Abstract
Brassica species (tribe Brassiceae) belonging to U's triangle—B. rapa (AA), B. nigra (BB), B. oleracea (CC), B. juncea (AABB), B. napus (AACC) and B. carinata (BBCC)—originated via two polyploidization rounds: a U event producing the three allopolyploids, and a more ancient b genome-triplication event giving rise to the A-, B-, and C-genome diploid species. Molecular mapping studies, in situ hybridization, and genome sequencing of B. rapa support the genome triplication origin of tribe Brassiceae, and suggest that these three diploid species diversified from a common hexaploid ancestor. Analysis of plastid DNA has revealed two distinct lineages—Rapa/Oleracea and Nigra—that conflict with hexaploidization as a single event defining the tribe Brassiceae. We analysed an R-block region of A. thaliana present in six copies in B. juncea (AABB), three copies each on A- and B-genomes to study gene fractionation pattern and synonymous base substitution rates (Ks values). Divergence time of paralogues within the A and B genomes and homoeologues between the A and B genomes was estimated. Homoeologous R blocks of the A and B genomes exhibited high gene collinearity and a conserved gene fractionation pattern. The three progenitors of diploid Brassicas were estimated to have diverged approximately 12 mya. Divergence of B. rapa and B. nigra, calculated from plastid gene sequences, was estimated to have occurred approximately 12 mya, coinciding with the divergence of the three genomes participating in the b event. Divergence of B. juncea A and B genome homoeologues was estimated to have taken place around 7 mya. Based on divergence time estimates and the presence of distinct plastid lineages in tribe Brassiceae, it is concluded that at least two independent triplication events involving reciprocal crosses at the time of the b event have given rise to Rapa/Oleracea and Nigra lineages.
Introduction
The evolution of land plants, particularly angiosperms, has been characterised by repeated rounds of polyploidization [1] and reticulation [2]. Both of these processes appear to have played a major role in the evolution of the Brassicaceae [3], [4]. Taxonomically, Brassicaceae is a well-defined family with a much conserved floral structure. Classification within the family, in contrast, has been contentious and has undergone many revisions [4]–[6]. The most recent taxonomic treatment of the family, which also takes into consideration evidence from molecular systematics, identifies 49 tribes, 321 genera and 3,600 species [6]. Relationship between many taxa, however, remains poorly resolved as a consequence of processes such as convergent evolution and reticulation.
In this study, we focused on evolution within the tribe Brassiceae, which contains some of the world's most extensively grown vegetable and oilseed crops. The most recent polyploidization event in the tribe Brassiceae was described by U [7] based on cytogenetic analysis of three allopolyploid species, B. juncea (AABB; 2n = 36), B. napus (AACC; 2n = 38) and B. carinata (BBCC; 2n = 34), and their three diploid progenitor species B. rapa (AA; 2n = 20), B. nigra (BB; 2n = 16) and B. oleracea (CC; 2n = 18). This round of polyploidy has been termed the U event [4]. The three allopolyploids behave as strict diploids, in all probability because of a mechanism that suppresses homoeologous chromosome pairing [8], [9].
A more intriguing case of polyploidization is an earlier genome triplication event, referred to as event b [4], which gave rise to the three diploid Brassica species of U's triangle, i.e., B. rapa, B. nigra and B. oleracea, as well as other species included in tribe Brassiceae. While earlier molecular mapping studies of B. nigra suggested the possibility of genome triplication [10], [11], the first clear evidence for this past event came from comparative mapping of the A and C genomes of B. napus (AACC) with genic markers from the model crucifer—Arabidopsis thaliana [12]. Based on extensive gene collinearity between A. thaliana and the A and C genomes of B. napus (AACC), the A. thaliana genome has been divided into blocks A through X [13]. Each A. thaliana gene block is present in at least three copies in each of the two constituent genomes, A and C, of B. napus [12]. Similar results have been observed between the A and B genomes of B. juncea (AABB) and A. thaliana in regard to triplication of each A. thaliana gene block [14]. Unequivocal evidence for gene triplication was also uncovered by in situ hybridization of A. thaliana genomic bacterial artificial chromosomes (BACs) with pachytene-stage chromosome preparations of some key Brassiceae taxa [15].
Sequencing of the B. rapa genome has provided strong evidence for genome triplication [16]. The triplication event involving three ancestors (each one with n = 7) leading to the formation of a hexaploid (2n = 42) was followed by gene fractionation [16], [17], chromosome reshuffling and reduction in chromosome number [18], leading to the evolution of present-day diploid taxa of tribe Brassiceae. Based on gene fractionation patterns, it has been proposed that the hexaploidy was the result of two independent events: two diploid genomes first coming together to produce a tetraploid, followed by crosses with a third diploid species [16], [17]. Early-entry genomes (termed MF1 and MF2) are the most heavily fractionated, with the last genome to enter (designated as LF) experiencing the lowest gene loss [16], [17], [19].
In taxonomic classifications, tribe Brassiceae has generally been considered to be monophyletic based on three conserved morphological traits—conduplicate cotyledons, segmented fruits and simple or no trichomes [5], [6], [20]–[22]. Molecular analysis of the nuclear genome has uncovered evidence of ancient genome triplication across tribal members, supporting a monophyletic origin for Brassiceae [12], [14]–[16]. Analysis of plastid DNA, however, reveals the presence of two distinct lineages—the Nigra (also called Sinapis) lineage and the Rapa/Oleracea lineage [23]–[27]. The presence of these two lineages was first established by RFLP analysis of plastid DNA from various genera and species [23], [24]. These observations have since been confirmed in a number of studies on plastid genes [26] and plastid non-coding sequences [15], [25], [27]. The findings of major studies on plastid DNA divergence confirming the presence of two distinct lineages are summarized in Table S1. The number of distinct lineages has increased to eight through analysis of more number of species [25], [27].
Two competing hypotheses can be put forth to explain the evolution of the Brassica A and B genomes: (a) A and B genomes diverged after a single b event, or (b) A and B genomes arose independently from different reciprocal crosses. Three types of data—gene fractionation patterns, gene block arrangements and synonymous nucleotide substitution rates in homologous genes—are the most pertinent for assessing the validity of these two hypotheses.
In this study, we investigated the genomic structure of an R-block region of B. juncea (AABB). The R block is present in triplicate in genomes A (contributed by B. rapa) and B (contributed by B. nigra). R blocks are gene rich and fairly large [14]. Our initial interest was focused on the R block on linkage group (LG) A10 of B. juncea, as it contains some of the most important yield-related quantitative trait loci (QTLs) mapped in B. juncea using a doubled haploid (DH) population derived from a cross between the East European gene pool line Heera and the Indian gene pool cultivar Varuna [28]. We aligned genomic BACs in the six R blocks, sequenced the BACs, annotated the sequences and studied gene fractionation pattern in a syntenic region of the R blocks. Pairwise synonymous base substitution rates (Ks values) and divergence times were calculated for paralogous (within A and B genomes) and homoeologous (across A and B genomes) nuclear genes to estimate divergence times of the paralogues and homoeologues. Pairwise synonymous base substitution rates and divergence times were also calculated for the plastid genes matK and ndhF and compared with the nuclear gene divergence times.
We propose that a hypothesis of reciprocal crosses between progenitor genomes at the time of the entry of the third genome provides the most parsimonious explanation for the presence of two or more plastid lineages in tribe Brassiceae.
Results
Identification and sequencing of BACs mapped to the six R blocks of B. juncea
In A. thaliana, the R block is present as a single block between genes At5g01240 and At5g22030 on chromosome 5 [13]. The six R blocks of B. juncea have been previously mapped in DH lines developed from a cross between two lines, Heera and Varuna, from well-defined B. juncea East European and Indian gene pools, respectively [14], [29], [30]. The R block of A. thaliana is represented in the A genome of B. juncea on LGs A2 (∼20 cM), A3 (∼26 cM) and A10 (∼50 cM), and in the B genome on LGs B2 (∼24 cM), B3 (∼29 cM) and B8 (∼47 cM) [14]. Based on examination of gene fractionation patterns, R blocks of A2, A3 and A10 have been respectively identified as most fractionated (MF2), medium fractionated (MF1) and least fractionated (LF) [16]. Previous mapping work using intron-length polymorphism (IP) markers has shown that A2–B2, A3–B3 and A10–B8 R blocks are homoeologous [14].
For analysis of R blocks of the A and B genomes, we focused on a specific region between gene IDs At5g14660 and At5g15840 of the R block of LG A10, which contains major QTLs mapped in a cross between Heera and Varuna [28], [31]. Two BAC libraries developed using BamHI- and HindIII-digested Heera genomic DNA were used to construct a physical map for all six LGs in the targeted area. This region is syntenous with a 444-kb region of the R block of A. thaliana.
BACs corresponding to the above region were identified using region-specific IP markers available on each of the six LGs [14]. A total of 85 BAC clones for the target region in the six R blocks were identified using these markers (Table S2). The gene span of each BAC clone was determined by amplification of genes upstream and downstream of the IP markers used in the initial screening. Once the total gene span of each BAC clone was established, a minimum number of overlapping BAC clones were identified for the targeted region of each of the six R blocks. Contigs on R blocks of A2 and B2 were constructed from four overlapping BAC clones each. Contigs on R blocks of A3 and B3 were created from three overlapping BAC clones each, while contigs on R blocks of A10 and B8 were constructed from five and seven BAC clones, respectively (Figure S1).
A total of 26 BAC clones were sequenced, followed by assembly of BAC-specific contigs. Contigs of the overlapping BACs for each of the six R blocks were assembled into supercontigs (scaffolds) by establishing overlaps. BACs identified for LG A2 and LG B2 were assembled into single scaffolds. BACs of other LGs were assembled into multiple scaffolds. A list of sequenced BAC clones and assembled scaffolds is given in Table S3. A total of 2,470 kb of DNA covering all six R blocks was annotated using the Brassica BAC annotation pipeline (http://brassica.nbi.ac.uk/annotate.html).
Gene collinearity and fractionation patterns in the sequenced regions of B. juncea
Gene contents of the six R block regions of the homoeologous linkage groups A2–B2, A3–B3 and A10–B8 were compared with gene content information available for the R block of A. thaliana and syntenous regions of B. rapa in the BRAD database (http://brassicadb.org/brad/index.php) (Table S4). The syntenous region of A. thaliana on chromosome 5 contains 120 genes. The LF R blocks of B. juncea on A10 and B8 have retained about 63% of the genes found in A. thaliana, while the MF2 R blocks on A2 and B2 and MF1 R blocks on A3 and B3 have retained about 50% and 43% of these genes, respectively (Table 1). Contrary to the overall trend observed in the genome of B. rapa [16], MF2 has more gene retention than MF1 in the region sequenced here.
Table 1. Difference in gene content of R blocks in the A and B genomes of B. juncea, as compared with the gene content reported in B. rapa (A) and A. thaliana.
| A. thaliana | B. rapa MF2-R-A2 | B. juncea MF2-R-A2 | B. juncea MF2-R-B2 | B. rapa MF1-R-A3 | B. juncea MF1-R-A3 | B. juncea MF1-R-B3 | B. rapa LF-R-A10 | B. juncea LF-R-A10 | B. juncea LF-R-B8 | |
| Number of A. thaliana genes of R block | 120 | 56 ▴ | 61 | 60 | 51 ▴ | 53 | 51 | 69 ▴ | 75 | 78 |
| Number of Brassica lineage specific genes of R block | - | 5 | 3 | 2 | 10 | 3 | - | 9 | 9 | 6 |
| * Number of CDS showing similarity with proteins coded by the B. rapa genes from non-collinear blocks | - | - | 2 | 13 | - | 3 | 2 | - | 9 | 14 |
| * Number of CDS showing similarity with proteins from genera other than B. rapa | - | - | 3 | 8 | - | 1 | - | - | - | 4 |
| * Number of CDS showing similarity with transposon related proteins | - | - | - | 10 | - | 1 | - | - | 1 | 11 |
MF2 - Most fractionated subgenome; MF1 - Medium fractionated subgenome; LF - Least fractionated subgenome.
A - A genome; B - B genome; R - R block; LG - linkage group.
CDS - coding sequence.
*- The rows marked with an asterix indicate that the recorded CDS are unique to that LG.
-Difference in gene numbers between B. rapa and A genome of B. juncea is observed because these genes are present in scaffold sequences of Brassica database (BRAD) but have not been recorded.
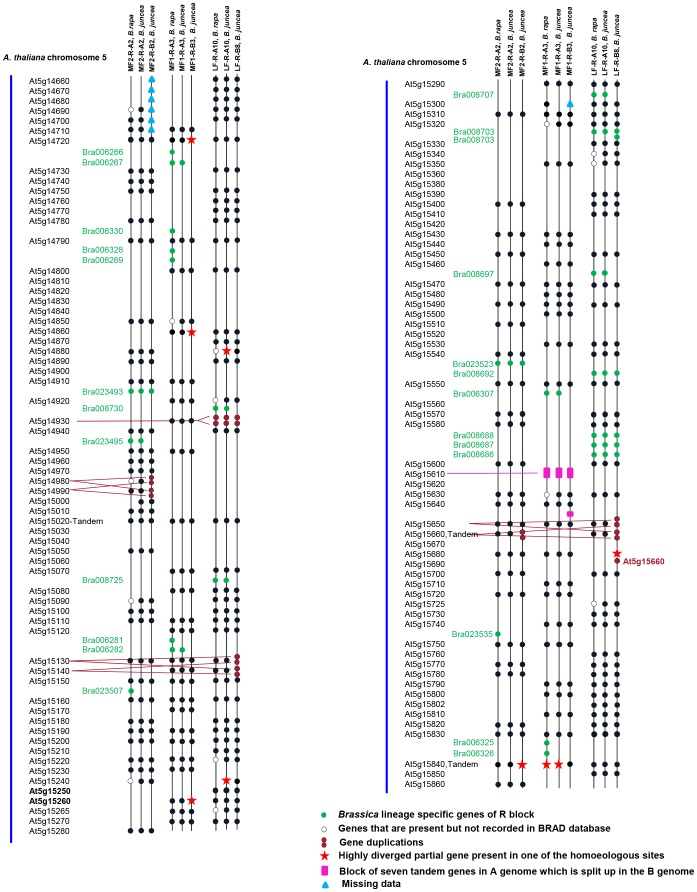
Because intergenic regions are prone to rapid evolutionary changes, we only examined the arrangement and fractionation pattern of coding sequences. Overall, the sequenced region in the A and B genomes of B. juncea showed strong collinearity in gene arrangement with that of A. thaliana (Figure 1). Sixteen out of 120 genes present in the syntenous region of A. thaliana were missing from both A and B genomes of B. juncea. The pattern of fractionation between syntenous LGs was identical for 97 of the 104 retained A. thaliana orthologues. With respect to the seven other retained A. thaliana genes, fractionation patterns differed between A and B genome homoeologues of B. juncea: while one of the homoeologues was present as a complete gene, the other was present either as a partial genic fragment or as a sequence showing only low-level of identity. In particular, A. thaliana At5g14720, At5g14860 and At5g15260 orthologues were observed in the A genome, while their homoeologues showing low-identity were observed at collinear positions in the B genome. Likewise, At5g14880 and At5g15240 orthologues were observed in the B genome, while their homoeologues in the A genome showed low-identity. At5g15840, a transcription factor-encoding gene, had one orthologue present in MF1 R block of B and another one in MF2 R block of A genome; their respective homoeologues at collinear positions in A and B genomes showed only remnants of identity. The A. thaliana gene At5g15680 was absent from the LF block of the A genome, while a partial well conserved sequence of the gene was identified in the LF block of the B genome (Figure 1).
Figure 1. Comparison of the gene organisation in the six R blocks of B. juncea.
Gene organisation in R blocks of the constituent A and B genomes of B. juncea has been compared with the gene organisation in the syntenous region of the R block of A. thaliana in chromosome 5 and R blocks of B. rapa.
We compared the coding sequence (CDS) content of B. rapa available in the BRAD database [32] with predicted CDSs of the B. juncea A-genome sequence assembly, and observed a disparity in the number of retained genes (Table 1). However, a BLASTN search for missing genes not recorded in the B. rapa BRAD database showed that most were actually present in the B. rapa scaffold assembly (Figure 1). High gene collinearity is thus evident between the B. juncea A genome and the B. rapa genome, in terms of CDS arrangement and the gene content is also similar.
In addition to A. thaliana gene orthologues identified in the sequenced region of B. juncea, other CDSs were detected by the annotation pipeline. A BLASTN search revealed 23 of these CDSs to be Brassica lineage-specific R-block genes. Brassica lineage-specific genes from the R block of B. rapa described in the BRAD database, i.e., Bra023493, Bra008703, Bra023523, Bra008692, Bra008688, Bra008687 and Bra008686, were observed on homoeologous LGs of both A and B genomes of B. juncea (Figure 1). This result is strongly suggestive of A- and B-genome common ancestry. Some Brassica lineage-specific genes identified in the A genome of B. juncea, i.e., Bra006267, Bra023495, Bra008725, Bra006282, Bra008730, Bra008707, Bra008697 and Bra006307, did not have homoeologues in the B genome (Figure 1). This difference can be attributed to gene losses in the Nigra lineage after the Rapa-Nigra split. A few lineage-specific genes described in the BRAD database, i.e., Bra023507, Bra023535, Bra006266, Bra006330, Bra006328, Bra006269, Bra006281, Bra006325 and Bra006326, were not present in either A or B genomes of B. juncea. It is interesting to observe that out of 10 lineage-specific genes on the LG-A3 R block (MF1) of B. rapa, only 3 have been retained on LG A3 of B. juncea, whereas all 9 lineage-specific genes on the LG-A10 R block (LF) of B. rapa are present on LG A10 of B. juncea.
A total of 82 CDSs showing no identity with A. thaliana orthologues at collinear positions were recognised by the annotation pipeline. These inserted sequences were always specific to a single R block, and appeared as clusters predominantly on LGs B2 and B8. BLASTX comparison of the predicted CDSs with protein databases using a threshold of E<e−5 revealed significant similarity with protein(s) in the UniProt database. Forty-three CDSs showed similarity with functional proteins encoded by B. rapa genes from other blocks, 16 CDSs showed similarity with proteins encoded by genera other than B. rapa, and 23 CDSs were associated with transposon-related proteins (Table S5). Transposon-related CDSs were not included in subsequent analyses. A BLASTN search of CDSs against unpublished B. juncea, B. rapa and B. nigra transcriptome databases with an E<e−10 threshold revealed that 62% were represented in the B. juncea transcriptome database. More than 80% of the CDSs expressed in the transcriptome were highly divergent from B. rapa gene transcripts and appeared to be ancient insertions. Most of these insertions were observed in the B genome, with a few specific to the A genome. The remaining 20% of CDSs were mostly Brassica lineage-specific genes or gene fragments that appear to have recently transposed from noncollinear regions. Most of these transpositions were specific to the A genome. It can be concluded that the A and B genomes of B. juncea contain many unique CDSs. This situation may be due to either loss of these CDSs from homoeologous sites after genome triplication or to divergence in the progenitor species contributing to evolution of the A and B genomes.
An interesting feature is the tandemly duplicated nature of several genes present in single copies in A. thaliana (Figure 1). Single-copy genes At5g14930 and At5g15610 were found to be present as tandem repeats in both the A and B genomes of B. juncea and the B. rapa genome. Arabidopsis thaliana gene At5g14930 showed tandem duplication on the LF blocks of LGs A10 and B8, while At5g15610, a proteasome component domain protein, was present in seven tandem copies on the MF1 block of LG A3; this latter gene also occurred in four copies on LG B3, although the fourth copy was found to have separated from the other three (Figure 1). Gene-pair duplications specific to the B genome of B. juncea were also observed: At5g14980-At5g14990, At5g15130-At5g15140 and At5g15650-At5g15660. In addition to the At5g15650-At5g15660 duplicated gene pair, a third copy of At5g15660 was observed on the LF R block of LG B8 (Figure 1).
Nuclear genome divergence
Nuclear genome divergence was studied by estimating the synonymous base substitution rates (Ks) for 11 genes (Table S6) retained in all six R blocks of B. juncea and their respective orthologues from A. thaliana. For each of the 11 genes, six CDSs of B. juncea annotated by the Brassica BAC annotation pipeline were aligned with A. thaliana sequences using the CLUSTAL W codons option of the Alignment explorer in MEGA5 [33]. After removing gaps and codons with ambiguous bases, only common aligned regions from all the sequences were considered for further analysis. Pairwise comparisons were used to estimate Ks values between B. juncea and A. thaliana genomes (orthologue divergence), and within A and B genomes (paralogue divergence) and between A and B genomes (homoeologue divergence) of B. juncea. Divergence times were calculated assuming a mutation rate of 1.5×10−8 substitutions per synonymous site per year [21].
Average Ks values for A. thaliana (AT)-B. juncea (BJ) ranged from 0.43±0.1 (for AT-BJ A3) to 0.49±0.25 (for AT-BJ A2) on the A genome, and from 0.45±0.15 (for AT-BJ B3) to 0.46±0.14 (for AT-BJ B8) on the B genome (Table 2). The mean synonymous base substitution rate, 0.46±0.15, calculated for Arabidopsis-Brassica indicates that the two genomes diverged about 15 mya (Figure 2). Analysis of variance showed no significant difference (F[5,60] = 0.17; p = 0.97) among Ks values derived from the different Arabidopsis-Brassica comparisons, indicating that all six subgenomes are almost equally diverged from A. thaliana.
Table 2. Nuclear genome divergence analysis based on synonymous nucleotide substitutions in the genes retained across all the six R blocks of B. juncea and their orthologues from A. thaliana.
| Orthologue divergence | Paralogue divergence | Homoeologue divergence | ||||||
| Between A. thaliana and B. juncea | Within the A and B genomes of B. juncea | Between the A and B genomes of B. juncea | ||||||
| Mean Ks value | Divergence time (mya) | Mean Ks value | Divergence time (mya) | Mean Ks value | Divergence time (mya) | |||
| A genome | A genome | 11 homoeologues | ||||||
| AT-BJ A2 | 0.49±0.25 | 16.3±8 | BJ A2-BJ A3 | 0.36±0.14 | 11.9±5 | BJ A2-BJ B2 | 0.27±0.14 | 8.9±5 |
| AT-BJ A3 | 0.43±0.10 | 14.5±3 | BJ A2-BJ A10 | 0.39±0.12 | 13.1±4 | BJ A3-BJ B3 | 0.18±0.07 | 6.1±2 |
| AT-BJ A10 | 0.48±0.12 | 15.9±4 | BJ A3-BJ A10 | 0.36±0.15 | 12.0±5 | BJ A10-BJ B8 | 0.19±0.08 | 6.5±3 |
| B genome | B genome | All homoeologues | ||||||
| AT-BJ B2 | 0.45±0.12 | 15.2±4 | BJ B2-BJ B3 | 0.33±0.08 | 11.0±3 | BJ A2-BJ B2 | 0.23±0.09 | 7.6±3 |
| AT-BJ B3 | 0.45±0.15 | 15.4±5 | BJ B2-BJ B8 | 0.34±0.13 | 11.2±4 | BJ A3-BJ B3 | 0.21±0.07 | 6.8±2 |
| AT-BJ B8 | 0.46±0.14 | 15.9±4 | BJ B3-BJ B8 | 0.38±0.13 | 12.6±4 | BJ A10-BJ B8 | 0.21±0.06 | 6.9±2 |
| Overall Mean | 0.46±0.15 | 15.3±4 | 0.36±0.12 | 12.0±4 | 0.21±0.10 | 7.1±3 | ||
Divergence time was calculated by assuming a mutation rate of 1.5×10−8 synonymous substitutions per site per year [21].
Overall mean values were calculated from the comparisons made for the 11 genes retained in all the six R blocks.
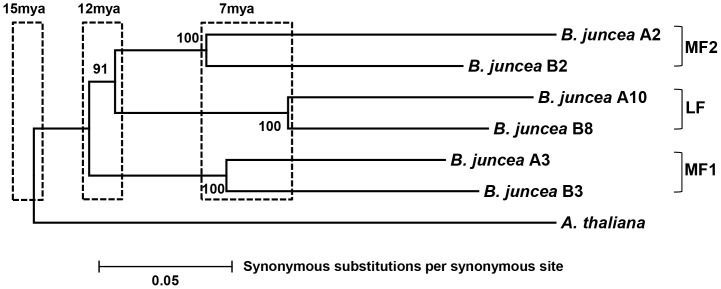
Figure 2. Neighbour-joining tree constructed on the basis of distances estimated from synonymous base substitutions of the concatenated nucleotide sequences of 11 genes retained in all the six R blocks of B. juncea.
The numbers on the nodes represent percentage from bootstrap analysis of 1000 replicates. A. thaliana-Brassica divergence time ∼15 mya; Brassica A genome paralogue and B genome paralogue divergence time ∼12 mya; A and B genome homoeologue divergence time ∼7 mya.
To decipher the divergence order of the three paralogues within the A and B genomes of B. juncea, mean Ks values were estimated by pairwise comparison of genes from these regions. Average Ks values ranged from 0.36±0.14 (for BJ A2-BJ A3) to 0.39±0.12 (for BJ A2-BJ A10) within the A genome, and from 0.33±0.08 (for BJ B2-BJ B3) to 0.38±0.13 (for BJ B3-BJ B8) within the B genome (Table 2). No significant variation was found among Ks values calculated from pairwise comparisons of paralogues on the A genome (F [2], [30] = 0.27; p = 0.77) and B genome (F [2], [30] = 0.54; p = 0.58), indicating that the three progenitors contributing to the A and B genomes diverged at the same time, about 12 mya (Figure 2). The mean Ks value of 0.36±0.12 (n = 66) obtained from paralogue comparison within A and B genomes is significantly different (p<0.0001, unpaired Student's t-test) from the mean of 0.46±0.15 (n = 66) calculated from Arabidopsis-Brassica comparisons. These results indicate that the three progenitors of Brassica A and B genomes diverged after the Arabidopsis-Brassica split.
To examine divergence of the A and B genomes of B. juncea, the rate of synonymous substitutions was calculated for homoeologous gene pairs. Average Ks values ranged from 0.18±0.07 (for BJ A3-BJ B3) to 0.27±0.14 (for BJ A2-BJ B2) (Table 2) with no significant variation (F [2], [30] = 2.2; p = 0.12). The mean Ks value of 0.21±0.1 (n = 33) calculated from the homoeologue comparisons implies a divergence time of approximately 7 mya for the A–B genome split (Figure 2). This calculated Ks value is significantly different (p<0.0001, unpaired Student's t-test) from the mean of 0.36±0.12 (n = 66) derived from comparisons of paralogues within the A and B genomes. This difference suggests that the divergence of the three paralogues of A and B genomes has more antiquity than the divergence between homoeologues of A and B genomes i.e the divergence of B. rapa and B. nigra.
We constructed a neighbour-joining tree taking into consideration the evolutionary distances estimated from synonymous base substitution rates, using a 9,399-bp long data set comprising concatenated sequences of the 11 genes (Figure 2).
The above-described homoeologue divergence analysis between A and B genomes considered only genes retained across all six R blocks. We carried out an additional analysis that included every possible homoeologous gene pair i.e. 44 gene pairs for BJ A2-BJ B2, 45 for BJ A3-BJ B3 and 54 for BJ A10-BJ B8. Average Ks values ranged from 0.21±0.1 for BJ A3-BJ B3, BJ A10-BJ B8 and 0.23±0.1 for BJ A2-BJ B2 (Table 2). Even with the larger number of genes included in this analysis, the results were similar to the previous analysis, with no significant variation observed in Ks values (F[2,140] = 1.2; p = 0.30) between the three homoeologous LGs.
Pairwise divergence was estimated for seven Brassica lineage-specific genes present in the homoeologous LGs—two from MF2 (BJ A2-BJ B2) and five from LF (BJ A10-BJ B8) (Figure 1). Calculated Ks values ranged from 0.12 for Bra008703 to 0.23 for Bra023493, with a mean of 0.19±0.04, almost identical to estimated levels of divergence between homoeologous gene pairs of A and B genomes. This result indicates that these genes were present in the progenitors before the A-B genome split.
The antiquity of Brassica tandem duplications was studied by calculating pairwise divergence levels between the duplicated gene copies. A Ks value of 0.6 was estimated between copies of At5g14930, a tandemly duplicated gene in both BJ A10 and BJ B8; this value was much higher than the Ks value of 0.2 calculated for the corresponding homoeologues (i.e., Ks between A genome and B genome copies). This result suggests that the gene duplication predated the divergence of A and B genomes and occurred in the progenitor(s) contributing to the b event. Orthologues of A. thaliana genes At5g14980 and At5g14990 are present as tandem duplicates on LG B2, and At5g15130, At5g15140 and At5g15650 orthologues are found on LG B8. Ks values of these duplicates were between 0.01 and 0.15, indicating that the associated duplication events were recent and specific to the B genome.
Plastid genome divergence
A phylogeny was reconstructed for the six Brassica species of U's triangle from plastid maturase K (matK) gene sequences available in the NCBI database (www.ncbi.nlm.nih.gov/nuccore). Sequence of A. thaliana matK was included as an outgroup. The aligned set of matK sequences of the six Brassica species was 1,572 bp in length, which was 6 bp shorter than the matK sequence of A. thaliana. After removing gaps, pairwise distances were calculated by the Kimura two-parameter method [34], and a neighbour-joining tree was constructed. The allotetraploid B. carinata and its cytoplasm donor, the diploid species B. nigra, formed one lineage; allotetraploids B. juncea and B. napus, which acquired cytoplasm from B. rapa, and diploid species B. rapa and B. oleracea clustered together in another lineage (Figure S2). These results are in congruence with earlier studies reporting the presence of the two lineages Rapa/Oleracea and Nigra in the genus Brassica (Table S1) [23]–[27].
Results obtained with the matK gene were confirmed by similar analysis using another plastid gene, ndhF (NADH dehydrogenase subunit F). Partial gene sequences of the three Brassica diploid species and B. napus were downloaded from the NCBI database (www.ncbi.nlm.nih.gov/nuccore) and aligned with the 2,241-bp complete gene sequence of A. thaliana. The reading frame of partial sequences was determined using A. thaliana as a reference. The phylogenetic tree constructed from the 732-bp common aligned sequence set revealed two lineages, with B. rapa, B. oleracea and B. napus clustering into one lineage, and B. nigra comprising the other.
To estimate the divergence time of chloroplast genomes of B. rapa and B. nigra, the two progenitors of B. juncea, Ks values were calculated for matK and ndhF genes. Divergence times were estimated assuming a mutation rate of 1.7×10−9 substitutions per synonymous site per year previously calculated for Brassicaceae chloroplast DNA [35]. Based on the Ks value of 0.044 calculated for matK, Rapa/Oleracea and Nigra plastid lines were estimated to have diverged from one another 12.0 mya; using the Ks value of 0.036 calculated for ndhF, a divergence time of 11.6 mya was inferred for these lineages. The calculated plastid genome divergence time for B. rapa and B. nigra, approximately 12 mya, coincides with the estimated nuclear divergence time of the three progenitors of present-day Brassica species A and B genomes.
Discussion
The best evidence for independent origins of A and B genomes is derived from divergence times estimated from levels of synonymous nucleotide sequence divergence in plastid and nuclear genes. The evidence presented here, as well as extensive data available from earlier studies (Table S1), clearly reveals that A and B genomes represent two independent plastid lineages in tribe Brassiceae—the Rapa/Oleracea lineage and the Nigra lineage. Separation of these two lineages dates back to around 12 mya based on Ks values for plastid matK and ndhF gene sequences and the plastid DNA mutation rate estimate given by Koch et al. [35].
Previous research involving mapping and in situ hybridization have clearly established that Brassica A, B and C genomes are triplicated genomes [10]–[12], [14], [15]. If the three constituent genomes of B. rapa (AA) are represented as X, Y and Z and those of B. nigra (BB) as X′, Y′ and Z′, our synonymous nucleotide substitution data indicate that the X-X′, Y-Y′ and Z-Z′ (the A and B genome homoeologues in B. juncea) split occurred around 7 mya, and that the paralogues X-Y-Z and X′-Y′-Z′ separated about 12 mya. Ks values between the paralogues of the A genome, X-Y-Z, and the paralogues of the B genome, X′-Y′-Z′ are significantly different from levels of divergence between X-X′, Y-Y′ and Z-Z′ (Table 2).
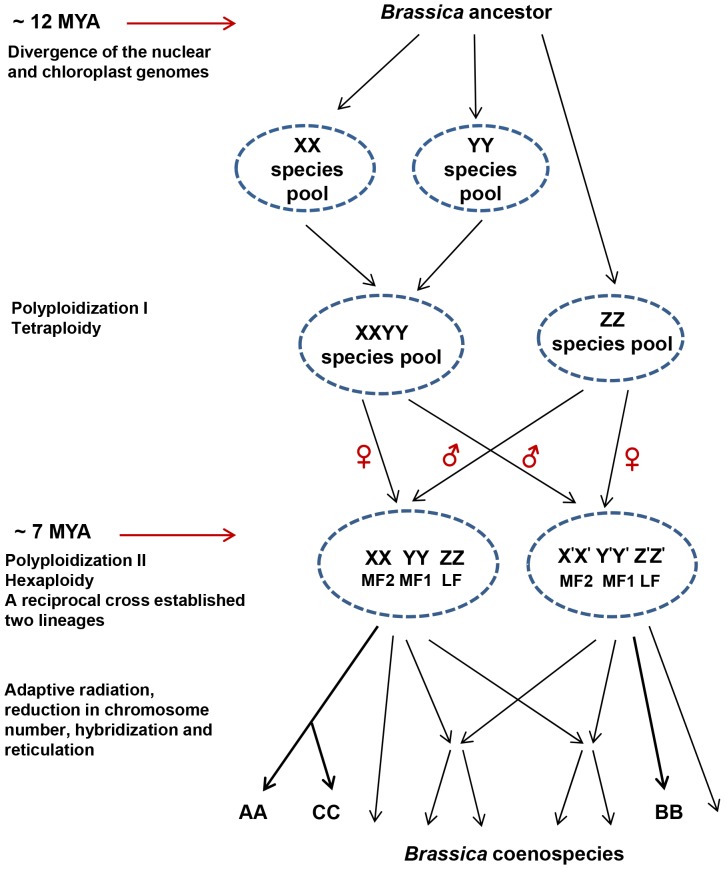
Based on the rate of synonymous substitutions among R-block nuclear genes and the overwhelming evidence supporting plastid Rapa/Oleracea and Nigra lineages, we propose the following model to explain the evolution of A and B genomes (Figure 3):
Figure 3. A model depicting the evolutionary events leading to divergence of the Rapa/Oleracea and Nigra lineages of Brassiceae.
Length of the arrows does not represent evolutionary distance.
The Brassica lineage split from the Arabidopsis lineage around 15 mya. Other studies have dated this separation to anywhere between 14.5 to 20 mya [36]–[38]. Nevertheless, all reported estimates show that the split between the Brassica and Arabidopsis lineages happened earlier than diversification of the Brassica lineage.
The Brassica progenitor evolved into a number of species. In Figure 3, we show only three—X, Y and Z—to explain A and B genome evolution. It may be more appropriate, however, to describe X, Y and Z as X, Y and Z species pools.
X, Y and Z have similar Ks values, and are therefore of similar antiquity. Based on gene fractionation patterns in B. rapa, it has been proposed recently that X and Y first came together to form an allotetraploid, followed by entry of the third genome, Z [16], [17]. Our model (Figure 3) agrees with this sequence of events.
Recently generated B. rapa nuclear genome sequence data reveals that the Z genome is the least fractionated (LF), X is the most fractionated (MF2) and Y is medium fractionated (MF1) [16]. Our model agrees with this fractionation pattern.
A minimum of two independent genome triploidization events, involving reciprocal crosses between an XY tetraploid and a Z diploid, and thereby establishing two plastid lineages, must have occurred about 7 mya. The ages of the plastid lineages date back to the independent formation of X, Y and Z species pools, approximately 12 mya.
Stochastically, two reciprocal crosses would not survive in a panmictic population. If one cross had a selective advantage over the other, the latter would be rapidly eliminated. The two reciprocal crosses leading to the establishment of hexaploids (XXYYZZ) must have therefore occurred under reproductive isolation, and may have involved somewhat differentiated X, Y and Z species pools.
Our model allows for gene fractionation [16], [17], chromosomal rearrangement [18], [39], hybridization and reticulation [3] during subsequent evolution of tribe Brassiceae (Figure 3).
A number of studies on family Brassicaceae have calculated Ks values and divergence times using different set of nuclear genes (Table S7). All the studies show that Arabidopsis-Brassica split has more antiquity than divergence within the tribe Brassiceae. The evolutionary divergence between the ancestral genomes—X, Y, Z—that took part in genome triplication has been reported to be more than divergence within each genome i.e between X-X′, Y-Y′, Z-Z′ (Table S7). Divergence time figures show the same trend in all the studies but have been interpreted to propose a single event of hexaploidy followed by divergence of the Nigra and Rapa/Oleracea lineages. This does not fit with the antiquity of plastid genome divergence of 12 mya calculated using the synonymous mutation rate estimates given by Koch et al. [35]. The model presented here reconciles both nuclear and plastid genome divergence times (Figure 3).
Brassica species have poor reproductive barriers, and hybrids could have readily formed at hexaploidy and subsequently between the descendants. Cytogenetic studies have clearly demonstrated the high intercrossability of Brassica species and significant pairing between homoeologous chromosomes [40]–[43]. As a consequence, a large number of species belonging to the tribe Brassiceae have been referred to as Brassica coenospecies.
A study of the Brassiceae U polyploidization event clearly demonstrates that reciprocal crosses can occur and survive. Brassica napus (AACC) and B. juncea (AABB) have A-genome (B. rapa) plastid genomes, whereas B. carinata (BBCC) has a B-genome (B. nigra) plastid genome [44]. The three polyploids originated in isolation. Further, sufficient data exists from plastid and nuclear genome studies of B. napus lines to suggest that this allopolyploid had two or more independent origins [9], [45]. Thus, the U polyploidization event involved both reciprocal and multiple crossing events. It is therefore entirely plausible that the b event involved multiple crosses that included reciprocal crossing.
The main difference between b and U events is the chromosomal number of the resulting allopolyploids. The U event gave rise to strict allotetraploids with chromosome numbers exactly equal to the sum of any two of the three U-triangle species—B. rapa (AA), B. oleracea (CC) and B. nigra (BB). In comparison, the b polyploidization event led to the evolution of mesoploids with chromosome numbers much reduced from the initial hexaploid chromosome number (2n = 6x = 42) [18].
Does the R-block gene fractionation pattern reported in this study support the hypothesis of independent origin of the two mesoploid genomes A and B? The six R blocks studied here (three from the A genome and three from the B genome) show very high gene collinearity and a conserved gene fractionation pattern with respect to the R block of A. thaliana and homoeologous LGs. The fractionation pattern is different between B. rapa and B. nigra in only seven of the 104 Arabidopsis genes (Figure 1). Gene redundancy exists in all these cases, as paralogues are available. Thus differential gene content could be explained either way—as evidence of independent origin or gradual gene loss over time.
Many gene duplications are specific to the B genome. Because these duplicated genes are of relatively recent origin, they do not provide any support for independent genome origins. The duplicated genes common to both A and B genomes may have either independent origins or shared lineages, and therefore are not rigorous evidence for the two-independent-lineage hypothesis. An interesting observation is the presence of many reading frames on LG B2 (MF2-R) and LG B8 (LF-R). These CDSs are unique to the B genome. Such unique CDSs were also observed on LG A2 (MF2-R) and LG A10 (LF-R) (Table 1). Some of these CDSs show identity with genes from distantly related plant families. It is possible that these genes were present in the putative Brassica ancestor but have been selectively lost in some of the descendants which constituted diverged species pools X, Y and Z. The differential loss of these CDS can be cited as an evidence for independent origin of A and B lineages.
A third piece of evidence can be gleaned from block arrangements on the A, B and C genomes. Based on the arrangement of 24 genomic blocks [13], some previous mapping studies using genic markers [12], [14] uncovered identical block arrangements between three LGs of A and C genomes (A1–C1, A2–C2 and A3–C3) and significant similarity between three LGs of the A and B genomes (A4–B4, A5–B5 and A6–B6). Our recent mapping work (unpublished) using transcriptome-based SNPs, however, has revealed much more extensive reshuffling on most LGs of the B genome. None of the LGs of the B genome seem to have the same block arrangement as those of the A genome. These differences are further suggestive of independent A and B genome origins.
Based on the divergence times estimated from synonymous base substitution rates in nuclear and plastid genes and their somewhat similar gene fractionation patterns, we conclude that reciprocal crossing between similar genomes at the time of the b event most plausibly explains the origin of Brassica A and B genomes. A recently published phylogenetic analysis of 89 Brassiceae species using sequences from four plastid intergenic regions—rpl32-trnL, atpI-atpH, psbD-trnT and ycf6-psbM—uncovered eight lineages: Rapa-Oleracea, Savignya, Nigra, Cakile, Crambe, Henophyton, Zilla and Vella [27]. Data was analysed by three different algorithms—maximum parsimony, maximum likelihood and Bayesian inference; all three methods yielded similar results confirming the eight independent lineages. Just like the Rapa/Oleracea and Nigra lineages, these additional lineages may also have resulted from independent triplication events involving species that were descendants of a putative ancestral species. Thus even though tribe Brassiceae is monophyletic – hexaploidization, the b event, was not a single event but a number of independent events involving hybridization between closely related or somewhat divergent species.
Materials and Methods
Plant materials and B. juncea BAC libraries
BAC library development was performed by Amplicon Express (Pullman, Washington, USA) using the B. juncea East European line Heera. Partial genomic digests of nuclear DNA were cloned into a pECBAC1 vector at a BamHI site and into a pCC1BAC vector at a HindIII site for the construction of two independent BAC libraries. PCR screening of the BAC libraries was carried out using the Matrix Pool and Superpool strategy of Amplicon Express (http://ampliconexpress.com/products-services/screening-services/pools-and-superpools). Each B. juncea BAC library consisted of 55,296 clones with an average insert size of 120–130 kb distributed over 144 384-well plates, thus representing approximately five genomic equivalents of the B. juncea haploid genome estimated to be 1,068 Mb in size [46].
Screening of BAC libraries
Dominant or co-dominant IP markers [14] specific to the target R-block region and the Heera line were used to screen the BAC libraries. Superpools were PCR-screened, with DNA from parents Varuna and Heera included as positive controls. A second PCR round for the specific superpool(s) identified in the first PCR round was then performed on the matrix pools. PCR conditions were the same as those previously used for genetic mapping of the IP markers onto a B. juncea linkage map [14]. The identified clones were streaked on LB agar plates, with five isolated colonies from each clone selected for plasmid isolation. Plasmid isolation was carried out using 3-ml LB cultures by the standard alkaline lysis method [47]. Clone confirmation of the isolated plasmid DNA was performed by PCR amplification of the IP marker region; the HindIII digestion pattern of the plasmid DNA was checked to detect any contamination.
Construction of contigs across six R blocks in B. juncea
To construct a contig across the targeted region of a particular R block, anchor BACs were initially identified using IP markers mapped to that region. The gene span of the BAC clone was determined by amplification of the BAC DNA using primers designed from genes upstream and downstream of the IP marker used to select that clone. In addition, B. juncea, B. rapa and B. nigra DNAs were included as controls. To extend the contigs into regions where mapped markers were not available, suitable primers amplified by the last identified BAC clone in that region were used to screen more BAC clones. Because each LG's amplification pattern was unique in terms of fragment size, the correct assignment of new BACs to a particular region was confirmed by comparing their amplification patterns in the overlapping region with those of previous BACs.
Suitable mapped markers were not available in the target region of LG A3. Amplification with the IP primer for the target R-block region of A. thaliana gene At5g15320 generated four bands, three of which were confirmed to belong to LGs A10, B8 and B3. Based on gene fractionation patterns, which are usually identical for homoeologous LGs, the fourth band was assumed to belong to LG A3. This band was sequenced and BLASTN-searched against the A3 chromosome sequence of B. rapa [48]. Based on sequence similarities, the fourth band was confirmed to belong to chromosome A3 of B. rapa. This primer was then used for BAC clone screening to construct the B. juncea LG A3 contig.
Sequencing of BAC clones and sequence analysis
Solexa-based next-generation sequencing of BAC clones was carried out by Amplicon Express. Annotation of the assembled scaffolds for the six R blocks was performed using the Brassica BAC annotation pipeline (http://brassica.nbi.ac.uk/annotate.html). Annotation details are available by scaffold-ID searching at http://brassica.nbi.ac.uk/cgi-bin/gbrowse/diy_brassica.
Estimation of Ks and genome divergence times
The rate of synonymous substitutions (Ks value) was estimated by computing pairwise distances using the Nei-Gojobori [49] method as implemented in MEGA5. Differences among Ks values for the six orthologue comparisons (AT-BJ), six paralogue comparisons (three each in A and B genome) and three homoeologue comparisons (between A and B genomes) were statistically analysed using single-factor ANOVA as implemented in Microcal Origin version 6.0 (Microcal, Northampton, MA, USA). Significance of variation between mean Ks values for orthologue divergence (AT-BJ), paralogue divergence (within A and B genomes) and homoeologue divergence (between A and B genomes) was analysed statistically by unpaired Student's t-tests using Microcal Origin. For both statistical analyses, p<0.05 was considered to be statistically significant. Nuclear genome divergence times were calculated from Ks values using the equation T = Ks/(2×[1.5×10−8]), where 1.5×10−8 substitutions per site per year is the synonymous mutation rate [21].
Chloroplast divergence times were calculated by the same formula used for nuclear genome divergence times by adopting the synonymous mutation rate of 1.7×10−9 substitutions per site per year [35].
Phylogenetic analyses
To reconstruct a nuclear genome phylogeny, alignments of 11 nuclear genes retained across all six R blocks were concatenated using the Phyutility software program [50]. A neighbour-joining tree was constructed using MEGA5 taking into consideration the synonymous base substitution rates and the distances were computed by Kimura two-parameter method [34]. The significance of nodes was tested by bootstrap analysis with 1,000 replicates.
Plastid genome phylogenies were reconstructed separately from full-length matK and partial ndhF sequences downloaded from the NCBI database (www.ncbi.nlm.nih.gov/nuccore) for the diploid and allotetraploid Brassica species and from the TAIR database (www.arabidopsis.org) for A. thaliana. Sampled taxa and their GenBank/TAIR accession numbers are as follows: for matK sequences, B. rapa (NC_015139), B. nigra (AB354272.1), B. oleracea (AB354271), B. napus (JF807904), B. juncea (AB354274.1), B. carinata (AB354275.1) and A. thaliana (ATCG00040), and for ndhF sequences, B. rapa (DQ200044), B. nigra (DQ200031), B. oleracea (AF064647), B. napus (DQ200041) and A. thaliana (ATCG01010). Neighbour-joining trees were constructed in MEGA5 based on nucleotide substitutions, with distances estimated by the Kimura two-parameter method [34].
Supporting Information
Contigs assembled using overlapping BAC clones in the target R block region in the six linkage groups of B. juncea . Mapping information is from Panjabi et al. [14]. BACs with H prefix are from HindIII library and those with B prefix are from BamHI library. Colour code in the contigs constructed from overlapping BAC clones indicates the A. thaliana genes used to design primers to pick up the corresponding BAC clone.
(PPTX)
Neighbour joining tree of six Brassica crop species constructed based on the chloroplast gene matK sequence. The numbers on the nodes represent percentage from bootstrap analysis of 1000 replicates.
(PPTX)
Assignment of different species of tribe Brassiceae to the Rapa/Oleracea or the Nigra lineages based on chloroplast genome analysis. Haploid chromosome number of the species as well as the species in which genome triplication has been shown by in situ hybridization are also recorded [51], [52].
(XLSX)
List of BAC clones identified for the target region in six R blocks of B. juncea . BACs with H prefix are from HindIII library and those with B prefix are from BamHI library.
(DOCX)
List of the BAC clones sequenced for the targeted regions of the six R blocks of B. juncea .
(DOCX)
Comparison of gene organisation in the targeted R block regions on six linkage groups of B. juncea with syntenous regions of B. rapa and A. thaliana .
(XLSX)
BLASTX analysis of the coding sequences (CDS) in the B. juncea R blocks from noncollinear regions.
(DOCX)
List of 11 genes considered for the nuclear divergence analysis with their assigned functions in A. thaliana .
(XLSX)
Nuclear genome divergence time estimates in Arabidopsis / Brassica evolution from different studies [53]–[56] .
(XLSX)
Acknowledgments
We would like to thank Dr. Martin Trick from John Innes Centre for helpful suggestions in the annotation of BAC clones using Brassica BAC Annotation Pipeline. We would also like to thank Prof. Geeta from Department of Botany, University of Delhi for her comments on the manuscript.
Funding Statement
This work was funded by the National Dairy Development Board (NDDB) and Centre of Excellence on Mustard Breeding by the Department of Biotechnology (DBT), Government of India. DP acknowledges a J. C. Bose Fellowship from the Department of Science and Technology (DST), Government of India. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, et al. (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- 2. McBreen K, Lockhart PJ (2006) Reconstructing reticulate evolutionary histories of plants. Trends Plant Sci 11: 398–404. [DOI] [PubMed] [Google Scholar]
- 3. Marhold K, Lihová J (2006) Polyploidy, hybridization and reticulate evolution: lessons from the Brassicaceae. Plant Syst Evol 259: 143–174. [Google Scholar]
- 4.Lysak MA, Koch MA (2011) Phylogeny, genome and karyotype evolution of crucifers (Brassicaceae). In: Schmidt R, Bancroft I, editors. Genetics and genomics of the Brassicaceae. Springer.pp. 1–31.
- 5. Al-Shehbaz IA, Beilstein MA, Kellogg EA (2006) Systematics and phylogeny of the Brassicaceae (Cruciferae): An overview. Plant Syst Evol 259: 89–120. [Google Scholar]
- 6. Al-Shehbaz IA (2012) A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon 61: 931–954. [Google Scholar]
- 7. U N (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jap J Bot 7: 389–452. [Google Scholar]
- 8. Liu Z, Adamczyk K, Manzanares-Dauleux M, Eber F, Lucas MO, et al. (2006) Mapping PrBn and other quantitative trait loci responsible for the control of homeologous chromosome pairing in oilseed rape (Brassica napus L.) haploids. Genetics 174: 1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cifuentes M, Eber F, Lucas MO, Lode M, Chèvre AM, et al. (2010) Repeated polyploidy drove different levels of crossover suppression between homoeologous chromosomes in Brassica napus allohaploids. Plant Cell 22(7): 2265–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lagercrantz U, Lydiate DJ (1996) Comparative genome mapping in Brassica. Genetics 144: 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lagercrantz U (1998) Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150: 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parkin IA, Gulden SM, Sharpe AG, Lukens L, Trick M, et al. (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana . Genetics 171: 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genome. Trends Plant Sci 11: 535–542. [DOI] [PubMed] [Google Scholar]
- 14. Panjabi P, Jagannath A, Bisht NC, Padmaja KL, Sharma S, et al. (2008) Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics 9: 113–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae. Genome Res 15: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Wang H, Wang J, Sun R, Wu J, et al. (2011) The genome of the mesopolyploid crop species Brassica rapa . Nat Genet 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- 17. Tang H, Woodhouse MR, Cheng F, Schnable JC, Pedersen BS, et al. (2012) Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics 190: 1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng F, Mandáková T, Wu J, Xie Q, Lysak MA, et al. (2013) Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa . Plant Cell 25: 1541–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang H, Lyons E (2012) Unleashing the genome of Brassica rapa . Front Plant Sci 3: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulz OE (1936) Cruciferae. In: Engler A, Prantl K, editors. Die natürlichen Pflanzenfamilien. Engelmann: Leipzig, Germany. Vol 17B: pp. 227–658.
- 21. Koch MA, Haubold B, Mitchell-Olds T (2000) Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis and related genera (Brassicaceae). Mol Biol Evol 17: 1483–1498. [DOI] [PubMed] [Google Scholar]
- 22. Bailey CD, Koch MA, Mayer M, Mummenhoff K, O' Kane SL Jr, et al. (2006) Toward a global phylogeny of the Brassicaceae. Mol Biol Evol 23: 2142–2160. [DOI] [PubMed] [Google Scholar]
- 23. Warwick SI, Black LD (1991) Molecular systematics of Brassica and allied genera (subtribe Brassinae, Brassiceae)-chloroplast genome and cytodeme congruence. Theor Appl Genet 82: 81–92. [DOI] [PubMed] [Google Scholar]
- 24. Pradhan AK, Prakash S, Mukhopadhyay A, Pental D (1992) Phylogeny of Brassica and allied genera based on variation in chloroplast and mitochondrial DNA patterns: molecular and taxonomic classifications are incongruous. Theor Appl Genet 85: 331–340. [DOI] [PubMed] [Google Scholar]
- 25. Warwick SI, Sauder CA (2005) Phylogeny of tribe Brassiceae (Brassicaceae) based on chloroplast restriction site polymorphisms and nuclear ribosomal internal transcribed spacer and chloroplast trnL intron sequences. Can J Bot 83: 467–483. [Google Scholar]
- 26. Hall JC, Tisdale TE, Donohue K, Wheeler A, Al-Yahya MA, et al. (2011) Convergent evolution of a complex fruit structure in the tribe Brassiceae (Brassicaceae). Am J Bot 98: 1989–2003. [DOI] [PubMed] [Google Scholar]
- 27. Arias T, Pires C (2012) A fully resolved chloroplast phylogeny of the brassica crops and wild relatives (Brassicaceae: Brassiceae): Novel clades and potential taxonomic implications. Taxon 61(5): 980–988. [Google Scholar]
- 28. Ramchiary N, Padmaja KL, Sharma S, Gupta V, Sodhi YS, et al. (2007) Mapping of yield influencing QTL in Brassica juncea: implications for breeding of major oilseed crop of dryland areas. Theor Appl Genet 115: 807–817. [DOI] [PubMed] [Google Scholar]
- 29. Srivastava A, Gupta V, Pental D, Pradhan AK (2001) AFLP-based genetic diversity assessment amongst agronomically important natural and some newly synthesized lines of Brassica juncea . Theor Appl Genet 102: 193–199. [Google Scholar]
- 30. Pradhan AK, Gupta V, Mukhopadhyay A, Arumugam N, Sodhi YS, et al. (2003) A high-density linkage map in Brassica juncea (Indian mustard) using AFLP and RFLP markers. Theor Appl Genet 106: 607–614. [DOI] [PubMed] [Google Scholar]
- 31. Yadava SK, Arumugam N, Mukhopadhyay A, Sodhi YS, Gupta V, et al. (2012) QTL mapping of yield-associated traits in Brassica juncea: meta-analysis and epistatic interactions using two different crosses between east European and Indian gene pool lines. Theor Appl Genet 125: 1553–1564. [DOI] [PubMed] [Google Scholar]
- 32. Cheng F, Liu S, Wu J, Fang L, Sun S, et al. (2011) BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol 11: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 35. Koch MA, Haubold B, Mitchell-Olds T (2001) Molecular systematics of the Brassicaceae: Evidence from coding plastidic matK and nuclear Chs sequences. Am J Bot 88: 534–544. [PubMed] [Google Scholar]
- 36. Yang YW, Lai KN, Tai PY, Li WH (1999) Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J Mol Evol 48: 597–604. [DOI] [PubMed] [Google Scholar]
- 37. Cho K, O'Neill CM, Kwon SJ, Yang TJ, Smooker AM, et al. (2010) Sequence-level comparative analysis of the Brassica napus genome around two stearoyl-ACP desaturase loci. Plant J 61: 591–599. [DOI] [PubMed] [Google Scholar]
- 39. Mandáková T, Lysak MA (2008) Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae). Plant Cell 20: 2559–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harberd DJ, McArthur ED (1980) Meiotic analysis of some species and genus hybrids in the Brassiceae. In: Tsunoda S, Hinata K, Gómez-Campo C, editors. Brassica crops and wild allies.Japan Scientific Societies Press, Tokyo. pp. 65–87.
- 41.Mizushima U (1980) Genome analysis in Brassica and allied genera. In: Tsunoda S, Hinata K, Gómez-Campo C, editors. Brassica crops and wild allies. Japan Scientific Societies Press, Tokyo. pp. 89–106.
- 42. Takahata Y, Hinata K (1983) Studies on cytodemes in subtribe Brassicinae (Cruciferae). Tohoku J Agr Res 33: 111–124. [Google Scholar]
- 43.Prakash S, Takahata Y, Kirti PB, Chopra VL (1999) In: Gomez-Campo C, editor. Biology of Brassica coenospecies. Cytogenetics. Elsevier: Amsterdam, Netherlands. pp. 59–105.
- 44. Erickson LR, Straus NA, Beversdorf WD (1983) Restriction patterns reveal origins of chloroplast genomes in Brassica amphiploids. Theor Appl Genet 65: 201–206. [DOI] [PubMed] [Google Scholar]
- 45. Allender CJ, King GJ (2010) Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Johnston JS, Pepper AE, Hall AE, Chen ZJ, Hodnett G, et al. (2005) Evolution of genome size in Brassicaceae. Ann Bot 95: 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Russell DW (2001) Molecular cloning, A laboratory manual 3rd edition. Woodbury, NY: Cold Spring Harbor Laboratory Press.
- 48. Mun JH, Kwon SJ, Seol YJ, Kim JA, Jin M, et al. (2010) Sequence and structure of Brassica rapa chromosome A3. Genome Biol 11: R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418–426. [DOI] [PubMed] [Google Scholar]
- 50. Smith SA, Dunn CW (2008) Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 24(5): 715–716. [DOI] [PubMed] [Google Scholar]
- 51. Warwick SI, Al-Shehbaz IA (2006) Brassicaceae: chromosome number index and database on CD-ROM. Plant Syst Evol 259: 237–248. [Google Scholar]
- 52. Lysak MA, Cheung K, Kitschke M, Bures P (2007) Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol 145: 402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang TJ, Kim JS, Kwon SJ, Lim KB, Choi SB, et al. (2006) Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa . Plant Cell 18: 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang J, Long Y, Wu B, Liu J, Jiang C, Shi L, et al. (2009) The evolution of Brassica napus FLOWERING LOCUS T paralogues in the context of inverted chromosomal duplication blocks. BMC Evol Biol 9: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Inaba R, Nishio T (2002) Phylogenetic analysis of Brassiceae based on the nucleotide sequences of the S-locus related gene, SLR1. Theor Appl Genet 105: 1159–1165. [DOI] [PubMed] [Google Scholar]
- 56. Cheung F, Trick M, Drou N, Lim YP, Park JY, et al. (2009) Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell 21: 1912–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contigs assembled using overlapping BAC clones in the target R block region in the six linkage groups of B. juncea . Mapping information is from Panjabi et al. [14]. BACs with H prefix are from HindIII library and those with B prefix are from BamHI library. Colour code in the contigs constructed from overlapping BAC clones indicates the A. thaliana genes used to design primers to pick up the corresponding BAC clone.
(PPTX)
Neighbour joining tree of six Brassica crop species constructed based on the chloroplast gene matK sequence. The numbers on the nodes represent percentage from bootstrap analysis of 1000 replicates.
(PPTX)
Assignment of different species of tribe Brassiceae to the Rapa/Oleracea or the Nigra lineages based on chloroplast genome analysis. Haploid chromosome number of the species as well as the species in which genome triplication has been shown by in situ hybridization are also recorded [51], [52].
(XLSX)
List of BAC clones identified for the target region in six R blocks of B. juncea . BACs with H prefix are from HindIII library and those with B prefix are from BamHI library.
(DOCX)
List of the BAC clones sequenced for the targeted regions of the six R blocks of B. juncea .
(DOCX)
Comparison of gene organisation in the targeted R block regions on six linkage groups of B. juncea with syntenous regions of B. rapa and A. thaliana .
(XLSX)
BLASTX analysis of the coding sequences (CDS) in the B. juncea R blocks from noncollinear regions.
(DOCX)
List of 11 genes considered for the nuclear divergence analysis with their assigned functions in A. thaliana .
(XLSX)
Nuclear genome divergence time estimates in Arabidopsis / Brassica evolution from different studies [53]–[56] .
(XLSX)