Abstract
Asthma originates from genetic and environmental factors with about half the risk of disease attributable to heritable causes. Genome-wide association studies, mostly in populations of European ancestry, have identified numerous asthma-associated single nucleotide polymorphisms (SNPs). Studies in populations with diverse ancestries allow both for identification of robust associations that replicate across ethnic groups and for improved resolution of associated loci due to different patterns of linkage disequilibrium between ethnic groups. Here we report on an analysis of 745 African-American subjects with asthma and 3,238 African-American control subjects from the Candidate Gene Association Resource (CARe) Consortium, including analysis of SNPs imputed using 1,000 Genomes reference panels and adjustment for local ancestry. We show strong evidence that variation near RAD50/IL13, implicated in studies of European ancestry individuals, replicates in individuals largely of African ancestry. Fine mapping in African ancestry populations also refined the variants of interest for this association. We also provide strong or nominal evidence of replication at loci near ORMDL3/GSDMB, IL1RLML18R1, and 10pl4, all previously associated with asthma in European or Japanese populations, but not at the PYHIN1 locus previously reported in studies of African-American samples. These results improve the understanding of asthma genetics and further demonstrate the utility of genetic studies in populations other than those of largely European ancestry.
Introduction
Asthma is a disease characterized by bronchial hyper-responsiveness, reversible airflow limitation, and airway inflammation. In the United States, the prevalence of asthma varies between ethnic groups, with estimates ranging from 8.2 % in European Americans to 14.6 % in African-Americans and 18.4 % in Latino Americans (Moorman et al. 2011). The prevalence of asthma, as well as asthma morbidity and mortality in the United States remains high, with certain ethnic groups disproportionately affected by this trend (Scirica and Celedon 2007). The striking ethnic disparities in asthma prevalence cannot be explained entirely by measured environmental, social, cultural, or economic factors (Boudreaux 2003; Ray et al. 1998; Scirica and Celedon 2007). It remains difficult, however, to identify specific causal genes underlying ethnic differences and to determine whether genetic control contributes to the observed ethnic disparities for this complex disease.
Multiple recent genome-wide association (GWA) studies have confirmed the genetic contribution to asthma with heritability estimated at around 75 % (Ferreira et al. 2011; Gudbjartsson et al. 2009; Hancock et al. 2009; Himes et al. 2009; Mathias et al. 2010 Hirota et al. 2011; Li et al. 2010; Moffatt et al. 2010; Sleiman et al. 2010; Thomsen et al. 2010; Torgerson et al. 2011; Willemsen et al. 2012). These studies have primarily been conducted in populations of European origin and have demonstrated increasingly robust results with multiple signals at several loci. While far fewer genome-wide studies have focused on asthma in individuals with different ancestries, several loci discovered in European ancestry samples have been replicated in diverse ethnic populations (Galanter et al. 2008; Liu et al. 2011; Mathias et al. 2010; Torgerson et al. 2011), with some of these loci demonstrating population-specific associations (Galanter et al. 2011; Torgerson et al. 2012). At least one locus near PYHIN1 has been specifically associated with asthma in subjects of African decent (Torgerson et al. 2011). Together, these results suggest that some asthma susceptibility loci are associated with disease risk across a range of ancestries, but that population-specific loci may play a role in explaining ethnic differences in asthma phenotypes.
Studies of diverse populations allow for the discovery of robust associations that replicate across ethnic groups, as well as unique associations that contribute to the heterogeneity in disease prevalence amongst groups with different ancestries (Cooper et al. 2008). Moreover, studying populations with diverse ancestries has been shown to increase the resolution of associated regions as a result of different patterns of linkage disequilibrium (LD) between ethnic groups (N’Diaye et al. 2011; Teo et al. 2010). Investigating genetic risk factors for asthma in ethnically diverse populations is complicated because population samples are small, thus limiting the power of discovery. One approach to increasing sample size involves examining population-based cohorts for which basic clinical information about asthma has been collected and for which genome-wide genotype data exists (Ramasamy et al. 2012). The Candidate Gene Association Resource (CARe) assembled phenotype and genotype data from 9 NHLBI cohorts with a specific emphasis on genome-wide genotype data in individuals of African ancestry from 5 of the cohorts (Lettre et al. 2011; Musunuru et al. 2010). To date, this study has contributed to understanding the genetics of cardiovascular disease, quantitative traits in African ancestry populations, and the effects of ancestry on disease risk (Lettre et al. 2011; N’Diaye et al. 2011; Pasaniuc et al. 2011). We performed an analysis of 3,983 African-Americans in the CARe Project with an emphasis on replicating and fine mapping previously identified signals associated with asthma.
Materials and methods
Ethics statement
All participants gave written informed consent. The CARe project is approved by the institutional review boards of the participating study centers and the Massachusetts Institute of Technology.
Studies
African-American participants for the GWA study were drawn from four population-based studies: Atherosclerosis Risk in Communities (ARIC; n = 2,789), Coronary Artery Risk Development in young Adults (CARDIA; n = 1,037), Jackson Heart Study (JHS; n = 2,162), and Multi-Ethnic Study of Atherosclerosis (MESA; n = 1636). The Cleveland Family Study (CFS) had too few individuals with asthma to generate stable regression estimates, and was not included in the analyses. Information collected at the time of recruitment as well as longitudinal information was used in this study. See Table 1 and Supplemental Tables 1 and 2 for descriptions of study demographics. Individual study descriptions have been published previously (Lettre et al. 2011) and are briefly summarized in the supplemental section.
Table 1.
Demographics of the CARe African-American cohorts
| Phenotype | ARIC | CARDIA | JHS | MESA | CARe totals |
|---|---|---|---|---|---|
| Asthma | |||||
| Age | 53.9 ± 6.0 | 24.1 ± 3.8 | 47.9 ± 11.3 | 60.4 ± 10.8 | - |
| Female | 99 | 101 | 139 | 150 | 489 |
| Male | 61 | 56 | 80 | 59 | 256 |
| Total | 160 | 157 | 219 | 209 | 745 |
| Control | |||||
| Age | 53.0 ± 5.8 | 24.5 ±3.9 | 49.2 ± 12.0 | 62.4 ± 10.0 | - |
| Female | 601 | 108 | 476 | 683 | 1,868 |
| Male | 339 | 84 | 366 | 581 | 1,370 |
| Total | 940 | 192 | 842 | 1,264 | 3,238 |
Individuals with asthma include those with self-reported and physician-diagnosed asthma and with no evidence of other lung disease. Non-asthmatic controls are those without evidence of lung disease or low lung function (see “Materials and methods” for details)
Asthma-associated loci used in replication/fine mapping
Asthma-associated SNPs identified from GWA studies were determined from the NIH GWAS database and from published literature. We focused on those loci that approached or exceeded genome-wide significance (p < 5 × 10−8), as well as top results from studies that included predominantly African-American individuals. The following is a list of the 21 asthma-associated loci used in this replication/fine mapping (see Supplemental Table 3): PDE4D (Himes et al. 2009), DENND1B (Sleiman et al. 2010), RAD50/IL13 (Li et al. 2010; Moffatt et al. 2010), IL1RL1-IL18R1 (Gudbjartsson et al. 2009; Moffatt et al. 2010; Torgerson et al. 2011), GSDMB/ORMDL3 (Moffatt et al. 2010; Torgerson et al. 2011), IL33 (Moffatt et al. 2010; Torgerson et al. 2011), RORA (Moffatt et al. 2010), SMAD3 (Moffatt et al. 2010), IL2RB (Moffatt et al. 2010), IL6R (Ferreira et al. 2011), 11q13.5 (C11orf30-LRRC32) (Ferreira et al. 2011), 10p14 (gene desert) (Hirota et al. 2011), 4q31 (USP38-GAB1) (Hirota et al. 2011), 5q22 (TSLP-WDR36) (Hirota et al. 2011; Torgerson et al. 2011), 6p21 (HLA-DR/DQ/NOTCH4) (Moffatt et al. 2010), ch 12ql3 (IKZF4/EOS) (Hirota et al. 2011), HLA-DP (Noguchi et al. 2011), PYHIN1 (Torgerson et al. 2011), ADRA1B (Mathias et al. 2010), DPP10 (Mathias et al. 2010), PRNP (Mathias et al. 2010).
Phenotype definition
Subjects with asthma were defined based on at least one positive response to study questions and included both physician-diagnosed and self-reported asthma. Examples of study questions include “Have you ever been diagnosed with asthma?” or “Have you ever had asthma?” (exact questions varied between studies—see Supplemental Table 6 for details). The remaining subjects served as healthy controls if they did not report any of the following conditions: wheezing, chronic obstructive pulmonary disease, emphysema, chronic bronchitis, chronic cough, chronic sputum production, or other lung disease (see Supplemental Tables 7 and 8 for details). Individuals were excluded from the control group if they had <70 % of the predicted FEV1 based on race- and sex-specific NHANES III prediction equations adjusting for age and height, or FEV1/FVC less than lower limits of normal for age, race and sex (based on Hankinson criteria) (Hankinson et al. 1999). Individuals with reports of chronic obstructive pulmonary disease, emphysema, chronic bronchitis, or other lung diseases were also excluded from the case group.
Genotyping and SNP imputation
All discovery samples (GWA study) were genotyped on the Affymetrix Genome-Wide Human SNP array 6.0 according to the manufacturer’s protocol. SNPs were filtered using standard QC metrics (Lettre et al. 2011). To increase coverage and facilitate comparison with other datasets, we imputed genotype data using MACH version 1.0.16 (Kang et al. 2010; Lettre et al. 2011). For this study, we created a set of haplotypes containing all 1,000 Genome samples with reported European or African ancestry contained within the December 2010 haplotype release. The resulting set of SNPs was conservatively filtered to remove low imputation quality (RSQ <0.4) or minor allele frequency SNPs (MAF <0.01), for which association results would be less reliable.
Genome-wide association study analyses
Association studies with asthma used an additive genetic model and were adjusted for sex, ancestry-informative principal components and age using PLINK v1.07 (Purcell et al. 2007). We considered >8.5 million SNPs. The first ten principal components generated from EIGENSTRAT were added to the logistic model to control for global ancestry and population stratification (Price et al. 2006). No inflation of the resulting test statistic was detected, so genomic control adjustment was not performed. Metaanalysis of the four cohorts was performed using METAL under the inverse normal model (Willer et al. 2010). To test for local effects of admixture, a separate GWA study was conducted by adding an additional covariate at each SNP containing the local ancestry estimate for the SNP generated by HAPMIX (Price et al. 2009). A detailed description of the analysis methods and the phenotypic definitions used can be found in the supplemental data.
Replication analyses
Because of differing LD patterns between African and European ancestry individuals, a lead “European” SNP may not be in LD with the causal SNP in African ancestry populations. Thus, it was necessary to test multiple SNPs within each locus for association, which we did by considering SNPs that are in LD in African ancestry reference panels (r2 > 0.8) with the lead SNP. In addition, we were searching over 21 different loci for signals of replication. Thus, a stringent p value threshold for replication must take into account two types of multiple testing: testing multiple correlated SNPs within each locus, and testing 21 loci. Simply counting the total number of SNPs tested is overly conservative, as many of the additional SNPs within each locus are still partially correlated. To determine a more appropriate empirical threshold, we generated 250 “null” sets of 21 SNPs by matching each of the previously published associated 21 SNPs on allele frequency, distance to nearest gene, and number of genes in linkage disequilibrium, and by requiring the resulting SNPs within each list to be uncorrelated (simulating 21 independent loci). For each list, we then repeated the same search for replication signals as was performed on the true data, and recorded the most significant p value for each set of SNPs. The Bonferroni threshold for the test was set at the top 5 % (12/250) of best p values for the simulations. The actual replication test thus showed significant replication of multiple European signals at a Bonferroni threshold of significance, and all loci with more significant p values than this threshold were reported. Because we expect multiple replications, this Bonferroni threshold is actually overly conservative, but replications exceeding this threshold are highly likely to be correct.
Results
The meta-analysis included results from four population-based studies: ARIC, CARDIA, JHS, and MESA. Overall, a total of 3,983 African-Americans with adequate phenotype information, and with high genotype quality were available for analysis (see “Materials and methods”). After all exclusion criteria were applied, there were 745 individuals in the asthma category, and 3,238 individuals in the control category from the CARe cohorts.
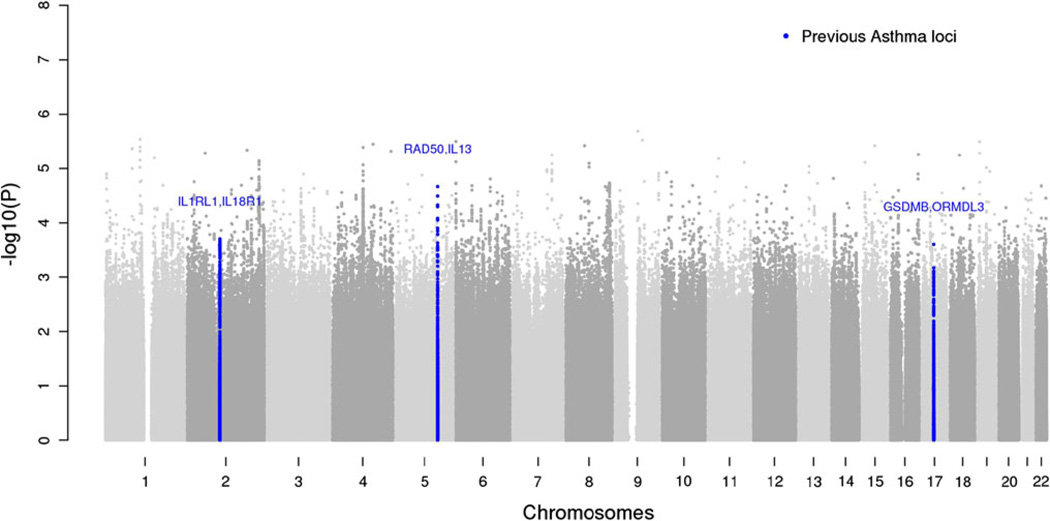
Quantile–quantile (QQ) plots of the meta-analyses show that the test statistics follow the null expectations (Supplemental Figure 1 and Supplemental Figure 2). No SNPs in the analysis surpassed genome-wide significance with p < 5 × 10−8 (Fig. 1 and Supplemental Table 3). Additional analyses of asthma phenotypes stratified by severity (determined from lung function data), age of onset (age of onset <16 years and age of onset >16 years), and allergic status (presence or absence of self-reported hay fever) were performed but did not yield genome-wide significant results (data not shown). We also asked whether the top 100 most significant SNPs in our analysis were associated with asthma in GABRIEL (Moffatt et al. 2010), a large-scale consortium-based GWA study of asthma, but found no evidence of associations beyond that expected by chance.
Fig. 1.
Manhattan plot of study results. Observed log-linear p values on the logarithmic scale are sorted by physical location on the 22 autosomes. Blue dots indicate loci with strong evidence of replication in African ancestry populations shown in Table 2 (color figure online)
Although no loci reached genome wide significance, we hypothesized that we might still be able to provide evidence of replication and help localize signals for associations that had previously been described, mainly in European ancestry populations. This analysis is particularly relevant in African ancestry populations because replication across diverse ethnic groups is an important determinant of the robustness of association at a given locus, and because differing patterns of linkage disequilibrium may permit fine mapping of well-established associations (Cooper et al. 2008). Of the 21 genomic loci previously reported from the GABRIEL consortium, the EVE consortium, and other GWA studies of asthma, the effect estimates of 17 loci were in the same direction and nominally associated with asthma in the African-American meta-analysis (Supplemental Table 3). Thus, it is likely that the loci discovered thus far (mostly in samples of European ancestry) are relevant across multiple ethnicities. The strongest signals of replication in our African- American population were at variants near ORMDL3, IL18RL1-ILRL1, and RAD50/IL13 (see Table 2; all p < 0.0002, the Bonferroni-corrected threshold for the number of loci and variants tested). We also observed nominal evidence of replication at 10p14 (see Table 2) (Hirota et al. 2011), but not at the PYHIN1 locus previously reported as genome-wide significant in African-Americans (see Supplemental Table 3) (Torgerson et al. 2011). To account for a potential “winner’s curse” (Lohmueller et al. 2003; Skol et al. 2006) that could have inflated the effect size estimates in the original report, we estimated our power to replicate the PYHIN association using the originally described lower end of the 95 % confidence interval for effect size (Torgerson et al. 2011). We calculated that we had approximately 74 % power to reach a significance level of p < 0.05 for PYHIN1 in our sample. The higher prevalence of asthma in African-Americans raises the possibility that asthma cases might have a higher percentage of African ancestry than controls. However, this was not the case in subjects from a large meta-analysis of asthma in African-Americans and African-Caribbeans (Torgerson et al. 2011).
Table 2.
Replication of European asthma-associated SNPs in the CARe African-American meta-analyses
| Locus | Reported SNP |
References | EAF-CARe | CARe SNP | Ancestry specific r2 |
African ancestry r2 |
EAF |
p value-one tailed |
Effect direction |
|---|---|---|---|---|---|---|---|---|---|
| RAD50/IL13 | rs2244012 | Moffatt et al. (2010) | 0.5943 | rs17622991 | 0.86971 | 0.117662 | 0.195 | 1.07E–05 | + |
| IL1RL1/IL18R1 | rsl0173081 | Moffatt et al. (2010) | 0.7394 | rs17027029 | 0.773331 | 0.511213 | 0.688 | 9.97E–05 | + |
| GSDMB/ORMDL3 | rs2305480 | Moffatt et al. (2010) | 0.8424 | rs869402 | 0.79411 | 0.570927 | 0.801 | 1.25E–04 | + |
| 10pl4 (gene desert) | rs10508372 | Hirota et al. (2011) | 0.8016 | rs11255975 | 0.985498 | 0.793969 | 0.777 | 8.49E–04 | + |
Reported SNP refers to the top SNP at the particular locus from the referenced GWA studies, and CARe SNP refers the top SNP in the CARe African-American cohort. EAF-CARe is the effect allele frequency for the reported SNP in CARe. EAF is the effect allele frequency for the “better” SNP (i.e., the SNP with the strongest signal of replication) in CARe. Effect direction: + indicates that the CARe SNP effect was in the same direction as the reported SNP
Admixture between populations results in genome-wide ancestry variation that can confound the results of association studies by producing associations involving variants that are distant from the causal variant (Seldin et al. 2011). To account for the potentially confounding effects of admixed populations, we adjusted for local ancestry using HAPMIX, but this did not significantly change the above associations (see Supplemental Table 4). Despite the lack of replication at some individual loci, these results in aggregate indicate good overall evidence of replication in our meta-analysis of African-American individuals for SNPs previously associated with asthma in individuals of European ancestry, confirming a substantial shared genetic basis for asthma across populations with differing genetic ancestry.
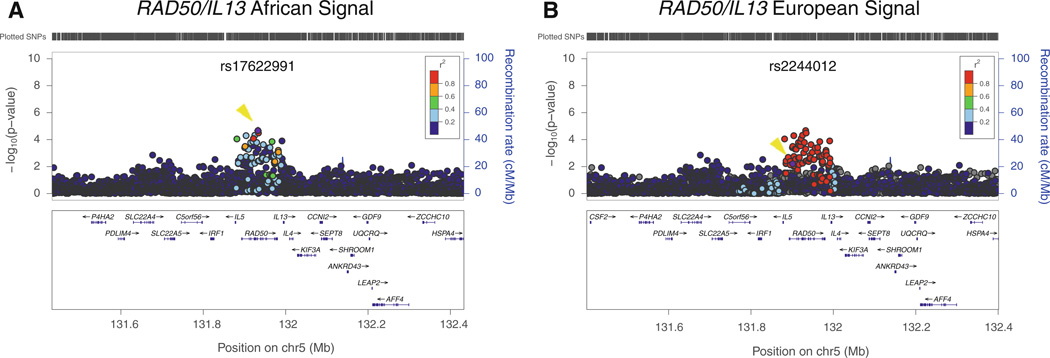
Taking advantage of the different LD patterns between African and European ancestry populations (LD breaks down over shorter distances in African-derived chromosomes), we assessed whether we could fine-map the loci that showed strong evidence of replication in our sample. For this analysis, we evaluated the association with asthma in our African-American sample of those SNPs that were correlated with the index SNP in 1,000 Genomes European ancestry samples (r2 > 0.8). This approach helped refine the association signal for the RAD50/IL13 locus. The “European-ancestry SNP” (rs2244012) and the SNP identified by fine mapping in the African-American metaanalysis (rs17622991) are in strong LD with each other in European ancestry individuals (r2 = 0.87). However, in African-Americans, LD between these two SNPs is weaker (r2 = 0.12) and the association with asthma is stronger for rs17622991 than for rs2244012 (p = 0.000021 and 0.0062, respectively; see Table 2 and Fig. 2). Thus, variants correlated in African ancestry samples with rs17622991 rather than with rs2244012 are more likely to be causal at this locus. We also examined how many SNPs were in strong LD (r2 = 0.8) with rs17622991 in YRI and compared them to the number of SNPs in strong LD with rs2244012 in CEU. We found 76 proxy SNPs in YRI and 77 proxy SNPs in CEU, with only 4 SNPs present in both panels.
Fig. 2.
Fine mapping of association results at RAD50/IL13 locus. Association results in the CARe African-American samples with LD estimated either in African ancestry individuals (a) or European individuals (b) plotted using LocusZoom (Pruim et al. 2010) against the position on chromosome 5. The purple dot (also marked with a yellow arrowhead) represents the most significantly associated SNP after meta-analysis (rs17622991 for African-Americans and rs2244012 for Europeans). The SNPs surrounding the most significant SNP are color coded to reflect their LD with this SNP. Estimated recombination rates are plotted in cyan to reflect the local LD structure. Genes, the position of exons, and the direction of transcription from the UCSC genome browser are noted. Hashmarks represent SNP positions available in the meta-analysis (color figure online)
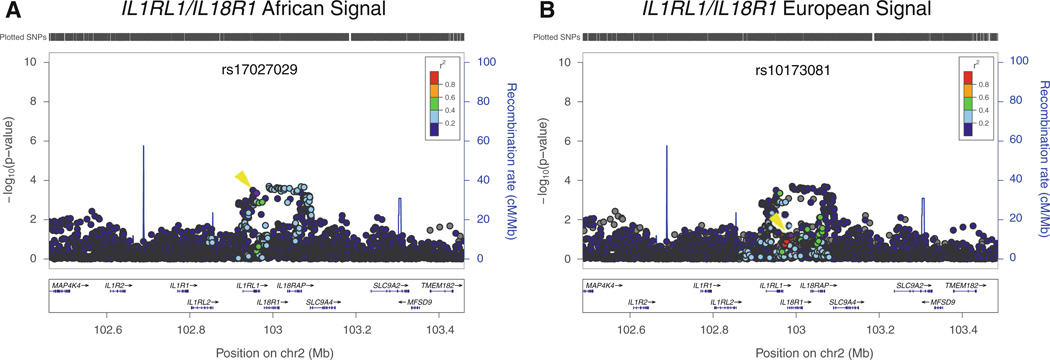
Similarly, at the IL1RL1/IL18R1 locus on chromosome 2, the SNP with the strongest association in our sample, rs17027029, is more strongly associated with asthma than the “European-ancestry SNP” rs10173081, and also defines a narrower genomic interval than was implicated in analysis of European-derived samples (Fig. 3). Examining SNPs in strong LD with rs17027029 and rs10173081, we found that were 37 proxies in YRI and 26 proxies in CEU with only one SNP present in both panels. Thus, both the RAD50/IL13 locus and the IL1RL1/IL18R1 locus illustrate the utility of fine-mapping association signals in other ethnic groups to limit the list of potential causal variants. For variants near GDSMB/ORMDL3 the most strongly associated SNPs in African-Americans and the SNPs implicated from studies of European-derived samples were not distinguishable in our data (Supplemental Figure 3).
Fig. 3.
Fine mapping of association results at ILlRLl/IL18R1 locus. Association results in the CARe African-American samples with LD estimated either in African ancestry individuals (a) or European individuals (b) plotted using LocusZoom (Pruim et al. 2010) against the position on chromosome 2. The purple dot (also marked with a yellow arrowhead) represents the most significantly associated SNP after meta-analysis (rs17027029 for African-Americans and rsl0173081 for Europeans). The SNPs surrounding the most significant SNP are color coded to reflect their LD with this SNP. Estimated recombination rates are plotted in cyan to reflect the local LD structure. Genes, the position of exons, and the direction of transcription from the UCSC genome browser are noted. Hashmarks represent SNP positions available in the meta-analysis (color figure online)
We next performed a conditional analysis of the signals in the RAD50/IL13 region in our African-American sample in an effort to distinguish the association data of the “European ancestry SNP” rs224012 from the “African ancestry SNP” rs17622991 (Supplemental Table 5). Conditioning for the effect of rs17622991 eliminated evidence for association for rs224012 (conditioned p value >0.1). By contrast, conditioning for the effect of rs224012 did not abolish the association signal of rs17622991 (conditioned p value 2.9 × 10−4). We also performed a conditional analysis of the signals in IL1RL1/IL18R1, but this analysis did not distinguish the signal at the European ancestry SNP from the signal at the African ancestry SNP (Supplemental Table 5) (Price et al. 2009).
Discussion
The majority of large-scale genetic efforts to identify risk factors for asthma have focused on populations of European ancestry, despite the fact that in the US, this disease disproportionately affects minority populations. In this study, we found strong overall evidence of replication in our meta-analysis of African-Americans for SNPs previously associated with asthma in individuals of European ancestry. In particular, we demonstrate that the association of variation near RAD50/IL13, first discovered in individuals of European ancestry, replicates in individuals of African ancestry (Li et al. 2010; Moffatt et al. 2010). We also demonstrate strong evidence of replication at loci near ORMDL3/GSDMB and IL1RL1/IL18R1, each previously associated with asthma in European populations. Further, we show for the first time that variation near 10p14, which has been associated with asthma in Japanese populations, is also nominally associated with asthma in African ancestry populations. The overall replication of many of the asthma loci in African ancestry populations adds to the robustness of these results, and suggests that many association signals are relevant across different populations and caused by shared genetic factors.
In a still early application of the approach, we also use imputation based on the 1000 Genomes Project data to fine map known association signals. In addition, the analysis of the RAD50/IL13 and IL1RL1/IL18R1 loci in African ancestry populations in the present study and the differences in LD structure between European and African ancestry populations have delimited the list of potentially causal SNPs at this locus. Our results also provide further evidence for the utility of a self-reported asthma phenotype, which is widely available, in genetic association studies of asthma (Moffatt et al. 2010; Ramasamy et al. 2012).
The size of the current GWA study is quite modest compared to consortia focusing on European-derived individuals, and as a consequence of the limited discovery power, we did not identify novel loci specifically associated with asthma that reached the level of genome-wide significance. In addition, despite the fact that we found strong overall evidence of replication of SNPs previously associated with asthma, there were many loci originally found in European- and African-derived populations that were not formally replicated in the CARe meta-analyses presented in this manuscript (Supplemental Table 3). The small sample size of this analysis and the statistical thresholds used to control the false positive rate together provide the most likely explanation for why many loci largely showed directional consistency but did not formally replicate in the CARe African-Americans. As such, the lack of replication at certain loci (in particular near PYHIN1) in this study should not be interpreted as definitively disproving the association between those loci and asthma. Taken together, our results suggest that asthma risk in African-Americans is influenced by multiple variants with modest effects, as has been observed for asthma in European-derived samples, and that many of these variants are likely shared across populations. This and future analyses of larger samples with diverse populations will allow for the identification of robust associations that replicate across ethnic groups and will add to the understanding of the genetic architecture of asthma across populations.
Supplementary Material
Acknowledgments
This work is supported by Contract HHSN26800625226C from the National Heart, Lung, and Blood Institute, T32 5T32HD040128-09 from the National Institutes of Health, the American Asthma Foundation, the American Medical Association Seed Grant Research Program, and the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. The authors wish to acknowledge the support of the National Heart, Lung, and Blood Institute and the contributions of the research institutions, study investigators, field staff, and study participants in creating the CARe resource for biomedical research. The following studies, funded by the listed National Institutes of Health grants, were utilized in this study. Atherosclerosis Risk in Communities (ARIC): University of North Carolina at Chapel Hill (N01-HC-55015, N01-HC-55018), Baylor Medical College (N01-HC-55016), University of Mississippi Medical Center (N01-HC-55021), University of Minnesota (N01-HC-55019), Johns Hopkins University (N01-HC-55020), University of Texas, Houston (N01-HC-55022). Coronary Artery Risk in Young Adults: University of Alabama at Birmingham (N01-HC-48047, N01-HC- 95095), University of Minnesota (N01-HC-48048), Northwestern University (N01-HC-48049), Kaiser Foundation Research Institute (N01-HC-48050), Tufts-New England Medical Center (N01-HC- 45204), Wake Forest University (N01-HC-45205), Harbor-UCLA Research and Education Institute (N01-HC-05187), University of California, Irvine (N01-HC-45134, N01-HC-95100); Jackson Heart Study: Contracts N01-HC-95170, N01-HC-95171, N01-HC-95172 from the National Heart, Lung, and Blood Institute and the National Institute on Minority Health and Health Disparities, with additional support from the National Institute on Biomedical Imaging and Bioengineering. Multi-Ethnic Study of Atherosclerosis: University of Washington (N01-HC-95159), University of California, Los Angeles (N01-HC-95160), Columbia University (N01-HC-95161), Johns Hopkins University (N01-HC-95162, N01-HC-95168), University of Minnesota (N01-HC-95163), Northwestern University (N01-HC-95164), Wake Forest University (N01-HC-95165), University of Vermont (N01-HC-95166), New England Medical Center (N01-HC-95167), Harbor-UCLA Research and Education Institute (N01-HC-95169), Cedars-Sinai Medical Center (R01-HL-071205), University of Virginia (subcontract to R01-HL-071205). Pulmonary phenotyping and analyses in MESA are funded by NIH grants R01-HL077612, RC1-HL100543 and R01-HL093081.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-013-1310-7) contains supplementary material, which is available to authorized users.
Contributor Information
David B. Kantor, Email: david.kantor@childrens.harvard.edu, Division of Critical Care Medicine, Department of Anesthesiology, Perioperative and Pain Medicine, Boston Children’s Hospital, 300 Longwood Avenue, Boston, MA 02115, USA.
Cameron D. Palmer, Program in Medical and Population Genetics, Broad Institute, 7 Cambridge Center, Cambridge, MA 02142, USA
Taylor R. Young, Program in Medical and Population Genetics, Broad Institute, 7 Cambridge Center, Cambridge, MA 02142, USA
Yan Meng, Program in Medical and Population Genetics, Broad Institute, 7 Cambridge Center, Cambridge, MA 02142, USA.
Zofia K. Gajdos, Program in Medical and Population Genetics, Broad Institute, 7 Cambridge Center, Cambridge, MA 02142, USA
Helen Lyon, Program in Medical and Population Genetics, Broad Institute, 7 Cambridge Center, Cambridge, MA 02142, USA.
Alkes L. Price, Departments of Epidemiology and Biostatistics, Harvard School of Public Health, 665 Huntington Avenue, Boston, MA 02115, USA
Samuela Pollack, Departments of Epidemiology and Biostatistics, Harvard School of Public Health, 665 Huntington Avenue, Boston, MA 02115, USA.
Stephanie J. London, Division of Intramural Research, DHHS, National Institute of Environmental Health Sciences, National Institutes of Health, PO Box 12233, Research Triangle Park, NC 27709, USA
Laura R. Loehr, Department of Epidemiology, University of North Carolina Gillings School of Global Public Health, Chapel Hill, NC 27599, USA
Lewis J. Smith, Northwestern University, Feinberg School of Medicine, Division of Pulmonary and Critical Care Medicine, 240 E. Huron Ave, Chicago, IL 60611, USA
Rajesh Kumar, Ann & Robert H. Lurie Children’s Hospital of Chicago, 225 E Chicago Avenue, Chicago, IL 60611, USA.
David R. Jacobs, Jr, Division of Epidemiology and Community Health, University of Minnesota, 1300 S. 2nd Street Suite 300, Minneapolis, MN 55454, USA.
Marcy F. Petrini, Division of Pulmonary, Critical Care and Sleep Medicine, University of Mississippi Medical Center, 2500 North State St, Jackson, MS 39216-4505, USA
George T. O’Connor, Department of Medicine, Pulmonary Center, Boston University School of Medicine, Room R 304, 72. E. Concord St, Boston, MA 02118, USA
Wendy B. White, Tougaloo College and Jackson Heart Study, 500 West County Line Road, Tougaloo, MS 39174, USA
George Papanicolaou, Division of Prevention and Population Sciences, National Heart, Lung and Blood Institute, National Institutes of Health, Two Rockledge Center, Suite 10018 6701 Rockledge Drive, MSC 7936, Bethesda, MD 20892-7936, USA.
Kristin M. Burkart, Division of Pulmonary, Allergy and Critical Care, College of Physicians and Surgeons, Columbia University, 622 West 168th Street, PH 8 East, Room 101, New York, NY 10032, USA
Susan R. Heckbert, University of Washington, Department of Epidemiology, School of Public Health, Cardiovascular Health Research Unit, 1730 Minor Avenue, Suite 1360, Seattle, WA 98101-1466, USA
R. Graham Barr, Department of Medicine, College of Physicians and Surgeons, Department of Epidemiology, Mailman School of Public Health, Columbia University, PH 9 East, Room 105, New York, NY 10032, USA.
Joel N. Hirschhorn, Email: joelh@broadinstitute.org, Program in Medical and Population Genetics, Broad Institute, 7 Cambridge Center, Cambridge, MA 02142, USA; Division of Endocrinology, Boston Children’s Hospital, 300 Longwood Avenue, Boston, MA 02115, USA; Departments of Genetics and Pediatrics, Harvard Medical School, 25 Shattuck Street, Boston, MA 02115, USA.
References
- Boudreaux ED. Acute asthma among adults presenting to the emergency department: the role of race/ethnicity and socioeconomic status. Chest. 2003;124:803–812. doi: 10.1378/chest.124.3.803. [DOI] [PubMed] [Google Scholar]
- Cooper RS, Tayo B, Zhu X. Genome-wide association studies: implications for multiethnic samples. Hum Mol Genet. 2008;17:R151–R155. doi: 10.1093/hmg/ddn263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MAR, Matheson MC, Duffy DL, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanter J, Choudhry S, Eng C, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177:1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanter JM, Torgerson D, Gignoux CR, et al. Cosmopolitan and ethnic-specific replication of genetic risk factors for asthma in 2 Latino populations. J Allergy Clin Immunol. 2011;128(37–43):e12. doi: 10.1016/j.jaci.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Bjornsdottir US, Halapi E, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- Hancock DB, Romieu I, Shi M, et al. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in Mexican children. PLoS Genet. 2009;5:e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159:179–187. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- Himes BE, Hunninghake GM, Baurley JW, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Takahashi A, Kubo M, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43:893–896. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Chiang CWK, Palmer CD, et al. Genome-wide association of anthropometric traits in African- and African-derived populations. Hum Mol Genet. 2010;19:2725–2738. doi: 10.1093/hmg/ddq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Palmer CD, Young T, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Howard TD, Zheng SL, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010;125:328–335. doi: 10.1016/j.jaci.2009.11.018. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Rogers L, Cheng Q, et al. Genetic Variants of TSLP and asthma in an admixed urban population. PLoS ONE. 2011;6:e25099. doi: 10.1371/journal.pone.0025099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Pearce CL, Pike M, et al. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- Mathias RA, Grant AV, Rafaels N, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125:336–346. doi: 10.1016/j.jaci.2009.08.031. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman JE, Zahran H, Truman BI, et al. Current asthma prevalence—United States, 2006–2008. MMWR Surveill Summ. 2011;(60 Suppl):84–86. [PubMed] [Google Scholar]
- Musunuru K, Lettre G, Young T, et al. Candidate Gene Association Resource (CARe): design, methods, and proof of concept. Circ Cardiovasc Genet. 2010;3:267–275. doi: 10.1161/CIRCGENETICS.109.882696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N’Diaye A, Chen GK, Palmer CD, et al. Identification, replication, and fine-mapping of loci associated with adult height in individuals of African ancestry. PLoS Genet. 2011;7:e1002298. doi: 10.1371/journal.pgen.1002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Sakamoto H, Hirota T, et al. Genome-wide association study identifies HLA-DP as a susceptibility gene for pediatric asthma in Asian populations. PLoS Genet. 2011;7:e1002170. doi: 10.1371/journal.pgen.1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B, Zaitlen N, Lettre G, et al. Enhanced statistical tests for GWAS in admixed populations: assessment using African Americans from CARe and a Breast Cancer Consortium. PLoS Genet. 2011;7:e1001371. doi: 10.1371/journal.pgen.1001371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A, Kuokkanen M, Vedantam S, et al. Genome-wide association studies of asthma in population-based cohorts confirm known and suggested loci and identify an additional association near HLA. PLoS ONE. 2012;7:e44008. doi: 10.1371/journal.pone.0044008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray NF, Thamer M, Fadillioglu B, Gergen PJ. Race, income, urbanicity, and asthma hospitalization in California: a small area analysis. Chest. 1998;113:1277–1284. doi: 10.1378/chest.113.5.1277. [DOI] [PubMed] [Google Scholar]
- Scirica CV, Celedon JC. Genetics of asthma: potential implications for reducing asthma disparities. Chest. 2007;132:770S–781S. doi: 10.1378/chest.07-1905. [DOI] [PubMed] [Google Scholar]
- Seldin MF, Pasaniuc B, Price AL. New approaches to disease mapping in admixed populations. Nat Rev Genet. 2011;12:523–528. doi: 10.1038/nrg3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Sleiman PMA, Flory J, Imielinski M, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- Teo Y-Y, Small KS, Kwiatkowski DP. Methodological challenges of genome-wide association analysis in Africa. Nat Rev Genet. 2010;11:149–160. doi: 10.1038/nrg2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen SF, van der Sluis S, Kyvik KO, et al. Estimates of asthma heritability in a large twin sample. Clin Exp Allergy. 2010;40:1054–1061. doi: 10.1111/j.1365-2222.2010.03525.x. [DOI] [PubMed] [Google Scholar]
- Torgerson DG, Ampleford EJ, Chiu GY, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson DG, Gignoux CR, Galanter JM, et al. Case-control admixture mapping in Latino populations enriches for known asthma-associated genes. J Allergy Clin Immunol. 2012 doi: 10.1016/j.jaci.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen G, van Beijsterveldt TCEM, van Baal CGCM, et al. Heritability of self-reported asthma and allergy: a study in adult Dutch twins, siblings and parents. Twin Res Hum Genet. 2012;11:132–142. doi: 10.1375/twin.11.2.132. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.