Abstract
Among four vertebrate arrestins, only two are ubiquitously expressed. Arrestins specifically bind active phosphorylated G protein-coupled receptors (GPCRs), thereby precluding further G protein activation. Recent discoveries suggest that the formation of the arrestin-receptor complex initiates the second round of signaling with comparable biological importance. Despite having virtually no recognizable sequence motifs known to mediate protein-protein interactions, arrestins bind a surprising variety of signaling proteins with mind-boggling range of functional consequences. High conformational flexibility allows arrestins to show many distinct “faces” to the world, which allows these relatively small ~45 kDa proteins to bind various partners under different physiological conditions, organizing multi-protein signaling complexes and localizing them to distinct sub-cellular compartments.
Introduction
Exquisitely timed precisely regulated protein-protein interactions determine every aspect of cell behavior. If we clearly understood what makes proteins bind each other and what prevents their interactions, we would be able to tell the cell what to do in a language it cannot ignore. Arrestins are a family of only four proteins in mammals with remarkably similar structures [1–4]. Their only function in life is to bind an amazing variety of partners at appropriate times, organizing multi-protein complexes and localizing them to proper sub-cellular compartments, thereby orchestrating cell signaling [5,6]. Because of unimpressive size (~45 kDa), an arrestin molecule can accommodate no more than 5–6 other proteins simultaneously, suggesting that at any given moment it interacts only with select few out of hundreds of possible partners [7]. Thus, arrestins provide a perfect model for elucidating how the structural changes in proteins make them “decide” which partners to bind and in what combinations. Extensive crystallographic, biophysical, and mutagenesis studies of arrestin proteins are beginning to shed light on the molecular mechanisms underlying these critical choices.
Basal does not mean inactive
The protein that specifically binds active phosphorylated rhodopsin was discovered less than 30 years ago [8]. The fact that receptor binding induces a global conformational change in this protein was established soon thereafter [9]. This idea was one of the cornerstones of the first mechanistic model (which still holds water) coherently explaining how arrestin can bind with high affinity to the receptor that is active and phosphorylated at the same time, but neither to inactive phosphorylated nor active unphosphorylated form of the same protein [10]. Therefore, subsequently solved remarkably similar crystal structures of all four arrestins [1–4,11,12] were believed to represent their basal conformation, which was deemed “inactive”. In essence, arrestins were assumed to exist in this “waiting” state, ready to be “activated” by receptor binding [13], whereupon they would acquire the ability to interact with various non-receptor partners [14]. This view was largely shaped by the pioneering reports in 1999–2001 that arrestin-assisted activation of Src [15,16], MAP kinases JNK3 [17] and ERK1/2 [15,16,18,19], and arrestin ubiquitination [20] happened only in response to receptor activation. Although the first evidence contradicting this view emerged as early as 2001, when Miller at al reported receptor-independent facilitation of JNK3 phosphorylation by arrestin-3 [21], the idea that only GPCR-bound arrestin can interact with non-receptor partners is still popular. In fact, it is an example of unwarranted generalization: it is correct in some cases, but misleading in others. Careful comparison of ERK1/2 and JNK3 activation in the same cell showed that while ERK1/2 phosphorylation is strictly dependent on receptor stimulation and is only mediated by arrestins that can bind receptors, the activation of JNK3 is not affected by receptor activity or the ability of arrestin to interact with GPCRs [22]. This finding was consistent with previous demonstration that ERK1/2 preferentially binds receptor-associated arrestins [23], whereas JNK3 binds free arrestins in their basal state perfectly well [24], and its activation is facilitated even by arrestin-3 mutant with the deletion in the inter-domain hinge that precludes receptor binding [22,25]. Admittedly, cell-based assays cannot yield unambiguous answers, because every cell expresses thousands of different proteins that can mediate indirect interactions and significantly affect outcomes. Recently the ability of arrestin-3 to facilitate JNK3 activation by its upstream kinases was demonstrated in the system reconstituted from purified proteins in the absence of receptors [26,27], proving beyond reasonable doubt that this is a function of free arrestin-3.
In fact, remarkably few signaling proteins were shown to preferentially interact with receptor-bound arrestins: in addition to ERK1/2 [23], this short list includes clathrin and AP2 [28,29], E3 ubiquitin ligase AIP4 [30], and cRaf1 [23]. Several partners strongly prefer the basal conformation of arrestins: microtubules [31], calmodulin [32], E3 ubiquitin ligases Mdm2 [24] and parkin [33]. Some proteins apparently bind arrestins in both basal and “active” conformations comparably: JNK3 [24], ASK1, MKK4, MEK1 [25], and PDE4D [34] (Table 1). Unfortunately, we do not know the conformational preferences (if any) of the great majority of arrestin-binding proteins. This issue must be addressed experimentally for each partner, and it is safe to predict that for various proteins the answers will be very different. It seems logical that if the interaction site of a particular partner overlaps with the receptor footprint, this partner would only bind free arrestins. This prediction is supported by the evidence in case of calmodulin [32] and microtubules [31]: both interact with the concave sides of the two domains engaged by GPCRs, and therefore receptors successfully outcompete them [32,35]. Unfortunately, arrestin residues mediating the interaction were identified in very few cases (Fig. 1), which makes the usability of this logic very limited. To complicate things further, different functional forms of the same receptor apparently interact with distinct arrestin elements, as has been recently shown by comparing the residues engaged by active unphosphorylated and inactive phosphorylated rhodopsin on arrestin-1 [36] (Fig. 2). Thus, it is entirely possible that certain partners would compete with some forms of the receptor, but not others. Precise identification of the binding sites of different functional forms of GPCRs and other signaling proteins on arrestins is yet another unmet need in arrestin biology.
Table 1.
Known conformational preferences of arrestin binding partners.
| Arrestin Binding Partner |
Arrestin isoform |
Preferred Arrestin Conformation |
References |
|---|---|---|---|
| ERK1/2 | Arrestin-2, arrestin-3 |
receptor-bound | [23] |
| clathrin | Arrestin-2, arrestin-3 |
receptor-bound | [28,29] |
| AP-2 | Arrestin-2, arrestin-3 |
receptor-bound | [28,29] |
| E3 ubiquitin ligase AIP4 |
Arrestin-2 | receptor-bound | [30] |
| cRaf1 | Arrestin-2 | receptor-bound | [23] |
| microtubules | Arrestin-1, arrestin-2, arrestin-3 |
basal | [31,59] |
| calmodulin | arrestin2 | basal | [32] |
| E3 ubiquitin ligase Mdm2 |
Arrestin-1, Arrestin-2, arrestin-3 |
basal | [24] |
| E3 ubiquitin ligase parkin |
Arrestin-2, arrestin-3 |
basal | [33] |
| JNK3 | Arrestin-1, arrestin-2, arrestin-3 |
either | [24] |
| ASK1 | Arrestin-2, arrestin-3 |
either | [25] |
| MKK4 | Arrestin-3 | either | [25] |
| MEK1 | Arrestin-2, arrestin-3 |
either | [25,66] |
| c-Raf1 | Arrestin-3 | either | [23] |
| PDE4 | Arrestin-3 | either | [34] |
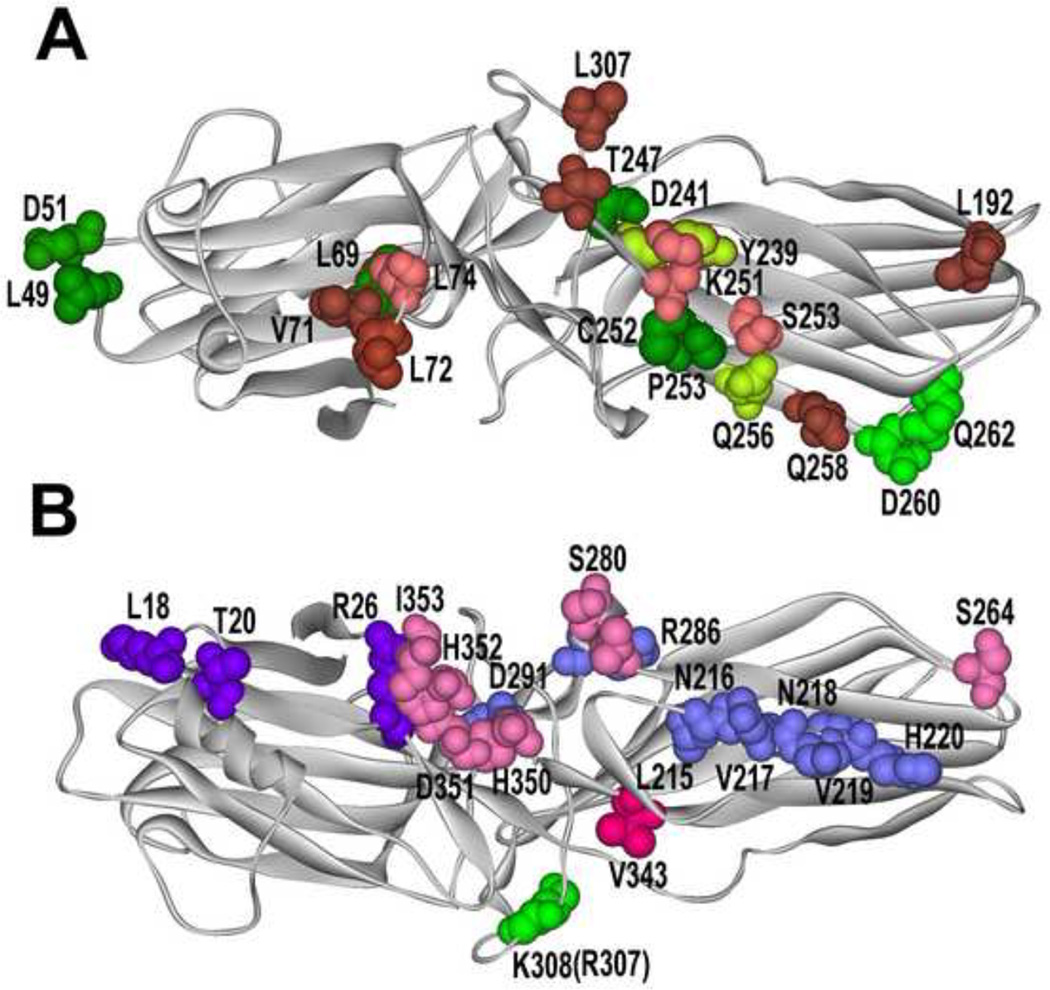
Fig. 1. What arrestin elements do GPCRs and other partners engage?
A. Arrestin from “receptor viewpoint”. Arrestin residues that determine receptor specificity [46,64] are shown in different shades of green: yellow-green, those where mutations (Y239T and Q256Y) result in reduced dopamine D2 receptor binding; bright green, those where mutations (D260K and Q262P) result in high preference for other GPCRs over β2-adrenergic receptor. Elements engaged by Ca2+-liganded calmodulin are shown in pink and brown (darker color indicates greater immobilization by calmodulin [32]). Residues in green with red border (L69 and T239) are shared by receptors and calmodulin. B. Non-receptor- binding side of arrestins. Residues engaged by all forms of PDE4D are shown in blue, those specific for PDE4D5 in bright violet [34], the residue critical cRaf1-binding is in bright green (mutation R307A in arrestin-3 prevents cRaf1 interaction; homologous mutation K308A in arrestin-3 does not) [65], residues responsible for the ability of arrestin-3 to promote JNK3 activation are shown in bright red (critical V343) and pink (supporting residues) [67]. Arrestin-3 structure 3P2D [4] was used to generate this figure (since calmodulin-binding elements were identified in arrestin-2 [32], homologous arrestin-3 residues are highlighted).
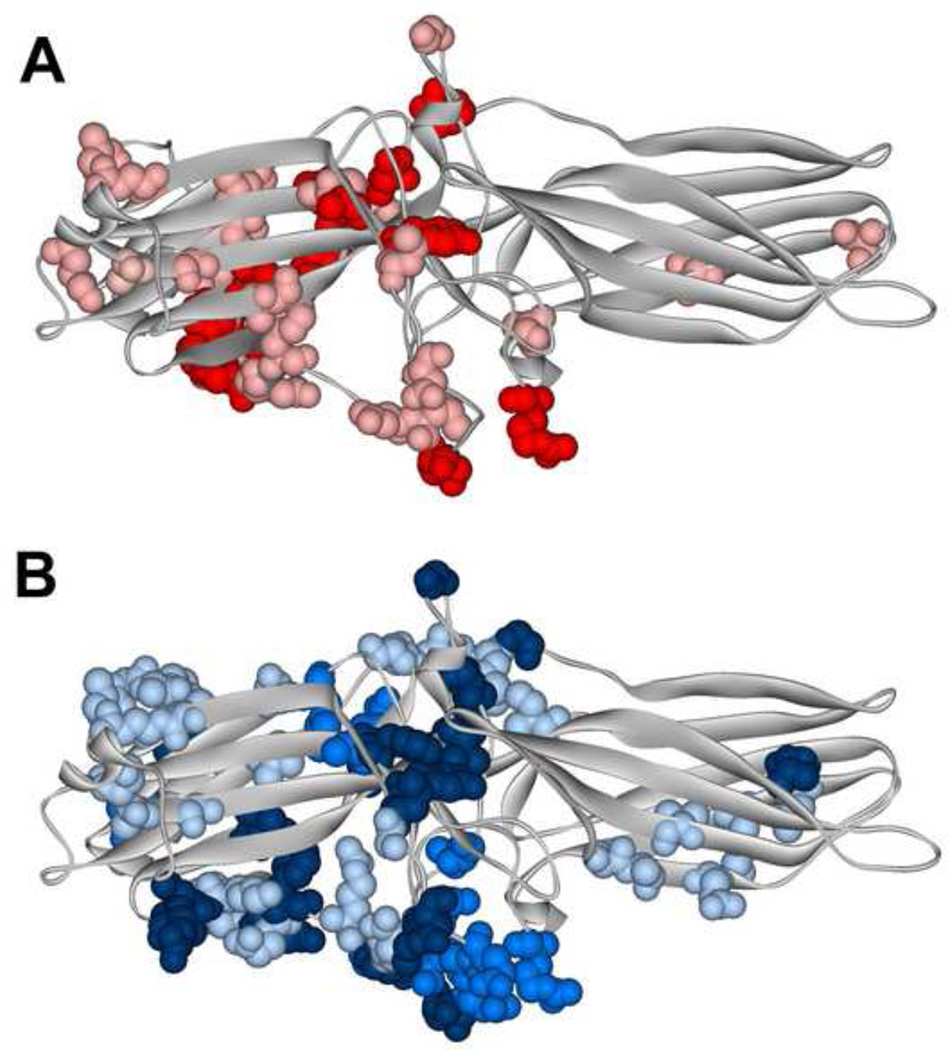
Fig. 2. Different functional forms of the same receptor engage distinct arrestin elements.
Arrestin-1 residues demonstrating significant chemical shift changes in NMR spectra upon the binding of light-activated unphosphorylated (A) or inactive phosphorylated (B). A. The magnitude of binding-induced changes is shown by color coding: bright red, shifts greater than 0.01; pink, shifts greater than 0.005, but smaller than 0.01. B. The magnitude of binding-induced changes is shown by color coding: dark blue, peaks disappeared; blue, shifts greater than 0.01; light blue, shifts greater than 0.005, but smaller than 0.01. The two forms of rhodopsin affect distinct arrestin-1 elements that only partially overlap [36].
Receptor-bound conformation: what does it look like?
It became clear that the shape of receptor-bound arrestins must be quite different from the basal state long before any crystal structures were solved [9]. The release of the C-tail was the first receptor-induced conformational rearrangement documented [37]. However, the finding that the deletion of the C-tail does not make arrestin bind all forms of the receptor indiscriminately [10] suggested that additional changes are likely. Recently several attempts to determine receptor-bound “active” conformation of arrestins were made using various methods, such as NMR [36], EPR [38], and crystallography [39,40] (Fig. 3). Each of these approaches has obvious caveats: biophysical studies of arrestin complexes with real receptor cannot yield atomic resolution [36,38], whereas real receptors are absent in beautiful crystal structures of “active” arrestins [39,40]. Nonetheless, collectively these attempts proved very informative. As could be expected, every study detected significant conformational rearrangements in arrestins. EPR documented the movement of four flexible loops on the receptor-binding side [38], some of which were predicted, while others were quite unexpected (Fig. 3B). NMR confirmed the engagement of distinct arrestin elements by inactive phosphorylated and active unphosphorylated forms of the receptor [36] (Fig. 2), which was predicted earlier based on mutagenesis and EPR data [10,41]. Unexpectedly, NMR study also revealed global increase in arrestin flexibility upon the binding to active phosphorylated receptor [36]. General “melting” of arrestin structure upon receptor binding was never predicted, but biologically it makes perfect sense: receptor-bound arrestin interacts with numerous signaling proteins, and protein-protein interactions are often mediated by unstructured elements, where the binding to an appropriate partner promotes folding [42,43]. The global conformational change induced by receptor binding was hypothesized to include the movement of the two arrestin domains relative to each other [13], partially based on the finding that the deletions in the 12-residue inter-domain hinge impair receptor binding of all arrestins [31,44]. However, proposed clam-like movement of the two domains was not detected by EPR [38] (Fig. 3B). Recently two crystal structures of presumably “active” arrestins were reported: one of truncated arrestin-2 (previously shown to be constitutively active [45]) associated with multi-phosphorylated C-terminus of vasopressin V2 receptor [40](Fig. 3C) and the other of short arrestin-1 splice variant p44 also lacking the C-tail [39] (Fig. 3D). These very similar structures suggest that the conformational changes include the rotation of the two domains relative to each other by 20–21° [39,40] (Fig. 3C,D). Both also revealed a large movement of the “139 loop” in the central crest of the receptor-binding surface, away from the “finger loop” implicated in receptor binding [41,46] (Fig. 3). This movement was earlier discovered using EPR [38] and confirmed by mutagenesis [47]. Because the receptor is not present in the “active” structures, we do not know how much arrestin domains actually move upon GPCR binding, but domain rotation allows to rationalize hinge deletion results [31,44], which distance measurements by EPR [38] left unexplained.
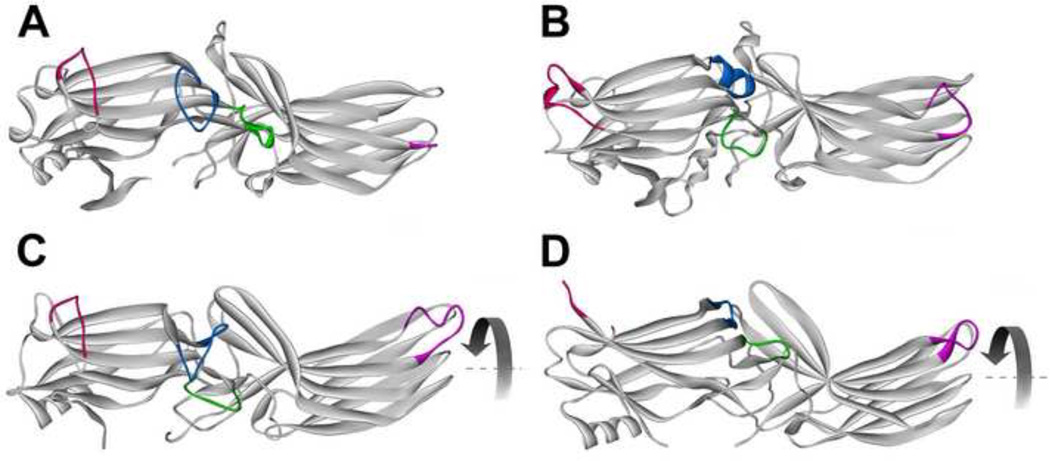
Fig. 3. Receptor binding-induced structural changes in arrestins.
A. Basal arrestin conformation (based on arrestin-2 structure 1G4M, which has the highest resolution among published arrestin structures [2]). B. A model of arrestin-1 bound to lightactivated phosphorylated rhodopsin based on intra-molecular distance measurements in free and bound arrestin-1 [38]. C. Crystal structure 4JQI of truncated arrestin-2-(1–382) in complex with multi-phosphorylated C-terminus of vasopressin V2 receptor receptor [40]. D. Crystal structure of p44, truncated splice variant of arrestin-1. Arrestin elements that change conformation upon receptor binding are color-coded, as follows: blue, the “finger loop” (G68-S78 in arrestin-1; G65-T74 in arrestin-2); green, “139 loop” (P134-S142 in arrestin-1; P131-A139 in arrestin-2); red, “157 loop” (H155-P165 in arrestin-1; E152-H159 in arrestin-2); magenta, “344 loop” (S336-S344 in arrestin-1; S330-S340 in arrestin-2). Residue numbers for corresponding bovine proteins are given. The rotation of the two domains relative to each other by 20–21° is indicated by curved arrow in C and D.
It is worth noting that back in 2006 this type of domain movement was proposed as a possibility due to hydrophobic nature of the inter-domain interface [48] and even suggested as the most likely based on molecular modeling [49]. The extent of domain rotation was estimated at ~20° based the length of the hinge in WT arrestins [49]. The coincidence is remarkable, even though this model was originally proposed to explain how two GPCRs can fit a single arrestin [49]. A series of recent studies proved that monomeric class A (rhodopsin-like) receptor is necessary and sufficient to perform all known signaling functions: activate cognate G protein [50–52], become phosphorylated by G protein-coupled receptor kinases [53,54], and bind arrestins [53–56]. Interestingly, while the high-affinity binding invariably involves one-to-one arrestin-receptor stoichiometry, an alternative mode, where a single arrestin-1 molecule apparently engages two rhodopsins, one with high and one with low affinity, was recently reported in the visual system [57,58]. It remains to be elucidated whether this is a special adaptation in rods, where half of the surface of disc membranes is occupied by rhodopsin, with the concentration in rod outer segments of ~ 3 mM [6], or reflects the capability of all arrestin proteins.
Although the true shape of “active” arrestins will be revealed only by the structure of the arrestin-receptor complex, recent studies provided interesting clues (Fig. 3). Unless the real structure proves otherwise, domain rotation appears the most likely global conformational change, accompanied by the movement of several loops on the receptor-binding side. This model, along with the identification of the docking sites of nonreceptor partners, which is sadly missing in most cases, should help us understand their conformational preferences.
Microtubule-bound state: the third face of Janus?
All arrestins bind microtubules [31,35], and this interaction apparently triggers a conformational change different from that induced by GPCRs [59], because hinge deletion mutants impaired in receptor binding showed higher affinity for microtubules [31]. Microtubule binding was proposed to determine the localization of visual arrestin-1 in dark-adapted photoreceptors [35] and to play a role in sub-cellular distribution and activity of signaling proteins associated with non-visual arrestins [31]. Several proteins, including ERK1/2 and Mdm2, interact with arrestins in microtubule-bound conformation (and/or hinge deletion mutants mimicking this form) better than with free arrestins [23,24,31]. This was particularly surprising in case of ERK1/2 with strong preference for the receptor-bound form [23]. We know even less about the actual shape of microtubule-bound arrestins than about their receptor-bound conformation. Since in this case crystallization is not an option, only careful biophysical studies can give additional clues. This information is biologically important, because it might give us tools to direct signaling proteins to the cytoskeleton and/or specifically regulate their activity in this compartment.
Elusive transitions and signaling bias
Among many arrestin conformations we know well only the free state, with one structure affording better than 2Å resolution [2]. Several recent studies [36,38–40] provided clues about receptor-induced conformational changes, although the structure of the arrestin-receptor complex, preferably several revealing different “flavors” of these complexes that likely exist [60], would be most welcome. The shape of the microtubule-bound arrestin remains to be elucidated. Recent finding that arrestin-3 binding to JNK3α2 differentially affects its affinity for the upstream kinases, enhancing the interaction with MKK4, while decreasing MKK7 binding [27], strongly suggests that many partners induce conformational changes in arrestins, so that GPCRs and microtubules are not unique in this regard. Thus, the shape of arrestins in complex with different partners must be investigated. Most importantly, we know virtually nothing about the process of conformational transitions in arrestins, which can be only elucidated by biophysical methods revealing protein dynamics, because it is virtually impossible to deduce them from inherently static crystal structures.
Identification of arrestin elements engaged by various partners is another challenge. Mapping of the receptor-binding site on arrestins [41,46] (Fig. 1A) and even partial elucidation of the mechanics of the arrestin-receptor interactions [1–4,61] led to the construction of enhanced arrestins that bind unphosphorylated active GPCRs, change receptor trafficking pattern [62], and even compensate for the lack of receptor phosphorylation in vivo [63]. Reengineering of the receptor-binding surface of inherently promiscuous non-visual arrestin-3 yielded mutants with >50-fold preference for some GPCRs over others [64] (Fig. 1A). Similarly, even imperfect identification of MAP kinase interaction sites led to the construction of arrestin-2 that does not facilitate ERK1/2 activation [65] (Fig. 1B), as well as arrestin-3 mutant that acts as a dominant-negative silent scaffold, recruiting the kinases of JNK activation cascade away from productive scaffolds and suppressing JNK activity in the cell [22] (Fig. 1B). Considering how many different proteins arrestins bind [7], these pioneering attempts show that there are many possible ways of constructing arrestins with signaling bias [60], which can be used to change cell signaling in the desired manner.
Conclusions
Arrestins are typical scaffolds that bind numerous partners, organizing multi-protein signaling complexes and ensuring their proper sub-cellular localization via interactions with “anchoring” proteins, such as GPCRs and microtubules. Arrestins exist in at least three (very likely more) distinct conformations: free, GPCR-associated, and microtubule-bound. This flexibility underlies their functional versatility: each conformational state preferentially recruits particular partners. Current challenge is to elucidate the shape of arrestins in each state, the mechanism of their transition from one state to another, and the sites where other proteins bind consistent with their conformational preferences. This will pave the way to engineering arrestins that bind the partners we want and recruit them to sub-cellular locations of our choosing in order to channel cell signaling in desired directions.
Acknowledgements
Supported in part by NIH grants GM077561, EY011500, and GM081756 (VVG), NS065868 and DA030103 (EVG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Different systems of arrestin names are used in the field. We use the systematic names of arrestin proteins: arrestin-1 (historic names S-antigen, 48 kDa protein, visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin; for unclear reasons its gene is called “arrestin 3” in the HUGO database).
References
- 1. Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 A crystal structure of visual arrestin: a model for arrestin's regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. **This paper provided the first comprehensive functional interpretation of arrestin structure and coined most of arrestin terminology.
- 2. Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. *This is the highest resolution arrestin structure in existence. It gave the first clues regarding the structural basis of receptor specificity of arrestin proteins.
- 3.Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. Crystal Structure of Cone Arrestin at 2.3Å: Evolution of Receptor Specificity. J Mol Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 4. Zhan X, Gimenez LE, Gurevich VV, Spiller BW. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two nonvisual arrestins. J Mol Biol. 2011;406:467–478. doi: 10.1016/j.jmb.2010.12.034. *The comparison of this structure with other arrestins and follow-up mutagenesis revealed the feature enabling relatively promiscuous binding of arrestin-3 to different GPCR subtypes and functional forms of receptors.
- 5. Shukla AK, Xiao K, Lefkowitz RJ. Emerging paradigms of β-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem Sci. 2011;36:457–469. doi: 10.1016/j.tibs.2011.06.003. *This is a very comprehensive review of arrestin-mediated signaling pathways.
- 6. Gurevich VV, Hanson SM, Song X, Vishnivetskiy SA, Gurevich EV. The functional cycle of visual arrestins in photoreceptor cells. Prog Retin Eye Res. 2011;30:405–430. doi: 10.1016/j.preteyeres.2011.07.002. *This review gives a comprehensive synthesis of the information on arrestin-1 structure, functional cycle, and its biological role in photoreceptor cells.
- 7. Xiao K, McClatchy DB, Shukla AK, Zhao Y, Chen M, Shenoy SK, Yates JR, Lefkowitz RJ. Functional specialization of beta-arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci U S A. 2007;104:12011–12016. doi: 10.1073/pnas.0704849104. *This work reports an amazing variety of cellular proteins that directly interact with non-visual arrestins.
- 8. Kuhn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett. 1984;176:473–478. doi: 10.1016/0014-5793(84)81221-1. *The fact that arrestin specifically binds rhodopsin that is active and phosphorylated at the same time was first described in this paper.
- 9. Schleicher A, Kuhn H, Hofmann KP. Kinetics, binding constant, and activation energy of the 48-kDa protein-rhodopsin complex by extrametarhodopsin II. Biochemistry. 1989;28:1770–1775. doi: 10.1021/bi00430a052. *This is the first report indicating that receptor binding induces a global conformational change in the arrestin molecule.
- 10. Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin: Sequential multisite binding ensures strict selectivity towards lightactivated phosphorylated rhodopsin. J. Biol. Chem. 1993;268:11628–11638. **This paper proposed the first (and thus far the only) coherent model proposing the mechanism ensuring that arrestin binds active phosphorylated receptor with high affinity, while interacting with active unphosphorylated and inactive phosphorylated forms with fairly low affinity.
- 11.Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, Buldt G. X-ray crystal structure of arrestin from bovine rod outer segments. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- 12.Milano SK, Pace HC, Kim YM, Brenner C, Benovic JL. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry. 2002;41:3321–3328. doi: 10.1021/bi015905j. [DOI] [PubMed] [Google Scholar]
- 13. Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. *This review gives a concise structure-based summary of the mechanism whereby active phosphorylated receptors induce global conformational changes in arrestins.
- 14.Gurevich VV, Gurevich EV. The new face of active receptor bound arrestin attracts new partners. Structure. 2003;11:1037–1042. doi: 10.1016/s0969-2126(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 15. Luttrell LM, Ferguson SS, Daaka Y, Miller WE, Maudsley S, Della Rocca GJ, Lin F, Kawakatsu H, Owada K, Luttrell DK, et al. Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. **This is the first report of the signaling cascade specifically initiated by the formation of the arrestin-receptor complex. This paper is the first description of the recruitment of signaling proteins to the receptor-bound arrestins.
- 16. DeFea KA, Vaughn ZD, O'Bryan EM, Nishijima D, Déry O, Bunnett NW. The proliferative and antiapoptotic effects of substance P are facilitated by formation of a beta -arrestin-dependent scaffolding complex. Proc Natl Acad Sci U S A. 2000;97:11086–11091. doi: 10.1073/pnas.190276697. *This is an important independent confirmation that arrestins can facilitate ERK1/2 activation via Src (compare to ref 15) .
- 17. McDonald PH, Chow CW, Miller WE, Laporte SA, Field ME, Lin FT, Davis RJ, Lefkowitz RJ. Beta-arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science. 2000;290:1574–1577. doi: 10.1126/science.290.5496.1574. *This is one of the two pioneering reports (with ref 19) of MAP kinase cascade being scaffolded by arrestin. This study proposed the first model of the complex of arrestin with three MAP kinases constituting a signaling module.
- 18.Luttrell LM, Roudabush FL, Choy EW, Miller WE, Field ME, Pierce KL, Lefkowitz RJ. Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proc Natl Acad Sci U S A. 2001;98:2449–2454. doi: 10.1073/pnas.041604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. DeFea KA, Zalevsky J, Thoma MS, Déry O, Mullins RD, Bunnett NW. betaarrestin- dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J Cell Biol. 2000;148:1267–1281. doi: 10.1083/jcb.148.6.1267. *This is one of the two pioneering reports (with ref 17) of MAP kinase cascade being scaffolded by arrestins. It emphasized two distinct pathways, one via G protein and the other via arrestins, connecting active GPCRs with ERK1/2 phosphorylation.
- 20. Shenoy SK, McDonald PH, Kohout TA, Lefkowitz RJ. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and betaarrestin. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. *This is the first study documenting that the arrestin-receptor interaction triggers the ubiquitination of both receptor and arrestin.
- 21. Miller WE, McDonald PH, Cai SF, Field ME, Davis RJ, Lefkowitz RJ. Identification of a motif in the carboxyl terminus of beta -arrestin2 responsible for activation of JNK3. J Biol Chem. 2001;276:27770–27777. doi: 10.1074/jbc.M102264200. **This is the first report of receptor-independent facilitation of MAP kinase activity by an arrestin.
- 22. Breitman M, Kook S, Gimenez LE, Lizama BN, Palazzo MC, Gurevich EV, Gurevich VV. Silent scaffolds: inhibition of c-Jun N-terminal kinase 3 activity in the cell by a dominant-negative arrestin-3 mutant. J Biol Chem. 2012;287:19653–19664. doi: 10.1074/jbc.M112.358192. *This is the first report describing the use of reengineered arrestin to suppress MAP kinase signaling in the cell. Described dominant-negative mutant recruts all three kinases in the ASK1-MKK4-JKN3 cascade away from productive scaffolds and holds them in a configuration that does not facilitate signaling.
- 23.Coffa S, Breitman M, Hanson SM, Callaway K, Kook S, Dalby KN, Gurevich VV. The Effect of Arrestin Conformation on the Recruitment of c-Raf1, MEK1, and ERK1/2 Activation. PLoS One. 2011;6:e28723. doi: 10.1371/journal.pone.0028723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song X, Raman D, Gurevich EV, Vishnivetskiy SA, Gurevich VV. Visual and both non-visual arrestins in their "inactive" conformation bind JNK3 and Mdm2 and relocalize them from the nucleus to the cytoplasm. J Biol Chem. 2006;281:21491–21499. doi: 10.1074/jbc.M603659200. *This is the first evidence that arrestins in their free “inactive” conformation bind non-receptor partners.
- 25. Song X, Coffa S, Fu H, Gurevich VV. How does arrestin assemble MAPKs into a signaling complex? J Biol Chem. 2009;284:685–695. doi: 10.1074/jbc.M806124200. * This study proposed an alternative model of the complex of arrestin with the three MAP kinases constituting a signaling module, where all three kinases directly bind both arrestin domains.
- 26. Zhan X, Kaoud TS, Dalby KN, Gurevich VV. Non-visual arrestins function as simple scaffolds assembling MKK4- JNK3α2 signaling complex. Biochemistry. 2011;50:10520–10529. doi: 10.1021/bi201506g. *This is the first study where arrestin-MAP kinase signaling complex was reconstituted from purified proteins. These experiments unambiguously proved that arrestin-dependent JNK3 activation does not require arrestin binding to a GPCR.
- 27. Zhan X, Kaoud TS, Kook S, Dalby KN, Gurevich VV. JNK3 binding to arrestin-3 differentially affects the recruitment of upstream MAP kinase kinases. J Biol Chem. 2013;288:28535–28547. doi: 10.1074/jbc.M113.508085. *This is the first evidence that MAP kinases affect each other's interaction with arrestin, providing the first clue of the mechanisms arrestins employ to “select” the combination of partners they bind simulatneously (out of an enormous number of possible combinations, see ref 7).
- 28.Xiao K, Shenoy SK, Nobles K, Lefkowitz RJ. Activation-dependent conformational changes in {beta}-arrestin 2. J Biol Chem. 2004;279:55744–55753. doi: 10.1074/jbc.M409785200. [DOI] [PubMed] [Google Scholar]
- 29.Kim YM, Benovic JL. Differential roles of arrestin-2 interaction with clathrin and adaptor protein 2 in G protein-coupled receptor trafficking. J Biol Chem. 2002;277:30760–30768. doi: 10.1074/jbc.M204528200. [DOI] [PubMed] [Google Scholar]
- 30.Bhandari D, Trejo J, Benovic JL, Marchese A. Arrestin-2 interacts with the ubiquitin-protein isopeptide ligase atrophin-interacting protein 4 and mediates endosomal sorting of the chemokine receptor CXCR4. J Biol Chem. 2007;282:36971–36979. doi: 10.1074/jbc.M705085200. [DOI] [PubMed] [Google Scholar]
- 31. Hanson SM, Cleghorn WM, Francis DJ, Vishnivetskiy SA, Raman D, Song X, Nair KS, Slepak VZ, Klug CS, Gurevich VV. Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J Mol Biol. 2007;368:375–387. doi: 10.1016/j.jmb.2007.02.053. *This is the first report that microtubule-bound arrestins specifically recruit signaling proteins to the cytoskeleton with distinct functional consequences (compare to arrestin-mediated recruitment of signaling proteins to GPCRs, ref 15).
- 32.Wu N, Hanson SM, Francis DJ, Vishnivetskiy SA, Thibonnier M, Klug CS, Shoham M, Gurevich VV. Arrestin binding to calmodulin: a direct interaction between two ubiquitous signaling proteins. J Mol Biol. 2006;364:955–963. doi: 10.1016/j.jmb.2006.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ahmed MR, Zhan X, Song X, Kook S, Gurevich VV, Gurevich EV. Ubiquitin ligase parkin promotes Mdm2-arrestin interaction but inhibits arrestin ubiquitination. Biochemistry. 2011;50:3749–3763. doi: 10.1021/bi200175q. *This is the first demonstration that non-receptor partners affect each other's interaction with arrestins.
- 34.Baillie GS, Adams DR, Bhari N, Houslay TM, Vadrevu S, Meng D, Li X, Dunlop A, Milligan G, Bolger GB, et al. Mapping binding sites for the PDE4D5 cAMPspecific phosphodiesterase to the N- and C-domains of beta-arrestin using spot-immobilized peptide arrays. Biochem J. 2007;404:71–80. doi: 10.1042/BJ20070005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nair KS, Hanson SM, Mendez A, Gurevich EV, Kennedy MJ, Shestopalov VI, Vishnivetskiy SA, Chen J, Hurley JB, Gurevich VV, et al. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. *This study proposed the first model of light-dependent arrestin-1 translocation within rod photoreceptors that explains how arrestin-1 can move without expending energy by virtue of binding to microtubules in the dark and rhodopsin in the light.
- 36. Zhuang T, Chen Q, Cho M-K, Vishnivetskiy SA, Iverson TI, Gurevich VV, Hubbell WL. Involvement of Distinct Arrestin-1 Elements in Binding to Different Functional Forms of Rhodopsin. Proc Nat Acad Sci USA. 2013;110:942–947. doi: 10.1073/pnas.1215176110. *This NMR study of the arrestin-1 interactions with different functional forms of rhodopsin was the first to reveal receptor binding-induced “loosening up” of the arrestin structure.
- 37. Palczewski K, Pulvermuller A, Buczylko J, Hofmann KP. Phosphorylated rhodopsin and heparin induce similar conformational changes in arrestin. J Biol Chem. 1991;266:18649–18654. *This was the first direct demonstration that receptor binding induces a conformational change in arrestin.
- 38. Kim M, Vishnivetskiy SA, Van Eps N, Alexander NS, Cleghorn WM, Zhan X, Hanson SM, Morizumi T, Ernst OP, Meiler J, et al. Conformation of receptor-bound visual arrestin. Proc Nat Acad Sci USA. 2012;109:18407–18412. doi: 10.1073/pnas.1216304109. *This is the most comprehensive comparison of the confromation of free and receptor-bound arrestin based on the measurements of intra-molecular distances in free and rhodopsin-bound arrestin-1 using pulse EPR technique.
- 39. Kim YJ, Hofmann KP, Ernst OP, Scheerer P, Choe HW, Sommer ME. Crystal structure of pre-activated arrestin p44. Nature. 2013;497:142–146. doi: 10.1038/nature12133. *This study (along with ref 40) reported the first crystal structure of an arrestin conformation other than basal.
- 40. Shukla AK, Manglik A, Kruse AC, Xiao K, Reis RI, Tseng WC, Staus DP, Hilger D, Uysal S, Huang LY, et al. Structure of active β-arrestin-1 bound to a Gprotein- coupled receptor phosphopeptide. Nature. 2013;497:137–141. doi: 10.1038/nature12120. *This study (along with ref 39) reported the first crystal structure of an arrestin conformation other than basal.
- 41. Hanson SM, Francis DJ, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, Gurevich VV. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc Natl Acad Sci U S A. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. *This is the first comrehensive elucidation of the receptor “footprint” on arrestin.
- 42.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci U S A. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 44.Vishnivetskiy SA, Hirsch JA, Velez M-G, Gurevich YV, Gurevich VV. Transition of arrestin in the active receptor-binding state requires an extended interdomain hinge. J Biol Chem. 2002;277:43961–43968. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- 45.Kovoor A, Celver J, Abdryashitov RI, Chavkin C, Gurevich VV. Targeted construction of phosphorylation-independent b-arrestin mutants with constitutive activity in cells. J Biol Chem. 1999;274:6831–6834. doi: 10.1074/jbc.274.11.6831. [DOI] [PubMed] [Google Scholar]
- 46. Vishnivetskiy SA, Gimenez LE, Francis DJ, Hanson SM, Hubbell WL, Klug CS, Gurevich VV. Few residues within an extensive binding interface drive receptor interaction and determine the specificity of arrestin proteins. J Biol Chem. 2011;286:24288–24299. doi: 10.1074/jbc.M110.213835. *This study identified surprisingly few residues on an extensive receptor-binding surface of arrestins that drive the interaction and determine receptor specificity of arrestin proteins.
- 47.Vishnivetskiy SA, Baameur F, Findley KR, Gurevich VV. Critical role of the central 139-loop in stability and binding selectivity of arrestin-1. J Biol Chem. 2013;288:11741–11750. doi: 10.1074/jbc.M113.450031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G protein-coupled receptors. Pharm Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Modzelewska A, Filipek S, Palczewski K, Park PS. Arrestin interaction with rhodopsin: conceptual models. Cell Biochem Biophys. 2006;46:1–15. doi: 10.1385/CBB:46:1:1. *This molecular modeling study contains a remarkable prediction that receptor binding induces the rotation of the two arrestin domains relative to each other (revealed by crystal structures reported in refs 39 and 40)
- 50. Whorton MR, Bokoch MP, Rasmussen SG, Huang B, Zare RN, Kobilka B, Sunahara RK. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. **This study (along with ref. 52) unambiguosly demonstrated that monomeric class A GPCR effectively couples to its cognate G protein.
- 51.Whorton MR, Jastrzebska B, Park PSH, Fotiadis D, Engel A, Palczewski K, Sunahara RK. Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J. Biol. Chem. 2008;283:4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J. Biol. Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. **This study (along with ref. 50) unambiguosly demonstrated that monomeric class A GPCR effectively couples to its cognate G protein.
- 53. Bayburt TH, Vishnivetskiy SA, McLean M, Morizumi T, Huang C-c, Tesmer JJ, Ernst OP, Sligar SG, Gurevich VV. Rhodopsin monomer is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. **This study unambiguosly demonstrated that monomeric class A GPCR is effectively phosphorylated by its cogante G protein-coupled receptor kinase and binds arrestin with physiologically relevant affinity.
- 54.Vishnivetskiy SA, Ostermaier MK, Singhal A, Panneels V, Homan KT, Glukhova A, Sligar SG, Tesmer JJ, Schertler GF, Standfuss J, et al. Constitutively active rhodopsin mutants causing night blindness are effectively phosphorylated by GRKs but differ in arrestin-1 binding. Cell Signal. 2013;25:2155–2162. doi: 10.1016/j.cellsig.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsukamoto H, Sinha A, Dewitt M, Farrens DL. Monomeric Rhodopsin Is the Minimal Functional Unit Required for Arrestin Binding. J Mol Biol. 2010;399:501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Singhal A, Ostermaier MK, Vishnivetskiy SA, Panneels V, Homan KT, Tesmer JJ, Veprintsev D, Deupi X, Gurevich VV, Schertler GF, et al. Insights into congenital night blindness based on the structure of G90D rhodopsin. EMBO Rep. 2013;14:520–526. doi: 10.1038/embor.2013.44. *This is the first structure of a disease-associated GPCR mutant, demonstrating how mutation-induced structural perturbations affect receptor function.
- 57. Sommer ME, Hofmann KP, Heck M. Arrestin-rhodopsin binding stoichiometry in isolated rod outer segment membranes depends on the percentage of activated receptors. J Biol Chem. 2011;286:7359–7369. doi: 10.1074/jbc.M110.204941. *This report (along with ref. 58) documents for the first time that a single arrestin molecule can simulatenously engage two GPCRs.
- 58. Sommer ME, Hofmann KP, Heck M. Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin. Nat Commun. 2012;3:995. doi: 10.1038/ncomms2000. *This report (along with ref. 57) documents for the first time that a single arrestin molecule can simulatenously engage two GPCRs and proposes possible biological role of this binding mode.
- 59. Hanson SM, Francis DJ, Vishnivetskiy SA, Klug CS, Gurevich VV. Visual arrestin binding to microtubules involves a distinct conformational change. J Biol Chem. 2006;281:9765–9772. doi: 10.1074/jbc.M510738200. *This is the first report of the conformational change in arrestin induced by microtubule binding.
- 60.Gurevich VV, Gurevich EV. Synthetic biology with surgical precision: Targeted reengineering of signaling proteins. Cell Signal. 2012;24:1899–1908. doi: 10.1016/j.cellsig.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vishnivetskiy SA, Paz CL, Schubert C, Hirsch JA, Sigler PB, Gurevich VV. How does arrestin respond to the phosphorylated state of rhodopsin? J Biol Chem. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. *This is a comprehensive structure-function study of the phosphate sensor in arrestins. Proposed mechanism explained for the first time how the phosphates attached to structurally diverse receptors can similarly “activate” arrestins. This explained how very few arrestins that we have can “serve” hundreds of GPCR subtypes.
- 62.Pan L, Gurevich EV, Gurevich VV. The nature of the arrestin x receptor complex determines the ultimate fate of the internalized receptor. J Biol Chem. 2003;278:11623–11632. doi: 10.1074/jbc.M209532200. [DOI] [PubMed] [Google Scholar]
- 63. Song X, Vishnivetskiy SA, Gross OP, Emelianoff K, Mendez A, Chen J, Gurevich EV, Burns ME, Gurevich VV. Enhanced Arrestin Facilitates Recovery and Protects Rod Photoreceptors Deficient in Rhodopsin Phosphorylation. Curr Biol. 2009;19:700–705. doi: 10.1016/j.cub.2009.02.065. **This is the first demonstration that enhanced phosphorylation-independent arrestin mutants can compensate for the defects in receptor phosphorylation in living animals.
- 64. Gimenez LE, Vishnivetskiy SA, Baameur F, Gurevich VV. Manipulation of very few receptor discriminator residues greatly enhances receptor specificity of non-visual arrestins. J Biol Chem. 2012;287:29495–29505. doi: 10.1074/jbc.M112.366674. **This is the first demonstration that non-visual arestins with high preference for particular GPCR subtypes can be constructed on the basis of naturally promiscuous arrestin-3 by mutagenesis of very few residues.
- 65.Coffa S, Breitman M, Spiller BW, Gurevich VV. A single mutation in arrestin-2 prevents ERK1/2 activation by reducing c-Raf1 binding. Biochemistry. 2011;50:6951–6958. doi: 10.1021/bi200745k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meng D, Lynch MJ, Huston E, Beyermann M, Eichhorst J, Adams DR, Klusmann E, Houslay MD, Baillie GS. MEK1 binds directly to betaarrestin1, influencing both its phosphorylation by ERK and the timing of its isoprenalinestimulated internalization. J Biol Chem. 2009;284:11425–11435. doi: 10.1074/jbc.M806395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seo J, Tsakem EL, Breitman M, Gurevich VV. Identification of arrestin-3- specific residues necessary for JNK3 activation. J Biol Chem. 2011;286:27894–27901. doi: 10.1074/jbc.M111.260448. [DOI] [PMC free article] [PubMed] [Google Scholar]