Abstract
Alzheimer’s disease (AD) is a neurodegenerative disorder associated with amyloid accumulation and autophagic changes. Parkin is an E3 ubiquitin ligase involved in proteasomal and autophagic clearance. We previously demonstrated decreased parkin solubility and interaction with the key autophagy enzyme Beclin-1 in AD, but tyrosine kinase inhibition restored parkin-Beclin-1 interaction. In the current studies we determined the mechanisms of Nilotinib-induced parkin-Beclin-1 interaction, which leads to amyloid clearance. Nilotinib increased endogenous parkin levels and ubiquitination, which may enhance parkin recycling via the proteasome, leading to increased activity and interaction with Beclin-1. Parkin solubility was decreased and autophagy was altered in amyloid expressing mice, suggesting that amyloid stress affects parkin stability, leading to failure of protein clearance via the lysosome. Isolation of autophagic vacuoles revealed amyloid and parkin accumulation in autophagic compartments but Nilotinib decreased insoluble parkin levels and facilitated amyloid deposition into lysosomes in wild type, but not parkin−/− mice, further underscoring an essential role for endogenous parkin in amyloid clearance. These results suggest that Nilotinib boosts the autophagic machinery, leading to increased level of endogenous parkin that undergoes ubiquitination and interacts with Beclin-1 to facilitate amyloid clearance. These data suggest that Nilotinib-mediated autophagic changes may trigger parkin response via increased protein levels, providing a therapeutic strategy to reduce Aβ and Tau in AD.
Keywords: Ubiquitination, parkin, autophagy, Tau, amyloid, Alzheimer’s
Introduction
Alzheimer’s disease (AD) is an aging disorder characterized by deposition of β-amyloid (Aβ) plaques and hyper-phosphorylated Tau (p-Tau). Amyloid precursor protein (APP) is sequentially cleaved by secretases to yield C-terminal APP fragments (CTFs) and amyloid peptides Aβ1–40 and Aβ1–42 [1–4] giving rise to extracellular plaques. The tyrosine kinase Abelson (Abl) is distributed in the nucleus and cytoplasm and involved in a wide range of functions, including apoptosis [5]. Abl phosphorylation at tyrosine 412 (T412) is elevated in the hippocampus and entorhinal cortex in AD brains [6, 7], and Abl phosphorylates Tau at several tyrosine residues [6, 8]. In primary neuronal culture, Abl inhibition prevents Aβ1–42 fibrils and cell death [9], and hippocampal injection of Aβ fibrils results in up-regulation of Abl levels [10]. Furthermore, Abl is also increased in Parkinson’s Disease (PD) brains, while Abl inhibition leads to autophagic α-Synuclein degradation in PD models [11].
It was previously reported that Abl inhibition may activate parkin and improve dopaminergic neuron survival in PD models [12]. Decreased parkin solubility is associated with defects in autophagic clearance of β-amyloid and p-Tau in post-mortem AD brains [13], indicating that aging may alter parkin function. Parkin mediates mitophagy [14–17], and clears autophagic vacuoles (AVs) in AD mice [13, 18]. These data led to the hypothesis that tyrosine kinase inhibition (TKI) will activate parkin and facilitate autophagic clearance of β-amyloid and p-Tau. Nilotinib is a second-generation TKI, which was approved by the US Food and Drug Administration (FDA) in 2007 for the treatment of adult chronic myelogenous leukemia (CML) [19–21]. We previously demonstrated that Nilotinib increases parkin-beclin-1 interaction, leading to clearance of β-amyloid and p-Tau in AD models [22]. The current studies further examined the effects of Nilotinib on endogenous parkin function to mediate autophagic clearance. Here we report that TKI via Nilotinib increases endogenous parkin levels and self-ubiquitination, which may enhance physiological re-cycling via the proteasome, leading to increased activity and interaction with Beclin-1. Together these data suggest that decreased parkin solubility in AD and sporadic PD [13, 23] may be due to inefficient ubiquitination, leading to parkin instability and decreased parkin-Beclin-1 interaction. Nilotinib alters autophagy, triggering an increase in parkin level, perhaps enhancing parkin re-cycling. The current studies provide novel mechanistic evidence that the FDA-approved drug, Nilotinib, can increase endogenous parkin level and function in response to autophagic changes to clear amyloid, suggesting therapeutic benefits for AD.
Materials and methods
Human postmortem brain tissues
Human postmortem samples were obtained from John’s Hopkins University brain bank. Patients’ description and sample preparation are summarized in [13]. Data were analyzed as mean±SEM, using Two-tailed t-test (P<0.05).
Stereotaxic injection
Lentiviral constructs encoding LacZ, or Aβ1–42 [24] were stereotaxically injected 1×106 multiplicity of infection (m.o.i) bilaterally into the CA1 hippocampus of 1 year old male C57BL/6 or parkin−/−. All procedures were approved by the Georgetown University Animal Care and Use Committee (GUACUC). Nilotinib was dissolved in DMSO and a total volume of 30μl were IP injected once a day for 3 weeks. Half the animals received DMSO and the other half received Nilotinib in DMSO.
Western blot
Brain tissues were homogenized in 1× STEN buffer [13], centrifuged at 10,000 × g for 20 min at 4°C, and the supernatants containing the soluble fraction were collected. To extract the insoluble fraction, pellets were re-suspended in either 4M urea or 30% formic acid and centrifuged at 10,000 × g for 20 min at 4°C. Parkin was probed (1:1000) with PRK8 antibody as indicated [25]. Rabbit polyclonal antibodies anti-beclin-1 (1:1000). Total Abl was probed with (1:500) rabbit polyclonal (K12) antibody (Thermo Fisher), or p-Abl (T412) with (1:500) rabbit polyclonal antibody (Millipore). Phospho-tyrosine proteins were probed (1:1000) with rabbit polyclonal antibody (Cell signaling). β-actin was probed (1:1000) with polyclonal antibody (Cell Signaling Technology, Beverly, MA, USA). Rabbit polyclonal (1:1000) tubulin (Thermo Scientific) was used. MAP-2 was probed (1:1000) with mouse monoclonal antibody (Pierce).
Immunohistochemistry
was performed on 20 micron-thick 4% paraformaldehyde (PFA) fixed cortical brain sections. Aβ1–42 was probed (1:200) with rabbit polyclonal specific anti-Aβ1–42 antibody (Zymed) that recognizes a.a.1–42, and (1:200) mouse monoclonal antibody (4G8) that recognizes a.a. 17–24 (Covance) and counterstained with DAPI. Parkin was immunoprobed (1:200) with mouse anti-parkin (PRK8) antibody that recognizes a.a. 399–465 (Signet Labs, Dedham, MA) and rabbit polyclonal (1:200) anti-parkin (AB5112) antibody that recognizes a.a. 305–622 (Millipore) and counterstained with DAPI. Mouse monoclonal (6E10) antibody (1:100) with DAB were used (Covance) and thioflavin-S was performed according to manufacturer’s instructions (Sigma). Stereological methods- were applied by a blinded investigator using unbiased stereology analysis (Stereologer, Systems Planning and Analysis, Chester, MD) as described in [13, 24].
Aβ and p-Tau enzyme-linked immunosorbent assay (ELISA)
using specific p-Tau, Aβ1–40 and Aβ1–42 ELISA and caspase-3 activity were performed according to manufacturer’s protocol as described in [13, 24].
Subcellular fractionation to isolate autophagic vacuoles
Animal brains were homogenized at low speed (Cole-Palmer homogenizer, LabGen 7, 115 Vac) in 1×STEN buffer and centrifuged at 1,000g for 10 minutes to isolate the supernatant from the pellet. The pellet was re-suspended in 1×STEN buffer and centrifuged once to increase the recovery of lysosomes. The pooled supernatants were then centrifuged at 100.000 rpm for 1 hour at 4°C to extract the pellet containing autophagic vacuoles (AVs) and lysosomes. The pellet was then re-suspended in 10 ml (0.33 g/ml) 50% Metrizamide and 10 ml in cellulose nitrate tubes. A discontinuous Metrizamide gradient was constructed in layers from bottom to top as follows: 6 ml of pellet suspension, 10 ml of 26%; 5 ml of 24%; 5 ml of 20%; and 5 ml of 10% Metrizamide. After centrifugation at 100,000 rpm for 1 hour at 4°C, the fraction floating on the 10% layer (Lysosome) and the fractions banding at the 24%/20% (AV 20) and the 20%/10% (AV10) inter-phases were collected by a syringe and examined.
Ubiquitination assay
Parkin or ubiquitin were separately immunoprecipitated in 100 μl (100 μg of proteins) 1×STEN buffer using (1:100) anti-ubiquitin monoclonal antibody (Abnova) or (1:100) anti-parkin mouse monoclonal antibody (PRK8; Signet Labs; Dedham, MA), respectively. Following immunoprecipitation and normalization of the aliquots, 300 ng of each substrate protein (parkin and ubiquitin) were mixed in the presence of 1 μg recombinant human ubiquitin (Boston Biochem, MA), 100 mm ATP, 1 μg recombinant UbcH7 (Boston Biochem), 40 ng E1 recombinant enzyme (Boston Biochem) and incubated at 37°C in an incubator for 20 min. The reaction was heat inactivated by boiling for 5 min and the substrates were analyzed by WB.
Transmission Electron Microscopy
Brain tissue were fixed in (1:4, v:v) 4% paraformaldehydepicric acid solution and 25% glutaraldehyde overnight, then washed 3× in 0.1M cacodylate buffer and osmicated in 1% osmium tetroxide/1.5% potassium ferrocyanide for 3h, followed by another 3× wash in distilled water. Samples were treated with 1% uranyl acetate in maleate buffer for 1 h, washed 3 × in maleate buffer (pH 5.2), then exposed to a graded cold ethanol series up to 100% and ending with a propylene oxide treatment. Samples are embedded in pure plastic and incubated at 60°C for 1–2 days. Blocks are sectioned on a Leica ultracut microtome at 95 nm, picked up onto 100 nm formvar-coated copper grids, and analyzed using a Philips Technai Spirit transmission EM. All images were collected by a blind investigator.
Cell culture and transfection
Rat neuroblastoma B35 cells were grown in 24 well dishes (Falcon) as previously described [13, 24]. Transient transfection was performed with 3 μg Aβ1–42 cDNA, or 3 μg LacZ cDNA for 24hr. Cells were treated with 10 μM Nilotinib (AMN-107, Selleck Chemical, LLC, USA). for 24 hr and harvested 48 hr after transfection.
Parkin ELISA
was performed on brain soluble brain lysates (in STEN buffer) or insoluble brain lysates (4M urea) using parkin kit (MYBioSource) in 50 μl (1μg/μl) of brain lysates detected with 50μl primary antibody (3h) and 100 μl anti-rabbit antibody (30 min) at RT. Extracts were incubated with stabilized Chromogen for 30 minutes at RT and solution was stopped and read at 450nm, according to manufacturer’s protocol.
Parkin E3 ubiquitin ligase activity
An E3LITE Customizable Ubiquitin Ligase Kit (Life Sensors, UC#101), which measures the mechanisms of E1-E2-E3 activity in the presence of different ubiquitin chains was used. UbcH7 was used as an E2 that provides maximum activity with parkin E3 ligase and added E1 and E2 in the presence of recombinant ubiquitin, including WT or no lysine mutant (K0) to determine the lysine-linked type of ubiquitin. E3 was added as IP parkin (normalized to make sure equal amount of protein loaded) to an ELISA microplate that captures poly-ubiquitin chains formed in the E3-dependent reaction, which was initiated with ATP at RT for 60 min. Controls included, E1-E2-E3, no parkin as an E3 or only IgG were used and poly-ubiquitin chain read in addition to E1, E2 and Aβ1–42 without parkin and assay buffer. Plates were washed 3 times and incubated with streptavidin-HRP for 5 min and read on a chemiluminecense plate reader.
20S proteasome activity assay
Brain extracts (100 μg) were incubated with 250 μM of the fluorescent 20S proteasome specific substrate Succinyl-LLVY-AMC at 37°C for 2h. The medium was discarded and proteasome activity was measured in tissue homogenates as previously described [13, 24].
Results
Nilotinib increases parkin level and ubiquitination
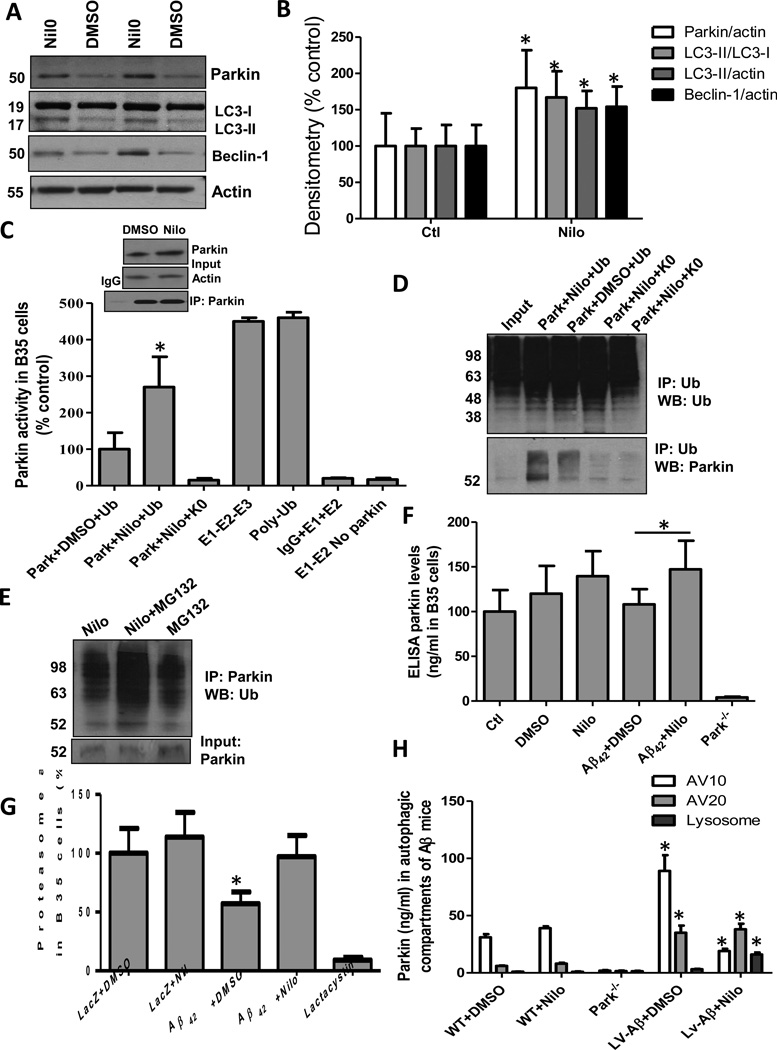
We previously demonstrated decreased parkin-Beclin-1 interaction in AD brains [22], which display decreased parkin solubility and autophagic changes [13]. TKI reversed the loss of parkin-Beclin-1 interaction and led to increased autophagic clearance of Aβ and p-Tau [13, 22]. To determine how TKI alters endogenous parkin response to interact with Beclin-1, 2-months old wild type C57BL/6 mice were treated with intraperotineal (I.P) injection of 10mg/kg Nilotinib or DMSO for 3 weeks [22]. Western blot analysis of total brain lysates showed an increase (80%) in parkin levels (Fig.1A&B, p<0.05, n=9) and significant LC3-II accumulation relative to LC3-I (67%) and actin (52%) in Nilotinib treated mice (Fig. 1A&B) compared to DMSO, suggesting autophagic alteration via autophagosome formation. A significant increase (54%) in Beclin-1 was also observed (Fig. 1A&B), further suggesting autophagic changes in Nilotinib treated mice. To determine parkin modification in response to Nilotinib, rat B35 neuroblastoma cells were treated with 10μM Nilotinib or DMSO (1μl) for 24hr and in vitro ubiquitination assays were performed to determine whether parkin was ubiquitinated, which may affect its re-cycling and prevent its accumulation as observed in AD [13] and PD [23] brains. Parkin (E3) was immuno-precipitated from transfected B35 cells, which had increased parkin levels upon Nilotinib treatment, but the parkin IPs were normalized after protein assays (Fig. 1C, Inset) to ensure equal amounts of parkin are used in the ELISA. An enzymatic mix of E1, E2 (UbcH7) and ATP were added with either WT ubiquitin containing all seven lysines or no lysine (K0) ubiquitin [26] to parkin, and IgG and no parkin were used as controls. Nilotinib increased parkin auto-ubiquitination compared to DMSO or K0 control (Fig. 1C, 170%, n=5, p<0.05). A control E1-E2-E3 mix and polyubiquitin chains were also used to control for chemiluminscence reading. We previously demonstrated using the same ubiquitination assays that parkin can self-ubiquitinate via K48 and K63 ubiquitin linkages [26]. To ascertain that Nilotinib promotes parkin ubiquitination, another in-vitro assay was performed as we previously described [25, 26]. Parkin was immunoprecipitated from B35 cells treated with DMSO or Nilotinib (42hr) and 20μM proteasome inhibitor MG132 for 6hr, and recombinant E1, E2 (UbcH7), ubiquitin and ATP were added at 60°C for 1hr. Immunoprecipitation of ubiquitin showed higher levels of ubiquitinated parkin in Nilotinib treated cells compared to DMSO with WT but not K0 ubiquitin (Fig. 1D, n=5), suggesting that Nilotinib increases parkin ubiquitination. Additionally, to ascertain that Nilotinib increased parkin ubiquitination, parkin was immunoprecipitated and showed a higher smear with ubiquitin antibodies (Fig. 1E, n=5), suggesting that Nilotinib increased the level of parkin ubiquitination. These results suggest that ubiquitinated parkin may increase recognition by the proteasome for degradation.
Fig. 1. Nilotinib increases parkin level and induces protein clearance.
A) WB of total brain lysates on 4–12% SDS-NuPAGE gel showing the levels of parkin (1st blot), LC3-II (2nd blot) and Beclin-1 (3rd blot) relative to actin in C57BL/6 mice treated with Nilotinib; and B) graphs represent densitometry analysis. C) Parkin E3 ubiquitin ligase function in B35 neuroblastoma cells treated with DMSO or Nilotinib for 24hr, insert shows parkin input and IP after normalization. D) WB of immunoprecipitated ubiquitin (1st blot, input) probed with parkin antibody (2nd blot) on 4–12% SDS-NuPAGE gel. E) WB of immunoprecipitated parkin probed with ubiquitin antibody (2nd blot) on 4–12% SDS-NuPAGE gel. F) Parkin levels as measured by ELISA in B35 expressing Aβ42 and treated with Nilotinib. G). Proteasome activity via Chymotrypsin-like assays in human neuroblastoma cells (n=12) ± Nilotinib. H) Parkin ELISA in AVs isolated from 1 year old male C57BL/6 or parkin−/− mice injected 10mg/kg for 3 weeks. * significantly different to control or as indicated, Mean±SEM, ANOVA with Neumann Keuls multiple comparison, p<0.05.
To determine whether parkin levels are also increased in response to amyloid stress, rat B35 neuroblastoma cells were transfected with 3μg human Aβ1–42 cDNA (or LacZ) for 24hr, and treated with 10μM for an additional 24hr. Parkin levels were measured via ELISA using parkin−/− brain extracts as a specificity control (Fig. 1F). An increase (39%) in parkin level was observed with Aβ1–42+Nilotinib (47%) compared to Aβ1–42+DMSO or control (Fig. 1F, n=12, p<0.05), suggesting that Nilotinib increases parkin levels in response to Aβ1–42 stress, which alone did not alter parkin level. To further determine whether alteration of parkin levels is associated with proteasome function, the Chymotrypsin-like assay was used with the 20S proteasome inhibitor lactacystin as a specificity control (Fig. 1G). Proteasome activity was decreased (43%, p<0.05, n=12) in Aβ1–42 cells, and Nilotinib reversed these effects compared to LacZ cells (Fig. 1F).
To determine Nilotinib effects on parkin fate in vivo, we stereotaxically injected 1×106 m.o.i lentiviral Aβ1–42 bilaterally into the hippocampus of 1 year old male C57BL/6 (WT) or parkin−/− mice (generated on a C57BL/6 background) [27] and 3 weeks later we injected 10mg/kg Nilotinib I.P. once a day for 3 additional weeks. Autophagic vacuoles (AVs) were isolated [13] and parkin levels were measured by ELISA. Parkin was significantly increased (2.8 fold, Fig. 1H, p<0.05, n=5) in AV10 (10% gradient) and AV20 (6 fold) in lentiviral Aβ1–42 brains compared to control (DMSO). Nilotinib significantly reduced parkin level (1.25 fold) in AV10 and increased it in the lysosomal fraction (5 fold) compared to control, suggesting parkin transfer from AV10 to lysosomes for degradation [28].
Nilotinib is a non-specific Abl inhibitor that increases parkin level, leading to Aβ clearance
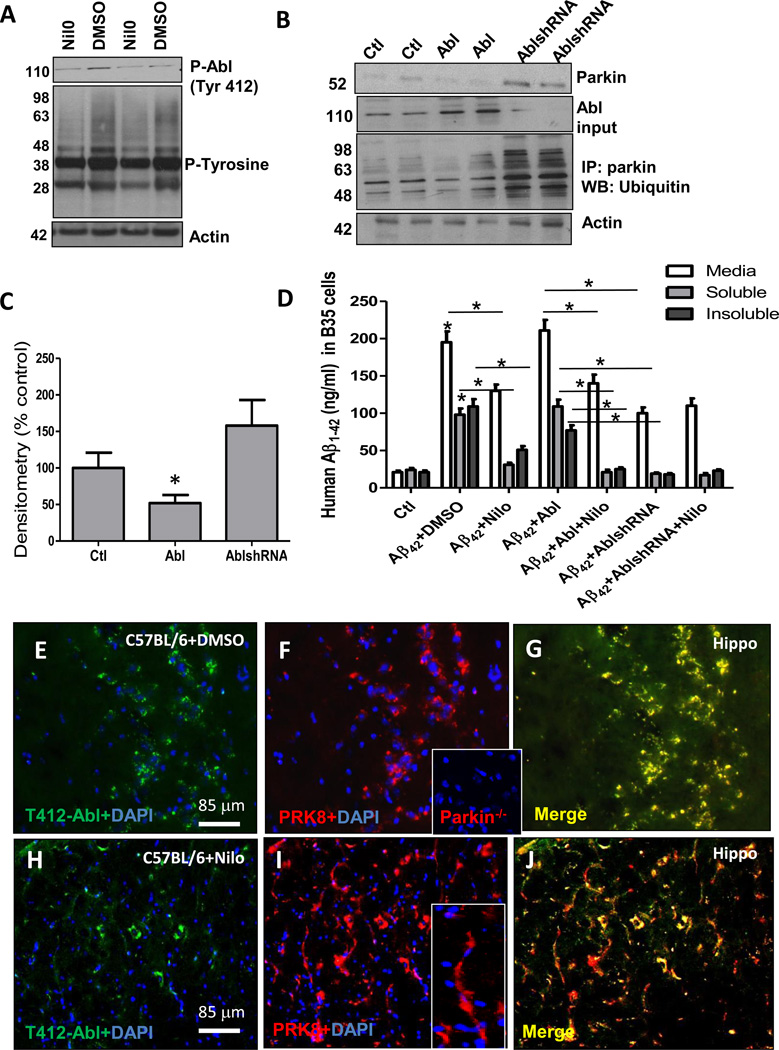
To determine whether Abl affects parkin activity via phosphorylation as previously reported [12], we immunoprecipitated parkin from Nilotinib treated mice or B35 cells, but we did not observe any tyrosine phosphorylation with the pan-phosphotyrosine antibody, suggesting that parkin is not a substrate for phosphorylation by Abl. However, Nilotinib decreased the levels of phosphorylated T412 Abl (Fig. 2A, 46%, p<0.05, n=9), and pan-phosphotyrosine (Fig. 1A, 2nd blot) relative to actin in WT mice, suggesting that Nilotinib is not a specific Abl inhibitor. To further examine the effects of TKI on parkin modification, rat neuroblastoma B35 cells were co-transfected with 3μg human Aβ1–42 and Abl cDNAs or Aβ1–42 cDNA and Abl shRNA (ThermoFisher) for 24hr and the proteasome was inhibited with 20μM MG132 for 6hr to allow accumulation of ubiquitinated parkin. Parkin levels were significantly decreased when Abl was over-expressed (Fig. 2B&C, n=12, p<0.05), compared to control, but parkin levels significantly increased (58%) when Abl expression was blocked relative to actin compared to control. Parkin was immunoprecipitated (Fig. 2B) and probed with ubiquitin, which showed an increase in ubiquitinated parkin smear when Abl expression was blocked with shRNA (Fig. 2B, 2nd blot, n=12). Additionally, the 50kDa band that corresponds to parkin was decreased (41%, n=12) with Abl overexpression and increased with Abl shRNA (81%) relative to actin in comparison to control (Fig. 2B, p<0.05), suggesting that Abl affects parkin ubiquitination and level. Furthermore, to determine whether Nilotinib effects are mediated via Abl inhibition, transfected B35 cells were treated with 10μM Nilotinib for 24hr without proteasome inhibition. ELISA showed that Aβ1–42 expression resulted in significant increases in secreted (195ng/ml, media), soluble (98ng/ml, STEN extract) and insoluble (109ng/ml, 30% formic acid extract) Aβ1–42 compared to control (Fig. 2D, p<0.05, n=12), and Nilotinib reduced secreted (130ng/ml), soluble (31ng/ml) and insoluble (51ng/ml) Aβ1–42. Over-expression of Abl (Fig. 2B) with Aβ1–42 led to a non-significant increase in secreted (211ng/ml), soluble (109ng/ml) and insoluble (77ng/ml) Aβ1–42, and Nilotinib reversed secreted (140ng/ml), soluble (21ng/ml) and insoluble (25ng/ml) Aβ1–42. Importantly, blocking Abl expression with shRNA (Fig. 2D) significantly reduced the levels of secreted (100ng/ml), soluble (19ng/ml) and insoluble (18ng/ml) Aβ1–42 compared to both Abl over-expression and DMSO, and these effects were not affected by Nilotinib (Fig, 2D), suggesting that reduction of Abl levels may increase Aβ1–42 clearance. Furthermore, the level of secreted Aβ1–42 when Abl shRNA was blocked at the same time with Aβ1–42 expression was significantly lower (100ng/ml), compared to Nilotinib (140ng/ml), which started 24hr after Aβ1–42 transfection, suggesting that reduction in secreted Aβ1–42 happened in the first 24hr before Nilotinib, accounting to the difference between secreted and intracellular Aβ1–42. To further assess the relationship between Nilotinib and parkin in 2 months old C57BL/6 mice, 20μm thick hippocampal sections were stained with T412 Abl (Fig. 2E) and parkin (Fig. 2F, insert is staining of parkin−/− brains to PRK8 specificity), showing co-localization between parkin and phosphorylated T412 Abl (Fig. 2G). Nilotinib significantly decreased (62% by stereology) T412 Abl (Fig. 2H) and increased parkin (29% by stereology) levels (Fig. 2I&J, p<0.05, n=6). Nilotinib also affected parkin expression pattern in mice, showing more parkin in the cell bodies and cellular processes (Fig. 2I, insert).
Fig. 2. Nilotinib is a non-specific TKI.
WB in brain lysates in A) 1-year old male C57BL/6 mice treated IP with 10mg/kg Nilotinib for 3 weeks showing T412 Abl (1st blot) and phospho-tyrosine proteins (2nd blot) relative to actin, and B) WB in rat neuroblastoma B35 cells transfected with Abl cDNA or shRNA (2nd blot) showing total parkin level (1st blot), and immunoprecipitated parkin probed with ubiquitin antibody (2nd blot) relative to actin on 4–12% SDS-NuPAGE gel. C) Densitometry of total parkin levels. D). ELISA of Aβ1–42 levels in B35 cells co-transfected with either Aβ1–42 and Abl or Aβ1–42 and Abl shRNA. Staining of 20μm thick hippocampal sections in C57BL/6 mice treated IP with 10mg/Kg Nilotinib or DMSO for 3 weeks showing E) T412 Abl, F) parkin (insert is PRK8 staining in parkin−/− cortex showing antibody specificity. and G) parkin and T412 Abl co-localization in DMSO treated mice. H) T412 Abl, I) parkin and insert parkin staining in neuronal processes, J) parkin and T412 Abl co-localization in Nilotinib treated mice. * significantly different to control or as indicated, Mean±SEM, ANOVA with Neumann Keuls multiple comparison, p<0.05.
Decreased parkin solubility in transgenic AD models and Nilotinib increases soluble parkin level, leading to amyloid clearance
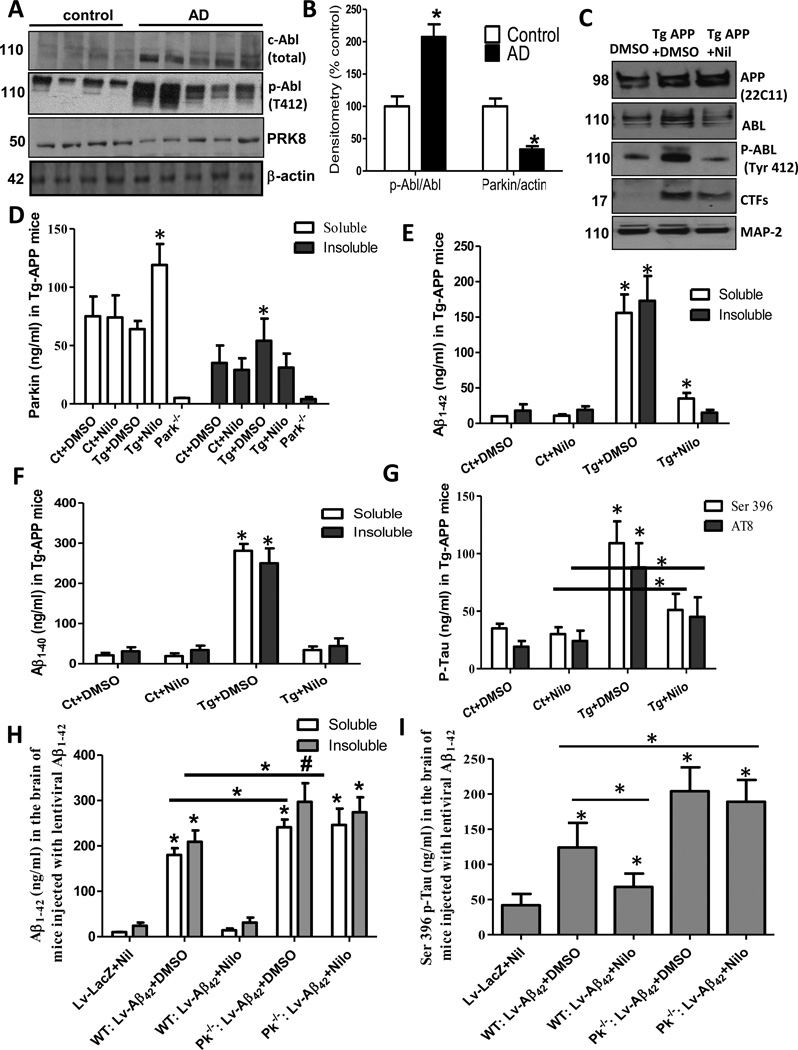
We previously showed that insoluble parkin levels were increased in post-mortem AD brains [13], so we examined the levels of Abl in these brain lysates (n=12 AD and 7 control, described in [13]). Significantly increased levels (90%) of total (Fig. 3A) and T412 (184%) Abl were detected in AD brains (Fig. 3A). The ratio of p-Abl over total Abl (Fig. 3B) was also increased (102%). In contrast, soluble parkin was decreased (70%) in AD cortex (Fig. 3A&B) relative to actin, suggesting that Abl activation and parkin inactivity may be a vulnerability to brain aging. To determine Nilotinib effects on parkin function in AD models in vivo, 8–12 months old male AD transgenic mice (Tg-APP) harboring the Swedish K670N/M671L, Dutch E693Q and Iowa D694N mutations [29] were treated with 10mg/kg IP injection of Nilotinib for 3 weeks. Significant increases in total (51%) and T412 Abl (64%) were detected in Tg-APP compared to control (Fig. 3C, p<0.05, n=11), while Nilotinib reversed these increases (Fig. 3E, p<0.05, n=11). Abl inhibition with Nilotinib also reduced the level of CTFs (44%, p<0.05, n=11) relative to MAP-2, with no effects on secretases as previously shown [22]. No changes in soluble or insoluble parkin were detected in control mice ± Nilotinib (Fig. 3, n=9). However, Nilotinib increased the level of soluble parkin from 64 ng/ml in Tg-APP+DMSO to 119 ng/ml (Fig. 3D, n=11, p<0.05) while it decreased insoluble parkin level from 54 ng/ml to 31 ng/ml in Nilotinib treated mice (Fig. 3D, p<0.05, n=11). Taken together, these data indicate that Nilotinib increases soluble parkin level and ubiquitination, suggesting increased activity and proteasomal recycling, leading to prevention of insoluble parkin accumulation.
Fig. 3. Nilotinib-induced parkin activation clears brain amyloid.
A) WB of post-mortem cortical extracts of AD patients (N=12 AD and 7 control) on 10% SDS Nu-PAGE and B) Graph is densitometry and ratio of Abl and p-Abl and parkin. C) WB analysis on 4–12% SDS Nu-PAGE gels of brain extracts from Tg-APP treated with Nilotinib or DMSO showing APP, Abl, p-Abl and CTFs and MAP-2 as control (n=11). D) Graph represents ELISA levels of soluble and insoluble mouse parkin in the brain of 8–12 months old Tg-APP mice (n=9) injected (IP) with 10mg/kg (daily for 3 weeks). Graph represents ELISA levels of soluble and insoluble E) Aβ1–42 and F) Aβ1–40 in the brain of 8–12 months old Tg-APP mice (n=9) injected (IP) with 10mg/kg once a day for 3 weeks. G) Graph represents ELISA levels of mouse p-Tau in the brain of 8–12 months old Tg-APP mice (n=9). Graph represents ELISA levels of H). soluble and insoluble human Aβ1–42, and I) ELISA levels of mouse p-Tau in the brain of mice (N=9) injected (IP) with 10mg/kg (3 weeks). * Significantly different to control or as indicated, Mean±SEM, ANOVA with Neumann Keuls multiple comparison, p<0.05.
Tg-APP mice expressed significantly higher levels of soluble (156ng/ml) and insoluble (173ng/ml) Aβ1–42 compared to 1-year old control ± Nilotinib (Fig. 3E, p<0.05, n=9) while Nilotinib reduced soluble Aβ1–42 (35ng/ml, which remained significantly higher than control) and reversed the increase in insoluble Aβ1–42. Significant increases in soluble (281ng/ml) and insoluble (250ng/ml) Aβ1–40 were also detected in Tg-APP mice compared to 1-year old control (Fig. 3F, p<0.05, n=9), and were reversed by Nilotinib. p-Tau was also increased at ser 396 (109ng/ml) and AT8 (288ng/ml) compared to 1-year old control (Fig. 2G, p<0.05, n=9). Nilotinib significantly reduced but did not completely reverse these increases in Tau. The decrease in parkin solubility may reflect loss of function, leading to failure of amyloid clearance, so we determined whether endogenous parkin can mediate clearance in lentiviral Aβ1–42 expressing WT and parkin−/− mice. Significant increases (p<0.05, n=12) in soluble (180ng/ml) and insoluble (209ng/ml) Aβ1–42 were observed in WT lentiviral Aβ1–42 mice (Fig. 3H), and Nilotinib completely reversed Aβ1–42 back to control level. Lentiviral Aβ1–42 in parkin−/− mice (Fig. 3H) increased soluble (241ng/ml) and insoluble (246ng/ml) Aβ1–42 compared to lentiviral Aβ1–42 in WT mice (n=12). Interestingly, Nilotinib failed to clear soluble (297ng/ml) and insoluble (274mg/ml) Aβ1–42 in parkin−/− mice, suggesting that endogenous parkin is required for Aβ1–42 clearance. Similarly, Nilotinib decreased p-Tau ser 396 (Fig. 3I) in WT mice (68ng/ml) compared to Aβ1–42 expression (124ng/ml) while p-Tau was increased (264ng/ml) in parkin−/− mice and Nilotinib did not lower p-Tau (189ng/ml) level (Fig. 3I, p<0.05, n=11). These data are consistent with our previous report showing that TKI, either via Nilotinib or Bosutinib, can increase amyloid clearance [22].
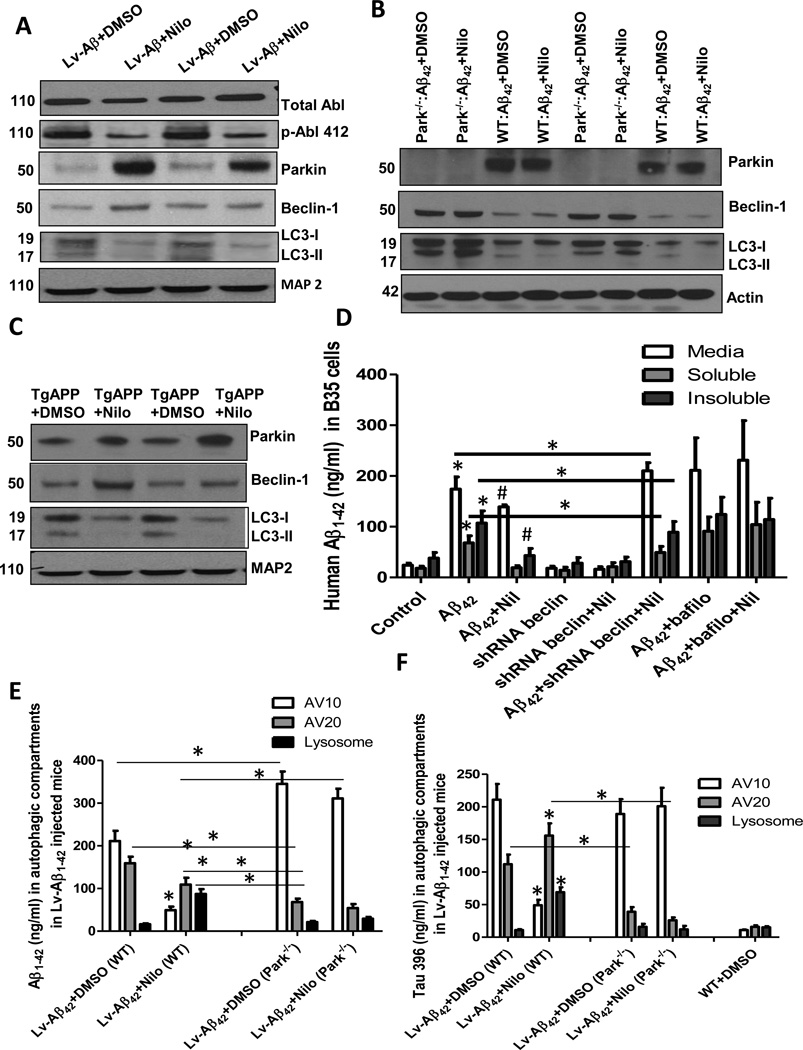
Nilotinib promotes autophagic clearance of amyloid in a parkin-Beclin-1-dependent manner
WB of total brain lysates in 1 year old WT mice injected with lentiviral Aβ1–42 showed a significant decrease in T412 (45%) relative to total Abl following daily treatment with 10mg/kg Nilotinib for 3 weeks (Fig. 4A, p<0.05, n=9). An increase in parkin level (62%) was associated with a similar increase in beclin-1 (53%) relative to MAP-2 (Fig 4A, p<0.05, n=9), consistent with the hypothesis that Abl inhibition may mediate clearance via increased parkin activity. LC3-II and its precursor LC3-I were both increased in lentiviral Aβ1–42 mice (DMSO) relative to MAP-2, but Nilotinib led to LC3-II disappearance and decreased levels of LC3-I relative to MAP-2 (Fig. 4A, 45%, p<0.05, n=9). Interestingly, parkin−/− mice had higher levels of beclin-1 relative to actin (Fig. 4B, 120%, n=9) compared to control but Nilotinib did not clear LC3-II in parkin−/− mice (Fig. 3B), suggesting that despite the increase in Beclin-1, Nilotinib cannot clear autophagosomes in parkin−/− mice. To verify the effects of Nilotinib on parkin level and autophagic markers Beclin-1 and LC3, TgAPP mice were treated I.P. with 10mg/kg Nilotinib once a day for 3 weeks. A significant increase in parkin (Fig. 4C, 41%, p<0.05, n=9), Beclin-1 (45%) and LC3-I (26%) relative to tubulin were observed in Nilotinib treated mice, which showed disappearance of LC3-II (Fig. 4C), suggesting autophagic clearance. To ascertain that autophagy is involved in Nilotinib-mediated amyloid clearance, Beclin-1 expression was blocked with shRNA (ThermoFisher) and autophagy was inhibited with 100nM Bafilomycin for 3hr in rat B35 neuroblastoma cells (Fig. 4D). Beclin-1 shRNA expression showed that Aβ1–42 levels were unaffected in the media with Nilotinib compared to Aβ1–42 expressing cells, and were significantly higher than Aβ1–42+Nilotinib (Fig. 4D). Soluble and insoluble Aβ1–42 were partially (42% and 21%, respectively) decreased compared to Aβ1–42 cells, but remained 2-fold higher compared to Aβ1-42+Nilotinib (Fig. 4D, p<0.05, n=12), indicating that Beclin-1 is required for complete Aβ1–42 clearance. Secreted Aβ1–42 (media) may have accumulated in the first 24hr after transfection, prior to Nilotinib treatment. Furthermore, inhibition of autophagy with Bafilomycin A1, led to Aβ1–42 accumulation and Nilotinib failed to reverse these effects (Fig. 4D, n=12).
Fig. 4. Nilotinib promotes autophagic clearance of amyloid.
WB of brain extracts on 4–12% Nu-Page SDS gels A) in lentiviral Aβ1–42 in WT mice±Nilotinib showing, Abl, p-Abl, LC3, parkin, and beclin-1 relative to MAP-2 (N=9) and B) in Tg-APP±Nilotinib showing, parkin, beclin-1 and LC3 relative to tubulin. C). WB of brain extracts on 4–12% Nu-Page SDS gels in lentiviral Aβ1–42 in WT and parkin−/− mice±Nilotinib showing parkin, LC3, and beclin-1 relative to actin (N=9). D) Human Aβ1–42 ELISA before and after Nilotinib treatment in B35 rat neuroblastoma cells (N=12) in media, soluble and insolublelysates in the presence and absence of shRNA beclin-1. E) ELISA level of human Aβ1–42 and F) mouse p-Tau in autophagic vacuoles isolated from lentiviral Aβ1–42 expressing WT and parkin−/− mice (N=5).* Significantly different or as indicated, # significantly different to Aβ1–42 expressing WT, Mean±SEM, ANOVA with Neumann Keuls multiple comparison, p<0.05.
To further determine whether autophagy mediates clearance of amyloid proteins in vivo, AVs were isolated [13] and amyloid was measured via ELISA. Aβ1–42 was detected in AV10 and AV20, but not lysosomes, in lentiviral Aβ1–42 WT brain (Fig. 4E, n=5) but Nilotinib decreased Aβ1–42 in AV10 (4.3 fold, p<0.05, n=5) and AV20 (1.45 fold) and increased (5.5 fold) in lysosomes. Lentiviral Aβ1–42 expression in parkin−/− mice increased Aβ1–42 in AV10 (0.6 fold) compared to WT but decreased it in AV20 compared to WT treated with DMSO (4.1 fold) or Nilotinib (2.3 fold), and no Aβ1–42 was detected in lysosomes (Fig. 4E). Nilotinib did not decrease Aβ1–42 levels in AV10, which remained higher than WT (6.3 fold) and Aβ1-42 did not change in all AVs compared to Aβ1–42 parkin−/− mice (DMSO), suggesting lack of autophagic Aβ1-42 degradation in parkin−/− mice. Moreover, mouse p-Tau was detected in AV10 and AV20, but not lysosomes, in lentiviral Aβ1–42 expressing WT brain (Fig. 4F, n=5) but Nilotinib decreased p-Tau levels in AV10 (4.3 fold, p<0.05, N=5) and increased it in AV20 (1.39 fold) and lysosomes (6.27 fold). Similar levels of p-Tau were detected in AV10 in parkin−/− compared to WT mice but p-Tau was decreased in AV20 compared to WT (2.8 fold), and no p-Tau was detected in lysosomes (Fig. 4F, n=5). Nilotinib did not decrease p-Tau in AV10 and p-Tau was decreased in AV20 of Aβ1–42 parkin−/− (6 fold) compared to Aβ1–42 WT mice (Fig. 4F, p<0.05).
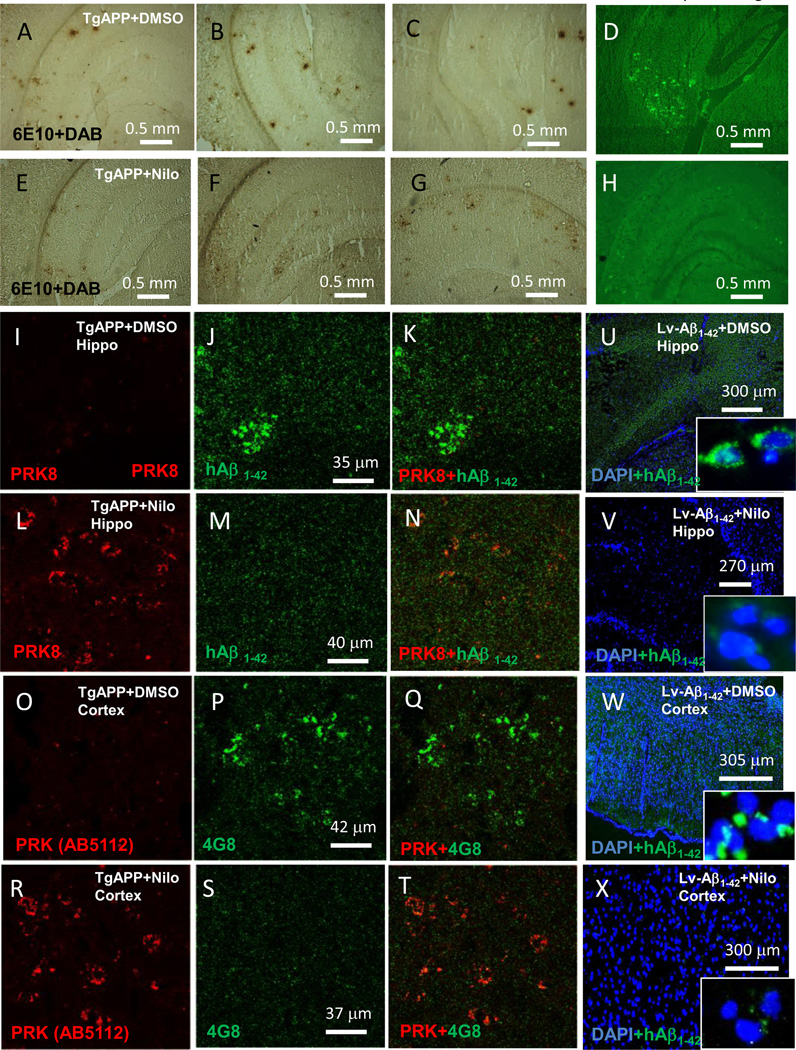
Nilotinib increases parkin level and decreases plaque load in Tg-APP mice
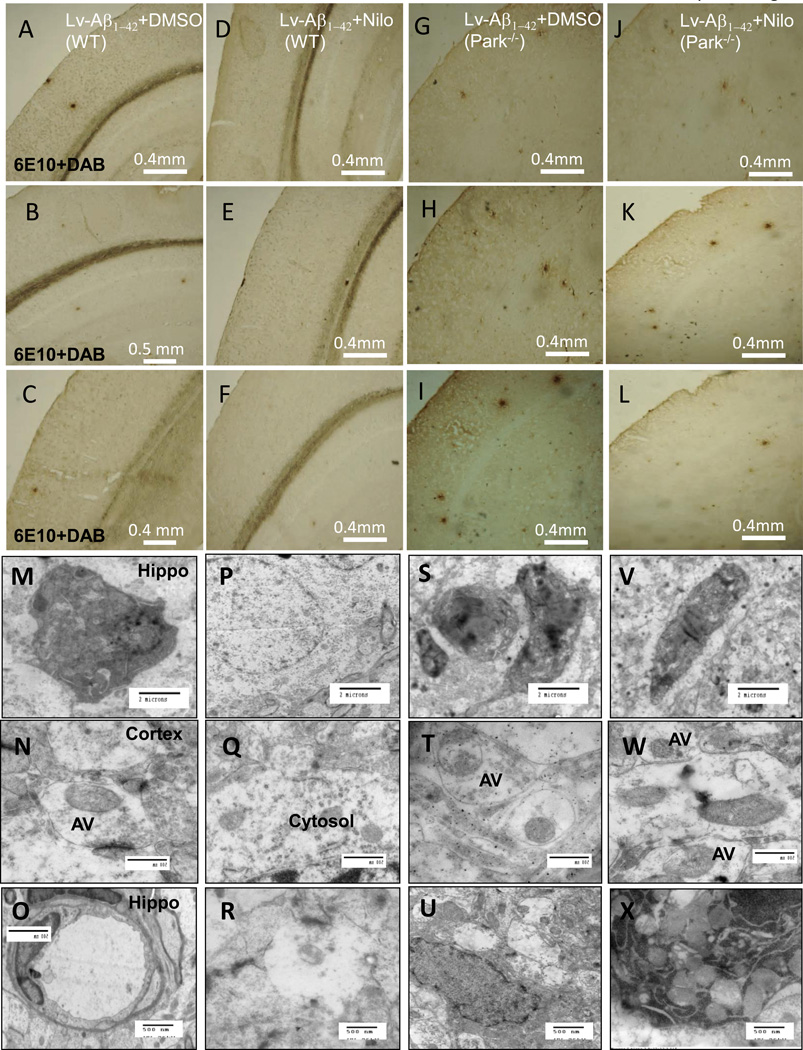
Staining of 20μm brain sections shows plaque formation in Tg-APP mice treated with DMSO (Fig. 5A-C representing different animals), though plaque staining disappeared in the Nilotinib group after 3-week treatment (Fig. 5E-G). Thioflavin-S staining was also decreased in Nilotinib treated Tg-APP mice (Fig. 5H) compared to DMSO (Fig. 5D). Higher magnification shows endogenous parkin (Fig. 5I) and plaque deposition (Fig. 5I &K) in Tg-APP mice hippocampus. Nilotinib increases endogenous parkin (Fig. 5L), resulting in plaque disappearance (Fig. 5M&N). Using different parkin antibodies to show parkin (Fig. 5O) and plaque (Fig. 5P&Q), Nilotinib increased parkin levels (Fig. 5R) and dissolved plaques (Fig. 5S&T). To determine whether parkin targets intracellular Aβ [24] to decrease extracellular plaque load, lentiviral injection was used to express intracellular Aβ1–42 within the hippocampus (Fig. 5U, inset higher magnification) and Nilotinib clearance of intracellular Aβ1–42 (Fig. 5V, inset is higher magnification). Lentiviral injection into the hippocampus led to intracellular Aβ1–42 expression throughout the cortex (Fig. 5W, inset higher magnification) and, again, Nilotinib eliminated Aβ1–42 accumulation (Fig. 5X, inset higher magnification). Lower magnification images show plaques in Aβ1–42 expressing mice 6 weeks post-injection (Fig. 6A-C). Nilotinib (daily for 3 weeks) eliminates plaques in Aβ1–42 WT mice (Fig. 6D-F). Aβ1–42 expression in parkin−/− mice showed more plaques (Fig. 6G-I) and Nilotinib did not reduce plaques (Fig. 6J-L).
Fig. 5. Nilotinib increases parkin level and decreases plaque load.
Staining of 20μm brain sections shows plaque formation within various brain regions in different A-C) Tg-APP+DMSO and E-G) Nilotinib group after 3-week treatment. Staining of 20μm thick brain sections shows D) thioflavin-S staining in Tg-APP+DMSO and Tg-APP+Nilotinib. Staining of 20μm thick brain sections shows I). parkin, J). Aβ1–42 and K) merged figure in hippocampus of Tg-APP mice after 3 weeks of DMSO treatment, and L). parkin, M). Aβ1–42 and N) merged figure in hippocampus of Tg-APP mice after 3 weeks of Nilotinib treatment. O). parkin, P). Aβ1–42 and Q) merged figure in cortex of Tg-APP mice after 3 weeks of DMSO treatment, and R). parkin, S). Aβ1–42 and T) merged figure in cortex of Tg-APP mice after 3 weeks of Nilotinib treatment. Intracellular Aβ1–42 within the U). hippocampus of lentiviral Aβ1–42 injected mice, inset higher magnification, and V) and Nilotinib clearance of intracellular Aβ1–42 (inset is higher magnification). Staining of 20μm brain sections shows intracellular Aβ1–42 within the W). cortex of lentiviral Aβ1–42 injected mice, inset higher magnification, and X) and Nilotinib clearance of intracellular Aβ1–42, inset is higher magnification.
Fig. 6. Nilotinib eliminates plaques in lentiviral Aβ1–42 injected WT but not parkin−/− mice.
Staining of 20μm brain sections shows plaque formation within various brain regions in different A-C) lentiviral Aβ1–42+DMSO WT mice and D-F) Nilotinib group after 3-week treatment. G-I) lentiviral Aβ1-42+DMSO in parkin−/− mice and J-L) Nilotinib group after 3-week treatment. Transmission EM shows autophagic defects in different lentiviral Aβ1–42+DMSO WT brains within M). hippocampus showing distrophic neurons, N) cortex showing accumulation of autophagic vacuoles, O). hippocampus showing enlarged lysosomes. And lentiviral Aβ1–42+Nilotinib WT brains within P). hippocampus, Q) cortex showing clearance of autophagic vacuoles, R). hippocampus. And lentiviral Aβ1–42±Nilotinib in parkin−/− brains within S&V). hippocampus showing distrophic neurons, T&W) cortex showing accumulation of autophagic vacuoles, U&X). hippocampus showing accumulation of autophagic vacuoles.
Transmission electron microscopy revealed (n=6 animals per treatment) autophagic defects in lentiviral Aβ1–42 expressing mice [13] hippocampal detection of dystrophic neurons (Fig. 6M), accumulation of undigested vacuoles in the cortex (Fig. 6N) and possible enlargement of hippocampal lysosomes (Fig. 6O), suggesting deficits in proteolytic degradation. Nilotinib reversed these effects in the hippocampus (Fig. 6P&R), where no such structures or defects were detected, and contributed to cortical clearance of vacuoles (Fig. 6Q). In contrast, Nilotinib failed to eliminate dystrophic neurons in the hippocampus of parkin−/− mice (Fig. 6S&V), and was unable to clear vacuoles and accumulating debris in cortex and hippocampus (Fig. 6T-X).
Discussion
These studies show that Nilotinib increases parkin levels and ubiquitination, leading to parkin-Beclin-1 interaction [23], and amelioration of amyloid pathology. We previously showed that parkin accumulates with intraneuronal Aβ1–42 in post-mortem AD [13] brains, suggesting that aging may alter parkin stability. Our results are in agreement with previous data showing decreased parkin solubility in Tg-APP mice [22], while Nilotinib increases parkin ubiquitination. WT and not mutant loss-of-function parkin increases proteasome activity [25, 30, 31], leading to degradation of ubiquitinated proteins, while amyloid stress alters parkin solubility [13]. Previous reports suggested that parkin insolubility is associated with alteration of its activity via increased phosphorylation by Abl [12, 32]. Although our results support the hypothesis that Abl activation leads to decreased parkin activity, we were unable to demonstrate that parkin is an Abl substrate. However, our data suggest that amyloid stress triggers autophagosome formation (LC3-II levels) in AD models, and Nilotinib-mediated endogenous parkin ubiquitination can lead to efficient proteasomal re-cycling and stability, and subsequent interaction with Beclin-1 to mediate autophagosome clearance [22]. Parkin activation via self-ubiquitination is consistent with parkin structure, which contains 9 potential ubiquitination sites on Lys residues [33], and its function as a hybrid RING (really interesting new gene) and HECT (homologous to the E6AP carboxyl terminus) enzyme [33–38], suggesting that ubiquitination may lead to parkin activation. Under normal physiological conditions, parkin is inactive [33, 39–41], but cellular stress, including amyloid and autophagosome accumulation may lead to its activation. We previously demonstrated that parkin is auto-ubiquitinated via both Lys48- and Lys63-linked ubiquitin chain, leading to ubiquitination of its substrate [26]. Exogenous parkin mediates autophagic clearance in Aβ−expressing animals [13]. Nilotinib-induced Aβ elimination was more efficient than p-Tau clearance, suggesting that p-Tau may affect autophagosome fusion with the lysosomes. The decrease in p-Tau levels in Nilotinib treated mice suggests clearance of free or unbound p-Tau, while the microtubule associated protein Tau remains intact. This is a novel mechanism involving Nilotinib-enhanced amyloid degradation to halt progression of AD-like pathology. Since Nilotinib is FDA approved, it could be used to test the validity of disease CSF and plasma biomarkers such as p-Tau and β-amyloid.
Although no parkin mutations are associated with AD, manipulation of parkin activity may serve as a disease modifying therapy that would provide an alternative approach to prevent progression from MCI to AD. Gene wide association studies (GWAS) do not associated Tau with AD, despite the fact that Tau pathology positively correlates with disease progression. Thus, loss of parkin stability, which results in lack of misfolded protein clearance, may have similar effects to Tau modification, which is associated with disease progression, independent of disease-causing mutations or genetic association with AD. Loss of parkin decreases amyloid toxicity in AD models [42, 43], while parkin deletion leads to more secretion and build-up of extracellular plaques, further underscoring the importance of intracellular Aβ in AD pathology. Parkin activation is associated with both K48 and K63-linked self-ubiquitination [26], indicating that ubiquitination may activate parkin to recruit its substrates. Parkin-mediated K63-linked poly-ubiquitination leads to autophagic clearance [44]. Decreased parkin solubility in association with autophagic or proteasomal defects suggest that aggregated proteins may overwhelm cellular quality control systems, leading to inclusion formation. Therefore, boosting parkin activation may increase autophagy and subsequent protein clearance via parkin-Beclin-1 interaction [22]. Although no dementia is reported with parkin mutations in early onset juvenile PD, loss of parkin function does not lead to cytosolic Lewy body inclusions [45], suggesting that functional parkin mediates cytosolic sequestration of misfolded proteins. In AD, Beclin-1 levels are altered [22, 46], suggesting inefficient execution of Beclin-1-dependent autophagy, leading to Aβ and p-Tau accumulation.
These studies provide novel insights into Nilotinib effects on parkin ubiquitination and activity, leading to amyloid clearance; and show the effects of an FDA-approved drug as a potential therapy for AD. Ubiquitination may lead to efficient re-cycling of parkin via the proteasome, leading to decreased parkin insolubility in human aging and AD models. Loss of parkin stability may also result in inefficiency of autophagic clearance due to decreased interaction with Beclin-1. These studies have significant translational and mechanistic impact on the AD field.
Key messages.
Parkin solubility (stability) is decreased in AD and APP transgenic mice.
Nilotinib-induced autophagic changes increase endogenous parkin level.
Increased parkin level leads to ubiquitination and proteasomal recycling.
Recycling decreases insoluble parkin and increases parkin-Beclin-1 interaction.
Beclin-1-parkin interaction enhances amyloid clearance.
Acknowledgments
Acknowledgements and disclosure statements: These studies were supported by NIH grant NIA 30378, Georgetown University funding and Merck & Co funds to Charbel E-H Moussa. The authors would like to thank Dr. Jim Driver from the University of Montana for his support in the EM studies.
Footnotes
Author Contributions:
Dr. Irina Lonskaya and Michaeline Hebron performed WB, IHC and ELISA and cell culture experiments. Dr. Joel Schachter provided analytic and intellectual input and proofread the manuscript; Dr. Charbel Moussa designed, oversaw the studies, performed gene transfer surgeries, analyzed the data and wrote the manuscript.
The authors have read the manuscript and declare no conflict of interest whatsoever.
Dr. Charbel Moussa has a pending application to use nilotinib and bosutinib as a treatment for neurodegenerative diseases. The PCT application number PCT/US13/039283 was filed on May 2, 2013 and claims priority to two provisional patent applications filed on May 2, 2012 and March 1, 2013. The title is “treating neural diseases with tyrosine kinase inhibitors”.
References
- 1.Cook DG, Forman MS, Sung JC, Leight S, Kolson DL, Iwatsubo T, Lee VM, Doms RW. Alzheimer's A beta(1–42) is generated in the endoplasmic reticulum/intermediate compartment of NT2N cells. Nat Med. 1997;3:1021–1023. doi: 10.1038/nm0997-1021. [DOI] [PubMed] [Google Scholar]
- 2.Greenfield JP, Tsai J, Gouras GK, Hai B, Thinakaran G, Checler F, Sisodia SS, Greengard P, Xu H. Endoplasmic reticulum and trans-Golgi network generate distinct populations of Alzheimer beta-amyloid peptides. Proc Natl Acad Sci U S A. 1999;96:742–747. doi: 10.1073/pnas.96.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skovronsky DM, Doms RW, Lee VM. Detection of a novel intraneuronal pool of insoluble amyloid beta protein that accumulates with time in culture. J Cell Biol. 1998;141:1031–1039. doi: 10.1083/jcb.141.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu H, Sweeney D, Wang R, Thinakaran G, Lo AC, Sisodia SS, Greengard P, Gandy S. Generation of Alzheimer beta-amyloid protein in the trans-Golgi network in the apparent absence of vesicle formation. Proc Natl Acad Sci U S A. 1997;94:3748–3752. doi: 10.1073/pnas.94.8.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang JY. Regulation of cell death by the Abl tyrosine kinase. Oncogene. 2000;19:5643–5650. doi: 10.1038/sj.onc.1203878. DOI 10.1038/sj.onc.1203878. [DOI] [PubMed] [Google Scholar]
- 6.Tremblay MA, Acker CM, Davies P. Tau phosphorylated at tyrosine 394 is found in Alzheimer's disease tangles and can be a product of the Abl-related kinase, Arg. J Alzheimers Dis. 2010;19:721–733. doi: 10.3233/JAD-2010-1271. DOI Q7H46J374V276467 [pii] 10.3233/JAD-2010-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlatterer SD, Acker CM, Davies P. c-Abl in Neurodegenerative Disease. J Mol Neurosci. 2011 doi: 10.1007/s12031-011-9588-1. DOI 10.1007/s12031-011-9588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derkinderen P, Scales TM, Hanger DP, Leung KY, Byers HL, Ward MA, Lenz C, Price C, Bird IN, Perera T, Kellie S, Williamson R, Noble W, Van Etten RA, Leroy K, Brion JP, Reynolds CH, Anderton BH. Tyrosine 394 is phosphorylated in Alzheimer's paired helical filament tau and in fetal tau with c-Abl as the candidate tyrosine kinase. J Neurosci. 2005;25:6584–6593. doi: 10.1523/JNEUROSCI.1487-05.2005. DOI 25/28/6584 [pii] 10.1523/JNEUROSCI.1487-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez AR, Sandoval PC, Leal NR, Castro PU, Kosik KS. Activation of the neuronal c-Abl tyrosine kinase by amyloid-beta-peptide and reactive oxygen species. Neurobiol Dis. 2004;17:326–336. doi: 10.1016/j.nbd.2004.06.007. DOI S0969-9961(04)00135-4 [pii] 10.1016/j.nbd.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Cancino GI, Toledo EM, Leal NR, Hernandez DE, Yevenes LF, Inestrosa NC, Alvarez AR. STI571 prevents apoptosis, tau phosphorylation and behavioural impairments induced by Alzheimer's beta-amyloid deposits. Brain. 2008;131:2425–2442. doi: 10.1093/brain/awn125. DOI awn125 [pii] 10.1093/brain/awn125. [DOI] [PubMed] [Google Scholar]
- 11.Hebron ML, Lonskaya I, Moussa CE. Nilotinib reverses loss of dopamine neurons and improves motor behavior via autophagic degradation of alpha-synuclein in Parkinson's disease models. Hum Mol Genet. 2013;22:3315–3328. doi: 10.1093/hmg/ddt192. DOI 10.1093/hmg/ddt192 ddt192 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imam SZ, Zhou Q, Yamamoto A, Valente AJ, Ali SF, Bains M, Roberts JL, Kahle PJ, Clark RA, Li S. Novel regulation of parkin function through c-Abl-mediated tyrosine phosphorylation: implications for Parkinson's disease. J Neurosci. 2011;31:157–163. doi: 10.1523/JNEUROSCI.1833-10.2011. DOI 31/1/157 [pii] 10.1523/JNEUROSCI.1833-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lonskaya I, Shekoyan AR, Hebron ML, Desforges N, Algarzae NK, Moussa CE. Diminished Parkin Solubility and Co-Localization with Intraneuronal Amyloid-beta are Associated with Autophagic Defects in Alzheimer's Disease. J Alzheimers Dis. 2012 doi: 10.3233/JAD-2012-121141. DOI V57263JK82055089 [pii] 10.3233/JAD-2012-121141. [DOI] [PubMed] [Google Scholar]
- 14.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkinmediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. DOI ncb2012 [pii] 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 15.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. DOI jcb.200809125 [pii] 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park J, Kim Y, Chung J. Mitochondrial dysfunction and Parkinson's disease genes: insights from Drosophila. Dis Model Mech. 2009;2:336–340. doi: 10.1242/dmm.003178. DOI 2/7-8/336 [pii] 10.1242/dmm.003178. [DOI] [PubMed] [Google Scholar]
- 17.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrane J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. DOI 0911187107 [pii] 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khandelwal PJ, Herman AM, Hoe HS, Rebeck GW, Moussa CE. Parkin mediates beclin-dependent autophagic clearance of defective mitochondria and ubiquitinated Abeta in AD models. Hum Mol Genet. 2011;20:2091–2102. doi: 10.1093/hmg/ddr091. DOI ddr091 [pii] 10.1093/hmg/ddr091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deremer DL, Ustun C, Natarajan K. Nilotinib: a second-generation tyrosine kinase inhibitor for the treatment of chronic myelogenous leukemia. Clin Ther. 2008;30:1956–1975. doi: 10.1016/j.clinthera.2008.11.014. DOI S0149-2918(08)00408-6 [pii] 10.1016/j.clinthera.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Skorski T. BCR-ABL1 kinase: hunting an elusive target with new weapons. Chem Biol. 2011;18:1352–1353. doi: 10.1016/j.chembiol.2011.11.001. DOI S1074-5521(11)00401-7 [pii] 10.1016/j.chembiol.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahon FX, Hayette S, Lagarde V, Belloc F, Turcq B, Nicolini F, Belanger C, Manley PW, Leroy C, Etienne G, Roche S, Pasquet JM. Evidence that resistance to nilotinib may be due to BCR-ABL, Pgp, or Src kinase overexpression. Cancer Res. 2008;68:9809–9816. doi: 10.1158/0008-5472.CAN-08-1008. DOI 68/23/9809 [pii] 10.1158/0008-5472.CAN-08-1008. [DOI] [PubMed] [Google Scholar]
- 22.Lonskaya I, Hebron ML, Desforges NM, Franjie A, Moussa CE. Tyrosine kinase inhibition increases functional parkin-Beclin-1 interaction and enhances amyloid clearance and cognitive performance. EMBO Mol Med. 2013 doi: 10.1002/emmm.201302771. DOI 10.1002/emmm.201302771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lonskaya I, Hebron ML, Algarzae NK, Desforges N, Moussa CE. Decreased parkin solubility is associated with impairment of autophagy in the nigrostriatum of sporadic Parkinson's disease. Neuroscience. 2012 doi: 10.1016/j.neuroscience.2012.12.018. DOI S0306-4522(12)01199-2 [pii] 10.1016/j.neuroscience.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rebeck GW, Hoe HS, Moussa CE. Beta-amyloid1–42 gene transfer model exhibits intraneuronal amyloid, gliosis, tau phosphorylation, and neuronal loss. J Biol Chem. 2010;285:7440–7446. doi: 10.1074/jbc.M109.083915. DOI M109.083915 [pii] 10.1074/jbc.M109.083915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burns MP, Zhang L, Rebeck GW, Querfurth HW, Moussa CE. Parkin promotes intracellular Abeta1–42 clearance. Hum Mol Genet. 2009;18:3206–3216. doi: 10.1093/hmg/ddp258. DOI ddp258 [pii] 10.1093/hmg/ddp258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hebron ML, Lonskaya I, Sharpe K, Weerasinghe PP, Algarzae NK, Shekoyan AR, Moussa CE. Parkin Ubiquitinates Tar-DNA Binding Protein-43 (TDP-43) and Promotes Its Cytosolic Accumulation via Interaction with Histone Deacetylase 6 (HDAC6) J Biol Chem. 2013;288:4103–4115. doi: 10.1074/jbc.M112.419945. DOI 10.1074/jbc.M112.419945 M112.419945 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, Meloni EG, Wu N, Ackerson LC, Klapstein GJ, Gajendiran M, Roth BL, Chesselet MF, Maidment NT, Levine MS, Shen J. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. DOI 10.1074/jbc.M308947200 M308947200 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Marzella L, Ahlberg J, Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol. 1982;93:144–154. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis J, Xu F, Deane R, Romanov G, Previti ML, Zeigler K, Zlokovic BV, Van Nostrand WE. Early-onset and robust cerebral microvascular accumulation of amyloid beta-protein in transgenic mice expressing low levels of a vasculotropic Dutch/Iowa mutant form of amyloid beta-protein precursor. J Biol Chem. 2004;279:20296–20306. doi: 10.1074/jbc.M312946200. DOI 10.1074/jbc.M312946200 M312946200 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Khandelwal PJ, Dumanis SB, Feng LR, Maguire-Zeiss K, Rebeck G, Lashuel HA, Moussa CE. Parkinsonrelated parkin reduces alpha-Synuclein phosphorylation in a gene transfer model. Mol Neurodegener. 2010;5:47. doi: 10.1186/1750-1326-5-47. DOI 1750-1326-5-47 [pii] 10.1186/1750-1326-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosen KM, Moussa CE, Lee HK, Kumar P, Kitada T, Qin G, Fu Q, Querfurth HW. Parkin reverses intracellular beta-amyloid accumulation and its negative effects on proteasome function. J Neurosci Res. 2010;88:167–178. doi: 10.1002/jnr.22178. DOI 10.1002/jnr.22178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ko HS, Lee Y, Shin JH, Karuppagounder SS, Gadad BS, Koleske AJ, Pletnikova O, Troncoso JC, Dawson VL, Dawson TM. Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin's ubiquitination and protective function. Proc Natl Acad Sci U S A. 2010;107:16691–16696. doi: 10.1073/pnas.1006083107. DOI 1006083107 [pii] 10.1073/pnas.1006083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, Shaler T, Walker D, Yang Y, Regnstrom K, Diep L, Zhang Z, Chiou S, Bova M, Artis DR, Yao N, Baker J, Yednock T, Johnston JA. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4:1982. doi: 10.1038/ncomms2982. DOI 10.1038/ncomms2982 ncomms2982 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S, Youle RJ. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol. 2013;200:163–172. doi: 10.1083/jcb.201210111. DOI 10.1083/jcb.201210111 jcb.201210111 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. DOI 10.1038/nature09966 nature09966 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iguchi M, Kujuro Y, Okatsu K, Koyano F, Kosako H, Kimura M, Suzuki N, Uchiyama S, Tanaka K, Matsuda N. Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. J Biol Chem. 2013;288:22019–22032. doi: 10.1074/jbc.M113.467530. DOI 10.1074/jbc.M113.467530 M113.467530 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spratt DE, Martinez-Torres RJ, Noh YJ, Mercier P, Manczyk N, Barber KR, Aguirre JD, Burchell L, Purkiss A, Walden H, Shaw GS. A molecular explanation for the recessive nature of parkin-linked Parkinson's disease. Nat Commun. 2013;4:1983. doi: 10.1038/ncomms2983. DOI 10.1038/ncomms2983 ncomms2983 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng X, Hunter T. Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res. 2013;23:886–897. doi: 10.1038/cr.2013.66. DOI 10.1038/cr.2013.66 cr201366 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, Kimura M, Komatsu M, Hattori N, Tanaka K. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. DOI 10.1083/jcb.200910140 jcb.200910140 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32(15):2099–2112. doi: 10.1038/emboj.2013.125. DOI 10.1038/emboj.2013.125 emboj2013125 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trempe JF, Sauve V, Grenier K, Seirafi M, Tang MY, Menade M, Al-Abdul-Wahid S, Krett J, Wong K, Kozlov G, Nagar B, Fon EA, Gehring K. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. DOI 10.1126/science.1237908 science.1237908 [pii] [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Navarro JA, Gomez A, Rodal I, Perucho J, Martinez A, Furio V, Ampuero I, Casarejos MJ, Solano RM, de Yebenes JG, Mena MA. Parkin deletion causes cerebral and systemic amyloidosis in human mutated tau over-expressing mice. Hum Mol Genet. 2008;17:3128–3143. doi: 10.1093/hmg/ddn210. DOI ddn210 [pii] 10.1093/hmg/ddn210. [DOI] [PubMed] [Google Scholar]
- 43.Perucho J, Casarejos MJ, Rubio I, Rodriguez-Navarro JA, Gomez A, Ampuero I, Rodal I, Solano RM, Carro E, Garcia de Yebenes J, Mena MA. The effects of parkin suppression on the behaviour, amyloid processing, and cell survival in APP mutant transgenic mice. Exp Neurol. 2010;221:54–67. doi: 10.1016/j.expneurol.2009.09.029. DOI S0014-4886(09)00416-6 [pii] 10.1016/j.expneurol.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 44.Olzmann JA, Chin LS. Parkin-mediated K63-linked polyubiquitination: a signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy. 2008;4:85–87. doi: 10.4161/auto.5172. DOI 5172 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuno Y, Hattori N, Mori H, Suzuki T, Tanaka K. Parkin and Parkinson's disease. Curr Opin Neurol. 2001;14:477–482. doi: 10.1097/00019052-200108000-00008. [DOI] [PubMed] [Google Scholar]
- 46.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. DOI 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]