Background: Dysregulation of Wnt signaling plays a key role in colorectal cancer (CRC).
Results: Thymine DNA glycosylase (TDG) enhances Wnt signaling and is required for CRC growth.
Conclusion: TDG is a novel positive regulator in the Wnt pathway and CRC.
Significance: TDG could be a novel biomarker and therapeutic target for CRC.
Keywords: Cell Growth, Colorectal Cancer, Protein-Protein Interactions, Transcription Coactivators, Wnt Signaling
Abstract
Wnt signaling plays an important role in colorectal cancer (CRC). Although the mechanisms of β-catenin degradation have been well studied, the mechanism by which β-catenin activates transcription is still not fully understood. While screening a panel of DNA demethylases, we found that thymine DNA glycosylase (TDG) up-regulated Wnt signaling. TDG interacts with the transcription factor TCF4 and coactivator CREB-binding protein/p300 in the Wnt pathway. Knocking down TDG by shRNAs inhibited the proliferation of CRC cells in vitro and in vivo. In CRC patients, TDG levels were significantly higher in tumor tissues than in the adjacent normal tissues. These results suggest that TDG warrants consideration as a potential biomarker for CRC and as a target for CRC treatment.
Introduction
Colorectal cancer (CRC)3 is the second leading cause of cancer deaths in the United States (1) and a growing concern across the globe. The mortality associated with CRC rests in part with the absence of convenient biomarkers for its early detection. More than 80% of CRCs contain adenomatous polyposis coli mutations, and most of the other CRCs contain β-catenin mutations (2). These mutations activate Wnt signaling, which, if deregulated and uncontrolled, lead to cancer (3). In this study, we found that thymine DNA glycosylase (TDG) is a novel positive regulator in the Wnt pathway, that TDG is required for CRC growth in vitro and in vivo, and that TDG holds promise as a potential biomarker and as a therapeutic target for CRC.
In normal cells without Wnt stimulation, β-catenin forms a degradation complex with tumor suppressors, adenomatous polyposis coli, and Axin, and undergoes sequential phosphorylation by CKIα and GSK3. The phosphorylated β-catenin is degraded by the ubiquitination/proteasome machinery (3–5). The binding of an external Wnt protein to the Frizzled and Lrp5/6 receptors causes the disruption of the Axin complex and prevents β-catenin phosphorylation and its subsequent degradation (6). Under conditions where a Wnt signal is present, β-catenin translocates to the nucleus, where it interacts with the T-cell factor/lymphocyte enhancer factor (TCF/LEF) transcription factors (7, 8) and recruits transcriptional coactivators, such as histone acetyltransferase p300/CREB-binding protein (CBP), that activate target gene expression (9). Mutations of β-catenin or adenomatous polyposis coli prevent phosphorylation and normal ubiquitination/degradation and result in abnormal β-catenin accumulation in the nucleus and aberrant gene expression. In the intestine, Wnt signaling promotes stem cell self-renewal and normal homeostasis, and mutations in this pathway in the intestinal stem cells represent the initiation step in CRC development (10).
TDG is a member of the TDG/mismatch uracil glycosylase DNA glycosylase family (11) and has multiple roles. It removes thymine moieties from G/T mismatches as a key role in mismatch repair (12–14). TDG also plays essential roles in epigenetic regulation. It interacts with activation-induced deaminase and the non-enzyme factor Gadd45 (growth-arrested and DNA damage-induced protein 45) and works with 5-methylcytosine hydroxylases TETs (ten-eleven translocation) to regulate active DNA demethylation (15–17). In addition to its function in DNA modification, TDG interacts with several transcription factors, including thyroid transcription factor 1 (TTF-1) (18), estrogen receptor (ERα) (19), retinoic acid receptor (RAR) and retinoid X receptor (RXR) (20). Finally, TDG also interacts with CBP and its paralog p300 in regulating DNA repair and gene transcription (21).
Because TDG is required for DNA repair and demethylation, it may play a role in tumor prevention, a role supported by a report that TDG acts as a coactivator of p53 (22, 23), which binds the TDG promoter and regulates TDG transcription (24). In the human genome, the TDG gene is located on chromosome 12q24.1. Recently, in a mutational profile study, a heterozygous mutation in the TDG gene was identified in a rectal cancer patient (25, 26), suggesting that TDG may function as a tumor suppressor. To examine the mechanism and function of TDG in CRC, we analyzed the effects of TDG on Wnt signaling. We now report that TDG interacts with TCF4 and acts as a positive regulator in the Wnt pathway. Furthermore, we found that TDG is up-regulated in human CRCs and is required for CRC growth. These findings suggest that TDG has multiple, heretofore unappreciated functions in CRC.
EXPERIMENTAL PROCEDURES
Plasmids and Cell Culture
HA-TCF4, HA-CBP, Myc-β-catenin, HA-β-catenin HA-CBP-HAT, and TOPFlash have been described previously (27–29). HA-Gadd45α was purchased from Addgene (catalog no. 80452). FLAG-TDG was generated by cloning the TDG (NM_003211.4) open reading frame by PCR. TDG 1–119 and TDG 119–410 truncations were generated by PCR and cloned into the pcDNA 3.1(+) vector. The TDG N140A and D133A mutations were introduced by site-directed mutagenesis (30). HEK293T, HT29, and HCT116 cells were grown in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. LS174T cells were grown in RPMI medium supplemented with 5% fetal bovine serum and 1% penicillin/streptomycin. Cell transfection was performed as reported previously (29, 31, 32).
Cell-based Assays
For Western blot and immunoprecipitation assays, cells were lysed in an appropriate volume of lysis buffer (50 mm HEPES, 100 mm NaCl, 2 mm EDTA, 1% glycerol, 50 mM NaF, 1 mm Na3VO4, and 1% Triton X-100 with protease inhibitors). Proteins were analyzed with the following antibodies: rabbit anti-β-catenin (Sigma, catalog no. C2206), rat anti-HA (Roche, catalog no. 3F10), mouse anti-HA (Cell Signaling Technology, catalog no. 6E2), rabbit anti-TCF4 (Epitomics, catalog no. 2114-1), mouse anti-FLAG (Sigma, catalog no. F1804), rabbit anti-TDG (Gene Tex, catalog no. GTX110473), and mouse anti-β-actin (Sigma, catalog no. A1978). For immunofluorescent staining, cells were grown on coverslips and fixed with 4% paraformaldehyde. Alexa Fluor 633-labeled anti-mouse immunoglobulin G (1:800) and Alexa Fluor 488-labeled anti-rabbit immunoglobulin G (1:800) were used for fluorescent analysis with an Olympus laser confocal microscope. The luciferase and cell proliferation assays have been described previously (29, 31, 32).
shRNA
TDG shRNA constructs were purchased from Sigma (catalog nos. TRCN0000150193 and TRCN0000285278). HEK293T cells were cotransfected with a shRNA plasmid, a psPAX2 packaging plasmid, and a pMD2.G envelope plasmid (33). Viruses were harvested 48 h after transfection and used to infect CRC cell lines HCT116, HT29, and LS174T. Stable cell lines were selected with 2 μg/ml puromycin 48 h after infection.
RT-PCR
Total RNA was isolated from cultured cells using an RNeasy mini kit (Qiagen, Valencia, CA). The RNA samples were converted into cDNA using random primer 6 (New England Biolabs, catalog no. S1230S,) and M-MULV Reverse transcriptase (New England Biolabs, catalog no. 0230908). Amplified products were analyzed on a 1.5% agarose gel. Primers were ordered from Eurofins MWG Operon. Primer sequences were as follows: TDG, 5′-GGCTAATTGAGAGCGTGGAG-3′ (forward) and 5′-GCATGGCTTTCTTCTTCCTG-3′ (reverse); β-actin, 5′-CAACCGCGAGAAGATGAC-3′ (forward) and 5′-AGGAAGGCTGGAAGAGTG-3′ (reverse); survivin, 5′-CATTCGTCCGGTTGCGCTTTCC-3′ (forward) and 5′-GCGCACTTTCTCCGCAGTTTCC-3′ (reverse); and c-myc, 5′-TGGGCTGTGAGGAGGTTTG-3′ (forward) and 5′-TATGTGGAGCGGCTTCTCG-3′ (reverse).
ChIP
ChIP assays were performed according to the protocol developed by Nowak et al. (28) with some modifications. The cells were cross-linked with disuccimidyl glutarate (Pierce, catalog no. 20593) and formaldehyde at room temperature. Cells were collected and resuspended in L1 buffer (50 mm Tris, 2 mm EDTA, 0.1% IGEPAL, 10% glycerol, 1mM dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (pH 8.0)). After centrifugation, the nuclear pellet was resuspended in ChIP lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris (pH 8.0), and protease inhibitor mixture). Cell lysate was sonicated with a Branson Sonifier 150. The lysate was incubated with appropriate antibodies overnight, followed by incubation with a protein A-agarose/salmon sperm DNA 50% slurry (Upstate, catalog no. 16-157) for 3 h at 4 °C. Beads were then washed, and bound DNA-protein complexes were eluted and de-cross-linked. DNA was then purified by phenol/chloroform extraction and ethanol precipitation. DNA pellets were resuspended in Tris-EDTA buffer, and 1 μl was used for PCR. The following primers were used: c-myc, 5′-TATGTGGAGCGGCTTCTCG-3′ (forward) and 5′-TGGGCTGTGAGGAGGTTTG-3′ (reverse). For ChIP assays, Abs used included preimmune rabbit IgG, anti-TDG Ab (Proteintech Group, catalog no. 13370-1-AP,) and anti-acetylated histone H3 (Millipore, catalog no. 06-599).
Xenograft Assay
HT29 stable cell lines (1 × 106) were injected subcutaneously into both flanks of athymic nude mice as described previously (32). Tumor xenografts were analyzed twice weekly. After 3 weeks, tumor xenografts were harvested and analyzed. The animal studies were approved by the Institutional Animal Care and Use Committee at the University of Kentucky.
Human Tissue Specimens
Discarded human CRC samples were provided by Nanfang Hospital in China. Tissues fixed in 10% formalin or 4% paraformaldehyde were paraffin-embedded and sectioned. The sections (5 μm) were stained with an anti-TDG antibody (Proteintech Group, catalog no. 13370-1-AP). Evaluation of the staining was on the basis of the percentage of positive cells in each tissue slide as well as the intensity of the positively stained cells (supplemental Table S1). The studies with discarded human tissues were approved by the Research Ethics Committee of Nanfang Hospital, Southern Medical University, and the University of Kentucky.
Statistical Analysis
Paired t tests were used to compare TDG expression in tumor versus adjacent and normal tissues for the immunohistochemistry analysis on the basis of the Nanfang Hospital study. Microarray and patient clinical data from two colon cancer studies were downloaded from the Oncomine database (34, 35). A two-sample t test was used to compare TDG expression in colon adenocarcinoma patients versus normal controls. A one-way analysis of variance model was used to compare TDG expression in different tumor stages. A similar method was also used to compare TDG expression in different tumor grades. Pearson's correlation coefficient was calculated to quantify the correlation between expressions of TDG and survivin. Pairwise comparisons were performed on the basis of the Tukey's Honestly Significant Difference (HSD) method. To assess the association between TDG expression and patient survival, two methods were used. One method stratified subjects into TDG high and low expression groups on the basis of the median expression level and compared survival time in these two groups on the basis of Kaplan-Meier curves and log-rank tests. The other method treated TDG expression as continuous and applied the proportional hazards model.
RESULTS
TDG Is Up-regulated in CRC
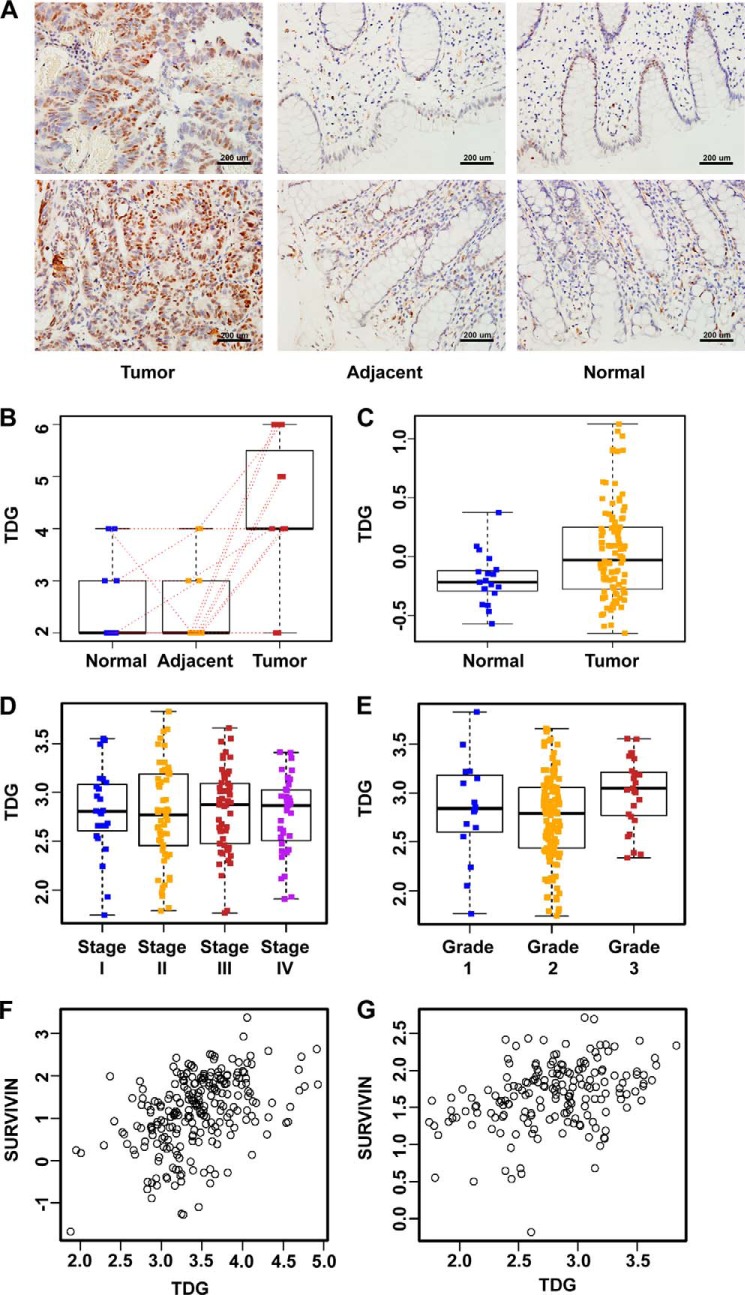
Recently, several enzymes involved in active DNA demethylation were identified (15–17). To understand the functions of these genes in CRC, we analyzed the expression of these genes in CRC cell lines and human tissues. One of the demethylation-related genes, TDG, is significantly increased in CRC. In this study, we first compared the expression of TDG in human CRC tissues versus adjacent and normal tissues. The difference in TDG expression on the basis of immunohistochemistry analysis (supplemental Table S1) of samples from Nanfang Hospital is illustrated in Fig. 1A. TDG is only weakly expressed in the nucleus of normal tissues but increased significantly in CRC (Fig. 1A). To further quantify the association between TDG levels and CRC, paired t tests were used to compare the immunohistochemistry scores from different tissue types. TDG expression was significantly higher in tumors than in adjacent tissues (p = 0.001) and in normal tissues (p = 0.007) (Fig. 1B).
FIGURE 1.
TDG is up-regulated in CRC. A, immunohistochemistry analysis of TDG in tumors, adjacent tissues, and normal tissues from the same patients. TDG expression was increased significantly in human colon cancer tissues. B, statistical analysis of TDG expression in tumor and normal tissues from Nanfang Hospital (supplemental Table S1). TDG expression was significantly higher in tumors than in adjacent tissues (p = 0.001) and normal tissues from the same patients (p = 0.007). Red dotted lines indicate scores from the same patient. C, TDG expression is significantly higher in colon adenocarcinoma patients than in normal controls, according to the TCGA database (276 tumor and normal pairs). D, TDG expression in patients with different tumor stages on the basis of data from Smith et al. (35). p = 0.92 (177 patients from the Moffitt Cancer Center as the independent dataset; stage I, 24 patients; stage II, 57 patients; stage III, 57 patients; stage IV, 39 patients). E, TDG expression in patients with different tumor grades on the basis of data from Smith et al. (35). p = 0.013. F and G, correlation between TDG expression and survivin expression in two human colon cancer microarray data sets. F, Smith et al. (35); Pearson's correlation coefficient = 0.36 (95% confidence interval, 0.22–0.48; p < 0.001). G, Pearson's correlation coefficient = 0.44 (95% confidence interval, 0.33–0.54, p < 0.001). (M. Bittner (Expression Project for Oncology, International Genomics Consortium)).
To further determine the clinical relevance of TDG in CRC, a series of statistical studies were performed on the basis of two publicly available microarray-based studies (34, 35). We also validated our findings using The Cancer Genome Atlas (TCGA) data (34). TDG expression was significantly higher in colon adenocarcinoma patients than in normal controls (Fig. 1C, two-sample t test, p = 0.001).
Next, we assessed the correlation between TDG expression and CRC stage and grade using the data from Smith et al. (35). There was no significant difference in TDG expression across different tumor stages (Fig. 1D, analysis of variance, p = 0.920). But there was a significant difference in TDG expression across different tumor grades (Fig. 1E, analysis of variance, p = 0.013). TDG expression was significantly higher in grade 3 patients than in grade 2 patients (Tukey's Honestly Significant Difference, p = 0.010).
TDG Activates Wnt Signaling
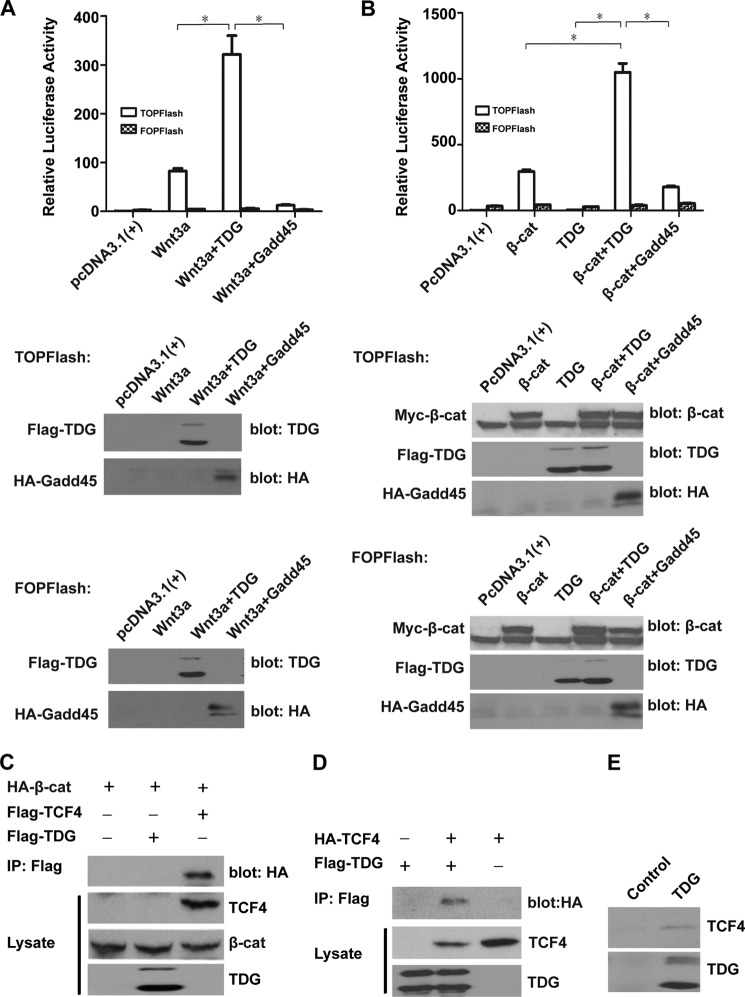
Given its function in DNA repair and demethylation, TDG should belong to a tumor suppressor family. It was unexpected to discover that TDG is up-regulated in CRC, suggesting that TDG may play other unappreciated roles in CRC. Because Wnt signaling has essential functions in CRC, we tested whether TDG plays any role in Wnt signaling. We analyzed the correlation between TDG and Wnt target gene survivin using the microarray data sets described above (Fig. 1, F and G). The expression of TDG is positively correlated with the expression of survivin, indicating a connection between TDG and Wnt signaling in colon cancer. We then analyzed the effects of TDG on Wnt signaling using the TOPFlash reporter, which can be activated by Wnt signaling (Fig. 2A). Expression of TDG significantly enhanced Wnt-induced TOPFlash reporter activity but not the control FOPFlash reporter activity. Another demethylation-related gene, Gadd45α, plays a negative role in Wnt signaling (36). As a control, expression of Gadd45α decreased Wnt-induced TOPFlash reporter activity (Fig. 2A). Similarly, TDG significantly enhanced β-catenin-induced TOPFlash reporter activity, whereas Gadd45 decreased this activity (Fig. 2B), suggesting that TDG and Gadd45α play different roles in Wnt signaling.
FIGURE 2.
TDG activates Wnt signaling. A, TDG enhanced Wnt3A-induced TOPFlash reporter activity. TOPFlash or FOPFlash was cotransfected with Wnt3A and Renilla plasmids into HEK293T cells with or without TDG and Gadd45. Top panel, relative luciferase reporter activity adjusted with Renilla. *, p < 0.05. Bottom panel, expression levels of TDG and Gadd45. B, TDG enhanced β-catenin-induced TOPFlash reporter activity. TOPFlash or FOPFlash was cotransfected with β-catenin (β-cat) and Renilla plasmids into HEK293T cells with or without TDG and Gadd45. Top panel, relative luciferase reporter activity adjusted with Renilla. *, p < 0.05. Bottom panel, expression levels of β-catenin, TDG, and Gadd45. C, β-catenin binds TCF4 but does not bind TDG. FLAG-tagged TDG or TCF4 and HA-tagged β-catenin (S33A mutant) were transfected into HEK293T cells. TDG or TCF4 was immunoprecipitated (IP) with an anti-FLAG Ab, and β-catenin was analyzed by Western blot analysis with an anti-HA Ab. TDG, TCF4, and β-catenin in the cell lysates were analyzed by Western blot analysis. D, TDG binds TCF4. FLAG-tagged TDG and HA-tagged TCF4 were transfected into HEK293T cells. TDG was immunoprecipitated with an anti-FLAG Ab, and TCF4 was analyzed by Western blot analysis with an anti-HA Ab. TDG and TCF4 in the cell lysates were analyzed by Western blot analysis. E, interaction between endogenous TDG and TCF4. Endogenous TDG was immunoprecipitated from HEK293T cells using an anti-TDG Ab. The presence of TCF4 was analyzed by Western blot analysis with an anti-TCF4 Ab. An unrelated Ab was used as a negative control for immunoprecipitation.
TDG Interacts with TCF4
TDG is a nuclear protein. To test whether TDG regulates Wnt signaling in the nucleus, we analyzed the binding between β-catenin and TDG, with TCF4 as a positive control. TCF4 strongly bound β-catenin, as expected, but TDG had no interaction with β-catenin in a coimmunoprecipitation assay (Fig. 2C). However, we found that TDG interacted with TCF4 in a coimmunoprecipitation assay (Fig. 2D). Endogenous TDG also interacted with endogenous TCF4 (Fig. 2E), suggesting that TDG plays a specific role in Wnt signaling by interacting with TCF4.
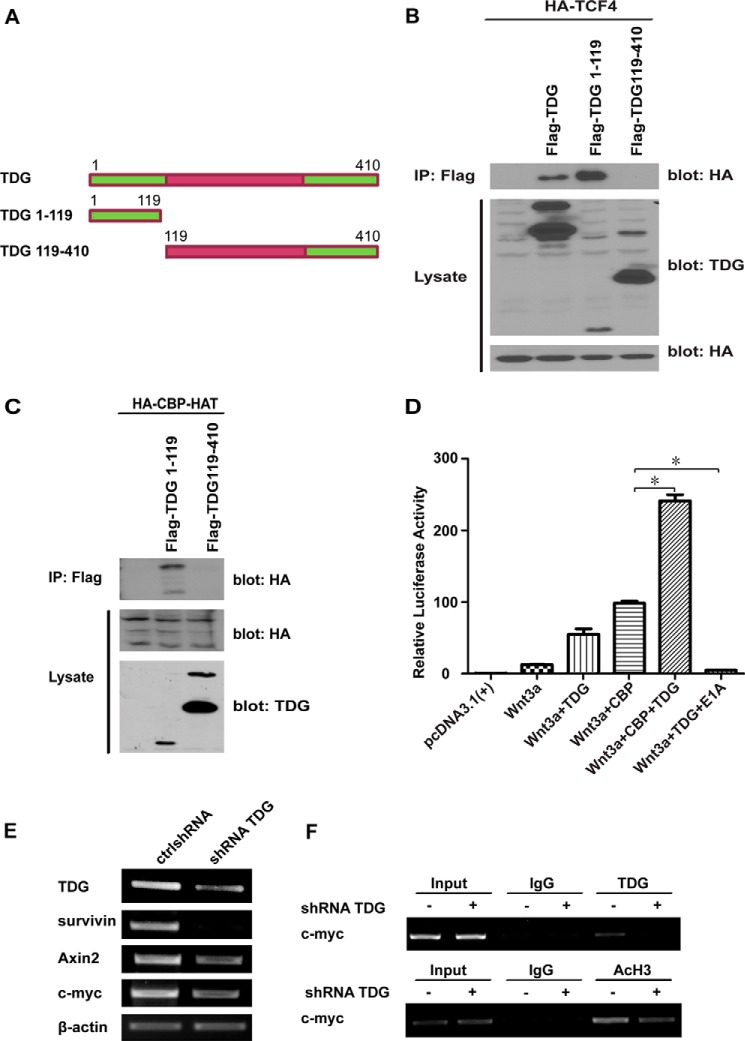
To determine which domain of TDG binds to TCF4, we generated several deletion constructs (Fig. 3A). The eukaryotic TDG proteins are composed of a conserved catalytic core domain in the middle of the protein sequence and non-conserved domains in the N and C termini (37). The N terminus of human TDG contains a DNA-binding domain required for base excision repair (38). We performed a coimmunoprecipitation experiment with FLAG-tagged TDG1–119 and TDG119–410. We found that the N terminus TDG1–119 is required for TCF4 binding (Fig. 3B).
FIGURE 3.
TDG interacts with TCF4 and CBP. A, TDG deletion constructs. B, the N terminus of TDG binds TCF4. HA-tagged TCF4 was cotransfected with FLAG-tagged TDG, TDG1–119, or TDG119–410 into HEK293T cells. Full-length and deleted TDG proteins were immunoprecipitated (IP) with an anti-FLAG Ab, and TCF4 was detected by Western blot analysis with an anti-HA Ab. Protein levels of TDG and TCF4 in the cell lysates were analyzed by Western blot analysis. C, the N terminus of TDG interacts with the HAT domain of CBP. HEK293T cells were transfected with HA-tagged HAT together with FLAG-tagged TDG1–119 or TDG119–410. The TDG fragments were immunoprecipitated with an anti-FLAG Ab, and the HAT fragment was analyzed by Western blot analysis with an anti-HA Ab (top blot). The expression levels of these proteins in the cell lysates were analyzed by Western blot analysis with anti-HA and anti-TDG Abs (center and bottom blots). D, TDG and CBP active TOPFlash reporter. Wnt3A was cotransfected with TOPFlash and Renilla plasmids into HEK293T cells with or without TDG and CBP. CBP enhanced TDG function in Wnt signaling. CBP inhibitor E1A abolished TDG-induced Wnt signaling. *, p < 0.05. E, TDG is required for Wnt target gene expression in colon cancer cells. HT29 colon cancer cells were infected with lentiviruses expressing a control shRNA (ctrlshRNA) or TDG shRNA. The mRNA levels of TDG and Wnt target genes, survivin, Axin2, and c-myc were analyzed by RT-PCR. β-actin was analyzed as a loading control. F, ChIP assay. HT 29 cells were treated with control shRNA or TDG shRNA. ChIP assays were performed using anti-TDG, anti-AcH3, and control Abs. The promoter sequence of the Wnt target gene, c-myc, was analyzed by PCR.
TDG Interacts with CBP
CBP/p300 is a transcriptional coactivator in the Wnt pathway. It has been found that CBP/p300 is associated with TDG in regulating DNA repair and transcription (21). To test whether TDG regulates Wnt signaling through CBP/p300, we analyzed the interaction between TDG and CBP. We found that TDG1–119 binds the HAT domain of CBP (Fig. 3C). HAT is the catalytic histone acetyltransferase domain of CBP, which modifies chromatin and regulates transcription (39).
To analyze the function of these interactions, we performed a TOPFlash reporter assay. Both TDG and CBP enhance Wnt3A-mediated TOPFlash reporter activation (Fig. 3D). When TDG and CBP were expressed together, the reporter activity was dramatically enhanced. In contract, E1A, a CBP/p300 inhibitor, abolished the reporter activity induced by Wnt3A and TDG (Fig. 3D). To determine whether TDG is required for Wnt target gene expression in colon cancer cells, we treated HT29 cells with control shRNA and TDG shRNA. TDG depletion decreased Wnt target gene expression (Fig. 3E). We then analyzed the binding between TDG and the Wnt target gene promoter using a ChIP assay. We found that TDG interacted with the c-myc promoter and that TDG shRNA inhibited the interaction (Fig. 3F, top panel). TDG shRNA also decreased the histone H3 acetylation at the c-myc promoter (Fig. 3F, bottom panel).
The SUMOylation Site of TDG Regulates Its Activity and Localization
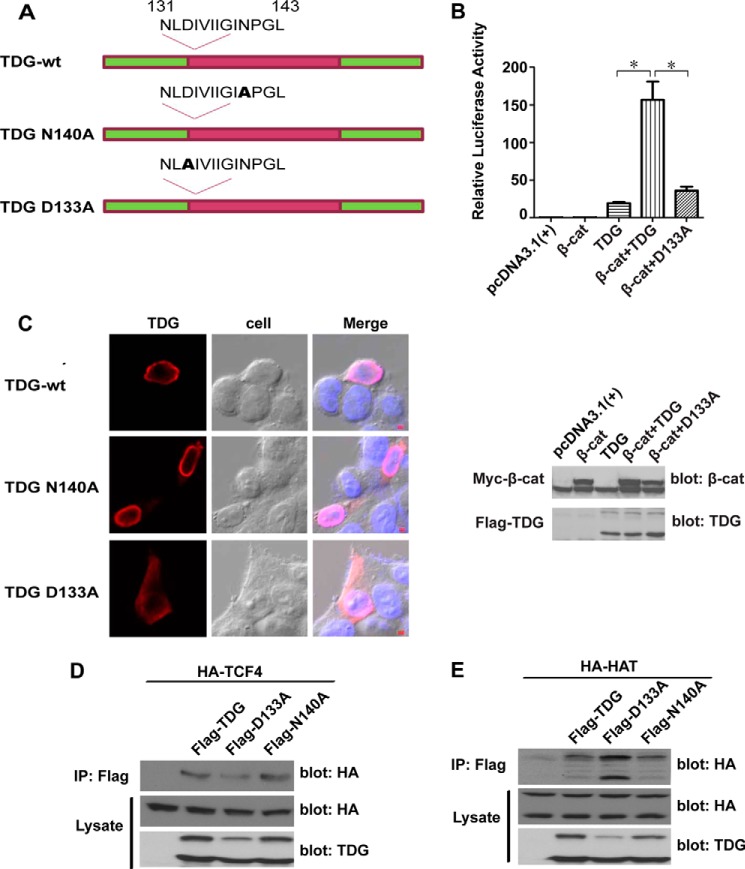
TDG is a DNA glycosylase that removes thymine generated by the deamination of 5-methylcytosine (16). The middle region of TDG contains the catalytic domain and asparagine 140 (Asn-140) and is required for its catalytic activity in DNA repair (40). To test whether the catalytic activity of TDG is required for Wnt signaling, we generated a point mutation, N140A (Fig. 4A). It is known that the N140A mutant is inactive in DNA repair assays (40). Surprisingly, N140A mutation has no significant effect on TDG activity in a TOPFlash reporter assay (data not shown), suggesting that the glycosylase activity of TDG is not essential for its role in Wnt signaling.
FIGURE 4.
SUMOylation of TDG regulates its activity in Wnt signaling. A, schematic of TDG mutations in the enzymatic activation site (N140A) and the SUMO conjugation site (D133A). B, the D133A mutation reduced TDG activity in enhancing β-catenin-mediated Wnt signaling. β-Catenin (β-cat), TOPFlash, and Renilla plasmids were cotransfected with TDG or TDG D133A into HEK293T cells. Top panel, relative luciferase reporter activity. *, p < 0.05. Bottom panel, expression levels of β-catenin and TDG. C, D133A mutation affects TDG cellular localization. FLAG-tagged TDG, TDG N140A, or TDG D133A was transfected into HCT116 cells. The cells were immunostained with an anti-FLAG Ab (red fluorescence). The nucleus was stained with DAPI (blue fluorescence). TDG and TDG N140A were detected in the nucleus, whereas D133A was detected in both the nucleus and the cytoplasm (>60% of cells). Scale bar = 100 μm. D, effects of TDG mutations on the interaction between TDG and TCF4. FLAG-tagged TDG, TDG D133A, or TDG N140A and HA-tagged TCF4 were cotransfected into HEK293T cells. TDG was immunoprecipitated (IP) with an anti-FLAG Ab, and TCF4 was analyzed by Western blot analysis with an anti-HA Ab. TCF4 and TDG in the cell lysates were analyzed by Western blot analysis. E, effects of TDG mutations on the interaction between TDG and HAT. FLAG-tagged TDG, TDG D133A, or TDG N140A and HA-tagged HAT were cotransfected into HEK293T cells. TDG was immunoprecipitated with an anti-FLAG Ab, and HAT was analyzed by Western blot analysis with an anti-HA Ab. HAT and TDG in the cell lysates were analyzed by Western blot analysis.
It has been reported that TDG can be conjugated with small ubiquitin-like modifiers (SUMOs) covalently and non-covalently (41) and that these SUMO modifications affect protein localization, stability, and function (14). SUMOylation of TDG, for example, affects its binding with DNA and other proteins (42). There are two SUMOylation sites in TDG: Asp-133 and Glu-310 (37). Because Asp-133 is close to Asn-140 and the N-terminal CBP-TCF4 binding domain, we generated a D133A point mutation in TDG (Fig. 4A). We compared the activity of the D133A mutant with the wild-type TDG using a TOPFlash reporter assay. The D133A mutation significantly reduced TDG ability to enhance Wnt signaling (Fig. 4B).
To test the effects of TDG mutations on TDG localization, FLAG-tagged TDG, TDG N140A, and TDG D133A were separately transfected into HCT116 CRC cells. The localization of TDG protein was analyzed by immunofluorescent staining and confocal microscopy. The wild-type TDG and the N140A mutant were localized in the nucleus, whereas a significant amount of the D133A mutant was detected in the cytoplasm (Fig. 4C), a result indicating that SUMOylation of TDG affects its nuclear translocation, which, in turn, regulates its activity in Wnt signaling. We next tested the binding between TDG mutants and TCF4. The D133A mutation, but not the N140A mutation, slightly reduced the interaction with TCF4 (Fig. 4D). Interestingly, the D133A mutation slightly increased the binding between TDG and the HAT domain of CBP (Fig. 4E).
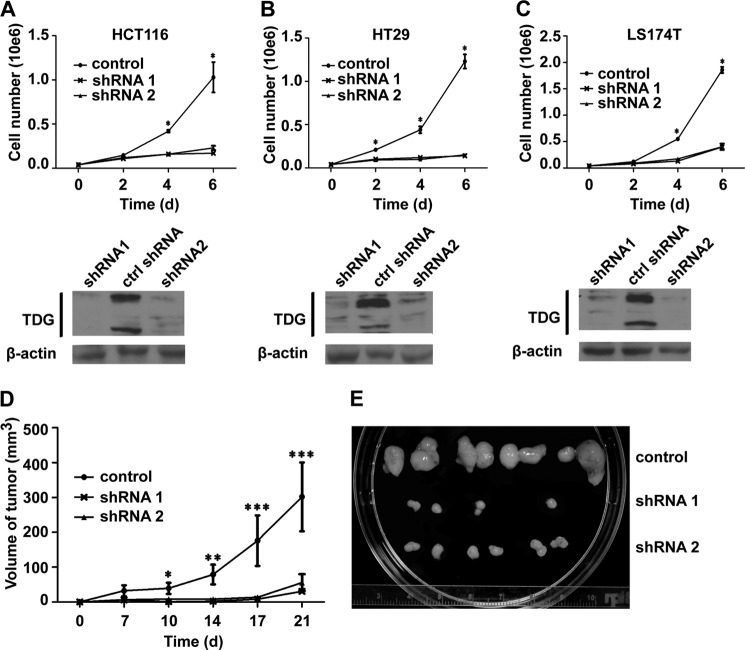
TDG Is Essential for CRC Cell Proliferation in Vitro and in Vivo
Because Wnt signaling is a key driver for CRC and because TDG regulates Wnt signaling, we analyzed the role of TDG in CRC cell growth. TDG shRNAs were transfected into different CRC cell lines using lentivirus infection. Cell proliferation assays indicated that TDG depletion significantly inhibited the growth of HCT116, HT29, and LS174T cells (Figs. 5, A–C).
FIGURE 5.
TDG regulates colon cancer cell growth in vitro and in vivo. A—C, TDG is required for the proliferation of colon cancer cells. Stable colon cell lines that expressed control shRNA (ctrlshRNA) and TDG shRNAs were established. Top panels, cell proliferation assays. *, p < 0.05. Bottom panels, TDG expression levels in the stable cell lines. D, deletion TDG inhibited colon cancer cell growth in vivo. Stable HT29 cell lines that expressed control shRNA and TDG shRNAs were inject subcutaneously into nude mice. Growth curves of these xenografted tumors were analyzed. *, p < 0.05; **, p < 0.01; ***, p < 0.001. E, xenografted tumors from control and TDG-depleted cells.
To determine the role of TDG on CRC growth in vivo, we generated stable HT29 cells that expressed control shRNA or TDG shRNAs. These cells were injected subcutaneously into the flanks of athymic nude mice. The xenografted tumors were measured twice weekly for 3 weeks. TDG depletion significantly inhibited xenografted tumor growth (Fig. 5, D and E), suggesting that TDG is required for CRC cell growth in vivo.
DISCUSSION
TDG plays a key role in DNA repair and an expanding role in a number of other events, including DNA demethylation. While searching for DNA demethylase enzymes that might regulate Wnt signaling, we identified TDG as a novel positive regulator in the Wnt pathway. The enzymatic activity of TDG was not essential for its function in Wnt signaling. Instead, it may act as an adaptor protein by interacting with transcription factor TCF4 and coactivator CBP/p300. TDG is overexpressed in human CRC and is required for CRC cell growth in vitro and in vivo, suggesting that TDG could be used as a novel marker and, possibly, even as a therapeutic target for CRC.
It has been well documented that Wnt signaling plays a central role in colon carcinogenesis and the majority of these cancers are initiated by mutations that affect the Wnt pathway. In normal cells, β-catenin levels are tightly controlled by Wnt signaling. However, in CRC, dysregulation of β-catenin leads to nuclear accumulation and unregulated gene expression. In the nucleus, β-catenin interacts with TCF and many coactivators, such as CBP/p300, pygopus, and Bcl9, to activate target gene expression. Our work points to TDG as a newly recognized coactivator for β-catenin/TCF in CRC. TDG has a catalytic domain in the middle of the protein sequence and a DNA binding domain at the N terminus (37). An N140A mutant at the enzymatic active site, however, still binds to the mismatched DNA but cannot catalyze base removal (40). We found that this N140A mutation has no significant effect on TDG activity in Wnt signaling. We tested several genes involved in DNA demethylation, including activation-induced deaminase and Gadd45α, and only TDG enhanced Wnt signaling in the reporter assay. Gadd45α even reduced Wnt signaling, supporting the previous report that Gadd45α down-regulates β-catenin (36). Although we cannot exclude the possibility that DNA demethylation is involved in Wnt signaling, our findings suggest that TDG regulates Wnt signaling through a mechanism independent of its DNA modification activity.
TDG interacts with both CBP and TCF4 (Fig. 3, B and C) in a manner that suggests that TDG regulates the assembly or function of the β-catenin·TCF·CBP complex. This finding is consistent with a previous report that TDG interacts with other transcriptional factors through its N-terminal domain (37). TDG is modified by SUMOylation at Asp-133, close to the Asn-140 activation site (Fig. 4A). Mutation of Asp-133 reduced nuclear levels of TDG. This could be another reason why TDG SUMOylation affects its interaction with CBP (43). The D133A mutation reduced TCF4 binding but increased CBP binding (Fig. 4, D and E), suggesting that TDG SUMOylation may differentially regulate TCF4 and CBP. Acetylation of TDG by CBP/p300 also regulates it function in DNA repair and transcription (21), suggesting that TDG function in Wnt signaling may be regulated by multiple posttranslational modifications, including SUMOylation and acetylation.
TDG is a key factor in DNA repair and active DNA demethylation. Approximately 25% of all cancers are related to mutations at the C-T transitions at the CpG sites, and this percentage rises to 50% in colon and gastric cancers (44). In addition, aberrant DNA methylation located in the CpG sites is associated with various human diseases, particularly cancer. It would be expected that TDG plays a tumor suppressor role in CRC by maintaining genetic and epigenetic integrity. We were surprised to find that TDG enhanced Wnt signaling and was required for CRC cell growth, an outcome that was inconsistent with previous reports that TDG is a potential tumor suppressor (22, 23). This inconsistency between our findings and previous reports is probably due to the fact that TDG has multiple functions in CRC. As a key enzyme in DNA repair and DNA demethylation, TDG may be required for CRC prevention. Loss of TDG may increase the rate of genetic mutation and induce abnormal DNA methylation associated with tumorigenesis. However, we have securely established that TDG is also required for Wnt signaling and probably other signals essential for CRC survival and growth. Thus, a cell with TDG mutation has no growth advantage and will be eliminated before it accumulates harmful genetic and epigenetic changes that lead to cancer. Consistent with this hypothesis, TDG deletion in mice is an embryonically lethal outcome (45). This can also explain why only a heterozygous TDG mutation, but no homozygous mutation, has been found in cancer patients (25, 26). This mechanism could significantly reduce the risk of tumorigenesis caused by loss of a DNA repair gene.
To understand the role of TDG in CRC, we analyzed the microarray and patient clinical data from the Oncomine database. TDG expression is significantly higher in CRC in comparison with adjacent normal tissues. Whether the TDG gene has point mutation in these tumor samples is not clear. However, the colon cancer cell lines used in this study express full-length TDG (Fig. 5, A–C). We evaluated the association between TDG expression and patient survival time on the basis of the data from Smith et al. (35) and TCGA data (34) (supplemental Fig. S1). TDG expression appears to be negatively correlated with patient survival, but the association is not statistically significant. More correlation studies need be performed to determine the true significance of TDG in CRC prognosis. Our findings suggest, however, that TDG could be used as a biomarker for CRC. Because knocking down TDG significantly inhibited xenografted CRC growth in nude mice, TDG may be a potential drug target for CRC treatment.
Supplementary Material
Acknowledgments
We thank Drs. Mark Evers and David Watt for critical reading of the manuscript and the Markey Bio-specimen and Tissue Procurement Shared Resource Facility for tissue embedding, sectioning, and H&E staining.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK071976 and R01 CA172379 (to C. L.).

This article contains supplemental Fig. S1 and Table S1.
- CRC
- colorectal cancer
- TDG
- thymine DNA glycosylase
- CBP
- CREB-binding protein
- CREB
- cAMP response element-binding protein
- Ab
- antibody
- HAT
- histone acetyltransferase
- SUMO
- small ubiquitin-like modifier.
REFERENCES
- 1. Siegel R., Naishadham D., Jemal A. (2013) Cancer Statistics, 2013. CA Cancer J. Clin. 63, 11–30 [DOI] [PubMed] [Google Scholar]
- 2. Giles R. H., van Es J. H., Clevers H. (2003) Caught up in a Wnt storm. Wnt signaling in cancer. Biochim. Biophys. Acta 1653, 1–24 [DOI] [PubMed] [Google Scholar]
- 3. Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., He X. (2002) Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108, 837–847 [DOI] [PubMed] [Google Scholar]
- 4. Hart M., Concordet J. P., Lassot I., Albert I., del los Santos R., Durand H., Perret C., Rubinfeld B., Margottin F., Benarous R., Polakis P. (1999) The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr. Biol. 9, 207–210 [DOI] [PubMed] [Google Scholar]
- 5. Amit S., Hatzubai A., Birman Y., Andersen J. S., Ben-Shushan E., Mann M., Ben-Neriah Y., Alkalay I. (2002) Axin-mediated CKI phosphorylation of β-catenin at Ser-45. A molecular switch for the Wnt pathway. Genes Dev. 16, 1066–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smalley M. J., Sara E., Paterson H., Naylor S., Cook D., Jayatilake H., Fryer L. G., Hutchinson L., Fry M. J., Dale T. C. (1999) Interaction of axin and Dvl-2 proteins regulates Dvl-2-stimulated TCF-dependent transcription. EMBO J. 18, 2823–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brunner E., Peter O., Schweizer L., Basler K. (1997) Pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature 385, 829–833 [DOI] [PubMed] [Google Scholar]
- 8. van de Wetering M., Cavallo R., Dooijes D., van Beest M., van Es J., Loureiro J., Ypma A., Hursh D., Jones T., Bejsovec A., Peifer M., Mortin M., Clevers H. (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88, 789–799 [DOI] [PubMed] [Google Scholar]
- 9. Wolf D., Rodova M., Miska E. A., Calvet J. P., Kouzarides T. (2002) Acetylation of β-catenin by CREB-binding protein (CBP). J. Biol. Chem. 277, 25562–25567 [DOI] [PubMed] [Google Scholar]
- 10. Barker N., van Es J. H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P. J., Clevers H. (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 [DOI] [PubMed] [Google Scholar]
- 11. Aravind L., Koonin E. V. (2000) The α/β fold uracil DNA glycosylases. A common origin with diverse fates. Genome Biol. 1, research0007.1-0007.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neddermann P., Jiricny J. (1993) The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem. 268, 21218–21224 [PubMed] [Google Scholar]
- 13. Wiebauer K., Jiricny J. (1989) In vitro correction of G.T mispairs to G.C pairs in nuclear extracts from human cells. Nature 339, 234–236 [DOI] [PubMed] [Google Scholar]
- 14. Cortázar D., Kunz C., Saito Y., Steinacher R., Schär P. (2007) The enigmatic thymine DNA glycosylase. DNA Repair 6, 489–504 [DOI] [PubMed] [Google Scholar]
- 15. Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D., Abramowitz L. K., Bartolomei M. S., Rambow F., Bassi M. R., Bruno T., Fanciulli M., Renner C., Klein-Szanto A. J., Matsumoto Y., Kobi D., Davidson I., Alberti C., Larue L., Bellacosa A. (2011) Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gong Z., Zhu J. K. (2011) Active DNA demethylation by oxidation and repair. Cell Res. 21, 1649–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu S. C., Zhang Y. (2010) Active DNA demethylation. Many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 11, 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Missero C., Pirro M. T., Simeone S., Pischetola M., Di Lauro R. (2001) The DNA glycosylase T:G mismatch-specific thymine DNA glycosylase represses thyroid transcription factor-1-activated transcription. J. Biol. Chem. 276, 33569–33575 [DOI] [PubMed] [Google Scholar]
- 19. Chen D., Lucey M. J., Phoenix F., Lopez-Garcia J., Hart S. M., Losson R., Buluwela L., Coombes R. C., Chambon P., Schär P., Ali S. (2003) T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor α. J. Biol. Chem. 278, 38586–38592 [DOI] [PubMed] [Google Scholar]
- 20. Um S., Harbers M., Benecke A., Pierrat B., Losson R., Chambon P. (1998) Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 273, 20728–20736 [DOI] [PubMed] [Google Scholar]
- 21. Tini M., Benecke A., Um S. J., Torchia J., Evans R. M., Chambon P. (2002) Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell 9, 265–277 [DOI] [PubMed] [Google Scholar]
- 22. Kim E. J., Um S. J. (2008) Thymine-DNA glycosylase interacts with and functions as a coactivator of p53 family proteins. Biochem. Biophys. Res. Commun. 377, 838–842 [DOI] [PubMed] [Google Scholar]
- 23. Sun W., Yang J. (2010) Functional mechanisms for human tumor suppressors. J. Cancer 1, 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. da Costa N. M., Hautefeuille A., Cros M. P., Melendez M. E., Waters T., Swann P., Hainaut P., Pinto L. F. (2012) Transcriptional regulation of thymine DNA glycosylase (TDG) by the tumor suppressor protein p53. Cell Cycle 11, 4570–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vasovcak P., Krepelova A., Menigatti M., Puchmajerova A., Skapa P., Augustinakova A., Amann G., Wernstedt A., Jiricny J., Marra G., Wimmer K. (2012) Unique mutational profile associated with a loss of TDG expression in the rectal cancer of a patient with a constitutional PMS2 deficiency. DNA Repair 11, 616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sjolund A. B., Senejani A. G., Sweasy J. B. (2012) MBD4 and TDG. Multifaceted DNA glycosylases with ever expanding biological roles. Mutat. Res. 743–744, 12–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhakat K. K., Hazra T. K., Mitra S. (2004) Acetylation of the human DNA glycosylase NEIL2 and inhibition of its activity. Nucleic Acids Res. 32, 3033–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Evans P. M., Zhang W., Chen X., Yang J., Bhakat K. K., Liu C. (2007) Krüppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J. Biol. Chem. 282, 33994–34002 [DOI] [PubMed] [Google Scholar]
- 29. Zhang W., Chen X., Kato Y., Evans P. M., Yuan S., Yang J., Rychahou P. G., Yang V. W., He X., Evers B. M., Liu C. (2006) Novel cross talk of Krüppel-like factor 4 and β-catenin regulates normal intestinal homeostasis and tumor repression. Mol. Cell Biol. 26, 2055–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Evans P. M., Liu C. (2005) SiteFind. A software tool for introducing a restriction site as a marker for successful site-directed mutagenesis. BMC Mol. Biol. 6, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang J., Zhang W., Evans P. M., Chen X., He X., Liu C. (2006) Adenomatous polyposis coli (APC) differentially regulates β-catenin phosphorylation and ubiquitination in colon cancer cells. J. Biol. Chem. 281, 17751–17757 [DOI] [PubMed] [Google Scholar]
- 32. Yu T., Chen X., Zhang W., Colon D., Shi J., Napier D., Rychahou P., Lu W., Lee E. Y., Weiss H. L., Evers B. M., Liu C. (2012) Regulation of the potential marker for intestinal cells, Bmi1, by β-catenin and the zinc finger protein KLF4. Implications for colon cancer. J. Biol. Chem. 287, 3760–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang W., Yang J., Liu Y., Chen X., Yu T., Jia J., Liu C. (2009) PR55 α, a regulatory subunit of PP2A, specifically regulates PP2A-mediated β-catenin dephosphorylation. J. Biol. Chem. 284, 22649–22656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cancer Genome Atlas Network (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith J. J., Deane N. G., Wu F., Merchant N. B., Zhang B., Jiang A., Lu P., Johnson J. C., Schmidt C., Bailey C. E., Eschrich S., Kis C., Levy S., Washington M. K., Heslin M. J., Coffey R. J., Yeatman T. J., Shyr Y., Beauchamp R. D. (2010) Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 138, 958–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ji J., Liu R., Tong T., Song Y., Jin S., Wu M., Zhan Q. (2007) Gadd45a regulates β-catenin distribution and maintains cell-cell adhesion/contact. Oncogene 26, 6396–6405 [DOI] [PubMed] [Google Scholar]
- 37. Smet-Nocca C., Wieruszeski J. M., Chaar V., Leroy A., Benecke A. (2008) The thymine-DNA glycosylase regulatory domain. Residual structure and DNA binding. Biochemistry 47, 6519–6530 [DOI] [PubMed] [Google Scholar]
- 38. Waters T. R., Gallinari P., Jiricny J., Swann P. F. (1999) Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 274, 67–74 [DOI] [PubMed] [Google Scholar]
- 39. Wang F., Marshall C. B., Ikura M. (2013) Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis. Structural and functional versatility in target recognition. Cell. Mol. Life Sci. 70, 3989–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hardeland U., Bentele M., Jiricny J., Schär P. (2000) Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. J. Biol. Chem. 275, 33449–33456 [DOI] [PubMed] [Google Scholar]
- 41. Smet-Nocca C., Wieruszeski J. M., Léger H., Eilebrecht S., Benecke A. (2011) SUMO-1 regulates the conformational dynamics of thymine-DNA glycosylase regulatory domain and competes with its DNA binding activity. BMC Biochem. 12, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hardeland U., Steinacher R., Jiricny J., Schär P. (2002) Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 21, 1456–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mohan R. D., Rao A., Gagliardi J., Tini M. (2007) SUMO-1-dependent allosteric regulation of thymine DNA glycosylase alters subnuclear localization and CBP/p300 recruitment. Mol. Cell Biol. 27, 229–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Greenblatt M. S., Bennett W. P., Hollstein M., Harris C. C. (1994) Mutations in the p53 tumor suppressor gene. Clues to cancer etiology and molecular pathogenesis. Cancer Res. 54, 4855–4878 [PubMed] [Google Scholar]
- 45. Cortázar D., Kunz C., Selfridge J., Lettieri T., Saito Y., MacDougall E., Wirz A., Schuermann D., Jacobs A. L., Siegrist F., Steinacher R., Jiricny J., Bird A., Schär P. (2011) Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470, 419–423 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.