Significance
In the interior of a cell, the volume accessible to each protein molecule is restricted by the presence of the large number of other macromolecules. Such a crowded environment is known to affect the stability and folding rates of proteins. In the case of intrinsically disordered proteins (IDPs), however, a class of proteins that lack stable structure, much less is known about the role of crowding effects. We have quantified the conformational changes occurring in IDPs in the presence of high concentrations of different polymers that act as crowding agents. Using single-molecule spectroscopy, we have identified effects that are typical of polymer solutions and have direct implications for the behavior of IDPs within the cell.
Keywords: single-molecule FRET, unfolded state collapse, excluded volume screening, Flory–Huggins theory
Abstract
Intrinsically disordered proteins (IDPs) are involved in a wide range of regulatory processes in the cell. Owing to their flexibility, their conformations are expected to be particularly sensitive to the crowded cellular environment. Here we use single-molecule Förster resonance energy transfer to quantify the effect of crowding as mimicked by commonly used biocompatible polymers. We observe a compaction of IDPs not only with increasing concentration, but also with increasing size of the crowding agents, at variance with the predictions from scaled-particle theory, the prevalent paradigm in the field. However, the observed behavior can be explained quantitatively if the polymeric nature of both the IDPs and the crowding molecules is taken into account explicitly. Our results suggest that excluded volume interactions between overlapping biopolymers and the resulting criticality of the system can be essential contributions to the physics governing the crowded cellular milieu.
A surprisingly large number of eukaryotic proteins either contain substantial unstructured regions or are entirely unfolded under physiological conditions (1, 2). These “intrinsically disordered proteins” (IDPs) are involved in many crucial cellular processes, such as transcription, translation, and signal transduction; their functional and conformational properties are thus of great interest for a wide range of biological questions. Important advances in understanding the structures of IDPs have been made over the past decade, especially with spectroscopic techniques, e.g., NMR (3, 4), single-molecule fluorescence (5–7), and with atomistic and coarse-grained molecular simulations (8–10). In contrast with the stable folded structures we are familiar with from 50 y of structural biology, IDPs comprise highly heterogeneous and dynamic ensembles of conformations, which either lack stable tertiary structure altogether or fold only on binding their cellular targets (4). Important components of the cellular environment that affect IDPs include not only specific cellular ligands, but also pH and the concentration of salts (11, 12). An additional contribution that has been difficult to investigate experimentally comes from the large number of different solutes present in a cell that do not interact with an IDP specifically, but result in an environment that is densely filled with macromolecules and metabolites (12–14). Given their lack of persistent structure, the conformations of IDPs are expected to be particularly sensitive to the effects of such molecular crowding. Indeed, first experiments indicate that some IDPs gain structure upon crowding (15), whereas others do not (16–18), but may change their dimensions (19–21). The question of how the conformational distributions of IDPs respond to crowded environments is of particular current interest because IDPs have a vital role in cellular compartments and regions with very high local concentrations of proteins and RNA, such as RNA granules and nuclear pore complexes (22–25). However, a quantitative comprehension of how the concentrations and sizes of the molecular crowding agents (or “crowders”) affect IDPs is currently incomplete (26), especially for polymeric crowders. Here we use single-molecule spectroscopy to investigate the influence of crowding on the conformational distributions of IDPs, as a step toward a quantitative framework of how the polydisperse cellular environment affects these highly flexible molecules.
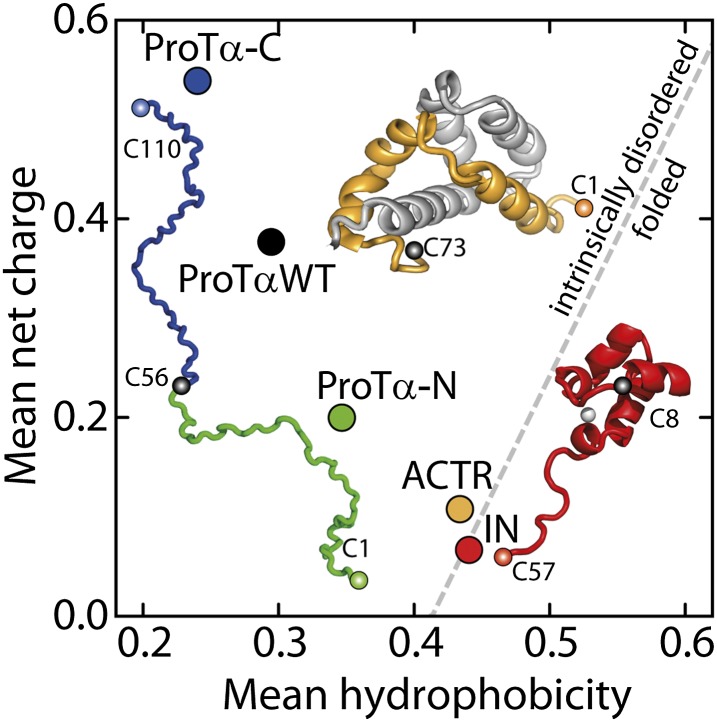
Single-molecule fluorescence detection in combination with Förster resonance energy transfer (FRET) is a method highly suited for addressing this question (5–7, 11, 27, 28) because it allows the heterogeneous structural ensemble of suitably labeled IDPs to be probed even in the presence of very large concentrations of unlabeled solutes. To investigate the physical principles underlying the crowding effects on IDPs, we study a selection of IDPs representative of the naturally occurring sequence compositions in combination with a broad range of molecular sizes of crowding agents. We primarily use polyethylene glycol (PEG) as a crowding agent. This uncharged polymer with high solubility in aqueous solution (29) (SI Appendix, Fig. S1) is available from monomeric ethylene glycol to degrees of polymerization of almost 1,000 (SI Appendix, Table S1) at sufficient purity for single-molecule experiments up to physiologically realistic volume fractions of crowder of ∼40% (30). PEG is widely used for biomedical applications (31) and for mimicking inert crowding agents (13, 26). Previous work has shown that the conformational properties of IDPs strongly depend on their amino acid sequence composition and charge patterning (8, 11, 27, 28, 32–34). Here we investigate the effect of crowding on four different IDP sequences that span a broad range of net charge per residue and average hydrophobicity (Fig. 1 and SI Appendix, Table S2): the N- and C-terminal segments of human prothymosin-α (ProTα-N and -C), the binding domain of the activator for thyroid hormones and retinoid receptors (ACTR), and the N-terminal domain of HIV-1 integrase (IN). Whereas ProTα is highly charged and does not assume a folded structure under any known conditions, ACTR and IN are representatives of the classes of IDPs that fold upon binding a protein or a small ligand, respectively.
Fig. 1.
Mean net charge versus mean hydrophobicity per residue for the four disordered protein sequences used in this study: the C- and N-terminal segments of prothymosin α, ProTα-C (blue) and ProTα-N (green), respectively (complete sequence: black), the activator for thyroid hormones and retinoid receptors, ACTR (orange), and the N-terminal domain of the HIV-1 integrase, IN (red). Folded structures refer to the conformations of ACTR and IN in presence of their ligands, NCBD (gray structure) and Zn2+ (light gray sphere). The FRET labeling sites (SI Appendix, Table S1) are indicated by colored spheres. The dashed gray line indicates the boundary between intrinsically disordered and folded proteins proposed by Uversky (32). Note that the contributions to the net charge from the fluorescent dyes are included (11).
Results
Quantifying Crowder-Induced Chain Compaction with Single-Molecule FRET.
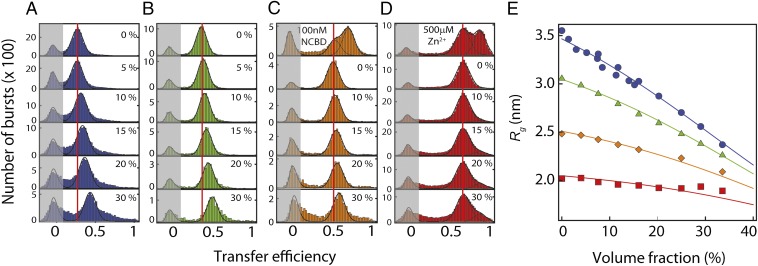
To probe the intramolecular distance distributions of the IDPs, we attached Alexa Fluor 488 and Alexa Fluor 594 as donor and acceptor fluorophores via cysteine residues introduced at suitable positions, with sequence separations of 55 (ProTα-N), 54 (ProTα-C), 72 (ACTR), and 49 residues (IN) (SI Appendix, Table S1). Fig. 2 shows examples of confocal single-molecule FRET experiments with the four different IDP sequences performed at increasing concentrations of PEG 6000 (i.e., PEG with a molecular mass of ∼6,000 Da; SI Appendix, Table S2). Up to three peaks are observed in the transfer efficiency (E) histograms from measurements of labeled IDPs freely diffusing in solution. The peak at E ∼ 0 results from molecules lacking an active acceptor dye and is not of interest here. The second peak at intermediate E corresponds to the disordered state. The appearance of a third peak at E ∼ 0.7 and E ∼ 0.9 for ACTR and IN, respectively, results from the formation of a folded structure in complex with their ligands, the nuclear coactivator binding domain (NCBD) and a Zn2+ ion, respectively (SI Appendix). This separation of subpopulations is essential for distinguishing the effects of solutes on the conformational distributions within the disordered state from a cooperative transition to a folded state. In the case of IN, our experiments indicate the formation of a small folded population at high PEG concentrations even in the absence of Zn2+ (Fig. 2D and SI Appendix, Fig. S2), but for all other proteins, only an unfolded population is present (Fig. 2 and SI Appendix, Fig. S2). However, with increasing concentration of PEG 6000, three of the four disordered sequences (ProTα-C, ProTα-N, and ACTR) exhibit a clear shift of the peak corresponding to the disordered state toward higher transfer efficiencies, indicating an overall tendency of these proteins to collapse in the presence of crowding agents. For the case of IN, which has the least charged and most hydrophobic sequence (Fig. 1), only very small changes in transfer efficiency are noticeable, clearly demonstrating that molecular crowding does not affect all IDPs equally. Given the importance of intramolecular electrostatic repulsion for their conformations (11, 33), it may seem surprising that the more highly charged IDPs exhibit a more pronounced collapse.
Fig. 2.
Single-molecule FRET can be used for quantifying the compaction of disordered proteins by molecular crowding. Representative FRET efficiency histograms at different volume fractions of polyethylene glycol (PEG) 6000 for ProTα-C (A, blue), ProTα-N (B, green), ACTR (C, orange), and IN (D, red). Histograms of ACTR and IN in the presence of their respective interaction partners, NCBD (C) and Zn2+ (D), are shown for comparison. Gaussian and lognormal distributions are used to fit the data (solid lines). The transfer efficiency peaks from molecules lacking an active acceptor dye are shaded in gray. At the highest volume fractions of PEG, some broadening of the peaks is observed due to the increasing fluorescent background. Only IN exhibits a small crowder-induced population at the transfer efficiency of the folded state (see SI Appendix, Fig. S2 for detailed controls). (E) The resulting radii of gyration (Rg) for ProTα-C (blue circles), ProTα-N (green triangles), ACTR (orange rhombi), and IN (red squares) illustrate the PEG-induced compaction. Fits (solid lines) are obtained using scaled-particle theory (SI Appendix, Eq. S5) with the size of PEG 6000 as a single, globally adjustable fitting parameter. The precision of the values of Rg as estimated from multiple measurements of selected data points is comparable to or smaller than the size of the symbols.
The changes in transfer efficiency of the IDPs induced by the crowding agents can be used to extract information on the corresponding changes in chain conformations. Following previous work on unfolded proteins (35) and IDPs (27, 28), we use a Flory–Fisk distribution, which provides a description of the underlying distance distributions, to quantify the dimensions of the polypeptide chains in terms of mean-squared intramolecular distances or the effective radii of gyration, Rg, of the segments probed by the FRET pair (SI Appendix). Note that the analysis is robust with respect to the polymer-physical model used and that the use of multiparameter detection allows us to exclude possible interfering artifacts, such as insufficient rotational averaging of the fluorophores or quenching of the dyes (SI Appendix).
Fig. 2E shows examples of the resulting changes in Rg as a function of the volume fraction φ of PEG 6000 for the four IDP sequences, all of which exhibit collapse upon crowding. Between 0% and 40% of crowder, the changes in Rg range from 0.2 nm (or ∼10%) for IN to ∼1 nm (or ∼30%) for ProTα-C. Qualitatively, this is the behavior expected even from a simple hard-sphere model for a crowding agent whose steric repulsion of the IDP chains leads to their compaction (13, 36). A commonly used quantitative framework for such effects is scaled-particle theory (37), which provides an estimate of the change in free energy required for creating a cavity equivalent to the size of the IDP in a solution of hard spheres with a radius corresponding to the size of the crowding agent, 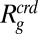 (SI Appendix). If we apply scaled-particle theory, a remarkably good fit is achieved with
(SI Appendix). If we apply scaled-particle theory, a remarkably good fit is achieved with 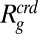 as a global fit parameter (Fig. 2E). However, the resulting value for
as a global fit parameter (Fig. 2E). However, the resulting value for 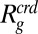 of (6.2 ± 0.1) nm is almost twice the measured radius of gyration of PEG 6000 (SI Appendix, Fig. S1), signifying that a hard-sphere description is not adequate for polymeric crowding agents such as PEG (38).
of (6.2 ± 0.1) nm is almost twice the measured radius of gyration of PEG 6000 (SI Appendix, Fig. S1), signifying that a hard-sphere description is not adequate for polymeric crowding agents such as PEG (38).
Crowder Size Variation Reveals the Importance of Polymer Effects.
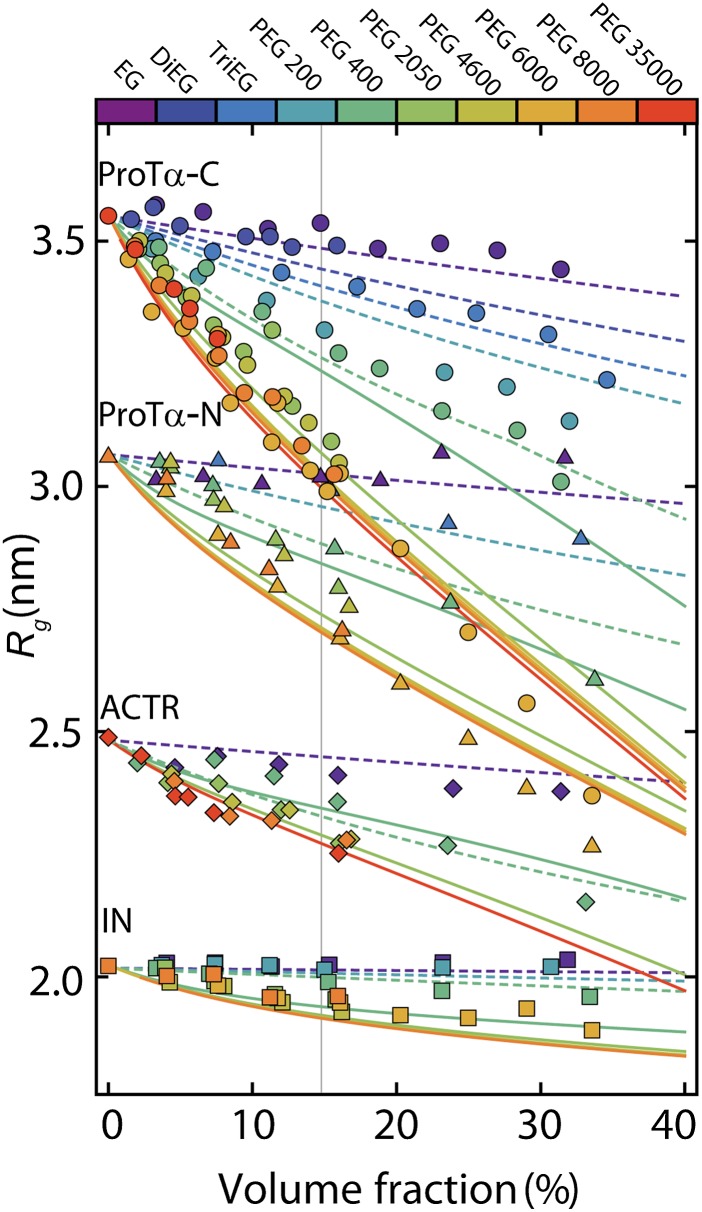
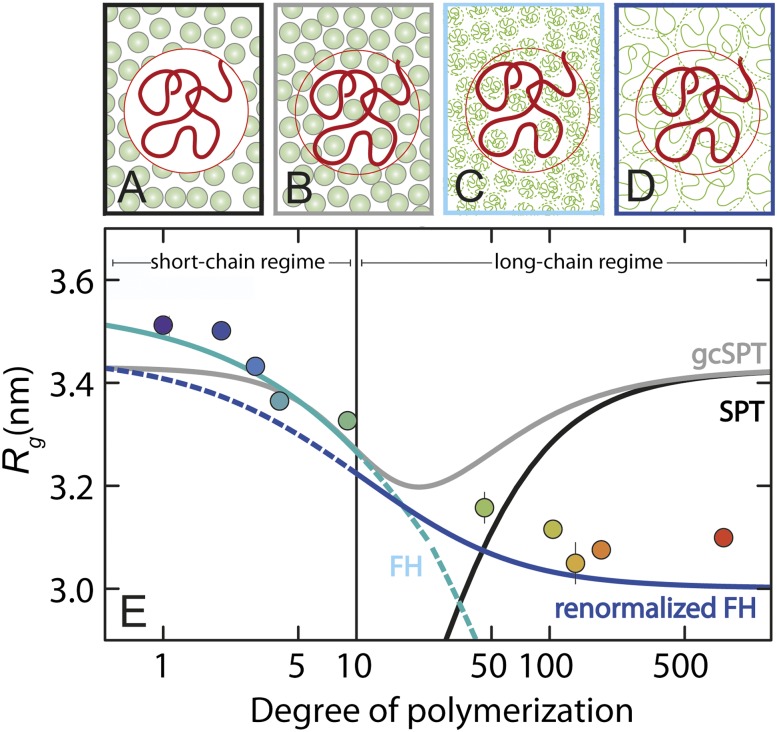
To identify the origin of this discrepancy, we choose a strategy orthogonal to varying the volume fractions of crowder and probe the influence of different sizes of crowding agents on the compaction of IDPs. Fig. 3 shows the complete data set for all four IDP sequences with PEGs of 10 different degrees of polymerization, P, at volume fractions from 0% to ∼40%. For all IDPs, we observe the tendency to collapse with increasing crowder concentration, but interestingly, the degree of compaction is highly dependent on crowder size. The characteristic behavior is most apparent if we consider the change in Rg of an IDP as a function of P at a fixed volume fraction of PEG, as illustrated in Fig. 4 for ProTα-C with φ = 15%. The IDPs collapse monotonically as the crowder size increases, but their Rg reaches a plateau for PEGs of more than ∼100 monomers. Notably, this behavior is the opposite of what we expect from scaled-particle theory because the free energy cost for creating a cavity of given size decreases with increasing crowder size (SI Appendix); in other words, larger solid-sphere crowding agents have larger interstitial cavities and would thus accommodate expanded IDPs more easily (Fig. 4A). To illustrate the discrepancy, Fig. 4E shows the resulting prediction for Rg(P) based on scaled-particle theory (solid black line, Fig. 4E).
Fig. 3.
Both increasing volume fraction and increasing crowder size lead to IDP compaction. Radii of gyration of ProTα-C (circles), ProTα-N (triangles), ACTR (rhombi), and IN (squares) as a function of the volume fraction of PEG obtained from single-molecule FRET experiments. Fits to the data corresponding to the short-chain regime (dashed lines, Eq. 1a) and the long-chain regime (solid lines, Eq. 1b) are shown. For the case of PEG 400, both types of fits are reported to illustrate the cross-over between the two regimes. The vertical dashed line indicates the volume fraction of 15% PEG used in Fig. 4.
Fig. 4.
Polymer concepts explain the compaction of IDPs by crowding agents of increasing size. Graphical representation of (A) scaled-particle theory (SPT), (B) Gaussian cloud model (gcSPT), (C) Flory–Huggins theory (FH) in the short-chain regime, and (D) renormalized Flory–Huggins theory (renormalized FH) in the long-chain regime. (E) Radius of gyration of ProTα-C as a function of the degree of polymerization of PEG at 15% volume fraction of crowding agent. The data points were obtained from linear interpolation of the volume fraction dependences shown in Fig. 3 (same color code for the PEG size). Fits according to the different theories are shown as black (SPT), gray (gcSPT), cyan (FH theory), and blue (renormalized FH theory) lines. Solid lines indicate the regime for which the respective theories were derived; outside of these regimes, dashed lines are used. Error bars reporting on the precision of the experiments are calculated as 1 SD from the linear fits of data for each PEG series in Fig. 3 (uncertainties are smaller than the size of the symbols unless shown explicitly).
An obvious deficit of scaled-particle theory for the treatment of unfolded proteins is the assumption that the crowders cannot penetrate the unfolded chain. To address this issue, Minton proposed the “Gaussian cloud” model (37) (Fig. 4B), where the unfolded protein is described in terms of a continuous Gaussian distribution of monomer density around the center of mass of the protein (SI Appendix, Fig. S3). Small solid-sphere crowders can pervade this protein cloud and thus have little effect on the density distribution of the chain. With increasing crowder size, the probability of accommodating the corresponding spheres without steric clashes with the chain decreases, leading to a compaction of the IDP, in agreement with experimental observation (solid gray line, Fig. 4E). For very large crowding agents, however, this penetration probability decreases further, and ultimately the limit of classic scaled-particle theory is recovered, in contrast with the experimental observation.
These results strongly suggest that we need to go a step further and take into account the polymeric nature of both IDP and crowding agent to explain the behavior observed experimentally. The simplest realistic model needs to comprise two polymers of different lengths in good solvent, i.e., a ternary system. Note that both the IDPs (24) and the crowder (SI Appendix, Fig. S2) (26) exhibit the scaling behavior characteristic of polymers in good solvent, which justifies this assumption.
We also need to take into consideration that, unlike the hard spheres assumed in scaled-particle theory, polymer chains can interpenetrate. This aspect becomes most relevant above a limiting volume fraction, referred to as the overlap concentration 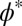 , where the solution can be thought of as being filled by nonintersecting spheres of the size of a single polymer chain. For volume fractions greater than
, where the solution can be thought of as being filled by nonintersecting spheres of the size of a single polymer chain. For volume fractions greater than 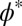 , the transition between dilute and semidilute regimes occurs, and the chains start to overlap, which will affect the conformations of the polymers (SI Appendix, Fig. S1). φ* depends only on the length P of the polymers and on the scaling exponent in the appropriate solvent regime (
, the transition between dilute and semidilute regimes occurs, and the chains start to overlap, which will affect the conformations of the polymers (SI Appendix, Fig. S1). φ* depends only on the length P of the polymers and on the scaling exponent in the appropriate solvent regime (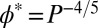 in good solvent; SI Appendix); for long chains, this semidilute regime is reached already at volume fractions of a few percent (SI Appendix, Fig. S1) and the interpenetration of the chains must thus be taken into account for the majority of our experimental conditions.
in good solvent; SI Appendix); for long chains, this semidilute regime is reached already at volume fractions of a few percent (SI Appendix, Fig. S1) and the interpenetration of the chains must thus be taken into account for the majority of our experimental conditions.
Within the framework of the commonly used Flory–Huggins theories, we therefore need to distinguish two scenarios under our experimental conditions: the short-chain regime (Fig. 4C) and the long-chain regime (Fig. 4D) (39). In the first case, the crowding polymer chains are short and consequently remain below the overlap concentration. The system can thus be depicted as a dilute (φ < φ*) solution of PEG chains of radius 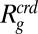 that do not overlap with each other but are able to pervade the volume explored by the IDP (Fig. 4C) (39). Inside this volume, the degrees of freedom of the crowders are reduced by the IDP, and the crowder chains will gain entropy by leaving this volume. A further increase in entropy of the crowder molecules results from reducing the volume occupied by the protein. In other words, the requisite equality of chemical potentials for crowders inside and outside the volume pervaded by the IDP predicts a collapse of the protein chain (39), similar to the Gaussian cloud model, and in good agreement with the experimental data (cyan line, Fig. 4E; also see SI Appendix). In the long-chain regime, however, this mean-field theory fails and diverges from the measured results. In this regime, the crowding polymers are often above their overlap concentrations, and their conformations are influenced by mutual interpenetration. In contrast with the case of a single chain in good solvent, where the dimensions are dominated by repulsive interactions between the monomers, the interpenetration by other crowders in the semidilute regime causes a screening of these repulsive interactions within each chain (40, 41). This excluded volume screening will also affect the conformations of the IDP. However, because the polymers have dimensions comparable to or larger than the protein, they will only partially penetrate the IDP. Under these conditions, the ternary system is close to a critical point and can exhibit density fluctuations over a broad range of length scales due to interactions within the protein, within the crowders, and between the crowders and the protein (41, 42). Many critical systems, ranging from the liquid–gas phase transition near the critical point to the magnetization near the Curie point of a ferromagnet and the Kondo effect of electrons in metals, have been successfully described by renormalization group theory (43). The same approach has provided fundamental insights into the scaling invariance for polymer solutions (41). Here we adopt a renormalized Flory–Huggins-type theory developed by Schäfer and Kappeler (44) for a multicomponent system in the long-chain regime.
that do not overlap with each other but are able to pervade the volume explored by the IDP (Fig. 4C) (39). Inside this volume, the degrees of freedom of the crowders are reduced by the IDP, and the crowder chains will gain entropy by leaving this volume. A further increase in entropy of the crowder molecules results from reducing the volume occupied by the protein. In other words, the requisite equality of chemical potentials for crowders inside and outside the volume pervaded by the IDP predicts a collapse of the protein chain (39), similar to the Gaussian cloud model, and in good agreement with the experimental data (cyan line, Fig. 4E; also see SI Appendix). In the long-chain regime, however, this mean-field theory fails and diverges from the measured results. In this regime, the crowding polymers are often above their overlap concentrations, and their conformations are influenced by mutual interpenetration. In contrast with the case of a single chain in good solvent, where the dimensions are dominated by repulsive interactions between the monomers, the interpenetration by other crowders in the semidilute regime causes a screening of these repulsive interactions within each chain (40, 41). This excluded volume screening will also affect the conformations of the IDP. However, because the polymers have dimensions comparable to or larger than the protein, they will only partially penetrate the IDP. Under these conditions, the ternary system is close to a critical point and can exhibit density fluctuations over a broad range of length scales due to interactions within the protein, within the crowders, and between the crowders and the protein (41, 42). Many critical systems, ranging from the liquid–gas phase transition near the critical point to the magnetization near the Curie point of a ferromagnet and the Kondo effect of electrons in metals, have been successfully described by renormalization group theory (43). The same approach has provided fundamental insights into the scaling invariance for polymer solutions (41). Here we adopt a renormalized Flory–Huggins-type theory developed by Schäfer and Kappeler (44) for a multicomponent system in the long-chain regime.
We thus analyzed the data in the short-chain and long-chain regimes according to
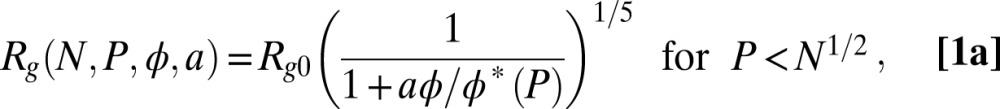 |
and
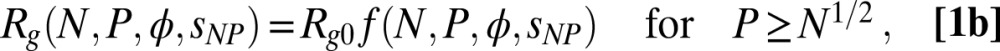 |
where 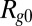 is the radius of gyration of the IDP in the absence of crowding; a is an empirical parameter that can account for differences in the solvent quality for the different proteins and interactions between protein and polymer (45) (SI Appendix);
is the radius of gyration of the IDP in the absence of crowding; a is an empirical parameter that can account for differences in the solvent quality for the different proteins and interactions between protein and polymer (45) (SI Appendix); 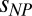 quantifies the interaction between the protein and the polymer chains; and f is a function that represents the renormalization mapping (SI Appendix). It is worth emphasizing that Eqs. 1a and 1b contain only a single adjustable parameter each, a and
quantifies the interaction between the protein and the polymer chains; and f is a function that represents the renormalization mapping (SI Appendix). It is worth emphasizing that Eqs. 1a and 1b contain only a single adjustable parameter each, a and 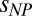 , respectively (SI Appendix, Table S4 and Fig. S4). The equations provide a good fit to the experimental data, including the approach to a limiting value of Rg for IDPs in very large crowders (Fig. 4E). In fact, the entire data set for all four IDP sequences is described remarkably well by a global fit (Fig. 3). The success of this approach supports the hypothesis that the polymeric properties of both IDP and PEG are essential for understanding the effect of molecular crowding, and that the criticality of the solution cannot be neglected. Considering the highly polydisperse cellular environment, it seems probable that related effects will be prominent in vivo and that mean-field descriptions are insufficient for a quantitative description of crowding in the cell.
, respectively (SI Appendix, Table S4 and Fig. S4). The equations provide a good fit to the experimental data, including the approach to a limiting value of Rg for IDPs in very large crowders (Fig. 4E). In fact, the entire data set for all four IDP sequences is described remarkably well by a global fit (Fig. 3). The success of this approach supports the hypothesis that the polymeric properties of both IDP and PEG are essential for understanding the effect of molecular crowding, and that the criticality of the solution cannot be neglected. Considering the highly polydisperse cellular environment, it seems probable that related effects will be prominent in vivo and that mean-field descriptions are insufficient for a quantitative description of crowding in the cell.
The Balance of Hard-Core Repulsion and Other Nonspecific Interactions.
Recent experimental results indicate that the presence of weak, nonspecific attractive interactions in the heterogeneous cellular environment can modulate or even dominate the effects of hard-core repulsion that are at the basis of molecular crowding (46–48). The role of such “chemical interactions” is a subject of debate also for proteins and PEG (13, 26). Notably, the approach presented here (Eq. 1b) allows the relative contributions of hard-core repulsion and other interactions to be quantified in terms of the interaction parameter 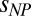 . In the cases investigated here, the analysis with Eq. 1b indicates that a small contribution of unfavorable interactions with PEG is present for ProTα and ACTR, and no such interactions are detected in the case of IN (SI Appendix, Table S4). We note, however, that even though interactions such as nonspecific attraction between crowder and IDP can modulate the amplitude of the change in Rg with crowder concentration (SI Appendix, Fig. S4), the polymeric effects dominate the overall behavior.
. In the cases investigated here, the analysis with Eq. 1b indicates that a small contribution of unfavorable interactions with PEG is present for ProTα and ACTR, and no such interactions are detected in the case of IN (SI Appendix, Table S4). We note, however, that even though interactions such as nonspecific attraction between crowder and IDP can modulate the amplitude of the change in Rg with crowder concentration (SI Appendix, Fig. S4), the polymeric effects dominate the overall behavior.
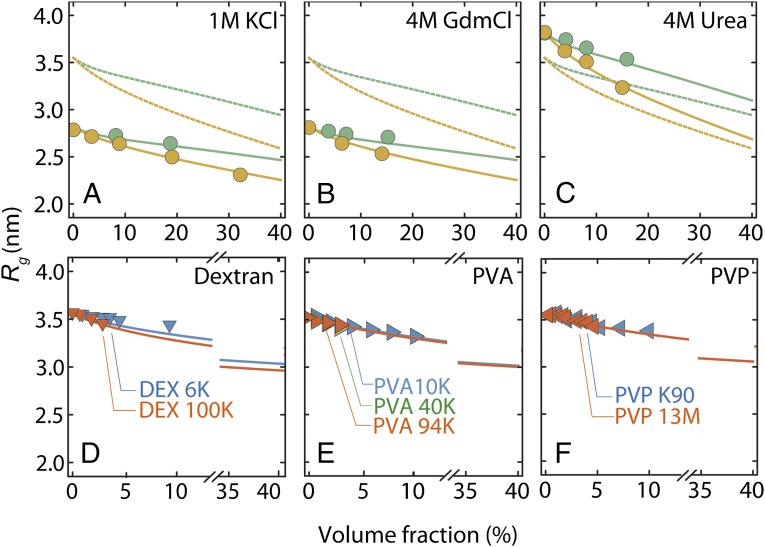
An independent means of interrogating the role of nonspecific charge and hydrophobic interactions is to add salt or denaturants to the solution. Fig. 5 shows that neither 1 M KCl nor 4 M guanidinium chloride (GdmCl) nor 4 M urea impedes the collapse of ProTα. The value of Rg0 depends on ionic strength and denaturant concentration owing to the known charge screening and/or denaturant-induced chain expansion (11). However, the dependence of Rg on the volume fraction of PEG is described by Eq. 1b with the same values of 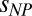 as in the absence of salt or denaturant, just by rescaling Rg0 to the value at the corresponding KCl, GdmCl, or urea concentrations without crowder, suggesting that the effect of additional interactions on the compaction of the IDP is small. Finally, we tested the influence of different chemical structures of the crowding polymer in experiments with dextran, polyvinyl alcohol (PVA), and polyvinylpyrrolidone (PVP) (Fig. 5). Even though we could measure these solutions only for volume fractions of crowder of up to 10% owing to fluorescent impurities, in all cases we observed a collapse of ProTα-C similar to that in PEG. The resulting values of
as in the absence of salt or denaturant, just by rescaling Rg0 to the value at the corresponding KCl, GdmCl, or urea concentrations without crowder, suggesting that the effect of additional interactions on the compaction of the IDP is small. Finally, we tested the influence of different chemical structures of the crowding polymer in experiments with dextran, polyvinyl alcohol (PVA), and polyvinylpyrrolidone (PVP) (Fig. 5). Even though we could measure these solutions only for volume fractions of crowder of up to 10% owing to fluorescent impurities, in all cases we observed a collapse of ProTα-C similar to that in PEG. The resulting values of 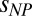 for dextran, PVA, and PVP are significantly lower than for PEG (SI Appendix, Table S5), indicating better compatibility––or less unfavorable interactions––with ProTα, but the collapse of the IDP is preserved. In summary, the polymeric crowding effects on IDPs observed here are dominated by hard-core repulsion between the monomers and the resulting excluded volume screening (40, 41), indicating a phenomenon of generic relevance. However, the analysis presented here does allow additional interactions to be included that can modulate the crowding effect.
for dextran, PVA, and PVP are significantly lower than for PEG (SI Appendix, Table S5), indicating better compatibility––or less unfavorable interactions––with ProTα, but the collapse of the IDP is preserved. In summary, the polymeric crowding effects on IDPs observed here are dominated by hard-core repulsion between the monomers and the resulting excluded volume screening (40, 41), indicating a phenomenon of generic relevance. However, the analysis presented here does allow additional interactions to be included that can modulate the crowding effect.
Fig. 5.
Variation of solution conditions and crowding agents suggest the importance of nonspecific effects on IDP compaction. Radius of gyration of ProTα-C versus the volume fraction of PEG 400 (green circles) and PEG 6000 (yellow circles) in (A) 1 M KCl solution, (B) 4 M GdmCl, and (C) 4 M urea. Fits according to Eq. 1b, assuming a different value of Rg0 but the same value of sNP as in Figs. 3 and 4, are shown as green and yellow solid lines for PEG 400 and PEG 6000, respectively. Fits for the same crowding agents in the absence of salt or denaturant (Fig. 3) are included as dashed lines with corresponding colors. The effects of molecular crowding with dextran (D), PVA (E), and PVP (F) on ProTα-C are shown for different sizes of these alternative crowders as indicated. Lines represent the fit to the renormalized FH theory (SI Appendix, Table S5) and are extrapolated up to 40% volume fraction for comparison with other polymers.
Discussion
Eqs. 1a and 1b can account for the dependence of Rg on crowder concentration and crowder size for all four IDPs investigated (Fig. 3). The question remains, however, why the extent of crowder-induced compaction is so different for the different IDPs. Polymer theory offers an interesting explanation. According to the Flory theorem, the chains in a melt (i.e., in the absence of solvent) of compatible polymers approach their Θ-state. Under these conditions, because of the screening of excluded volume interactions within and between the polymers, the dimensions of the chains scale approximately with the square root of the number of chain segments, and a characteristic radius of gyration RgΘ is observed (SI Appendix). Recent work indicates that RgΘ for the IDPs investigated here is in the range of ∼1.7–2.0 nm (27) (SI Appendix). The results in Fig. 3 for the larger PEGs are indeed consistent with asymptotic convergence of Rg for all of the IDP variants toward values in this range in the limit of very high volume fractions of crowder, i.e., under conditions that approach the situation of a melt. In other words, highly expanded IDPs with dimensions much greater than RgΘ (such as ProTα) are expected to undergo more pronounced compaction on polymeric crowding than those IDPs that are close to RgΘ already in the absence of crowders (such as IN). Based on the empirical relations between solvent quality and average net charge obtained previously (27), we estimate that ∼90% of all IDPs are above the θ-state in the absence of crowding (SI Appendix) and should thus be susceptible to compaction by polymeric crowders.
The observations reported here could thus have implications for the functional properties of many IDPs, e.g., for the capture radius for their cellular targets in the framework of a fly-casting mechanism (49, 50) and for the folding propensity of denatured ensembles in the crowded cellular environment (13). However, the balance of the different contributions may be subtle. Whereas a compaction of the chain by crowding will result in a decrease of the capture radius, it will increase the translational diffusion coefficient. These opposing effects will modulate the basic influence of crowding on solution viscosity and the concomitant changes in association rates (51). Similarly, the established effects of crowding on the stability of the folded and/or bound states of IDPs (13) may be affected by changes in unfolded state dimensions. Single-molecule experiments of the type presented here may help to dissect these contributions quantitatively. Complementary simulations of polymeric crowding could provide valuable insights into the underlying molecular mechanisms.
We note that a substantial fraction of crowding in the cell is due to polymeric molecules such as peptides, nucleic acids, polysaccharides, or other disordered proteins. However, the extent of crowding is strongly affected by the spatial organization of the cell. A remarkable example of very high local concentrations of IDPs are nucleoporins, which line the nuclear pore complexes (25). We estimate the volume fraction occupied by nucleoporins to be between 25% and 55% of the volume available in the pore, about an order of magnitude greater than the overlap concentration (SI Appendix). Similarly, IDPs involved in RNA granules (22, 24) or analogous nonmembrane-bound bodies with liquid-like properties (23, 24) are likely to exceed their overlap concentration locally (SI Appendix). Under these conditions, polymer effects characteristic of the semidilute regime will be highly relevant for the conformations of IDPs and for the occurrence of possible phase transitions. Interestingly, ProTα often colocalizes with dense speckles such as promyelocytic leukemia bodies (52). Given its abundance in the nucleus of mammalian cells and its high mobility within and near the nucleus (53), we expect that a compaction similar to what we observed here can occur in vivo. According to our results, the dense local environment resulting from liquid–liquid demixing (23, 24) or sol–gel transitions (22) should strongly influence the conformational distributions of IDPs, with consequent impact on the functional properties of the resulting assemblies and their mechanisms of formation. Flory–Huggins theories as used here might thus provide novel insights into the demixing of multicomponent polymeric systems (41). An interesting next step will be a direct comparison of experiments in vitro with intracellular measurements (14, 26), and the required quantitative tools are beginning to emerge (54–56).
Methods
Proteins were expressed, purified, and labeled similar to previous reports (11, 27, 28). Single-molecule measurements were performed using a MicroTime 200 confocal microscope equipped with a HydraHarp 400 counting module (PicoQuant). For details on experiments and theory, see SI Appendix.
Supplementary Material
Acknowledgments
We thank Rohit Pappu, Devarajan Thirumalai, and David Goldenberg for helpful discussions and comments on the manuscript. This work was supported by the Swiss National Science Foundation and a Starting Investigator Grant of the European Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322611111/-/DCSupplemental.
References
- 1.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18(6):756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Jensen MR, Ruigrok RW, Blackledge M. Describing intrinsically disordered proteins at atomic resolution by NMR. Curr Opin Struct Biol. 2013;23(3):426–435. doi: 10.1016/j.sbi.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19(1):31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreon AC, Moran CR, Gambin Y, Deniz AA. Single-molecule fluorescence studies of intrinsically disordered proteins. Methods Enzymol. 2010;472:179–204. doi: 10.1016/S0076-6879(10)72010-3. [DOI] [PubMed] [Google Scholar]
- 6.Schuler B, Müller-Späth S, Soranno A, Nettels D. Application of confocal single-molecule FRET to intrinsically disordered proteins. Methods Mol Biol. 2012;896:21–45. doi: 10.1007/978-1-4614-3704-8_2. [DOI] [PubMed] [Google Scholar]
- 7.Ferreon AC, Ferreon JC, Wright PE, Deniz AA. Modulation of allostery by protein intrinsic disorder. Nature. 2013;498(7454):390–394. doi: 10.1038/nature12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao AH, Lyle N, Pappu RV. Describing sequence-ensemble relationships for intrinsically disordered proteins. Biochem J. 2013;449(2):307–318. doi: 10.1042/BJ20121346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyle N, Das RK, Pappu RV. A quantitative measure for protein conformational heterogeneity. J Chem Phys. 2013;139(12):121907. doi: 10.1063/1.4812791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher CK, Stultz CM. Protein structure along the order-disorder continuum. J Am Chem Soc. 2011;133(26):10022–10025. doi: 10.1021/ja203075p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller-Späth S, et al. From the Cover: Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107(33):14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uversky VN. Intrinsically disordered proteins and their environment: Effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009;28(7-8):305–325. doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- 13.Zhou HX, Rivas GN, Minton AP. Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gershenson A, Gierasch LM. Protein folding in the cell: Challenges and progress. Curr Opin Struct Biol. 2011;21(1):32–41. doi: 10.1016/j.sbi.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dedmon MM, Patel CN, Young GB, Pielak GJ. FlgM gains structure in living cells. Proc Natl Acad Sci USA. 2002;99(20):12681–12684. doi: 10.1073/pnas.202331299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNulty BC, Young GB, Pielak GJ. Macromolecular crowding in the Escherichia coli periplasm maintains alpha-synuclein disorder. J Mol Biol. 2006;355(5):893–897. doi: 10.1016/j.jmb.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 17.Munishkina LA, Cooper EM, Uversky VN, Fink AL. The effect of macromolecular crowding on protein aggregation and amyloid fibril formation. J Mol Recognit. 2004;17(5):456–464. doi: 10.1002/jmr.699. [DOI] [PubMed] [Google Scholar]
- 18.Szasz CS, et al. Protein disorder prevails under crowded conditions. Biochemistry. 2011;50(26):5834–5844. doi: 10.1021/bi200365j. [DOI] [PubMed] [Google Scholar]
- 19.Hong JA, Gierasch LM. Macromolecular crowding remodels the energy landscape of a protein by favoring a more compact unfolded state. J Am Chem Soc. 2010;132(30):10445–10452. doi: 10.1021/ja103166y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikaelsson T, Adén J, Johansson LBA, Wittung-Stafshede P. Direct observation of protein unfolded state compaction in the presence of macromolecular crowding. Biophys J. 2013;104(3):694–704. doi: 10.1016/j.bpj.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansen D, Jeffries CM, Hammouda B, Trewhella J, Goldenberg DP. Effects of macromolecular crowding on an intrinsically disordered protein characterized by small-angle neutron scattering with contrast matching. Biophys J. 2011;100(4):1120–1128. doi: 10.1016/j.bpj.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han TW, et al. Cell-free formation of RNA granules: Bound RNAs identify features and components of cellular assemblies. Cell. 2012;149(4):768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 23.Li PL, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brangwynne CP. Soft active aggregates: Mechanics, dynamics and self-assembly of liquid-like intracellular protein bodies. Soft Matter. 2011;7(7):3052–3059. [Google Scholar]
- 25.Rout MP, et al. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J Cell Biol. 2000;148(4):635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elcock AH. Models of macromolecular crowding effects and the need for quantitative comparisons with experiment. Curr Opin Struct Biol. 2010;20(2):196–206. doi: 10.1016/j.sbi.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmann H, et al. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc Natl Acad Sci USA. 2012;109(40):16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soranno A, et al. Quantifying internal friction in unfolded and intrinsically disordered proteins with single-molecule spectroscopy. Proc Natl Acad Sci USA. 2012;109(44):17800–17806. doi: 10.1073/pnas.1117368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devanand K, Selser JC. Asymptotic-behavior and long-range interactions in aqueous-solutions of poly(ethylene oxide) Macromolecules. 1991;24(22):5943–5947. [Google Scholar]
- 30.Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 1991;222(3):599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 31.Harris JM. Poly(Ethylene Glycol) Chemistry: Biotechnical and Biomedical Applications. New York: Plenum; 1992. [Google Scholar]
- 32.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41(3):415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 33.Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107(18):8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das RK, Pappu RV. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc Natl Acad Sci USA. 2013;110(33):13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziv G, Haran G. Protein folding, protein collapse, and Tanford’s transfer model: Lessons from single-molecule FRET. J Am Chem Soc. 2009;131(8):2942–2947. doi: 10.1021/ja808305u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheung MS, Klimov D, Thirumalai D. Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc Natl Acad Sci USA. 2005;102(13):4753–4758. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minton AP. Models for excluded volume interaction between an unfolded protein and rigid macromolecular cosolutes: Macromolecular crowding and protein stability revisited. Biophys J. 2005;88(2):971–985. doi: 10.1529/biophysj.104.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mittal J, Best RB. Dependence of protein folding stability and dynamics on the density and composition of macromolecular crowders. Biophys J. 2010;98(2):315–320. doi: 10.1016/j.bpj.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joanny JF, Grant P, Pincus P, Turkevich LA. Conformations of polydisperse polymer-solutions - bimodal distribution. J Appl Phys. 1981;52(10):5943–5948. [Google Scholar]
- 40.Edwards SF. Theory of polymer solutions at intermediate concentration. Proc Phys Soc Lond. 1966;88(560P):265–280. [Google Scholar]
- 41.Schäfer L. Excluded Volume Effects in Polymer Solutions as Explained by the Renormalization Group. Berlin: Springer; 1999. [Google Scholar]
- 42.Tran HT, Pappu RV. Toward an accurate theoretical framework for describing ensembles for proteins under strongly denaturing conditions. Biophys J. 2006;91(5):1868–1886. doi: 10.1529/biophysj.106.086264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson KG. The renormalization-group and critical phenomena. Rev Mod Phys. 1983;55(3):583–600. [Google Scholar]
- 44.Schäfer L, Kappeler C. Interaction effects on the size of a polymer-chain in ternary solutions - a renormalization-group study. J Chem Phys. 1993;99(8):6135–6154. [Google Scholar]
- 45.Nose T. Chain dimension of a guest polymer in the semidilute solution of compatible and incompatible polymers. J Phys (Paris) 1986;47(3):517–527. [Google Scholar]
- 46.Minton AP. Quantitative assessment of the relative contributions of steric repulsion and chemical interactions to macromolecular crowding. Biopolymers. 2013;99(4):239–244. doi: 10.1002/bip.22163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarkar M, Li C, Pielak GJ. Soft interactions and crowding. Biophys. Rev. 2013;5(2):187–194. doi: 10.1007/s12551-013-0104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim YC, Mittal J. Crowding induced entropy-enthalpy compensation in protein association equilibria. Phys Rev Lett. 2013;110(20):208102-1–208102-5. doi: 10.1103/PhysRevLett.110.208102. [DOI] [PubMed] [Google Scholar]
- 49.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc Natl Acad Sci USA. 2000;97(16):8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trizac E, Levy Y, Wolynes PG. Capillarity theory for the fly-casting mechanism. Proc Natl Acad Sci USA. 2010;107(7):2746–2750. doi: 10.1073/pnas.0914727107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schreiber G, Haran G, Zhou HX. Fundamental aspects of protein-protein association kinetics. Chem Rev. 2009;109(3):839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vareli K, Frangou-Lazaridis M, van der Kraan I, Tsolas O, van Driel R. Nuclear distribution of prothymosin alpha and parathymosin: Evidence that prothymosin alpha is associated with RNA synthesis processing and parathymosin with early DNA replication. Exp Cell Res. 2000;257(1):152–161. doi: 10.1006/excr.2000.4857. [DOI] [PubMed] [Google Scholar]
- 53.Enkemann SA, Ward RD, Berger SL. Mobility within the nucleus and neighboring cytosol is a key feature of prothymosin-alpha. J Histochem Cytochem. 2000;48(10):1341–1355. doi: 10.1177/002215540004801005. [DOI] [PubMed] [Google Scholar]
- 54.Gelman H, Platkov M, Gruebele M. Rapid perturbation of free-energy landscapes: From in vitro to in vivo. Chemistry. 2012;18(21):6420–6427. doi: 10.1002/chem.201104047. [DOI] [PubMed] [Google Scholar]
- 55.Phillip Y, Kiss V, Schreiber G. Protein-binding dynamics imaged in a living cell. Proc Natl Acad Sci USA. 2012;109(5):1461–1466. doi: 10.1073/pnas.1112171109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakon JJ, Weninger KR. Detecting the conformation of individual proteins in live cells. Nat Methods. 2010;7(3):203–205. doi: 10.1038/nmeth.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.