific lipase (168, 172). Adipose-specific ablation of desnutrin/ATGL revealed that this enzyme is essential for adipocyte lipolysis and that it is phosphorylated by AMPK to increase its lipase activity (173). CGI-58, also known as α/β hydrolase domain-containing protein 5 (ABHD5), has been found to promote TG hydrolysis by coactivating ATGL (174). Moreover, other adipocyte lipases, such as TG hydrolases (175), adiponutrin, GS2 (176), and monoglyceride lipase (177) also may contribute to adipose tissue lipolysis.
Perilipin A and B on lipid droplets are believed to be a barrier that prevents adipocyte lipases from access to neutral lipid substrates (178). When the β-adrenergic receptor/adenylyl cyclase pathway is activated, perilipin A is phosphorylated by protein kinase A (PKA) and HSL is translocated to the lipid droplet surface to be able to meet neutral lipid substrates (179–183). The phosphorylation of perilipin A may also increase lipolysis by increasing the surface area of neutral lipid droplets to make more neutral lipid substrates accessible for lipases (184, 185).
Analysis of lipolysis can be accomplished by measuring basal, fasting, and β3 agonist-induced NEFA and glycerol release, as well as the activation of adipocyte lipases (186). In β3 agonist-induced lipolysis assays, note that the glycerol to NEFA ratio is an assessment of reesterification and cellular glyceroneogenesis (187). While a 1:3 molar ratio is predicted for the change, that ratio can be rather different and be anywhere from 1:0.1 to 1:2. Besides lipolysis, glucose and insulin levels during β3 agonist treatment are important reflections of whole-body glucose tolerance and β3 agonist-mediated insulin release, which is a direct effect linked to adipocyte function (188, 189).
TG clearance can be tested upon exposure of the mice to an oral TG gavage (137, 190). For example, ob/ob mice are expected to have high excursion/slow clearance of TGs from circulation (87). Though WAT is the major site of TG clearance, the impact of other tissues, such as BAT, cannot be ignored (191). Thus, injection of 3H-triolein to trace where the lipid has been cleared can further confirm tissue-specific responses (88). LPL hydrolyzes circulating TGs to fatty acids. In adipocytes, insulin activates LPL and its placement in the capillary endothelium. LPL activity assay can be measured enzymatically (192) to reveal the ability of adipose to take up TGs.
The liver was once considered the principle lipogenic organ; however, DNL in the WAT also plays a key role in energy storage and is controlled in response to nutritional, hormonal, and metabolic stimuli (193–195). In humans, WAT is likely to be the most important lipogenic organ; DNL contributes to approximately one-fifth of newly deposited TG in subcutaneous adipose tissue (196) with little DNL occurring in the liver (197–199). Thus, studying DNL in WAT of rodents has tremendous clinical relevance. A few groups reported that DNL in adipose tissue is positively correlated with insulin sensitivity (112, 200, 201) and is controlled by insulin in the brain (202). Leptin also appears to be a regulator of DNL through central STAT3-independent mechanisms (203). Lipogenic enzymes in the liver and epididymal adipose tissue are predominantly suppressed in response to high-fat diet (HFD) feeding; this occurs as a feedback mechanism of increased dietary lipid intake through increased leptin action. This feedback is gradually weakened by prolonged HFD-induced leptin resistance (203, 204). In contrast to HFD-induced obesity, DNL in db/db mice is dramatically upregulated in a liver-selective manner, accompanied by the abnormally elevated hepatic expression of PPARγ (205). Methods for analyzing DNL in adipose tissue include expression analyses of enzymes involved in DNL pathways, such as FAS, ACC, ACL, etc. (203, 204), and lipogenesis rate measurements in primary adipocytes isolated from adipose tissue (157).
Adipokines.
The endocrine action of adipocytes is critical for the maintenance of nutrient homeostasis and energy balance. Adipokines are critical to the systemic control of metabolic homeostasis; the measurement of the levels of some key adipokines is an excellent reflection of the physiological state of adipose tissue. Quantitative (q)PCR or Western blot analysis can determine the expression levels of adipokines, and there are a vast array of ELISA and RIA kits on the market to determine adipokine levels in circulation. Note that alterations in expression levels of adipokines are not always correlated with the changes in its secretion rate or concentrations in circulation, each stage is strictly regulated by distinct pathways and can be disturbed by specific manipulation of a component. Furthermore, the changes in adipokines should be very carefully interpreted because the exact function of most adipokines and their correlations with metabolic phenotypes are still being actively studied and are not yet fully understood. Taking adiponectin as an example, it is well established that the circulating adiponectin level is a reflection of metabolically healthy adipose tissue (206). Circulating adiponectin levels are generally negatively correlated with adipose tissue mass during high-fat diet-induced obesity; however, occasionally whole-body fat mass may decrease when there is a dramatic drop in circulating adiponectin levels, reflecting other physiological changes in adipose tissue (137). In other cases, if adipose tissue expansion occurs in a healthy manner, adiponectin levels can be positively correlated with adipose tissue mass (190).
BAT function.
Classical BAT, found most prominently in the interscapular region of rodents, is critical for TG clearance, glucose disposal, and for nonshivering thermogenesis. BAT has an enormous effect on metabolic rate; small alterations in BAT activity can impact multiple metabolic variables (207). Thus, understanding the effect of BAT is essential for complete metabolic phenotyping in mice. Excellent guidelines for the study of BAT function have recently been proposed and thus will not be discussed in detail here (208). Importantly, it is critical for investigators to note that standard housing conditions (22°C) may not be suitable for the analysis of BAT function in rodents; this “room temperature” setting actually poses a thermal (cold) challenge to most mice. The greatest activation of BAT is seen in mice maintained at thermoneutrality (∼30°C) prior to cold exposure. Furthermore, it is now appreciated that the thermogenic activity of BAT is under Rev-erbα-dependent circadian control; mice exposed to cold fare considerably better at Zeitgeber time 22 when Rev-erbα levels are low (81). Brown adipocyte activity can be assessed by calorimetry and through analysis of molecular markers as recently described (208). Importantly, the measurements of core body temperature during acute exposure to the cold only minimally reflect nonshivering thermogenesis of BAT; rather it is a reflection of muscle-based shivering thermogenesis.
Adipogenesis of beige fat cells.
The ectopic accumulation of UCP1+ multilocular adipocytes within the WAT depots has been well-documented for more than a decade; however, in recent years, these cells have garnered considerable interest for their potential role in rodent and human energy balance (210). Beige cell adipogenesis in mice occurs most rapidly in the subcutaneous inguinal WAT; however, prolonged cold exposure or stimulation with β3 agonists eventually trigger the formation of these cells in numerous WAT depots. As described above, the AdipoChaser model provides a clear approach to study beige fat cell neogenesis (73). However, in any rodent model, the abundance of inguinal beige adipocytes can be assayed either histologically (UCP1 immunostaining or localization of multilocular adipocytes) and molecularly by protein and mRNA analysis of thermogenic regulatory genes or markers [e.g., Pgc-1α, Prdm16, UCP1, Cidea, and Dio2; see a recent review by Harms and Seale (210) for complete information].
General considerations
In designing and interpreting results, it is essential to bear in mind that many of the pathological features of adipose tissue in obesity such as increased inflammation, fibrosis, or hypoxia will depend on age, strain, sex, and weight. Comparing weight- and sex-matched animals is often essential to understanding the primary defects caused by genetic manipulations. Furthermore, all of the potential phenotypes described above are intimately linked. Insight into the cell-autonomous functions of genes of interest can best be gained through tissue-specific gene ablation or cellular analyses as described below. In addition, if a given mouse strain is displaying a stunted overall growth (giving rise to a “runted” phenotype), care has to be used with respect to differentiating a developmental phenotype versus effects of a gene product on the functionality of the mature fat cell. Mice with disproportionately small growth can show improvements in insulin sensitivity and longevity, unrelated to the function of the protein in mature fat cells. Ultimately, inducible gain- or loss-of-function studies need to be performed to provide functional insights.
HOW DO I STUDY ADIPOGENESIS AND ADIPOCYTES USING CELLULAR MODELS?
Cellular models of adipogenesis remain tremendously helpful in understanding the cell-autonomous functions of any candidate regulator. In addition, they continue to serve as an excellent system to explore the biochemical properties and function of key adipogenic factors and pathways. Furthermore, given the limitations of the existing Cre lines, and the inability to target adipose progenitors in a specific manner, it remains essential to complement in vivo studies with cellular analysis. Below, we provide a general overview of how ex vivo and in vitro analyses of adipocytes can be conducted and highlight important markers that should be examined in order to properly interpret results.
In vitro adipogenesis assays
Adipogenic “cocktails”.
The commonly described standard protocol for differentiating immortalized or primary preadipose cell lines involves inducing differentiation of confluent (growth arrested) cultures with DMI for 48 h, followed by maintenance in media containing insulin for an additional 4–6 days. This protocol works well for well-maintained cell lines and primary SVCs; however, supplementation with a Pparγ agonist, such as the thiazolidinedione rosiglitazone or other adipogenic factors, is sometimes needed to enhance differentiation in vitro. Brown or beige cell adipogenesis can be induced using a similar protocol; triiodothyronine (T3) is often added to help initiate the thermogenic gene program via thyroid hormone signaling. It should be noted that very little is known about the bona fide signals of white adipogenesis in vivo; therefore, adherence to a strict standard protocol may be limiting. For example, members of the bone morphogenic protein (BMP) family can drive both white and brown adipocyte differentiation (211–214); the mechanisms required for this response may in fact be different than those that drive DMI-induced adipogenesis. In addition, some components of the standard differentiation cocktail may not be needed to stimulate differentiation. Primary Pdgfr+ cells from adipose SVF can differentiate in vitro spontaneously or with only the addition of insulin (37). 3T3-F442A cells, regarded as being a highly committed preadipose cell line, can differentiate in a similar manner (47). Several laboratories have differentiated the cultured mouse SVF with varying success; in our experience this likely reflects the varying degrees in which different lots of FBS can support adipogenesis. It is quite common for laboratories studying adipogenesis to identify FBS from different lots/sources that can best support primary preadipocyte differentiation under their desired conditions.
Assessing adipogenesis.
The extent and quality of adipocyte differentiation in vitro should be assessed by: 1) the morphological appearance of lipid-laden fat cells (determined by light microscopy and lipid staining), and 2) the expression of molecular markers of adipocytes. It is important to note that many cells can accumulate lipid; assaying lipid accumulation alone may not reveal the extent of adipocyte differentiation. As such, TG synthesis can often be confused with adipogenesis (Table 3). Adipocytes are hormone-producing cells with specialized functions. Thus, it is critical to assay the expression of adipocyte-selective genes that appear only in adipocytes and not in the undifferentiated progenitors. The most adipocyte-selective genes include those that encode adipokines, such as adipsin and adiponectin; however, as described above, some hormones such as leptin may not be expressed in cultured adipocytes. A list of some suitable markers can be found in Table 3.
TABLE 3.
Adipogenesis or lipogenesis? Reliable preadipocyte and adipocyte selective transcripts to assess in vitro adipogenesis
| Molecular Marker (mRNA levels) | Preadipocyte (in vitro) | Adipocyte (in vitro) |
| Stem cell/adipose progenitor genes: | ||
| CD34 | + (expression may be lost with prolonged culture) | − |
| Sca1 | ++ (expression may be lost with prolonged culture) | − |
| Pdgfrβ (CD140b) | ++ | − |
| Pdgfrα (CD140a) | +++ | − |
| Pref1 | +++ | − |
| Transcription Factors: | ||
| C/EBPα | + | + + + |
| Pparγ1 | + | + + + |
| Pparγ2 | − | + + + |
| Lipid/glucose metabolism: | ||
| Fabp4 | + | + + + + + |
| Glut4 | − | + + + |
| LpL | + | + + + |
| Adipokines: | ||
| Adipsin | − | + + + + + |
| Adiponectin | − | + + + + + |
The ability to store excess energy in the form of lipid is an ancient and fundamental requirement for growth and survival. Virtually all species have developed mechanisms for fat storage. Caenorhabditis elegans stores lipid in intestinal cells while Drosophila maintains a “fat body” which shares similarities to the mammalian liver. In vertebrates, many cell types can accumulate lipid; however, the evolution of adipocytes provided a specialized and safe compartment for this purpose. Furthermore, the identification of leptin, adiponectin, and other adipocyte-derived hormones and cytokines revealed the endocrine function of adipocytes. As such, an adipocyte must be viewed as being more than a mere lipid-accumulating cell. Studies of adipogenesis often rely on lipid-accumulation as a primary read-out for cellular differentiation. Our current knowledge of adipocytes demands a more precise definition of terminally differentiated adipocytes. The extent of adipocyte differentiation in vitro should be determined through quantitative measurements in the expression of preadipocyte-selective transcripts (downregulated during differentiation) and adipocyte-selective transcripts (activated during adipogenesis).
Gain- and loss-of-function analyses in vitro.
Most of our current knowledge regarding the mechanisms controlling adipocyte differentiation derives from functional studies in 3T3-L1 cells, primary SVCs, and mouse embryonic fibroblasts from various mouse models. These cellular systems can easily be manipulated in vitro using lentiviruses, retroviruses, or sometimes using simple transfection methods. Moreover, the effects of many small molecules on adipogenesis have been assessed using these cellular systems.
An important consideration in the design and interpretation of these cellular experiments is that many manipulations to primary or immortalized cell lines easily result in a loss of adipogenic capacity. Proteins or chemicals affecting cell growth or viability when introduced to preadipocytes may result in impaired adipogenesis; however, this may not reflect a true function in the differentiation process per se. While insights from these in vitro studies have proven to be very insightful and instrumental for our current understanding of adipogenesis, we have to be mindful of the limitations of the system. Conclusions based on the ability of stromal vascular isolates to undergo differentiation in vitro in support of a pro- or anti-adipogenic phenotype have to be performed on multiple isolates and may or may not be consistent with the in vivo phenotype. If the overall yield of differentiation of unsorted stromal vascular cell isolations is low, differences in differentiation efficiencies between genotypes are of limited value. The most convincing cellular studies of adipogenesis therefore utilize both gain- and loss-of-function analyses as well as multiple strains of preadipocyte cell lines and induction cocktails.
Cellular transplantation assays
As described above, an important caveat to the in vitro differentiation assays is that the full range of bona fide adipogenic signals present in vivo may not be represented under the standard culture conditions. A complementary approach that can be utilized is to perform cellular transplantation experiments. It has long been appreciated that among the various immortalized cell lines, the 3T3-F442A cells are capable of forming ectopic fat tissue when transplanted over the sternum of immune-deficient animals (48, 215). In numerous studies, these cells, along with other immortalized lines, have been engineered to overexpress or knock down genes of interest, and then are transplanted into recipient mice as a way to assess brown and white adipogenesis in vivo (13, 216, 217).
Transplantation assays have been utilized to characterize the properties of putative adipose progenitor populations (30, 37, 218). Primary APCs characterized by the expression of Pdgfrα or Pdgfrβ can form an ectopic fat pad following transplantation into isogenic or immunodeficient mice (34, 37). Interestingly, Pdgfrβ+ cells isolated from tissues other than adipose failed to generate a similar fat pad following transplantation (37); this suggests adipose-resident perivascular cells contain determined precursors. Furthermore, elegant transplantation studies from Berry and Rodeheffer (31) have revealed functional differences between distinct progenitor populations. The CD24+, but not the CD24−, population of adipose progenitors can reconstitute a lipodystrophic mouse and at least partially rescue the adipose deficiency when transplanted into the residual WAT of these animals (31). On the other hand, only the CD24− population contains cells that can be viewed as determined; this population is capable of forming an ectopic fat pad when transplanted into regions outside the normal WAT microenvironment (e.g., sternum). This suggests that the CD24+ progenitor population consists of more primitive adipose tissue stem cells while the CD24− population contains more committed preadipocytes. Importantly, these functional differences between the two populations could not be delineated in vitro using existing methods.
These recent studies utilizing cell transplantation assays provide a framework for future studies of adipogenesis. In principle, the aforementioned populations can be isolated from various mouse models of interest and then transplanted appropriately in wild-type animals as a means to assay the effect of specific genetic perturbations on adipogenesis. It should be noted, however, that these assays are not purely an assay of differentiation per se. Factors that influence the engraftment, proliferation, or survival of these cells may effect the formation of a fat pad. In addition, these assays remain inconsistent and the variability in the sizes of fat tissues formed makes quantifying the results difficult. Nevertheless, in the absence of a true preadipocyte-specific Cre driver, these assays provide a complementary approach to understanding the biology of APCs.
Cellular adipocyte function in a dish
The ability to isolate or derive cultures of adipocytes provides the opportunity to explore some of the functional aspects of fat cells in a cell autonomous manner. Classic adipocyte assays include measurements of β agonist-induced lipolysis and insulin-stimulated glucose uptake. These assays are routinely used with 3T3 adipocytes or primary adipocytes from various adipose depots (150–152) and combined with molecular analyses of downstream signaling cascades (156, 157).
Another aspect of adipocyte biology that has become easier to explore is mitochondrial function. Essential stages of lipid and glucose metabolism take place in mitochondria, and mitochondria are crucial for adipokine secretion and insulin sensitivity (190). Mitochondrial function or abundance in adipocytes can be determined by gene expression analysis, histology, or flow cytometry. For instance, mitochondrial membrane carrier (mDIC) expression can reflect higher mitochondrial potential (189). In recent years, profiling of mitochondrial function in vitro has become much easier and effective with the development of the Seahorse analyzer system. Mitochondrial respiration can be determined by measuring oxygen consumption rate; mitochondrial glycolysis can be determined by measuring extracellular acidification rate (88, 189).
Many of the functional differences between brown and white adipocytes are retained in cultured cells. Brown adipocytes derived from interscapular BAT have high baseline levels of UCP1, Pgc-1α, and other factors of thermogenic gene program when compared with cultured white adipocytes. Importantly, in response to forskolin or β3-adrenergic receptor agonists, cultured brown and beige adipocytes exhibit a more robust activation of UCP1 expression and ultimately, uncoupled respiration (10). Interestingly, 3T3-L1 adipocytes appear resistant to this “browning” response; primary adipocytes derived from the inguinal WAT appear to have the greatest propensity to activate the thermogenic gene program in vitro and in vivo. This cellular model of thermogenic function has been extremely useful in delineating the function of proteins in brown and beige fat biology.
OPPORTUNITIES AHEAD
The field of adipose biology has evolved rapidly in last several years (2). Progress is now being made in unraveling the identity of the elusive native APCs. The unique features of anatomically distinct brown and white adipocytes in rodents and humans are becoming apparent. Moreover, the therapeutic potentials of adipokines are increasingly evident. The uncovered cellular biology and physiology have led to refinements in our technical approach to study adipogenesis and adipocyte metabolism. Similarly, new and improved mouse models now provide the opportunity to address key biological questions that have yet to be explored. An immediate goal for the field is to understand how adipose tissue mass is regulated by physiological and dietary cues in a depot-specific manner; combining the AdipoChaser model with new tools for the study of APCs should help in this endeavor. Inducible Cre-loxP recombination systems will also allow us to examine the function of our genes of interest both during development as well as in adult animals.
Despite the progress we describe, a number of important obstacles remain. A true preadipocyte-selective Cre line does not currently exist; gene targeting specifically in APCs remains a huge challenge. Further profiling and characterization of the recently identified progenitor populations may help reveal a suitable marker. Furthermore, mouse models that enable investigators to genetically manipulate fat cells in a depot-specific manner would be extremely useful in understanding the biology of anatomically distinct brown/beige and white adipocytes. It has become certain that subcutaneous and visceral white adipocytes stably express a distinct set of genes; however, whether any of these genes can help lead to the derivation of WAT depot-specific Cre lines remains unclear, as the gene expression differences are merely quantitative in nature and hardly ever in the form of “all on” or “all off.”
For most of the 20th century, white adipocytes were viewed as mere energy-storing cells that serve as the reservoir for excess energy. However, it is now clear that adipocytes are quite complex and regulate many facets of energy metabolism, physiology, and disease. This extraordinary complexity creates many challenges for the field; however, it also provides great opportunities for unique therapeutic intervention. The tools and methodology available to adipose biologists are quickly evolving and it now seems certain that we will continue to rapidly gain new insight into this dynamic cell type.
Acknowledgments
The authors would like to thank Richard Howdy, Certified Medical Illustrator, at Visually Medical Inc. for help with the graphic arts.
Footnotes
Abbreviations:
- APC
- adipose progenitor cell
- ATGL
- adipose TG lipase
- ATM
- adipose tissue macrophage
- BAT
- brown adipose tissue
- DMI
- dexamethasone, iso-butyl-methyl-xanthine, and insulin
- DNL
- de novo lipogenesis
- ECM
- extracellular matrix
- FACS
- fluorescent-activated cell sorting
- HFD
- high-fat diet
- Hif-1α
- hypoxia-inducible factor-1α HSL, hormone-sensitive lipase
- Pgdfrα
- platelet-derived growth factor receptor α SVC, stromal vascular culture
- SVF
- stromal vascular fraction
- Ucp1
- uncoupling protein 1
- WAT
- white adipose tissue
This work was supported by National Institutes of Health Grants K01-DK090120-02 and R03-DK099428, and the Searle Scholars Program (Chicago, Illinois) (R.K.G.), National Institutes of Health Grants R01-DK55758, P01-DK088761, and R01-DK099110 (P.E.S.), and a postdoctoral fellowship from the American Diabetes Association (7-11-MN-47) (Q.A.W.). The authors declare that they have no competing financial interests.
REFERENCES
- 1.Green H., Kehinde O. 1974. Sublines of mouse 3T3 cells that accumulate lipid. Cell. 1: 113–116. [Google Scholar]
- 2.Rosen E. D., Spiegelman B. M. 2014. What we talk about when we talk about fat. Cell. 156: 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cristancho A. G., Lazar M. A. 2011. Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 12: 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannon B., Nedergaard J. 2004. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84: 277–359. [DOI] [PubMed] [Google Scholar]
- 5.Gesta S., Bluher M., Yamamoto Y., Norris A. W., Berndt J., Kralisch S., Boucher J., Lewis C., Kahn C. R. 2006. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc. Natl. Acad. Sci. USA. 103: 6676–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vidal H. 2001. Gene expression in visceral and subcutaneous adipose tissues. Ann. Med. 33: 547–555. [DOI] [PubMed] [Google Scholar]
- 7.Vohl M. C., Sladek R., Robitaille J., Gurd S., Marceau P., Richard D., Hudson T. J., Tchernof A. 2004. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes. Res. 12: 1217–1222. [DOI] [PubMed] [Google Scholar]
- 8.Kissebah A. H., Krakower G. R. 1994. Regional adiposity and morbidity. Physiol. Rev. 74: 761–811. [DOI] [PubMed] [Google Scholar]
- 9.Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scime A., Devarakonda S., Conroe H. M., Erdjument-Bromage H., et al. 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 454: 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J., Boström P., Sparks L. M., Ye L., Choi J. H., Giang A-H., Khandekar M., Virtanen K. A., Nuutila P., Schaart G., et al. 2012. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 150: 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J., Cohen P., Spiegelman B. M. 2013. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes Dev. 27: 234–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villanueva C. J., Vergnes L., Wang J., Drew B. G., Hong C., Tu Y., Hu Y., Peng X., Xu F., Saez E., et al. 2013. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARγ specifies lipid storage versus thermogenic gene programs. Cell Metab. 17: 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seale P., Kajimura S., Yang W., Chin S., Rohas L. M., Uldry M., Tavernier G., Langin D., Spiegelman B. M. 2007. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 6: 38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B. M. 2006. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 3: 333–341. [DOI] [PubMed] [Google Scholar]
- 15.Rajakumari S., Wu J., Ishibashi J., Lim H. W., Giang A. H., Won K. J., Reed R. R., Seale P. 2013. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 17: 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 92: 829–839. [DOI] [PubMed] [Google Scholar]
- 17.Cohen P., Levy J. D., Zhang Y., Frontini A., Kolodin D. P., Svensson K. J., Lo J. C., Zeng X., Ye L., Khandekar M. J., et al. 2014. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 156: 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee K. Y., Yamamoto Y., Boucher J., Winnay J. N., Gesta S., Cobb J., Blüher M., Kahn C. R. 2013. Shox2 is a molecular determinant of depot-specific adipocyte function. Proc. Natl. Acad. Sci. USA. 110: 11409–11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu L., Panel V., Ma X., Du C., Hugendubler L., Gavrilova O., Liu A., McLaughlin T., Kaestner K. H., Mueller E. 2013. The winged helix transcription factor foxa3 regulates adipocyte differentiation and depot-selective fat tissue expansion. Mol. Cell. Biol. 33: 3392–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaworski K., Sarkadi-Nagy E., Duncan R. E., Ahmadian M., Sul H. S. 2007. Regulation of triglyceride metabolism. IV. Hormonal regulation of lipolysis in adipose tissue. Am. J. Physiol. Gastrointest. Liver Physiol. 293: G1–G4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohm M., Sommerfeld A., Strzoda D., Jones A., Sijmonsma T. P., Rudofsky G., Wolfrum C., Sticht C., Gretz N., Zeyda M., et al. 2013. Transcriptional cofactor TBLR1 controls lipid mobilization in white adipose tissue. Cell Metab. 17: 575–585. [DOI] [PubMed] [Google Scholar]
- 22.Fujimoto Y., Nakagawa Y., Satoh A., Okuda K., Shingyouchi A., Naka A., Matsuzaka T., Iwasaki H., Kobayashi K., Yahagi N., et al. 2013. TFE3 controls lipid metabolism in adipose tissue of male mice by suppressing lipolysis and thermogenesis. Endocrinology. 154: 3577–3588. [DOI] [PubMed] [Google Scholar]
- 23.Eguchi J., Wang X., Yu S., Kershaw E. E., Chiu P. C., Dushay J., Estall J. L., Klein U., Maratos-Flier E., Rosen E. D. 2011. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 13: 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodbell M. 1964. Metabolism of isolated fat cells. I. Effects of hormones on glucose metabolism and lipolysis. J. Biol. Chem. 239: 375–380. [PubMed] [Google Scholar]
- 25.Hollenberg C. H., Vost A. 1969. Regulation of DNA synthesis in fat cells and stromal elements from rat adipose tissue. J. Clin. Invest. 47: 2485–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry R., Jeffery E., Rodeheffer M. S. 2014. Weighing in on adipocyte precursors. Cell Metab. 19: 8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crossno J. T., Jr, Majka S. M., Grazia T., Gill R. G., Klemm D. J. 2006. Rosiglitazone promotes development of a novel adipocyte population from bone marrow-derived circulating progenitor cells. J. Clin. Invest. 116: 3220–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majka S. M., Barak Y., Klemm D. J. 2011. Concise review: adipocyte origins: weighing the possibilities. Stem Cells. 29: 1034–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majka S. M., Fox K. E., Psilas J. C., Helm K. M., Childs C. R., Acosta A. S., Janssen R. C., Friedman J. E., Woessner B. T., Shade T. R., et al. 2010. De novo generation of white adipocytes from the myeloid lineage via mesenchymal intermediates is age, adipose depot, and gender specific. Proc. Natl. Acad. Sci. USA. 107: 14781–14786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodeheffer M. S., Birsoy K., Friedman J. M. 2008. Identification of white adipocyte progenitor cells in vivo. Cell. 135: 240–249. [DOI] [PubMed] [Google Scholar]
- 31.Berry R., Rodeheffer M. S. 2013. Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol. 15: 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takakura N., Yoshida H., Ogura Y., Kataoka H., Nishikawa S. 1997. PDGFR alpha expression during mouse embryogenesis: immunolocalization analyzed by whole-mount immunohistostaining using the monoclonal anti-mouse PDGFR alpha antibody APA5. J. Histochem. Cytochem. 45: 883–893. [DOI] [PubMed] [Google Scholar]
- 33.Joe A. W., Yi L., Natarajan A., Le Grand F., So L., Wang J., Rudnicki M. A., Rossi F. M. 2010. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 12: 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y. H., Petkova A. P., Mottillo E. P., Granneman J. G. 2012. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metab. 15: 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uezumi A., Fukada S., Yamamoto N., Takeda S., Tsuchida K. 2010. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 12: 143–152. [DOI] [PubMed] [Google Scholar]
- 36.Wassermann F. 1926. The fat organs of man: development, structure and systematic place of the so-called adipose tissue. Z. Zellforsch. Microskop. Anat. Abt. Histochem. 3: 325–329. [Google Scholar]
- 37.Tang W., Zeve D., Suh J. M., Bosnakovski D., Kyba M., Hammer R. E., Tallquist M. D., Graff J. M. 2008. White fat progenitor cells reside in the adipose vasculature. Science. 322: 583–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta R. K., Mepani R. J., Kleiner S., Lo J. C., Khandekar M. J., Cohen P., Frontini A., Bhowmick D. C., Ye L., Cinti S., et al. 2012. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 15: 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wassermann F. 1960. Electron microscopic investigation of the surface structures of the fat cell and of their changes during depletion of cell. Z. Zellforsch. Microsk. Anat. 52: 778–800. [Google Scholar]
- 40.Napolitano L. 1963. The differentiation of white adipose cells. an electron microscope study. J. Cell Biol. 18: 663–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shan T., Liu W., Kuang S. 2013. Fatty acid binding protein 4 expression marks a population of adipocyte progenitors in white and brown adipose tissues. FASEB J. 27: 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Armani A., Mammi C., Marzolla V., Calanchini M., Antelmi A., Rosano G. M., Fabbri A., Caprio M. 2010. Cellular models for understanding adipogenesis, adipose dysfunction, and obesity. J. Cell. Biochem. 110: 564–572. [DOI] [PubMed] [Google Scholar]
- 43.Poulos S. P., Dodson M. V., Hausman G. J. 2010. Cell line models for differentiation: preadipocytes and adipocytes. Exp. Biol. Med. (Maywood). 235: 1185–1193. [DOI] [PubMed] [Google Scholar]
- 44.Wolins N. E., Quaynor B. K., Skinner J. R., Tzekov A., Park C., Choi K., Bickel P. E. 2006. OP9 mouse stromal cells rapidly differentiate into adipocytes: characterization of a useful new model of adipogenesis. J. Lipid Res. 47: 450–460. [DOI] [PubMed] [Google Scholar]
- 45.Green H., Meuth M. 1974. An established pre-adipose cell line and its differentiation in culture. Cell. 3: 127–133. [DOI] [PubMed] [Google Scholar]
- 46.Green H., Kehinde O. 1975. An established preadipose cell line and its differentiation in culture II. Factors affecting the adipose conversion. Cell. 5: 19–27. [DOI] [PubMed] [Google Scholar]
- 47.Kuri-Harcuch W., Green H. 1977. Increasing activity of enzymes on pathway of triacylglycerol synthesis during adipose conversion of 3T3 cells. J. Biol. Chem. 252: 2158–2160. [PubMed] [Google Scholar]
- 48.Mandrup S., Loftus T. M., MacDougald O. A., Kuhajda F. P., Lane M. D. 1997. Obese gene expression at in vivo levels by fat pads derived from s.c. implanted 3T3-F442A preadipocytes. Proc. Natl. Acad. Sci. USA. 94: 4300–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farmer S. R. 2006. Transcriptional control of adipocyte formation. Cell Metab. 4: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta R. K., Arany Z., Seale P., Mepani R. J., Ye L., Conroe H. M., Roby Y. A., Kulaga H., Reed R. R., Spiegelman B. M. 2010. Transcriptional control of preadipocyte determination by Zfp423. Nature. 464: 619–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cristancho A. G., Schupp M., Lefterova M. I., Cao S., Cohen D. M., Chen C. S., Steger D. J., Lazar M. A. 2011. Repressor transcription factor 7-like 1 promotes adipogenic competency in precursor cells. Proc. Natl. Acad. Sci. USA. 108: 16271–16276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Todaro G. J., Green H. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17: 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siersbæk R., Nielsen R., John S., Sung M. H., Baek S., Loft A., Hager G. L., Mandrup S. 2011. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J. 30: 1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nielsen R., Pedersen T. A., Hagenbeek D., Moulos P., Siersbaek R., Megens E., Denissov S., Borgesen M., Francoijs K. J., Mandrup S., et al. 2008. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 22: 2953–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steger D. J., Grant G. R., Schupp M., Tomaru T., Lefterova M. I., Schug J., Manduchi E., Stoeckert C. J., Jr, Lazar M. A. 2010. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 24: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mikkelsen T. S., Xu Z., Zhang X., Wang L., Gimble J. M., Lander E. S., Rosen E. D. 2010. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 143: 156–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu E., Liang P., Spiegelman B. M. 1996. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J. Biol. Chem. 271: 10697–10703. [DOI] [PubMed] [Google Scholar]
- 58.Steppan C. M., Bailey S. T., Bhat S., Brown E. J., Banerjee R. R., Wright C. M., Patel H. R., Ahima R. S., Lazar M. A. 2001. The hormone resistin links obesity to diabetes. Nature. 409: 307–312. [DOI] [PubMed] [Google Scholar]
- 59.Scherer P. E., Williams S., Fogliano M., Baldini G., Lodish H. F. 1995. A novel serum protein similar to C1q, produced exclusively in adipocytes. J. Biol. Chem. 270: 26746–26749. [DOI] [PubMed] [Google Scholar]
- 60.Chawla A., Lazar M. A. 1994. Peroxisome proliferator and retinoid signaling pathways co-regulate preadipocyte phenotype and survival. Proc. Natl. Acad. Sci. USA. 91: 1786–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tontonoz P., Hu E., Spiegelman B. M. 1994. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 79: 1147–1156. [DOI] [PubMed] [Google Scholar]
- 62.Soukas A., Socci N. D., Saatkamp B. D., Novelli S., Friedman J. M. 2001. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J. Biol. Chem. 276: 34167–34174. [DOI] [PubMed] [Google Scholar]
- 63.Rentsch J., Chiesi M. 1996. Regulation of ob gene mRNA levels in cultured adipocytes. FEBS Lett. 379: 55–59. [DOI] [PubMed] [Google Scholar]
- 64.Van R. L., Roncari D. A. 1977. Isolation of fat cell precursors from adult rat adipose tissue. Cell Tissue Res. 181: 197–203. [DOI] [PubMed] [Google Scholar]
- 65.Poznanski W. J., Waheed I., Van R. 1973. Human fat cell precursors. Morphologic and metabolic differentiation in culture. Lab. Invest. 29: 570–576. [PubMed] [Google Scholar]
- 66.Van R. L., Bayliss C. E., Roncari D. A. 1976. Cytological and enzymological characterization of adult human adipocyte precursors in culture. J. Clin. Invest. 58: 699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deleted in proof.
- 68.Cawthorn W. P., Scheller E. L., MacDougald O. A. 2012. Adipose tissue stem cells meet preadipocyte commitment: going back to the future. J. Lipid Res. 53: 227–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fasshauer M., Klein J., Ueki K., Kriauciunas K. M., Benito M., White M. F., Kahn C. R. 2000. Essential role of insulin receptor substrate-2 in insulin stimulation of Glut4 translocation and glucose uptake in brown adipocytes. J. Biol. Chem. 275: 25494–25501. [DOI] [PubMed] [Google Scholar]
- 70.Tchkonia T., Giorgadze N., Pirtskhalava T., Thomou T., DePonte M., Koo A., Forse R. A., Chinnappan D., Martin-Ruiz C., von Zglinicki T., et al. 2006. Fat depot-specific characteristics are retained in strains derived from single human preadipocytes. Diabetes. 55: 2571–2578. [DOI] [PubMed] [Google Scholar]
- 71.Macotela Y., Emanuelli B., Mori M. A., Gesta S., Schulz T. J., Tseng Y. H., Kahn C. R. 2012. Intrinsic differences in adipocyte precursor cells from different white fat depots. Diabetes. 61: 1691–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maslowska M. H., Sniderman A. D., MacLean L. D., Cianflone K. 1993. Regional differences in triacylglycerol synthesis in adipose tissue and in cultured preadipocytes. J. Lipid Res. 34: 219–228. [PubMed] [Google Scholar]
- 73.Wang Q. A., Tao C., Gupta R. K., Scherer P. E. 2013. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med. 19: 1338–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karastergiou K., Smith S. R., Greenberg A. S., Fried S. K. 2012. Sex differences in human adipose tissues - the biology of pear shape. Biol. Sex Differ. 3: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shi H., Seeley R. J., Clegg D. J. 2009. Sexual differences in the control of energy homeostasis. Front. Neuroendocrinol. 30: 396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davis K. E., Neinast M. D., Sun K., Skiles W. M., Bills J. D., Zehr J. A., Zeve D., Hahner L. D., Cox D. W., Gent L. M., et al. 2013. The sexually dimorphic role of adipose and adipocyte estrogen receptors in modulating adipose tissue expansion, inflammation, and fibrosis. Mol. Metab. 2: 227–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tchkonia T., Morbeck D. E., Von Zglinicki T., Van Deursen J., Lustgarten J., Scrable H., Khosla S., Jensen M. D., Kirkland J. L. 2010. Fat tissue, aging, and cellular senescence. Aging Cell. 9: 667–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ando H., Yanagihara H., Hayashi Y., Obi Y., Tsuruoka S., Takamura T., Kaneko S., Fujimura A. 2005. Rhythmic messenger ribonucleic acid expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology. 146: 5631–5636. [DOI] [PubMed] [Google Scholar]
- 79.Zvonic S., Ptitsyn A. A., Conrad S. A., Scott L. K., Floyd Z. E., Kilroy G., Wu X., Goh B. C., Mynatt R. L., Gimble J. M. 2006. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 55: 962–970. [DOI] [PubMed] [Google Scholar]
- 80.Bray M. S., Young M. E. 2007. Circadian rhythms in the development of obesity: potential role for the circadian clock within the adipocyte. Obes. Rev. 8: 169–181. [DOI] [PubMed] [Google Scholar]
- 81.Gerhart-Hines Z., Feng D., Emmett M. J., Everett L. J., Loro E., Briggs E. R., Bugge A., Hou C., Ferrara C., Seale P., et al. 2013. The nuclear receptor Rev-erbα controls circadian thermogenic plasticity. Nature. 503: 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hirsch J., Han P. W. 1969. Cellularity of rat adipose tissue: effects of growth, starvation, and obesity. J. Lipid Res. 10: 77–82. [PubMed] [Google Scholar]
- 83.Stiles J. W., Francendese A. A., Masoro E. J. 1975. Influence of age on size and number of fat cells in the epididymal depot. Am. J. Physiol. 229: 1561–1568. [DOI] [PubMed] [Google Scholar]
- 84.Yang M. U., Presta E., Bjorntorp P. 1990. Refeeding after fasting in rats: effects of duration of starvation and refeeding on food efficiency in diet-induced obesity. Am. J. Clin. Nutr. 51: 970–978. [DOI] [PubMed] [Google Scholar]
- 85.Spalding K. L., Arner E., Westermark P. O., Bernard S., Buchholz B. A., Bergmann O., Blomqvist L., Hoffstedt J., Naslund E., Britton T., et al. 2008. Dynamics of fat cell turnover in humans. Nature. 453: 783–787. [DOI] [PubMed] [Google Scholar]
- 86.Sun K., Kusminski C. M., Scherer P. E. 2011. Adipose tissue remodeling and obesity. J. Clin. Invest. 121: 2094–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim J-Y., van de Wall E., Laplante M., Azzara A., Trujillo M. E., Hofmann S. M., Schraw T., Durand J. L., Li H., Li G., et al. 2007. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J. Clin. Invest. 117: 2621–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kusminski C. M., Holland W. L., Sun K., Park J., Spurgin S. B., Lin Y., Askew G. R., Simcox J. A., McClain D. A., Li C., et al. 2012. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat. Med. 18: 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jo J., Gavrilova O., Pack S., Jou W., Mullen S., Sumner A. E., Cushman S. W., Periwal V. 2009. Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth. PLOS Comput. Biol. 5: e1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Joe A. W., Yi L., Even Y., Vogl A. W., Rossi F. 2009. Depot-specific differences in adipogenic progenitor abundance and proliferative response to high-fat diet. Stem Cells. 27: 2563–2570. [DOI] [PubMed] [Google Scholar]
- 91.Dor Y., Brown J., Martinez O. I., Melton D. A. 2004. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 429: 41–46. [DOI] [PubMed] [Google Scholar]
- 92.Jopling C., Sleep E., Raya M., Marti M., Raya A., Izpisua Belmonte J. C. 2010. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 464: 606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun K., Asterholm I. W., Kusminski C. M., Bueno A. C., Wang Z. V., Pollard J. W., Brekken R. A., Scherer P. E. 2012. Dichotomous effects of VEGF-A on adipose tissue dysfunction. Proc. Natl. Acad. Sci. USA. 109: 5874–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perl A-K. T., Wert S. E., Nagy A., Lobe C. G., Whitsett J. A. 2002. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc. Natl. Acad. Sci. USA. 99: 10482–10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Soriano P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21: 70–71. [DOI] [PubMed] [Google Scholar]
- 96.Himms-Hagen J., Melnyk A., Zingaretti M. C., Ceresi E., Barbatelli G., Cinti S. 2000. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. Am. J. Physiol. Cell Physiol. 279: C670–C681. [DOI] [PubMed] [Google Scholar]
- 97.Barbatelli G., Murano I., Madsen L., Hao Q., Jimenez M., Kristiansen K., Giacobino J. P., De Matteis R., Cinti S. 2010. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am. J. Physiol. Endocrinol. Metab. 298: E1244–E1253. [DOI] [PubMed] [Google Scholar]
- 98.Rosenwald M., Perdikari A., Rülicke T., Wolfrum C. 2013. Bi-directional interconversion of brite and white adipocytes. Nat. Cell Biol. 15: 659–667. [DOI] [PubMed] [Google Scholar]
- 99.Rivers L. E., Young K. M., Rizzi M., Jamen F., Psachoulia K., Wade A., Kessaris N., Richardson W. D. 2008. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat. Neurosci. 11: 1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Spiegelman B. M., Flier J. S. 2001. Obesity and the regulation of energy balance. Cell. 104: 531–543. [DOI] [PubMed] [Google Scholar]
- 101.Kühn R., Torres R. M. 2002. Cre/loxP recombination system and gene targeting. Methods Mol. Biol. 180: 175–204. [DOI] [PubMed] [Google Scholar]
- 102.He W., Barak Y., Hevener A., Olson P., Liao D., Le J., Nelson M., Ong E., Olefsky J. M., Evans R. M. 2003. Adipose-specific peroxisome proliferator-activated receptor γ knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. USA. 100: 15712–15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barlow C., Schroeder M., Lekstrom-Himes J., Kylefjord H., Deng C-X., Wynshaw-Boris A., Spiegelman B. M., Xanthopoulos K. G. 1997. Targeted expression of Cre recombinase to adipose tissue of transgenic mice directs adipose-specific excision of loxP-flanked gene segments. Nucleic Acids Res. 25: 2543–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ross S. R., Graves R. A., Greenstein A., Platt K. A., Shyu H. L., Mellovitz B., Spiegelman B. M. 1990. A fat-specific enhancer is the primary determinant of gene expression for adipocyte P2 in vivo. Proc. Natl. Acad. Sci. USA. 87: 9590–9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Graves R. A., Tontonoz P., Platt K. A., Ross S. R., Spiegelman B. M. 1992. Identification of a fat cell enhancer: Analysis of requirements for adipose tissue-specific gene expression. J. Cell. Biochem. 49: 219–224. [DOI] [PubMed] [Google Scholar]
- 106.Lee K. Y., Russell S. J., Ussar S., Boucher J., Vernochet C., Mori M. A., Smyth G., Rourk M., Cederquist C., Rosen E. D., et al. 2013. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 62: 864–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mullican S. E., Tomaru T., Gaddis C. A., Peed L. C., Sundaram A., Lazar M. A. 2013. A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Mol. Endocrinol. 27: 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Makowski L., Boord J. B., Maeda K., Babaev V. R., Uysal K. T., Morgan M. A., Parker R. A., Suttles J., Fazio S., Hotamisligil G. S., et al. 2001. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat. Med. 7: 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Urs S., Harrington A., Liaw L., Small D. 2006. Selective expression of an aP2/fatty acid binding protein 4-Cre transgene in non-adipogenic tissues during embryonic development. Transgenic Res. 15: 647–653. [DOI] [PubMed] [Google Scholar]
- 110.Wang Z. V., Deng Y., Wang Q. A., Sun K., Scherer P. E. 2010. Identification and characterization of a promoter cassette conferring adipocyte-specific gene expression. Endocrinology. 151: 2933–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Deleted in proof.
- 112.Herman M. A., Peroni O. D., Villoria J., Schon M. R., Abumrad N. A., Bluher M., Klein S., Kahn B. B. 2012. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 484: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kleiner S., Mepani R. J., Laznik D., Ye L., Jurczak M. J., Jornayvaz F. R., Estall J. L., Chatterjee Bhowmick D., Shulman G. I., Spiegelman B. M. 2012. Development of insulin resistance in mice lacking PGC-1α in adipose tissues. Proc. Natl. Acad. Sci. USA. 109: 9635–9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zeve D., Seo J., Suh J. M., Stenesen D., Tang W., Berglund E. D., Wan Y., Williams L. J., Lim A., Martinez M. J., et al. 2012. Wnt signaling activation in adipose progenitors promotes insulin-independent muscle glucose uptake. Cell Metab. 15: 492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tallquist M. D., Weismann K. E., Hellstrom M., Soriano P. 2000. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 127: 5059–5070. [DOI] [PubMed] [Google Scholar]
- 116.Lepper C., Fan C. M. 2010. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis. 48: 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ohno H., Shinoda K., Ohyama K., Sharp L. Z., Kajimura S. 2013. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 504: 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sanchez-Gurmaches J., Hung C. M., Sparks C. A., Tang Y., Li H., Guertin D. A. 2012. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 16: 348–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schulz T. J., Huang P., Huang T. L., Xue R., McDougall L. E., Townsend K. L., Cypess A. M., Mishina Y., Gussoni E., Tseng Y. H. 2013. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature. 495: 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rudnicki M. A., Schnegelsberg P. N., Stead R. H., Braun T., Arnold H-H., Jaenisch R. 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 75: 1351–1359. [DOI] [PubMed] [Google Scholar]
- 121.Borycki A-G., Brunk B., Tajbakhsh S., Buckingham M., Chiang C., Emerson C. 1999. Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Development. 126: 4053–4063. [DOI] [PubMed] [Google Scholar]
- 122.Guerra C., Navarro P., Valverde A. M., Arribas M., Brüning J., Kozak L. P., Kahn C. R., Benito M. 2001. Brown adipose tissue-specific insulin receptor knockout shows diabetic phenotype without insulin resistance. J. Clin. Invest. 108: 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Imai T., Jiang M., Chambon P., Metzger D. 2001. Impaired adipogenesis and lipolysis in the mouse upon selective ablation of the retinoid X receptor α mediated by a tamoxifen-inducible chimeric Cre recombinase (Cre-ERT2) in adipocytes. Proc. Natl. Acad. Sci. USA. 98: 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sassmann A., Offermanns S., Wettschureck N. 2010. Tamoxifen-inducible Cre-mediated recombination in adipocytes. Genesis. 48: 618–625. [DOI] [PubMed] [Google Scholar]
- 125.Barros R. P., Gabbi C., Morani A., Warner M., Gustafsson J-Å. 2009. Participation of ERα and ERβ in glucose homeostasis in skeletal muscle and white adipose tissue. Am. J. Physiol. Endocrinol. Metab. 297: E124–E133. [DOI] [PubMed] [Google Scholar]
- 126.Huh W. J., Khurana S. S., Geahlen J. H., Kohli K., Waller R. A., Mills J. C. 2012. Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology. 142: 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Blüher M. 2013. Importance of estrogen receptors in adipose tissue function. Mol. Metab. 2: 130–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Reinert R. B., Kantz J., Misfeldt A. A., Poffenberger G., Gannon M., Brissova M., Powers A. C. 2012. Tamoxifen-induced Cre-loxP recombination is prolonged in pancreatic islets of adult mice. PLoS ONE. 7: e33529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tschöp M. H., Speakman J. R., Arch J. R., Auwerx J., Brüning J. C., Chan L., Eckel R. H., Farese R. V., Jr, Galgani J. E., Hambly C., et al. 2012. A guide to analysis of mouse energy metabolism. Nat. Methods. 9: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang J., Zhang Y., Sun T., Guo F., Huang S., Chandalia M., Abate N., Fan D., Xin H. B., Chen Y. E., et al. 2013. Dietary obesity-induced Egr-1 in adipocytes facilitates energy storage via suppression of FOXC2. Sci. Rep. 3: 1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Alekseev A. E., Reyes S., Yamada S., Hodgson-Zingman D. M., Sattiraju S., Zhu Z., Sierra A., Gerbin M., Coetzee W. A., Goldhamer D. J., et al. 2010. Sarcolemmal ATP-sensitive K(+) channels control energy expenditure determining body weight. Cell Metab. 11: 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fischer-Posovszky P., Wang Q. A., Asterholm I. W., Rutkowski J. M., Scherer P. E. 2011. Targeted deletion of adipocytes by apoptosis leads to adipose tissue recruitment of alternatively activated M2 macrophages. Endocrinology. 152: 3074–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Guilherme A., Virbasius J. V., Puri V., Czech M. P. 2008. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 9: 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ouchi N., Parker J. L., Lugus J. J., Walsh K. 2011. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 11: 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lumeng C. N., Bodzin J. L., Saltiel A. R. 2007. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 117: 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lumeng C. N., DelProposto J. B., Westcott D. J., Saltiel A. R. 2008. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes. 57: 3239–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sung H. K., Doh K. O., Son J. E., Park J. G., Bae Y., Choi S., Nelson S. M., Cowling R., Nagy K., Michael I. P., et al. 2013. Adipose vascular endothelial growth factor regulates metabolic homeostasis through angiogenesis. Cell Metab. 17: 61–72. [DOI] [PubMed] [Google Scholar]
- 138.Nishimura S., Manabe I., Nagasaki M., Eto K., Yamashita H., Ohsugi M., Otsu M., Hara K., Ueki K., Sugiura S., et al. 2009. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat. Med. 15: 914–920. [DOI] [PubMed] [Google Scholar]
- 139.Stolarczyk E., Vong C. T., Perucha E., Jackson I., Cawthorne M. A., Wargent E. T., Powell N., Canavan J. B., Lord G. M., Howard J. K. 2013. Improved insulin sensitivity despite increased visceral adiposity in mice deficient for the immune cell transcription factor T-bet. Cell Metab. 17: 520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Feng D., Tang Y., Kwon H., Zong H., Hawkins M., Kitsis R. N., Pessin J. E. 2011. High-fat diet-induced adipocyte cell death occurs through a cyclophilin D intrinsic signaling pathway independent of adipose tissue inflammation. Diabetes. 60: 2134–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sun K., Halberg N., Khan M., Magalang U. J., Scherer P. E. 2013. Selective inhibition of hypoxia-inducible factor 1alpha ameliorates adipose tissue dysfunction. Mol. Cell. Biol. 33: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Semenza G. L. 2000. Surviving ischemia: adaptive responses mediated by hypoxia-inducible factor 1. J. Clin. Invest. 106: 809–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ozcan U., Cao Q., Yilmaz E., Lee A-H., Iwakoshi N. N., Ozdelen E., Tuncman G., Gorgun C., Glimcher L. H., Hotamisligil G. S. 2004. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 306: 457–461. [DOI] [PubMed] [Google Scholar]
- 144.Hosogai N., Fukuhara A., Oshima K., Miyata Y., Tanaka S., Segawa K., Furukawa S., Tochino Y., Komuro R., Matsuda M., et al. 2007. Adipose tissue hypoxia in obesity and its impact on adipocytokine dysregulation. Diabetes. 56: 901–911. [DOI] [PubMed] [Google Scholar]
- 145.Huang G., Greenspan D. S. 2012. ECM roles in the function of metabolic tissues. Trends Endocrinol. Metab. 23: 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Khan T., Muise E. S., Iyengar P., Wang Z. V., Chandalia M., Abate N., Zhang B. B., Bonaldo P., Chua S., Scherer P. E. 2009. Metabolic dysregulation and adipose tissue fibrosis: role of collagen VI. Mol. Cell. Biol. 29: 1575–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Divoux A., Tordjman J., Lacasa D., Veyrie N., Hugol D., Aissat A., Basdevant A., Guerre-Millo M., Poitou C., Zucker J. D., et al. 2010. Fibrosis in human adipose tissue: composition, distribution, and link with lipid metabolism and fat mass loss. Diabetes. 59: 2817–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jonker J. W., Suh J. M., Atkins A. R., Ahmadian M., Li P., Whyte J., He M., Juguilon H., Yin Y. Q., Phillips C. T., et al. 2012. A PPARgamma-FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature. 485: 391–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Halberg N., Khan T., Trujillo M. E., Wernstedt-Asterholm I., Attie A. D., Sherwani S., Wang Z. V., Landskroner-Eiger S., Dineen S., Magalang U. J., et al. 2009. Hypoxia-inducible factor 1alpha induces fibrosis and insulin resistance in white adipose tissue. Mol. Cell. Biol. 29: 4467–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Miki T., Minami K., Zhang L., Morita M., Gonoi T., Shiuchi T., Minokoshi Y., Renaud J-M., Seino S. 2002. ATP-sensitive potassium channels participate in glucose uptake in skeletal muscle and adipose tissue. Am. J. Physiol. Endocrinol. Metab. 283: E1178–E1184. [DOI] [PubMed] [Google Scholar]
- 151.Mittendorfer B. 2005. Insulin resistance: sex matters. Curr. Opin. Clin. Nutr. Metab. Care. 8: 367–372. [DOI] [PubMed] [Google Scholar]
- 152.Abel E. D., Peroni O., Kim J. K., Kim Y. B., Boss O., Hadro E., Minnemann T., Shulman G. I., Kahn B. B. 2001. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 409: 729–733. [DOI] [PubMed] [Google Scholar]
- 153.Voshol P. J., Jong M. C., Dahlmans V. E. H., Kratky D., Levak-Frank S., Zechner R., Romijn J. A., Havekes L. M. 2001. In muscle-specific lipoprotein lipase-overexpressing mice, muscle triglyceride content is increased without inhibition of insulin-stimulated whole-body and muscle-specific glucose uptake. Diabetes. 50: 2585–2590. [DOI] [PubMed] [Google Scholar]
- 154.Jones J. R., Barrick C., Kim K-A., Lindner J., Blondeau B., Fujimoto Y., Shiota M., Kesterson R. A., Kahn B. B., Magnuson M. A. 2005. Deletion of PPARγ in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc. Natl. Acad. Sci. USA. 102: 6207–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Arkan M. C., Hevener A. L., Greten F. R., Maeda S., Li Z-W., Long J. M., Wynshaw-Boris A., Poli G., Olefsky J., Karin M. 2005. IKK-β links inflammation to obesity-induced insulin resistance. Nat. Med. 11: 191–198. [DOI] [PubMed] [Google Scholar]
- 156.Guo H., Jin D., Zhang Y., Wright W., Bazuine M., Brockman D. A., Bernlohr D. A., Chen X. 2010. Lipocalin-2 deficiency impairs thermogenesis and potentiates diet-induced insulin resistance in mice. Diabetes. 59: 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Macotela Y., Boucher J., Tran T. T., Kahn C. R. 2009. Sex and depot differences in adipocyte insulin sensitivity and glucose metabolism. Diabetes. 58: 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Le H. T., Ponniah S., Pallen C. J. 2004. Insulin signaling and glucose homeostasis in mice lacking protein tyrosine phosphatase α. Biochem. Biophys. Res. Commun. 314: 321–329. [DOI] [PubMed] [Google Scholar]
- 159.Yu X., Shen N., Zhang M. L., Pan F. Y., Wang C., Jia W. P., Liu C., Gao Q., Gao X., Xue B., et al. 2011. Egr-1 decreases adipocyte insulin sensitivity by tilting PI3K/Akt and MAPK signal balance in mice. EMBO J. 30: 3754–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sutanto M. M., Ferguson K. K., Sakuma H., Ye H., Brady M. J., Cohen R. N. 2010. The silencing mediator of retinoid and thyroid hormone receptors (SMRT) regulates adipose tissue accumulation and adipocyte insulin sensitivity in vivo. J. Biol. Chem. 285: 18485–18495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Unger R. H., Cherrington A. D. 2012. Glucagonocentric restructuring of diabetes: a pathophysiologic and therapeutic makeover. J. Clin. Invest. 122: 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Duncan R. E., Ahmadian M., Jaworski K., Sarkadi-Nagy E., Sul H. S. 2007. Regulation of lipolysis in adipocytes. Annu. Rev. Nutr. 27: 79–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Large V., Reynisdottir S., Langin D., Fredby K., Klannemark M., Holm C., Arner P. 1999. Decreased expression and function of adipocyte hormone-sensitive lipase in subcutaneous fat cells of obese subjects. J. Lipid Res. 40: 2059–2066. [PubMed] [Google Scholar]
- 164.Reynisdottir S., Langin D., Carlstrom K., Holm C., Rossner S., Arner P. 1995. Effects of weight reduction on the regulation of lipolysis in adipocytes of women with upper-body obesity. Clin. Sci. (Lond). 89: 421–429. [DOI] [PubMed] [Google Scholar]
- 165.Wang S. P., Laurin N., Himms-Hagen J., Rudnicki M. A., Levy E., Robert M. F., Pan L., Oligny L., Mitchell G. A. 2001. The adipose tissue phenotype of hormone-sensitive lipase deficiency in mice. Obes. Res. 9: 119–128. [DOI] [PubMed] [Google Scholar]
- 166.Holm C. 2003. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem. Soc. Trans. 31: 1120–1124. [DOI] [PubMed] [Google Scholar]
- 167.Vaughan M., Berger J. E., Steinberg D. 1964. Hormone-sensitive lipase and monoglyceride lipase activities in adipose tissue. J. Biol. Chem. 239: 401–409. [PubMed] [Google Scholar]
- 168.Villena J. A., Roy S., Sarkadi-Nagy E., Kim K. H., Sul H. S. 2004. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J. Biol. Chem. 279: 47066–47075. [DOI] [PubMed] [Google Scholar]
- 169.Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., et al. 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 306: 1383–1386. [DOI] [PubMed] [Google Scholar]
- 170.Haemmerle G., Zimmermann R., Hayn M., Theussl C., Waeg G., Wagner E., Sattler W., Magin T. M., Wagner E. F., Zechner R. 2002. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J. Biol. Chem. 277: 4806–4815. [DOI] [PubMed] [Google Scholar]
- 171.Lampidonis A. D., Rogdakis E., Voutsinas G. E., Stravopodis D. J. 2011. The resurgence of hormone-sensitive lipase (HSL) in mammalian lipolysis. Gene. 477: 1–11. [DOI] [PubMed] [Google Scholar]
- 172.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., et al. 2006. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 312: 734–737. [DOI] [PubMed] [Google Scholar]
- 173.Ahmadian M., Abbott M. J., Tang T., Hudak C. S., Kim Y., Bruss M., Hellerstein M. K., Lee H. Y., Samuel V. T., Shulman G. I., et al. 2011. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab. 13: 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3: 309–319. [DOI] [PubMed] [Google Scholar]
- 175.Soni K. G., Lehner R., Metalnikov P., O'Donnell P., Semache M., Gao W., Ashman K., Pshezhetsky A. V., Mitchell G. A. 2004. Carboxylesterase 3 (EC 3.1.1.1) is a major adipocyte lipase. J. Biol. Chem. 279: 40683–40689. [DOI] [PubMed] [Google Scholar]
- 176.Jenkins C. M., Mancuso D. J., Yan W., Sims H. F., Gibson B., Gross R. W. 2004. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem. 279: 48968–48975. [DOI] [PubMed] [Google Scholar]
- 177.Tornqvist H., Belfrage P. 1976. Purification and some properties of a monoacylglycerol-hydrolyzing enzyme of rat adipose tissue. J. Biol. Chem. 251: 813–819. [PubMed] [Google Scholar]
- 178.Brasaemle D. L., Rubin B., Harten I. A., Gruia-Gray J., Kimmel A. R., Londos C. 2000. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J. Biol. Chem. 275: 38486–38493. [DOI] [PubMed] [Google Scholar]
- 179.Souza S. C., Muliro K. V., Liscum L., Lien P., Yamamoto M. T., Schaffer J. E., Dallal G. E., Wang X., Kraemer F. B., Obin M., et al. 2002. Modulation of hormone-sensitive lipase and protein kinase A-mediated lipolysis by perilipin A in an adenoviral reconstituted system. J. Biol. Chem. 277: 8267–8272. [DOI] [PubMed] [Google Scholar]
- 180.Sztalryd C., Xu G., Dorward H., Tansey J. T., Contreras J. A., Kimmel A. R., Londos C. 2003. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J. Cell Biol. 161: 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tansey J. T., Huml A. M., Vogt R., Davis K. E., Jones J. M., Fraser K. A., Brasaemle D. L., Kimmel A. R., Londos C. 2003. Functional studies on native and mutated forms of perilipins. A role in protein kinase A-mediated lipolysis of triacylglycerols. J. Biol. Chem. 278: 8401–8406. [DOI] [PubMed] [Google Scholar]
- 182.Miyoshi H., Souza S. C., Zhang H. H., Strissel K. J., Christoffolete M. A., Kovsan J., Rudich A., Kraemer F. B., Bianco A. C., Obin M. S., et al. 2006. Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and -independent mechanisms. J. Biol. Chem. 281: 15837–15844. [DOI] [PubMed] [Google Scholar]
- 183.Brasaemle D. L. 2010. Lipolysis control: the plot thickens. Cell Metab. 11: 173–174. [DOI] [PubMed] [Google Scholar]
- 184.Marcinkiewicz A., Gauthier D., Garcia A., Brasaemle D. L. 2006. The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion. J. Biol. Chem. 281: 11901–11909. [DOI] [PubMed] [Google Scholar]
- 185.Moore H. P., Silver R. B., Mottillo E. P., Bernlohr D. A., Granneman J. G. 2005. Perilipin targets a novel pool of lipid droplets for lipolytic attack by hormone-sensitive lipase. J. Biol. Chem. 280: 43109–43120. [DOI] [PubMed] [Google Scholar]
- 186.Rohm M., Sommerfeld A., Strzoda D., Jones A., Sijmonsma T. P., Rudofsky G., Wolfrum C., Sticht C., Gretz N., Zeyda M., et al. 2013. Transcriptional cofactor TBLR1 controls lipid mobilization in white adipose tissue. Cell Metab. 17: 575–585. [DOI] [PubMed] [Google Scholar]
- 187.Mennes E., Dungan C. M., Frendo-Cumbo S., Williamson D. L., Wright D. C. Aging-associated reductions in lipolytic and mitochondrial proteins in mouse adipose tissue are not rescued by metformin treatment. J. Gerontol. A Biol. Sci. Med. Sci. Epub ahead of print. October 14, 2013; doi:10.1093/gerona/glt156. [DOI] [PubMed] [Google Scholar]
- 188.Grujic D., Susulic V. S., Harper M-E., Himms-Hagen J., Cunningham B. A., Corkey B. E., Lowell B. B. 1997. Beta3-adrenergic receptors on white and brown adipocytes mediate beta3-selective agonist-induced effects on energy expenditure, insulin secretion, and food intake. A study using transgenic and gene knockout mice. J. Biol. Chem. 272: 17686–17693. [DOI] [PubMed] [Google Scholar]
- 189.Asterholm I. W., Mundy D. I., Weng J., Anderson R. G., Scherer P. E. 2012. Altered mitochondrial function and metabolic inflexibility associated with loss of caveolin-1. Cell Metab. 15: 171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kusminski C. M., Scherer P. E. 2012. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol. Metab. 23: 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bartelt A., Bruns O. T., Reimer R., Hohenberg H., Ittrich H., Peldschus K., Kaul M. G., Tromsdorf U. I., Weller H., Waurisch C., et al. 2011. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 17: 200–205. [DOI] [PubMed] [Google Scholar]
- 192.Wei X., Wang D., Yang Y., Xia M., Li D., Li G., Zhu Y., Xiao Y., Ling W. 2011. Cyanidin-3-O-β-glucoside improves obesity and triglyceride metabolism in KK-Ay mice by regulating lipoprotein lipase activity. J. Sci. Food Agric. 91: 1006–1013. [DOI] [PubMed] [Google Scholar]
- 193.Khanade J. M., Nath M. C. 1961. Lipogenesis in adipose tissue of rats fed different diets. Am. J. Physiol. 201: 1041–1043. [DOI] [PubMed] [Google Scholar]
- 194.Lodhi I. J., Yin L., Jensen-Urstad A. P., Funai K., Coleman T., Baird J. H., El Ramahi M. K., Razani B., Song H., Fu-Hsu F., et al. 2012. Inhibiting adipose tissue lipogenesis reprograms thermogenesis and PPARγ activation to decrease diet-induced obesity. Cell Metab. 16: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Marcelino H., Veyrat-Durebex C., Summermatter S., Sarafian D., Miles-Chan J., Arsenijevic D., Zani F., Montani J-P., Seydoux J., Solinas G., et al. 2013. A role for adipose tissue de novo lipogenesis in glucose homeostasis during catch-up growth a Randle cycle favoring fat storage. Diabetes. 62: 362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Strawford A., Antelo F., Christiansen M., Hellerstein M. K. 2004. Adipose tissue triglyceride turnover, de novo lipogenesis, and cell proliferation in humans measured with 2H2O. Am. J. Physiol. Endocrinol. Metab. 286: E577–E588. [DOI] [PubMed] [Google Scholar]
- 197.Acheson K. J., Flatt J. P., Jequier E. 1982. Glycogen synthesis versus lipogenesis after a 500 gram carbohydrate meal in man. Metabolism. 31: 1234–1240. [DOI] [PubMed] [Google Scholar]
- 198.Hellerstein M. K. 1999. De novo lipogenesis in humans: metabolic and regulatory aspects. Eur. J. Clin. Nutr. 53(Suppl 1): S53–S65. [DOI] [PubMed] [Google Scholar]
- 199.Hellerstein M. K., Schwarz J. M., Neese R. A. 1996. Regulation of hepatic de novo lipogenesis in humans. Annu. Rev. Nutr. 16: 523–557. [DOI] [PubMed] [Google Scholar]
- 200.Hoffstedt J., Förster D., Löfgren P. 2007. Impaired subcutaneous adipocyte lipogenesis is associated with systemic insulin resistance and increased apolipoprotein B/AI ratio in men and women. J. Intern. Med. 262: 131–139. [DOI] [PubMed] [Google Scholar]
- 201.Ranganathan G., Unal R., Pokrovskaya I., Yao-Borengasser A., Phanavanh B., Lecka-Czernik B., Rasouli N., Kern P. A. 2006. The lipogenic enzymes DGAT1, FAS, and LPL in adipose tissue: effects of obesity, insulin resistance, and TZD treatment. J. Lipid Res. 47: 2444–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Scherer T., O'Hare J., Diggs-Andrews K., Schweiger M., Cheng B., Lindtner C., Zielinski E., Vempati P., Su K., Dighe S., et al. 2011. Brain insulin controls adipose tissue lipolysis and lipogenesis. Cell Metab. 13: 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Buettner C., Muse E. D., Cheng A., Chen L., Scherer T., Pocai A., Su K., Cheng B., Li X., Harvey-White J., et al. 2008. Leptin controls adipose tissue lipogenesis via central, STAT3-independent mechanisms. Nat. Med. 14: 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jiang L., Wang Q., Yu Y., Zhao F., Huang P., Zeng R., Qi R. Z., Li W., Liu Y. 2009. Leptin contributes to the adaptive responses of mice to high-fat diet intake through suppressing the lipogenic pathway. PLoS ONE. 4: e6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wang Q., Jiang L., Wang J., Li S., Yu Y., You J., Zeng R., Gao X., Rui L., Li W., et al. 2009. Abrogation of hepatic ATP-citrate lyase protects against fatty liver and ameliorates hyperglycemia in leptin receptor-deficient mice. Hepatology. 49: 1166–1175. [DOI] [PubMed] [Google Scholar]
- 206.Klöting N., Fasshauer M., Dietrich A., Kovacs P., Schön M. R., Kern M., Stümvoll M., Bluher M. 2010. Insulin-sensitive obesity. Am. J. Physiol. Endocrinol. Metab. 299: E506–E515. [DOI] [PubMed] [Google Scholar]
- 207.Nedergaard J., Bengtsson T., Cannon B. 2011. New powers of brown fat: fighting the metabolic syndrome. Cell Metab. 13: 238–240. [DOI] [PubMed] [Google Scholar]
- 208.Virtue S., Vidal-Puig A. 2013. Assessment of brown adipose tissue function. Front. Physiol. 4: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Deleted in proof.
- 210.Harms M., Seale P. 2013. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19: 1252–1263. [DOI] [PubMed] [Google Scholar]
- 211.Whittle A. J., Carobbio S., Martins L., Slawik M., Hondares E., Vázquez M. J., Morgan D., Csikasz R. I., Gallego R., Rodriguez-Cuenca S., et al. 2012. BMP8B increases brown adipose tissue thermogenesis through both central and peripheral actions. Cell. 149: 871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Taha M. F., Valojerdi M. R., Mowla S. J. 2006. Effect of bone morphogenetic protein-4 (BMP-4) on adipocyte differentiation from mouse embryonic stem cells. Anat. Histol. Embryol. 35: 271–278. [DOI] [PubMed] [Google Scholar]
- 213.Tseng Y. H., Kokkotou E., Schulz T. J., Huang T. L., Winnay J. N., Taniguchi C. M., Tran T. T., Suzuki R., Espinoza D. O., Yamamoto Y., et al. 2008. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 454: 1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tang Q. Q., Otto T. C., Lane M. D. 2004. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc. Natl. Acad. Sci. USA. 101: 9607–9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Green H., Kehinde O. 1979. Formation of normally differentiated subcutaneous fat pads by an established preadipose cell line. J. Cell. Physiol. 101: 169–171. [DOI] [PubMed] [Google Scholar]
- 216.Kajimura S., Seale P., Kubota K., Lunsford E., Frangioni J. V., Gygi S. P., Spiegelman B. M. 2009. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 460: 1154–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kang S., Akerblad P., Kiviranta R., Gupta R. K., Kajimura S., Griffin M. J., Min J., Baron R., Rosen E. D. 2012. Regulation of early adipose commitment by Zfp521. PLoS Biol. 10: e1001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Van R. L., Roncari D. A. 1982. Complete differentiation in vivo of implanted cultured adipocyte precursors from adult rats. Cell Tissue Res. 225: 557–566. [DOI] [PubMed] [Google Scholar]
- 219.Deleted in proof.
- 220.Asterholm I. W., Halberg N., Scherer P. E. 2007. Mouse models of lipodystrophy key reagents for the understanding of the metabolic syndrome. Drug Discov. Today Dis. Models. 4: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Shepherd P. R., Gnudi L., Tozzo E., Yang H., Leach F., Kahn B. B. 1993. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J. Biol. Chem. 268: 22243–22246. [PubMed] [Google Scholar]
- 222.Keller C., Hansen M. S., Coffin C. M., Capecchi M. R. 2004. Pax3: Fkhr interferes with embryonic Pax3 and Pax7 function: implications for alveolar rhabdomyosarcoma cell of origin. Genes Dev. 18: 2608–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lepper C., Conway S. J., Fan C-M. 2009. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 460: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]