Abstract
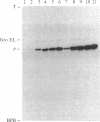
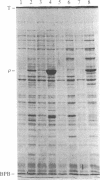
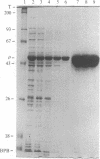
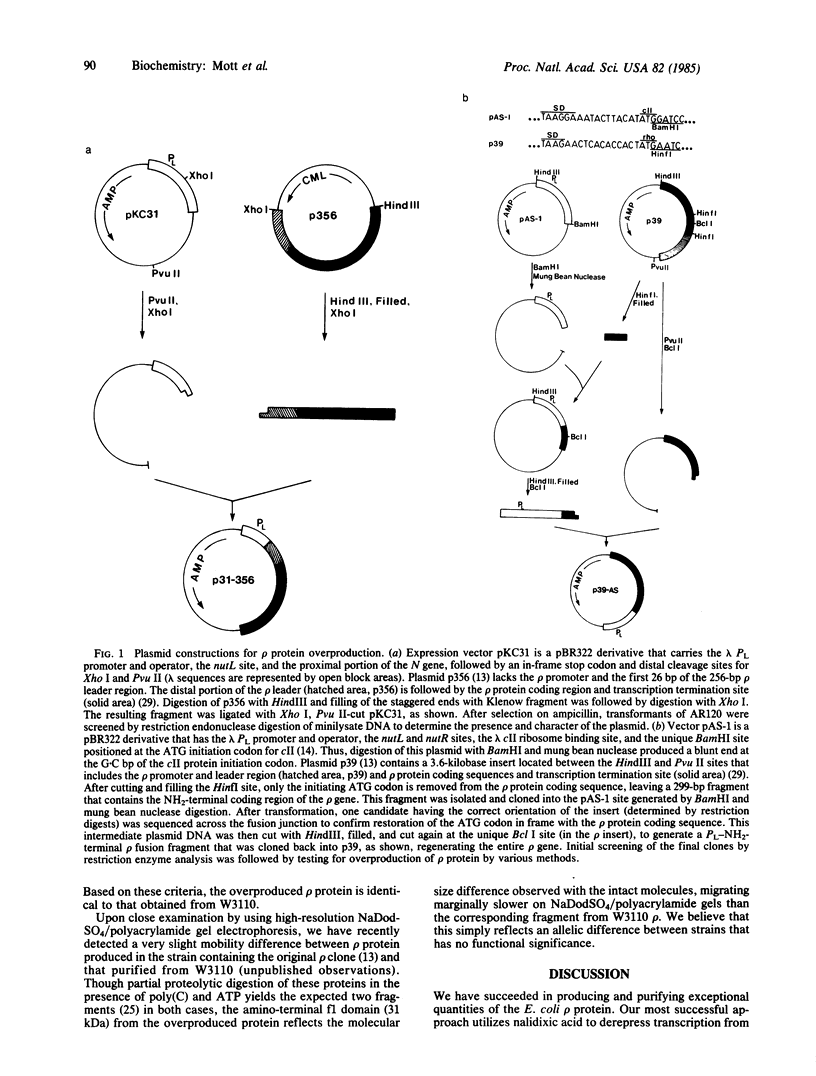
We have constructed two plasmids in which transcription of the rho gene from Escherichia coli K-12 is under the control of the lambda phage PL promoter. In p31-356, the normal rho promoter is deleted, but the remainder of the rho leader region, including the ribosome binding site, is present. In p39-AS, the rho leader is completely absent, and the lambda cII ribosome binding site replaces that of rho. Under noninducing conditions, expression of rho protein from these plasmids is repressed by the lambda cI protein in hosts carrying lambda cryptic prophage. Induction using mitomycin C or nalidixic acid in a cryptic lysogen carrying the cI+ repressor resulted in the overproduction of rho protein to levels of 3%-5% of the total cellular protein with p31-356, and to levels of approximately equal to 40% with p39-AS. The overproduced protein is functionally indistinguishable from the rho protein isolated from the K-12 strain W3110, and it can be obtained from cells harboring p39-AS in yields of up to 25 mg of rho per g of cells. In contrast to chemical induction, heat induction in four cryptic lambda lysogens carrying the thermolabile cI857 repressor failed to yield the same high levels of rho protein (with either plasmid). Our results show that chemical induction of PL-containing plasmid expression vectors can serve as a convenient and useful alternative to the commonly used method of heat induction.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S., Albrechtsen B., Pedersen S., Klemm P. Localization and regulation of the structural gene for transcription-termination factor rho of Escherichia coli. J Mol Biol. 1982 Dec 5;162(2):283–298. doi: 10.1016/0022-2836(82)90527-7. [DOI] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Merril C., Adhya S. Interaction of RNA polymerase and rho in transcription termination: coupled ATPase. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4828–4832. doi: 10.1073/pnas.75.10.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger L. R., Richardson J. P. Procedure for purification of Escherichia coli ribonucleic acid synthesis termination protein rho. Biochemistry. 1981 Mar 17;20(6):1640–1645. doi: 10.1021/bi00509a036. [DOI] [PubMed] [Google Scholar]
- Finger L. R., Richardson J. P. Stabilization of the hexameric form of Escherichia coli protein rho under ATP hydrolysis conditions. J Mol Biol. 1982 Mar 25;156(1):203–219. doi: 10.1016/0022-2836(82)90467-3. [DOI] [PubMed] [Google Scholar]
- Guarente L. P., Beckwith J. Mutant RNA polymerase of Escherichia coli terminates transcription in strains making defective rho factor. Proc Natl Acad Sci U S A. 1978 Jan;75(1):294–297. doi: 10.1073/pnas.75.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Restoration of termination by RNA polymerase mutations is rho allele-specific. J Mol Biol. 1979 Apr 5;129(2):295–304. doi: 10.1016/0022-2836(79)90283-3. [DOI] [PubMed] [Google Scholar]
- Hendrix R. W. Purification and properties of groE, a host protein involved in bacteriophage assembly. J Mol Biol. 1979 Apr 15;129(3):375–392. doi: 10.1016/0022-2836(79)90502-3. [DOI] [PubMed] [Google Scholar]
- Joyce C. M., Grindley N. D. Construction of a plasmid that overproduces the large proteolytic fragment (Klenow fragment) of DNA polymerase I of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1830–1834. doi: 10.1073/pnas.80.7.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung H., Bekesi E., Guterman S. K., Gray J. E., Traub L., Calhoun D. H. Autoregulation of the rho gene of Escherichia coli K-12. Mol Gen Genet. 1984;193(2):210–213. doi: 10.1007/BF00330669. [DOI] [PubMed] [Google Scholar]
- L'Italien J. J., Strickler J. E. Application of high-performance liquid chromatographic peptide purification to protein microsequencing by solid-phase Edman degradation. Anal Biochem. 1982 Nov 15;127(1):198–212. doi: 10.1016/0003-2697(82)90165-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Pleiotropic effects of a DNA adenine methylation mutation (dam-3) in Escherichia coli K12. Mutat Res. 1975 Apr;28(1):15–26. doi: 10.1016/0027-5107(75)90309-7. [DOI] [PubMed] [Google Scholar]
- Merrill B. M., Williams K. R., Chase J. W., Konigsberg W. H. Photochemical cross-linking of the Escherichia coli single-stranded DNA-binding protein to oligodeoxynucleotides. Identification of phenylalanine 60 as the site of cross-linking. J Biol Chem. 1984 Sep 10;259(17):10850–10856. [PubMed] [Google Scholar]
- Pinkham J. L., Platt T. The nucleotide sequence of the rho gene of E. coli K-12. Nucleic Acids Res. 1983 Jun 11;11(11):3531–3545. doi: 10.1093/nar/11.11.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Richardson J. P., Grimley C., Lowery C. Transcription termination factor rho activity is altered in Escherichia coli with suA gene mutations. Proc Natl Acad Sci U S A. 1975 May;72(5):1725–1728. doi: 10.1073/pnas.72.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. W. Termination factor for RNA synthesis. Nature. 1969 Dec 20;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Ho Y. S., Shatzman A. The use of pKc30 and its derivatives for controlled expression of genes. Methods Enzymol. 1983;101:123–138. doi: 10.1016/0076-6879(83)01009-5. [DOI] [PubMed] [Google Scholar]
- SUSSMAN R., JACOB F. [On a thermosensitive repression system in the Escherichia coli lambda bacteriophage]. C R Hebd Seances Acad Sci. 1962 Feb 19;254:1517–1519. [PubMed] [Google Scholar]
- Sharp J. A., Galloway J. L., Platt T. A kinetic mechanism for the poly(C)-dependent ATPase of the Escherichia coli transcription termination protein, rho. J Biol Chem. 1983 Mar 25;258(6):3482–3486. [PubMed] [Google Scholar]
- Sharp J. A., Platt T. Rho-dependent termination and concomitant NTPase activity requires a specific, intact RNA region. J Biol Chem. 1984 Feb 25;259(4):2268–2273. [PubMed] [Google Scholar]
- Shigesada K., Imai M. Studies on the altered rho factor in nitA mutants of Escherichia coli defective in transcription termination. II. Purification and molecular properties of the mutant rho. J Mol Biol. 1978 Apr 25;120(4):467–486. doi: 10.1016/0022-2836(78)90349-2. [DOI] [PubMed] [Google Scholar]
- Shigesada K., Tsurushita N., Matsumoto Y., Imai M. Overproduction of transcription termination factor Rho in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):199–209. doi: 10.1016/0378-1119(84)90180-x. [DOI] [PubMed] [Google Scholar]
- Tilly K., Murialdo H., Georgopoulos C. Identification of a second Escherichia coli groE gene whose product is necessary for bacteriophage morphogenesis. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1629–1633. doi: 10.1073/pnas.78.3.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A. M., Christie G. E., Platt T. Tandem termination sites in the tryptophan operon of Escherichia coli. Proc Natl Acad Sci U S A. 1981 May;78(5):2913–2917. doi: 10.1073/pnas.78.5.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen T. S., Webster R. E. Bacteriophage f1 gene II and X proteins. Isolation and characterization of the products of two overlapping genes. J Biol Chem. 1981 Nov 10;256(21):11259–11265. [PubMed] [Google Scholar]
- Yoakum G. H., Yeung A. T., Mattes W. B., Grossman L. Amplification of the uvrA gene product of Escherichia coli to 7% of cellular protein by linkage to the pL promoter of pKC30. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1766–1770. doi: 10.1073/pnas.79.6.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. F., Desselberger U., Palese P., Ferguson B., Shatzman A. R., Rosenberg M. Efficient expression of influenza virus NS1 nonstructural proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1983 Oct;80(19):6105–6109. doi: 10.1073/pnas.80.19.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]