Abstract
Following reports by ENCyclopedia Of DNA Elements (ENCODE; GENCODE) Consortium and others, it is now fairly evident that the majority (70–80%) of the mammalian genome has the potential to be transcribed into non-protein-coding RNAs (ncRNAs). Critical to our understanding of genetic processes is the mechanism by which ncRNAs exert their roles. Accordingly, ncRNAs are shown to regulate the expression of protein-coding loci (i.e., genes) at the transcriptional as well as post-transcriptional stages. We recently reported on a widespread transcription at the DNA enhancer elements in myogenic cells. In our study, we found certain enhancer RNAs (eRNAs) regulate chromatin accessibility of the transcriptional machinery at loci encoding master regulators of myogenesis (i.e., MyoD/MyoG), thus suggesting their significance and site-specific impact in cellular programming. Here, we examine recent discoveries pertinent to the proposed role(s) of eRNAs in regulating gene expression. We will highlight consistencies, discuss confounding observations, and consider a lack of critical information in a way to prioritize future objectives.
Keywords: eRNA, chromatin, transcription, mRNA synthesis, enhancers, histones
Introduction
With the advent of high-throughput sequencing, studies have taken a comprehensive approach in cataloguing regulatory genomic elements and the transcriptome in various cell types; as well as defining the relations between the global transcriptional activity and chromatin architecture.1-5 These studies have unraveled a complex and surprising description of the human genome that has challenged the views on the non-protein-coding compartment (equivalent to ~98% of the entire genome). Of note is the observation that the genome is pervasively transcribed with a greater proportion of long non-coding RNAs (lncRNAs), rather than protein-coding transcripts, showing cell type-specific expression.3,5,6 Subcategorized under lncRNAs are transcripts originating from regulatory enhancer elements (i.e., eRNAs). By knockdown approach, studies demonstrate the prominence of lncRNA as well as eRNAs in regulating gene expression and cellular programming.7-12 Genome-wide techniques are used to annotate enhancers, their connectivity, and mature transcripts. According to these studies (as they will be discussed herein), eRNAs display perplexing features, distinct from the rest of the transcriptome. These observations suggest that despite the collected data, there is much to be discovered about eRNAs that would precisely depict their molecular mechanism, including detailed biochemical characterization, processing (i.e., splicing, editing), co-factor identification, and genome-wide distribution.
Enhancers
Although we will briefly highlight key features of DNA enhancer elements, readers are referred to extensive reviews on this topic published elsewhere.13-17 Conventionally, transcriptional enhancers were shown to increase the expression of protein-coding genes in reporter-expression assays.18,19 Since then, endogenous enhancers, estimated to be in tens-of-thousands in metazoans, are being discovered through distinct chromatin signature, ablation, and cloning strategies. These enhancers control the expression of genes over a relatively large genomic distance, occasionally reaching megabases.13-17 Specifically, enhancers are binding sites for multiple transcription factors (TFs) that, to a large extent, are conserved, despite evolutionary divergence at regions flanking these binding sites.2,20,21 Furthermore, depending on their activity status, enhancer sites are more sensitive to endonucleases and are modified at distinct nucleosome residues (lysine-4 methylation and lysine-27 acetylation on histone H3), thus distinguishing them from surrounding regions and other transcribed loci.13,16,22-24 Active enhancers are also occupied by RNA polymerase II (PolII), though this observation was originally thought to be the result of indirect connectivity to proximal regulatory regions in chromatin immunoprecipitation (ChIP) studies. Nonetheless, enhancers are also sites of transcription,25-30 thus raising questions on the relevance of eRNAs in cellular processes.
Enhancer sites are further classified in two configurations: (1) Typical enhancers (< 1 kb) associated with housekeeping genes and (2) super-enhancers (enhancer-clusters or chromatin regulatory beacons; ranging in number from 200–1000 in certain cell types), which are generally confined near key developmental genes.31-33 Super-enhancers (~3–50 kb) are binding sites for unusually high levels of TFs, Mediator complex, and PolII. Segments of super-enhancers are found to be transcribed, whose eRNA levels correlate with the expression of nearby genes.8,31 As compared with typical enhancers where simple one-to-one promoter-connectivity is shown to drive gene expression,13,34 the significance of super-enhancer modules is not clear. Yet, given the high levels of Mediator complex at super-enhancers and the prominent role of Mediator complex in transmission of transcriptional instructions and 3D organization of the genome,35-38 it could be proposed that super-enhancers serve as genomic connectivity centers where appropriate regulatory networks are structurally organized in factories for coordinated expression. In fact, recent PolII-assisted chromatin connectivity mapping has revealed that, depending on the cellular context, SOX2 and OLIG1 loci (including their super-enhancers) are directly connected to distinct developmental networks.6,31,32 Overall, in addition to operating as information hubs,39 it may well be that super-enhancers are structural centers that stabilize the transcriptional architecture and perform as major ports for integrating developmental networks.
eRNA Synthesis and Biochemical Properties
eRNAs are detected in most cell types examined, and as mentioned above, originate from enhancers with a distinct chromatin signature.3,8,16 Aside from this distinction, molecular components governing the transcription of eRNAs thus far appear comparable to genes, including the contribution of transcriptional machinery (PolII), Mediator complex, nuclear receptors (estrogen receptor and Rev-Erbs), and transcription factors (Klf4; Egr1; FoxA1; MyoD, p53) in driving eRNAs transcription.8,9,11,12,40,41 To catalog nascent eRNAs and determine their rate of synthesis, scientists have used Global Run-On with Sequencing (GRO-Seq).11,12,40 Accordingly, nascent eRNAs contains a 7-methylguanylated cap with a rate of synthesis and levels comparable to the nearest protein-coding transcripts. Moreover, enhancers have the potential to maintain bidirectional RNA synthesis, much similar to occurrences found around transcriptional start sites (TSS) of genes.40,42 Still, the data regarding polyadenylation at 3′ end processing of eRNAs have been less clear. A report by Kim et al. (2010) suggests that rapidly inducible eRNAs are not polyadenylated (based on RNA-Seq data and circularization experiments), whereas studies examining steady-state and/or developmentally regulated eRNAs imply that eRNAs are subject to polyadenylation.8,10,29 Although it is quite conceivable that eRNAs differ in their post-transcriptional processing, generalization on this matter requires closer inspection as it will unravel key characteristics relevant for future studies.
There are reasons to suspect that the processing of eRNAs differs from the rest of transcriptome. One observation is that despite matching PolII occupancy signals at enhancers and genes in ChIP-Seq data, mature eRNAs are barely detectable in total RNA-Seq data sets, whereas nascent eRNAs levels (in GRO-Seq) are as high as nearby mRNAs.8,12,29,40 Another is the difficulty in cloning regulatory RNAs (including eRNAs) for characterization (our unpublished observations). These data suggest that eRNAs are either unstable for steady-state accumulation or not readily processed for sequencing/cloning with current protocols. Either way, with advances in molecular techniques, these questions should be addressed in the near future.
Proposed Molecular Mechanism(s)
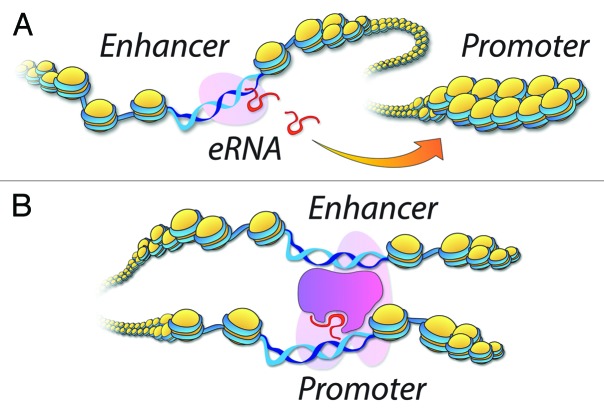
Recent discoveries support a role for eRNAs in promoting gene expression through chromatin accessibility, PolII recruitment, and enhancer-promoter contacts (EPCs). Common to these findings is the correlation between the levels of eRNAs and those of nearby mRNAs. Using RNA interference (RNAi), experiments show that targeted depletion of eRNAs results in significant reduction of nearby mRNAs in cultured cells and mice.8-12 Furthermore, this reduction occurs at the transcriptional stage, where PolII occupancy is noticeably reduced at genes; and eRNA depletion culminates in the loss of EPCs.8,9,12 In line with the latter observations, recent findings demonstrate that certain eRNAs directly associate with the Mediator complex to facilitate EPCs and augment mRNA transcription, whereas other experiments underscore the binding of specific eRNAs to Rad21+ Cohesin complex to stabilize chromatin looping and transcription.9,12 These observations are consistent with co-recruitment of the Mediator and Cohesin complexes for steady-state mRNA synthesis.37,43-45 Nonetheless, the model regarding the direct role(s) for eRNAs in shaping EPCs may not hold for several reasons. First, eRNA depletion by transcriptional inhibition indicates that PolII holoenzymes may reinforce EPCs.12,40 Second, a simple all-or-none contact does not explain the dose-response impact of eRNAs on gene expression observed in RNAi experiments. Perhaps analogous to an internal combustion system, proximity of the gas tank (i.e., enhancer) to the engine (i.e., promoter/gene) could only dictate agility in signal transmission. A “throttle” (i.e., eRNAs), which unlocks the gate for flow of reactive elements (i.e., PolII), dictates the rate of acceleration. In this context, designated enhancers have previously been shown to open chromatin for transcriptional activity at distinct promoters.46-49 In our recent published study, we asked whether the emanating eRNAs facilitate transcription by exposing proximal regulatory regions to PolII complex. Specifically, we focused on two eRNAs upstream of MYOD1, corresponding to a region classified as the abovementioned super-enhancer.8,31 By RNAi, we reported that while an eRNA (i.e., CERNA) controls chromatin accessibility and transcription at MYOD1, another (i.e., DRRRNA) operates to expose regulatory regions at another region, MYOG.8 Certainly, similar observations regarding transcriptional activity have been reported at other enhancer/gene combinations.9,10,12 Overall, these results demonstrate the specificity of enhancers/eRNAs and re-emphasize similarities to the “throttle” analogy, which satisfies the refractory nature of nucleosomes to transcription, role of enhancers in TSS specification, and dose-dependent enhancement of PolII occupancy and transcription.50-52 Third, interactome data reveal that although EPCs are developmentally dynamic, the connections in transient networks (e.g., TNFα) are static and stably formed even before the induction of eRNAs.12,40,53 Fourth, according to PolII-mediated chromatin connectivity mapping, a significant fraction of EPCs occur interchromosomally (i.e., between chromosomes), suggesting an interactome that is far more complex than a simple in cis contact afforded by eRNA-assisted looping model.6 Lastly, a comparison between evolutionarily divergent enhancers suggests an intricate regulatory function beyond the frequently observed higher-order chromatin configurations in metazoans.20,54-59 Given the above considerations, enhancers/eRNAs regulate transcription by establishing chromatin accessibility and PolII recruitment; and may not be directly responsible for the formation of EPCs, while it is PolII and its associated macromolecular complexes that may define the transcriptional architecture.38 These observations hint at a step-wise eRNA-mediated transcription activation events conceptualized in Figure 1.
Figure 1. Emerging roles of eRNAs in establishing chromatin accessibility and the subsequent formation of EPCs. (A) eRNA synthesis at an enhancer and its targeting to a defined regulatory region (i.e., Promoter). (B) eRNA-mediated chromatin accessibility and the subsequent recruitment of factors for transcription and the stabilization of EPCs.
Hierarchy Within Regulatory Networks
eRNA synthesis occurs prior to, and regulates the activation of genes in developmental regulatory networks. For example, chromatin marks associated with active enhancers are first observed at the core enhancer (CE) of MYOD1 before its transcriptional activation, and depletion of CERNA results in a significant reduction of MyoD transcript.8,44 Similarly, an eRNA (ncRNA-a7) activates SNAI1, a gene belonging to a family of TFs with roles in mesodermal determination and epithelial–mesenchymal transition (EMT).10 These data invoke an interesting hypothesis that certain eRNAs are at the top of the hierarchy within certain transcriptional regulatory networks. If so, this notion suggests that failure to activate, or mutations within, critical eRNAs leaves limited capabilities for TFs and results in disorders.33,60-64 Therefore, one investigative priority would be to resolve the targets of eRNAs genome-wide.
Conclusions and Perspectives
Latest data reaffirm the widespread transcription of the mammalian genomes, and that enhancers are among the transcribed regions. Thus far, evidence suggests that enhancers/eRNAs promote mRNA transcription by establishing chromatin accessibility at specified loci and over a large genomic space, resulting in PolII/Mediator/Cohesin complex recruitment and culminating in the formation of transcription networks. A question arising from these observations is “why would more complex biological systems evolve an enhancer-based regulatory system?” In the view of the positive relationship between the genome size and biological complexity, one can anticipate a superior governing and adaptive power of sequence-specific regulation rather than a more primitive TF-based system in multicellular and multilineage organisms.21,56,65-67 This way, coordinated spatiotemporal regulation of distinct transcriptional networks is organized and fine-tuned while utilizing similar protein components to drive complexity.6,57-59,68-70 Notwithstanding remarkable advances made thus far, a great deal is yet to be discovered about eRNAs, including their regulatory networks, biochemical characteristics, structure–activity relationship, and precise molecular mechanisms that make them key transcriptional integrators.71
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/27950
References
- 1.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 5.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–9. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Wong C-H, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–10. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–17. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam MTY, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–5. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–20. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–39. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ong C-T, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–93. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ong C-T, Corces VG. Enhancers: emerging roles in cell fate specification. EMBO Rep. 2012;13:423–30. doi: 10.1038/embor.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Natoli G, Andrau J-C. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 17.de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 18.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-X. [DOI] [PubMed] [Google Scholar]
- 19.Moreau P, Hen R, Wasylyk B, Everett R, Gaub MP, Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981;9:6047–68. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noonan JP. Regulatory DNAs and the evolution of human development. Curr Opin Genet Dev. 2009;19:557–64. doi: 10.1016/j.gde.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–16. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 22.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuan D, Kong S, Hu K. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proc Natl Acad Sci U S A. 1992;89:11219–23. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leach KM, Nightingale K, Igarashi K, Levings PP, Engel JD, Becker PB, Bungert J. Reconstitution of human β-globin locus control region hypersensitive sites in the absence of chromatin assembly. Mol Cell Biol. 2001;21:2629–40. doi: 10.1128/MCB.21.8.2629-2640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogan DF, Cousins DJ, Santangelo S, Ioannou PA, Antoniou M, Lee TH, Staynov DZ. Analysis of intergenic transcription in the human IL-4/IL-13 gene cluster. Proc Natl Acad Sci U S A. 2004;101:2446–51. doi: 10.1073/pnas.0308327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Black BL, et al. NISC Comparative Sequencing Program. National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program Authors. NISC Comparative Sequencing Program Authors Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–6. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annu Rev Biochem. 2000;69:729–49. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- 36.Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–22. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–72. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–95. doi: 10.1016/j.cell.2013.04.053. http://www.ncbi.nlm.nih.gov/pubmed/23706625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buecker C, Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 2012;28:276–84. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–23. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Léveillé N, Kalluri R, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–35. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 42.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–51. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taberlay PC, Kelly TK, Liu C-C, You JS, De Carvalho DD, Miranda TB, Zhou XJ, Liang G, Jones PA. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011;147:1283–94. doi: 10.1016/j.cell.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conaway RC, Conaway JW. Function and regulation of the Mediator complex. Curr Opin Genet Dev. 2011;21:225–30. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pikaart M, Feng J, Villeponteau B. The polyomavirus enhancer activates chromatin accessibility on integration into the HPRT gene. Mol Cell Biol. 1992;12:5785–92. doi: 10.1128/mcb.12.12.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim A, Dean A. A human globin enhancer causes both discrete and widespread alterations in chromatin structure. Mol Cell Biol. 2003;23:8099–109. doi: 10.1128/MCB.23.22.8099-8109.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jenuwein T, Forrester WC, Fernández-Herrero LA, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–72. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 49.Forrester WC, van Genderen C, Jenuwein T, Grosschedl R. Dependence of enhancer-mediated transcription of the immunoglobulin mu gene on nuclear matrix attachment regions. Science. 1994;265:1221–5. doi: 10.1126/science.8066460. [DOI] [PubMed] [Google Scholar]
- 50.Leimgruber E, Seguín-Estévez Q, Dunand-Sauthier I, Rybtsova N, Schmid CD, Ambrosini G, Bucher P, Reith W. Nucleosome eviction from MHC class II promoters controls positioning of the transcription start site. Nucleic Acids Res. 2009;37:2514–28. doi: 10.1093/nar/gkp116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boeger H, Bushnell DA, Davis R, Griesenbeck J, Lorch Y, Strattan JS, Westover KD, Kornberg RD. Structural basis of eukaryotic gene transcription. FEBS Lett. 2005;579:899–903. doi: 10.1016/j.febslet.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 52.Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133:716–26. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–4. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sholtis SJ, Noonan JP. Gene regulation and the origins of human biological uniqueness. Trends Genet. 2010;26:110–8. doi: 10.1016/j.tig.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 55.Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet. 2012;13:233–45. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- 56.Tautz D. Evolution of transcriptional regulation. Curr Opin Genet Dev. 2000;10:575–9. doi: 10.1016/S0959-437X(00)00130-1. [DOI] [PubMed] [Google Scholar]
- 57.Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, et al. Fine tuning of craniofacial morphology by distant-acting enhancers. Science. 2013;342:1241006. doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jónsson B, Schluter D, Kingsley DM. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–23. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 59.Chan YF, Marks ME, Jones FC, Villarreal G, Jr., Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–5. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He H, Li W, Wu D, Nagy R, Liyanarachchi S, Akagi K, Jendrzejewski J, Jiao H, Hoag K, Wen B, et al. Ultra-rare mutation in long-range enhancer predisposes to thyroid carcinoma with high penetrance. PLoS One. 2013;8:e61920. doi: 10.1371/journal.pone.0061920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakabe NJ, Savic D, Nobrega MA. Transcriptional enhancers in development and disease. Genome Biol. 2012;13:238. doi: 10.1186/gb-2012-13-1-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, Portnoy ME, Cutler DJ, Green ED, Chakravarti A. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–63. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 63.Iacopetta B, Grieu F, Joseph D, Elsaleh H. A polymorphism in the enhancer region of the thymidylate synthase promoter influences the survival of colorectal cancer patients treated with 5-fluorouracil. Br J Cancer. 2001;85:827–30. doi: 10.1054/bjoc.2001.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.French JD, Ghoussaini M, Edwards SL, Meyer KB, Michailidou K, Ahmed S, Khan S, Maranian MJ, O’Reilly M, Hillman KM, et al. GENICA Network. kConFab Investigators Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet. 2013;92:489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–99. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 66.Britten RJ. DNA sequence insertion and evolutionary variation in gene regulation. Proc Natl Acad Sci U S A. 1996;93:9374–7. doi: 10.1073/pnas.93.18.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rebeiz M, Jikomes N, Kassner VA, Carroll SB. Evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc Natl Acad Sci U S A. 2011;108:10036–43. doi: 10.1073/pnas.1105937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Werner T, Koshikawa S, Williams TM, Carroll SB. Generation of a novel wing colour pattern by the Wingless morphogen. Nature. 2010;464:1143–8. doi: 10.1038/nature08896. [DOI] [PubMed] [Google Scholar]
- 69.Cretekos CJ, Wang Y, Green ED, Martin JF, Rasweiler JJ, 4th, Behringer RR. Regulatory divergence modifies limb length between mammals. Genes Dev. 2008;22:141–51. doi: 10.1101/gad.1620408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crocker J, Tamori Y, Erives A. Evolution acts on enhancer organization to fine-tune gradient threshold readouts. PLoS Biol. 2008;6:e263. doi: 10.1371/journal.pbio.0060263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin F, Li Y, Ren B, Natarajan R. Enhancers: multi-dimensional signal integrators. Transcription. 2011;2:226–30. doi: 10.4161/trns.2.5.17712. [DOI] [PMC free article] [PubMed] [Google Scholar]