Abstract
Metastasis suppressors comprise a growing class of genes whose downregulation triggers metastatic progression. In contrast to tumor suppressors, metastasis suppressors are rarely mutated or deleted, and little is known regarding the mechanisms by which their expression is downregulated. Here, we demonstrate that the metastasis suppressor, NM23-H1, is degraded by lysosomal cysteine cathepsins (L,B), which directly cleave NM23-H1. In addition, activation of c-Abl and Arg oncoproteins induces NM23-H1 degradation in invasive cancer cells by increasing cysteine cathepsin transcription and activation. Moreover, c-Abl activates cathepsins by promoting endosome maturation, which facilitates trafficking of NM23-H1 to the lysosome where it is degraded. Importantly, the invasion- and metastasis-promoting activity of c-Abl/Arg is dependent on their ability to induce NM23-H1 degradation, and the pathway is clinically relevant as c-Abl/Arg activity and NM23-H1 expression are inversely correlated in primary breast cancers and melanomas. Thus, we demonstrate a novel mechanism by which cathepsin expression is upregulated in cancer cells (via Abl kinases). We also identify a novel role for intracellular cathepsins in invasion and metastasis (degradation of a metastasis suppressor). Finally, we identify novel crosstalk between oncogenic and metastasis suppressor pathways, thereby providing mechanistic insight into the process of NM23-H1 loss, which may pave the way for new strategies to restore NM23-H1 expression and block metastatic progression.
Keywords: oncogene, metastasis suppressor, c-Abl, Arg, NM23-H1, cathepsin
INTRODUCTION
For most cancers, death results from metastatic spread to distant organ sites. Since many cancers present without detectable metastatic disease, metastasis may be a targetable process. While the involvement of proto-oncogenes in metastasis is well documented, loss of metastasis suppressor expression also plays a significant role. NM23-H1 (encoded by NME1) was the first metastasis suppressor identified due its to decreased expression in metastatic breast cancer and melanoma cell lines (1-3). Interestingly, NM23-H1 has a dual role, as its expression is induced in primary cancers but suppressed at later stages during progression (4). NM23-H1 has multiple enzymatic functions, some of which are required for its metastasis suppressor activity (1, 3). Forced expression of NM23-H1 into invasive, metastatic cancer cells dramatically reduces migration, invasion, anchorage-independent growth, tumor dormancy, and metastasis (3, 5, 6). Given the importance of NM23-H1 loss during metastasis, agents that elevate NM23-H1 expression are being pursued in the clinic (2, 3, 7).
Protein degradation is also important for metastatic progression. Lysosomes, which contain proteases that function at an acidic pH, degrade old organelles (autophagy) as well as intracellular and membrane-bound proteins (8). Cysteine cathepsins degrade proteins within the lysosome but also have functions in the nucleus, plasma membrane, and in the extracellular environment (8). Cathepsins are synthesized as inactive proforms and routed to early endosomes, cleaved in late endosomes to form single chain intermediates, and transported to lysosomes for processing into double chain forms (8). In many cancer cells, cathepsins are overexpressed, and excess procathepsins are secreted, resulting in increased localization at the plasma membrane and in the extracellular compartment (8, 9). Secreted cathepsins promote invasion and metastasis by activating proteases and degrading extracellular matrix proteins (8). Intracellular cathepsins also contribute to invasion perhaps by inducing intracellular degradation of extracellular matrix; however, it is unclear whether they have additional roles (8).
The Abl family of non-receptor tyrosine kinases (c-Abl, Arg), encoded by Abl1 and Abl2 genes respectively, are proto-oncogenes known for their involvement in human leukemia (10). We showed that these kinases also are activated in solid tumors (breast cancer, melanoma, glioblastoma), and once activated, promote proliferation, survival during nutrient deprivation, anchorage-independent growth, invasion, and metastasis (11-14). Consistent with our findings, subsequent reports have shown that c-Abl/Arg are activated in other solid tumor types (e.g. lung, gastric, liver), and promote processes critical for progression (15); however, the molecular mechanisms by which they promote progression are only beginning to be elucidated.
Although NM23-H1 loss during metastasis was described over 20 years ago, the mechanism underlying its downregulation has not been adequately studied. Although a few reports noted an inverse relationship between NM23-H1 mRNA levels and metastatic propensity (16, 17), other articles have either shown no relationship (18-21) or demonstrate a positive correlation (22-24). Moreover, little is known regarding the regulation of NM23-H1 at the protein level. Here, we demonstrate that c-Abl and Arg induce NM23-H1 degradation by increasing expression and activation of cathepsin L and B, which directly cleave NM23-H1 in the lysosome. Importantly, this pathway has biological and clinical relevance, as c-Abl/Arg-dependent invasion and metastasis requires downregulation of NM23-H1, and c-Abl/Arg activity and NM23-H1 expression are inversely correlated in primary breast cancers and melanomas.
RESULTS
NM23-H1 is degraded by lysosomal cysteine cathepsins
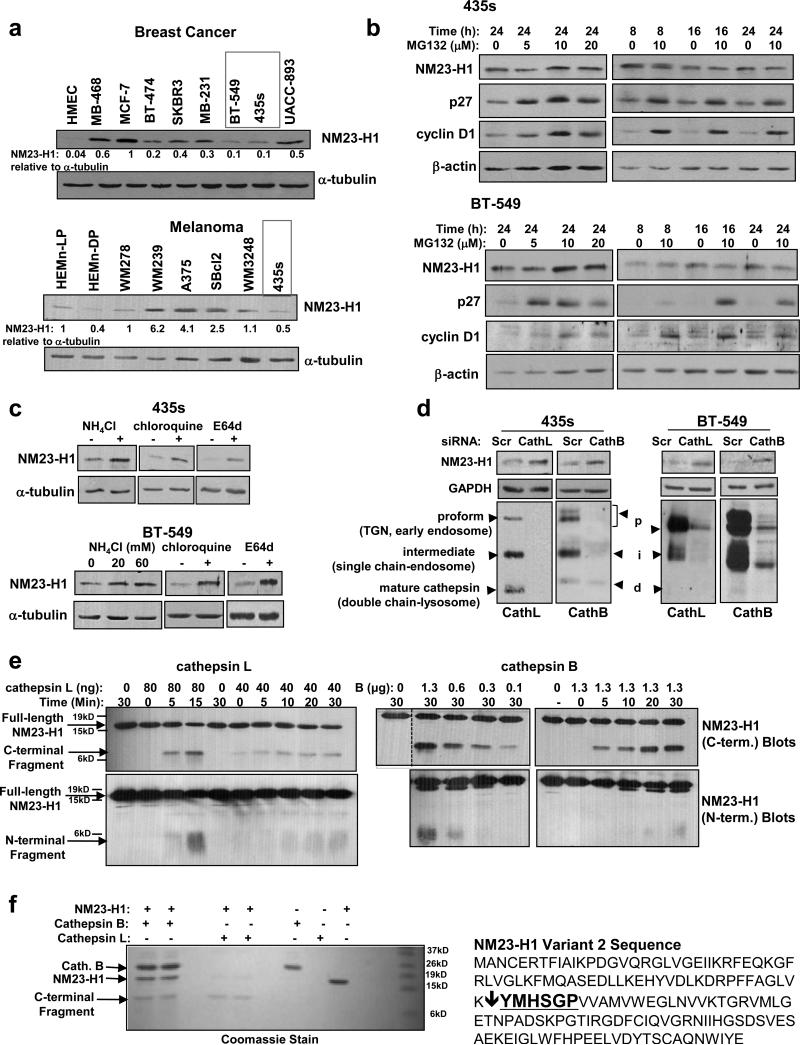
Since reduction in NM23-H1 expression is associated with breast cancer and melanoma progression, we screened a panel of breast cancer and melanoma cell lines to identify lines with extremely low levels of NM23-H1. MDA-MB-435s cells will be termed 435s cancer cells since their origin (breast vs. melanoma) is still under debate (14, 25, 26). Interestingly, we observed an induction in NM23-H1 protein in melanoma and breast cancer cell lines as compared to primary mammary epithelial cells (HMEC) or epidermal melanocytes (HEMn) (Figure 1a). However, a subset of highly invasive cancer lines had dramatically decreased NM23-H1 expression (e.g. 435s and BT-549; Figure 1a). These data confirm that NM23-H1 is induced in primary cancers but suppressed at later stages during progression (4). To identify the mechanism by which NM23-H1 is regulated, we first tested whether NM23-H1 mRNA levels correlate with protein expression using real-time RT-qPCR. We chose RSP13 as the reference gene since it had the least variation among the cell lines (Supplementary Figure 1a,d) (27). Primary cells (HMEC, HEMn) had very low NM23-H1 mRNA as compared to the cancer cell lines, indicating that NM23-H1 induction in primary cancers likely occurs at the mRNA level (Supplementary Figure 1b,c,e,f; Materials and Methods). However, we did not observe a correlation between mRNA and protein levels in the cancer lines (Supplementary Figure 1g), indicating that NM23-H1 loss during metastatic progression is not a result of decreased mRNA in this panel of cancer cell lines.
Figure 1. NM23-H1 is degraded by lysosomal cysteine proteases, cathepsins L and B.
(a) Western blot analysis of lysates from serum-starved breast cancer (top) and melanoma (bottom) cell lines. HMEC=human mammary epithelial cells. HEMn=human epidermal melanocytes. LP=light pigment, DP=dark pigment. (b) Lysates from detached and attached cells treated with the proteosomal inhibitor, MG132, were probed with antibodies. (c) Western blot analysis of lysates from cells treated with lysosomal inhibitors (ammonium chloride, 20mM (BT-549) or 60mM (435s, BT-549); chloroquine, 100μM) or the cysteine protease inhibitor, E64d (20μM) for 8h. (d) Lysates from cell lines transfected with siRNAs were blotted with antibodies 72h after the initial transfection. Mean±SEM for (a-d) are in Supplementary Figure S2. (e,f) Recombinant active human cathepsins L and B were incubated with recombinant NM23-H1, and reactions probed with antibodies for N- and C-termini of NM23-H1. Compared to cathepsin B, smaller amounts of cathepsin L were required to efficiently cleave NM23-H1. This is likely because the recombinant cathepsin B contained both inactive as well as active forms, and had 6X lower specific activity. (f) C-terminal cleavage products were sequenced by Edman Degradation. Input (left), and NM23-H1 sequence and cleavage site (bold; right) are shown. The site was identical for cathepsins B and L.
Next, we examined whether NM23-H1 protein is degraded by the proteasome in 435s and BT-549 cells, which contain very low levels of NM23-H1. Intriguingly, treatment with proteosomal inhibitors (MG132, PS1) effectively stabilized p27 and cyclin D1, known proteasome substrates, but either had no effect or decreased NM23-H1 protein (Figure 1b, Supplementary Figure 1h-j). In contrast, treatment with lysosomal inhibitors (ammonium chloride and chloroquine), a cell-permeable cysteine protease inhibitor (E64d), or silencing cysteine cathepsins L and B, increased NM23-H1 protein (Figure 1c,d and Supplementary Figure S2a-e). These data indicate that NM23-H1 is likely degraded in the lysosome.
To examine whether NM23-H1 is directly cleaved by cathepsins, recombinant, active cathepsins L,B were incubated with recombinant NM23-H1. NM23-H1 was cleaved in a dose- and time-dependent manner, resulting in ≈10kd C-terminal and ≈5-6kd N-terminal fragments (Figure 1e). Mass spectrometry analysis revealed 9.7kd and 6.7kd peaks as well as some smaller species for both cathepsin L and B reactions (Supplementary Figure S2f, not shown). The N-termini of the C-terminal fragments were sequenced and identified as “YMHSGP” (Figure 1f). If this were the only cleavage site, masses of 9.6kD and 7.6kD would be observed (on-line calculator). Since the N-terminal fragments were smaller than predicted (Figure 1e,f and Supplementary Figure S2f), they were likely cleaved into additional fragments, which were too small to visualize.
c-Abl/Arg promote cathepsin expression and activation
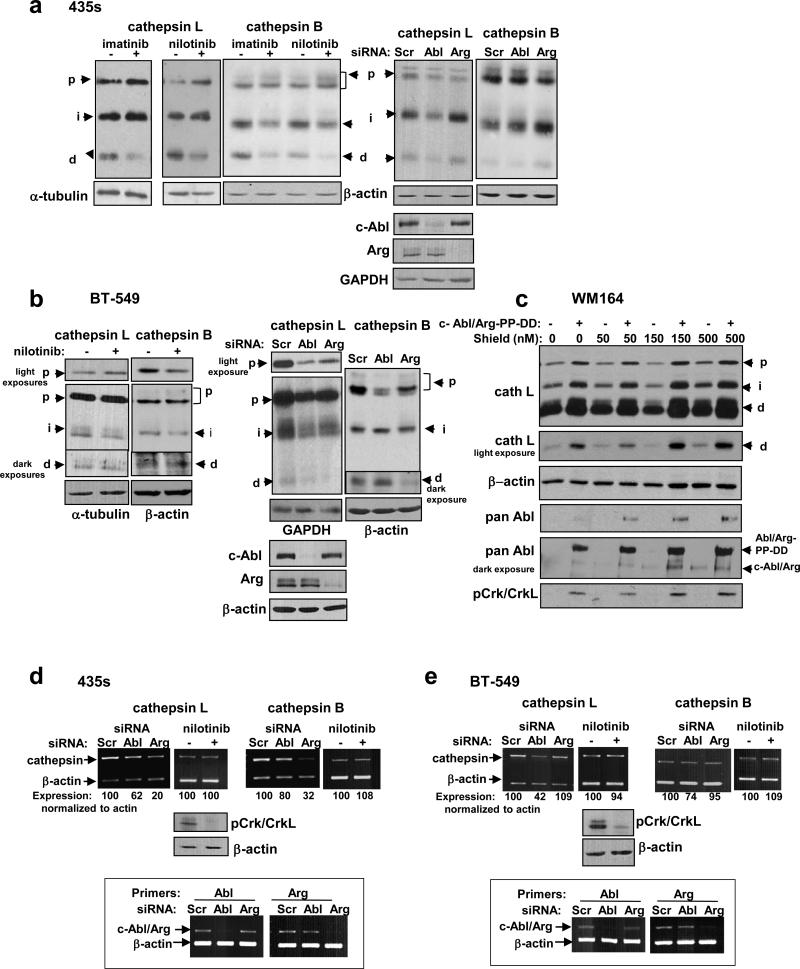
Previously, we showed that c-Abl and Arg non-receptor tyrosine kinases are activated in melanoma and breast cancer cell lines (including 435s and BT-549) (11, 14). Since c-Abl/Arg promote late stages of autophagy, a cathepsin-dependent process (28), we hypothesized that they may be upstream activators of the cathepsins, which in turn degrade NM23-H1. In 435s cells, treatment with the c-Abl/Arg inhibitors, imatinib or nilotinib, decreased cathepsin L,B activation, as the lysosomal double chain forms were reduced whereas proforms (golgi/early endosome) and intermediate forms (late endosome) accumulated (Figure 2a-left and Supplementary Figure S3a,b). In contrast, silencing c-Abl or Arg reduced cathepsin L,B proforms, and silencing c-Abl also reduced double-chain forms (Figure 2a-right and Supplementary Figure S3a,b). Thus, c-Abl and Arg promote cathepsin proform expression independent of their kinase activities, and c-Abl also activates the cathepsins in this line. In BT-549 cells, nilotinib inhibited cleavage of the proform to the intermediate form, silencing c-Abl or Arg inhibited cathepsin proform expression, and knockdown of Arg inhibited cleavage of cathepsin intermediates to their double chain forms (Figure 2b and Supplementary Figure S3c,d). Thus, like in 435s, c-Abl and Arg promote cathepsin proform expression independent of their kinase activities; however, unlike 435s, Arg plays a more prominent role in cathepsin activation in BT-549 cells. Importantly, activation of c-Abl and Arg also is sufficient to induce cathepsin expression and activation, as expression of constitutively active forms of c-Abl/Arg (PP) (29), into a low-invasive melanoma cell line (WM164), induced cathepsin expression (proform) and dramatically promoted activation (cleavage of intermediates to double chains; Figure 2c).
Figure 2. c-Abl and Arg promote cathepsin expression and activation.
(a,b) Western blot analysis of lysates from cells treated with imatinib or nilotinib for 48h (left) or transfected with siRNAs (right), and replated 24h prior to lysis such that densities were equivalent. (c) Western blot analysis of lysates from WM164 melanoma cells stably transfected with constitutively active c-Abl and Arg (PP) or vectors (Migr1/PK1). c-Abl/Arg-PP were DD-tagged in order to prevent high level expression, which is toxic. Significant leakiness was observed, and treatment with Shield1 (4h), which shields the DD tag from proteosomal degradation, further stabilized c-Abl/Arg-PP. Upper arrow=DD-tagged c-Abl/Arg, lower arrow indicates endogenous c-Abl/Arg. (d,e) Cathepsin mRNA levels in cells treated with nilotinib (48h) or transfected with siRNAs (72h) were examined by semi-quantitative RT-PCR. Knockdown efficiency is boxed below. pCrk/CrkL (c-Abl/Arg targets) blots indicate that nilotinib effectively inhibited c-Abl/Arg. p=proform, i=intermediate, d=double chain. Mean±SEM for all subfigures are in Supplementary Figure S3.
To determine the mechanism by which c-Abl/Arg increase procathepsin expression, we examined cathepsin mRNAs. Silencing c-Abl and/or Arg decreased cathepsin mRNA while nilotinib had no effect (Figure 2d,e and Supplementary Figure S3e-h). These data are consistent with the notion that c-Abl/Arg increase cathepsin mRNA and protein in a kinase-independent manner. Interestingly, Arg played a more prominent role in regulating cathepsin mRNA and proform expression in 435s cells, while c-Abl played a more prominent role in BT-549 cells (Fig. 2 and Supplementary Figure S3).
c-Abl and Arg induce downregulation of NM23-H1
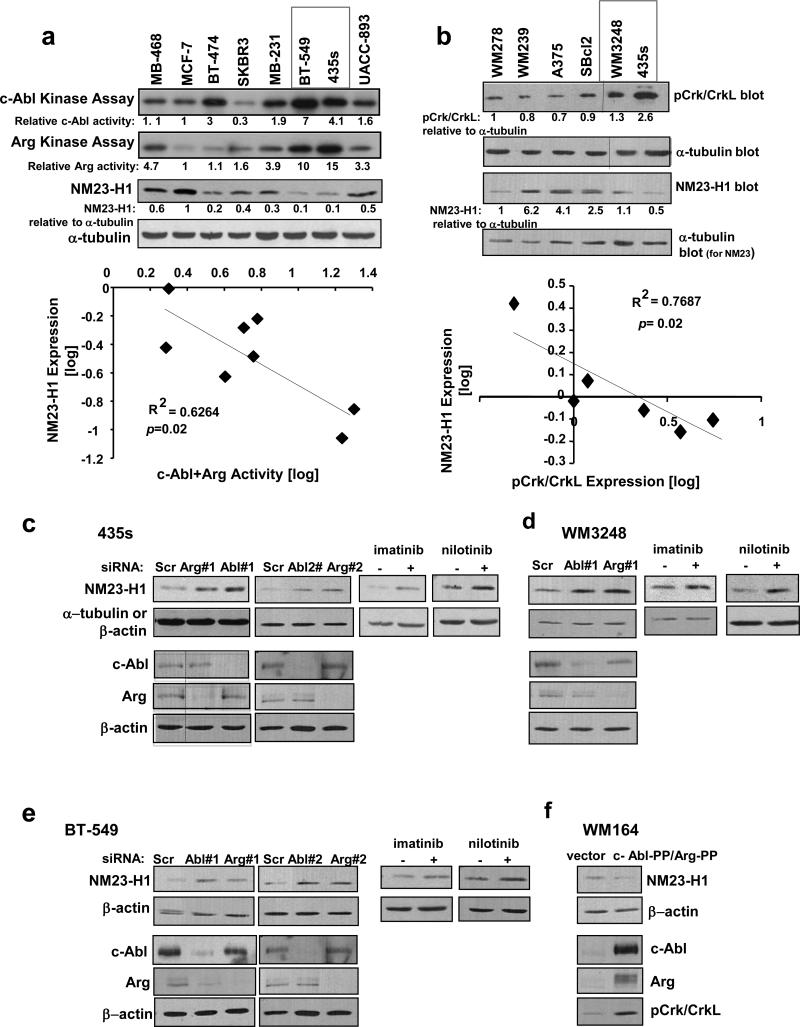
Since c-Abl and Arg activate cathepsins, we tested whether c-Abl/Arg regulate NM23-H1. Interestingly, c-Abl/Arg activities, as assessed by in vitro kinase assay or by phosphorylation of substrates Crk/CrkL, a reliable read-out of c-Abl/Arg activity (11, 14, 28), were inversely correlated with NM23-H1 expression in breast cancer and melanoma cell lines, and 435s and BT-549 cells contain highly active c-Abl and Arg and very low NM23-H1 (Figure 3a,b). Inhibiting or silencing c-Abl or Arg with two independent siRNAs induced NM23-H1 expression in 435s and BT-549 cells, as well as in a third line containing high c-Abl/Arg activity (WM3248; Figure 3c-e and Supplementary Figure S4a-c). Furthermore, expression of constitutively active forms of c-Abl/Arg into WM164 cells reduced NM23-H1 protein (Figure 3f and Supplementary Figure S4d). Thus, activation of c-Abl and Arg is necessary and sufficient for NM23-H1 downregulation in invasive cancer cells. Silencing c-Abl or Arg had no effect on NM23-H1 mRNA (Supplementary Figure S4e), confirming that c-Abl/Arg inhibit NM23-H1 protein expression rather than mRNA.
Figure 3. c-Abl and Arg activation induces loss of NM23-H1 expression.
(a,b) c-Abl/Arg activities were assessed directly by in vitro kinase assay (a) or indirectly via phosphorylation of substrates, Crk/CrkL (b), in breast cancer (a) or melanoma (b) cell lines, bands quantitated, and log-transformed values for c-Abl+Arg activity plotted against log-transformed NM23-H1 expression values. The inverse correlations were statistically significant. (a) Pearson's correlation coefficient =−0.79, 95% confidence interval −0.96 to −0.19, p=0.02. (b) Pearson's correlation coefficient =−0.85, 95% confidence interval −0.98 to −0.139, p=0.03. Kinase assays and pCrk/CrkL and tubulin blots were shown previously (11, 14). (c-e) Cell lines containing highly active c-Abl/Arg were transfected with c-Abl or Arg siRNAs (#1 and #2 are independent siRNAs) or treated with imatinib (10μM) or nilotinib (5μM) for 8h, and NM23-H1 expression examined by western blot. Bands were quantitated, and values expressed relative to loading controls and to scrambled- or vehicle-treated cells. Mean±SEM is in Supplementary Figure S4a-c. Similar results were obtained following 24-72h imatinib/nilotinib treatment (not shown). (f) Western blot analysis of lysates from WM164 melanoma cells transiently transfected with constitutively active c-Abl and Arg (PP) (72h). Mean±SEM is in Supplementary S4d.
c-Abl promotes endosome maturation, which facilitates trafficking of NM23-H1 to the lysosome
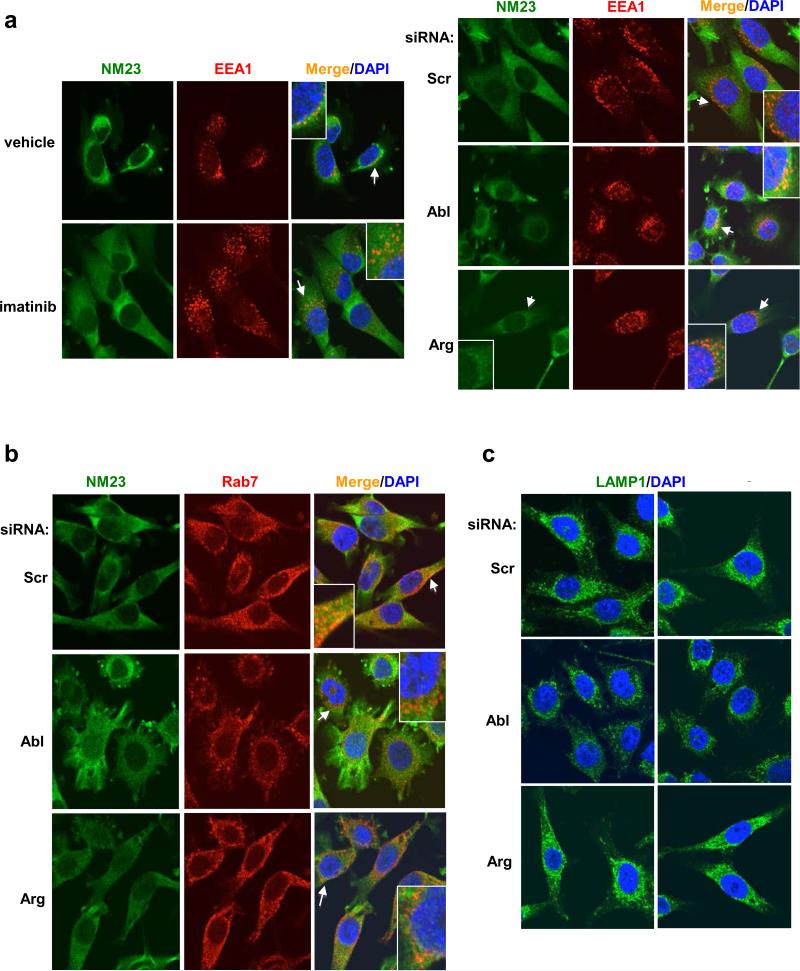
To determine the mechanism by which c-Abl promotes cathepsin cleavage and subsequent NM23-H1 degradation, we examined the effect of c-Abl/Arg on endocytic trafficking. Interestingly, imatinib treatment increased the size of perinuclear EEA1-positive vesicles (early endosome marker; Rab5 effector) (Figure 4a, Supplementary Fig. S5a-top). Moreover, silencing c-Abl induced clustering of EEA1- or Rab5-positive vesicles in a perinuclear region (Figure 4a and Supplementary Figure S5a-bottom, S5b). During endosome maturation, early endosomes are acidified, migrate towards the nucleus along microtubules, and are replaced with larger perinuclear-located endosomes (late endosomes), a process characterized by Rab5/Rab7 switching (30). Thus, we tested whether the clustered EEA1-positive perinuclear vesicles induced by silencing c-Abl express late endosome/multi-vesicular body markers (Rab7). Silencing c-Abl did not alter Rab7 distribution, but modestly reduced Rab7 staining intensity (Figure 4b, Supplementary Fig. S5c-top). Thus, the clumped perinuclear EEA1-positive vesicles are not late or hybrid endosomes. Silencing c-Abl also did not affect LAMP1 (lysosome) distribution, but significantly reduced LAMP1 staining intensity, indicating that there are likely fewer lysosomes present (Figure 4c and Supplementary Figure S5c-bottom). Thus, since c-Abl loss resulted in an accumulation of early endosomes and depletion of late endosomes/lysosomes, we hypothesized that c-Abl may promote endosome maturation. Interestingly, chloroquine treatment, which prevents endosome maturation by preventing lysosome acidification, did not produce the same effects as silencing c-Abl (Supplementary Figure S5d), indicating that c-Abl does not affect the acidification step of endosome maturation (31).
Figure 4. c-Abl promotes endosome maturation.
435s cells treated with vehicle/imatinib (4h) or transfected with siRNAs (72h) were stained with antibodies to markers for early endosomes (EEA1) (a), late endosomes (Rab7) (b), lysosomes (LAMP1) (c) and NM23-H1 (sc-465) (a,b), and counterstained with DAPI. Confocal microscopy pictures, using the brightest EEA1, Rab7 or LAMP1 plane as the focal plane are shown. Arrows denote areas used to create enlarged images. Fields from two independent LAMP1 experiments are shown (c). Quantitation is shown in Supplementary Fig. S5a,c.
Next, we tested whether silencing/inhibiting c-Abl/Arg induces NM23-H1 accumulation in endosomes. Silencing NM23-H1 completely abrogated all NM23-H1 (sc-465) immunofluorescence staining indicating that the antibody used for immunofluorescence is specific for NM23-H1 (Supplementary Figure S5e). In the majority of control cells, NM23-H1 localized to the cytoplasm, and some small NM23-H1-containing puncta appeared to partially colocalize with EEA1 (orange-yellow; Figure 4a). Silencing c-Abl appeared to increase NM23-H1 puncta size and EEA1 colocalization in some cells (orange-yellow vesicles; Figure 4a). In contrast, NM23-H1 colocalized with Rab7 within vesicles in the absence or presence of c-Abl/Arg inhibitors or siRNAs, although silencing c-Abl seemed to modestly reduce colocalization (Figure 4b). These data suggest that a pool of NM23-H1 likely resides in late endosomes, and inhibition/silencing c-Abl may promote its localization to early endosomes likely due to a block in endosome maturation (Supplementary Figure S6).
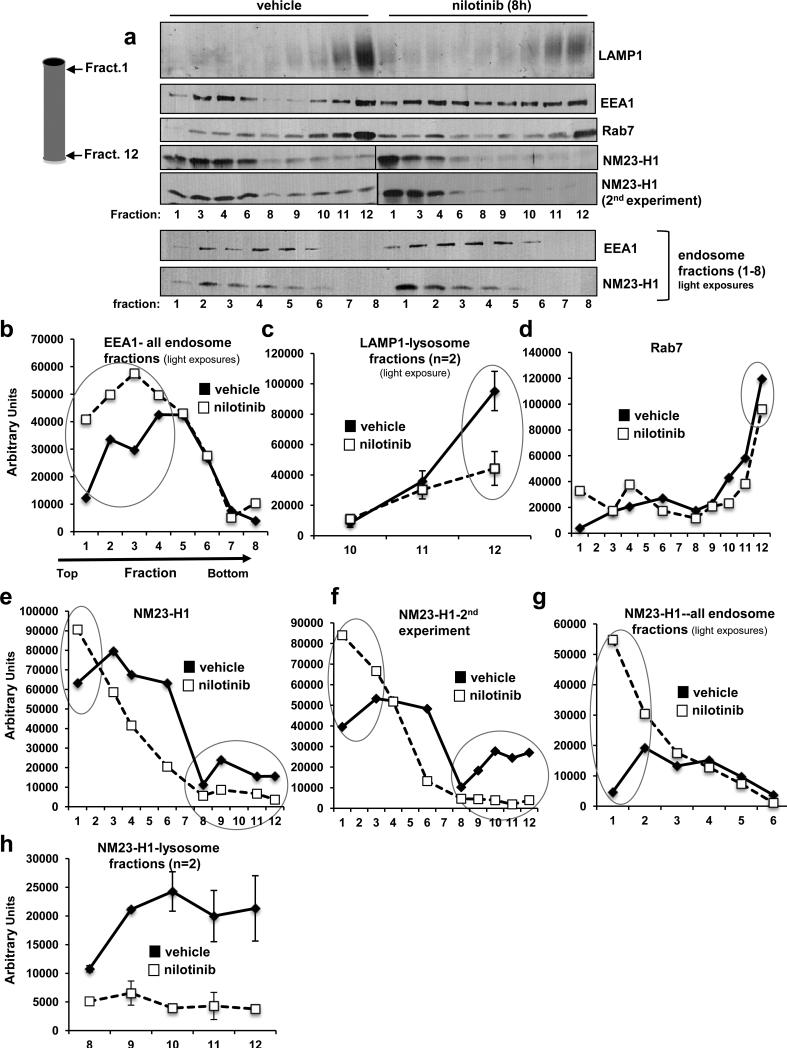
To confirm that NM23-H1 resides in endosomes and that c-Abl/Arg promote endosome->lysosome trafficking, we performed Percoll gradient subcellular fractionation (32). In vehicle-treated cells, Rab7- and LAMP1-positive vesicles were localized in high-density fractions (fractions 10-12), and EEA1-positive vesicles were located predominantly in low-density fractions (fractions 1-6); however, some EEA1-positive vesicles also were present in high-density fractions, likely indicating the presence of Rab5/Rab7 hybrid vesicles (Figure 5a). Nilotinib treatment increased the amount of EEA1-positive vesicles in low-density and intermediate-density fractions (fractions 1-4), decreased the amount of LAMP1 present in high-density fractions (fraction 12), and slightly reduced Rab7 expression in high-density fractions (Figure 5a-d). These data are consistent with the immunofluorescence data and indicate that nilotinib induces an accumulation of low-density early endosomes and decreases lysosome number. In vehicle-treated cells, NM23-H1 was enriched in low-density endosome (fractions 1-6) fractions, but also was present in late endosome/lysosome-enriched fractions (fractions 10-12), and nilotinib treatment increased NM23-H1 localization in low-density endosome fractions and reduced its presence in lysosome fractions (Figure 5a,e-h). Thus, these data are consistent with the notion that c-Abl induces NM23-H1 degradation by promoting its accumulation in lysosomes (Supplementary Figure S6).
Figure 5. Nilotinib inhibits endosome->lysosome trafficking, and induces NM23-H1 accumulation in endosomal fractions and depletion from lysosomal fractions.
(a) 435s cells treated with vehicle/nilotinib (8h) were subjected to Percoll gradient fractionation. Twelve fractions were collected, the indicated fractions (1,3,4,6,8,9-12) from vehicle- and nilotinib-treated cells were run on the same gel, and blotted with EEA1, Rab7 or LAMP-1 antibodies (top). Since all 24 fractions could not be run on one gel and fractions 2,5,7 were missing from the first gel, consecutive vehicle- and nilotinib-treated endosomal fractions (1-8) were rerun on a second gel. Western blots from one of three experiments are shown. Two independent experiments are shown for NM23-H1. (b-h) Bands were quantified and graphed. (c) Mean±SEM, n=2 for fractions 10-12 blotted with LAMP1 antibody. (h) Mean±SEM, n=2 for fractions 8-12 blotted with NM23-H1 antibody.
c-Abl/Arg promote invasion and metastasis by inducing NM23-H1 degradation
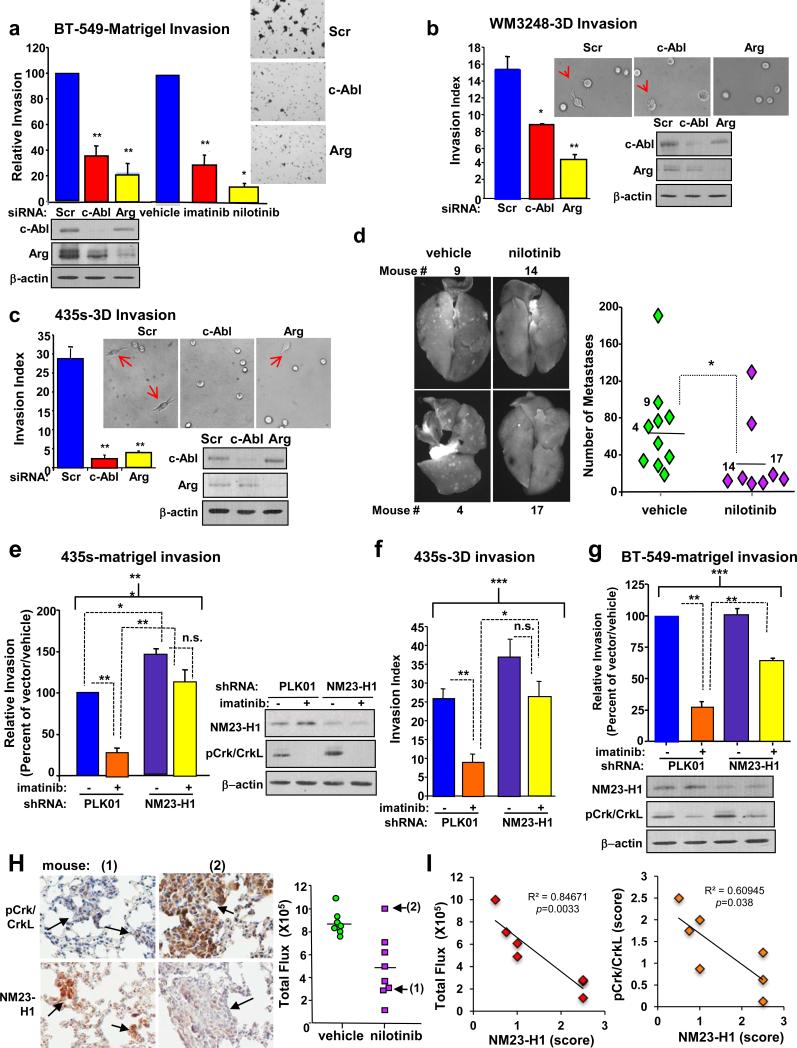
Previously, we demonstrated that activation of c-Abl/Arg drive matrigel invasion (435s, WM3248), anchorage-independent growth (435s, BT-549), and lung colonization/metastasis (435s) (11, 14). Here, we show that c-Abl/Arg also promoted matrigel invasion of BT-549 cells (Figure 6a), single-cell 3D-collagen invasion, which lacks a migratory component (Figure 6b,c) (33), and metastasis of WM3248 cells (Figure 6d). In contrast, expression of exogenous NM23-H1 into invasive cancer cells inhibited matrigel invasion, anchorage-independent growth, and metastasis (6, 34, 35). Thus, we tested whether NM23-H1 downregulation is required for c-Abl/Arg-mediated effects on these processes. NM23-H1 shRNA (or non-targeted control vector, PLK01) was stably expressed in order to prevent stabilization of NM23-H1 following c-Abl/Arg inhibition. In control vector-infected 435s cells, imatinib treatment significantly reduced matrigel and 3D invasion; however, this effect was rescued in cells expressing the NM23-H1 shRNA (Figure 6e,f), indicating that NM23-H1 upregulation is required for imatinib to inhibit invasion. In BT-549 cells, there was only a partial rescue, indicating that, in this cell type, c-Abl/Arg promote invasion via NM23-H1-dependent and -independent mechanisms (Figure 6g). Interestingly, silencing NM23-H1 led to increased activation of c-Abl/Arg (pCrk/CrkL) in both cell types, suggesting the presence of a positive feedback loop (Figure 6e,g). Although silencing NM23-H1 increased soft agar growth, it only modestly rescued the inhibitory effects of imatinib, indicating that c-Abl/Arg promote anchorage-independent growth via NM23-H1-dependent and -independent mechanisms (Supplementary Figure S7a).
Figure 6. c-Abl/Arg promote invasion and metastasis by inducing NM23-H1 loss.
(a) BT-549 cells transfected with siRNAs and serum-starved (16h) or treated with imatinib (10μM) or nilotinib (0.5μM) during starvation were utilized in matrigel boyden chamber invasion assays. Graphs are Mean±SEM for three independent experiments (n=2 for nilotinib); each experiment was normalized to scrambled or vehicle control. *p<0.05 or **p<0.01. Representative fields were photographed. Lysates from a representative experiment were probed with antibodies. (b,c) siRNA-transfected cells were incubated in a single-cell 3D-invasion assay for 5h (b) or 20h (c). Non-invasive and invasive cells were scored, and invasive index calculated (cells with invasive extensions versus the total number of cells multiplied by 100). Graphs are Mean±SEM for three independent experiments; representative fields were photographed. Arrows denote invading cells. *p<0.05, **p<0.01. (d) Nude mice, injected (i.v.) with WM3248 cells expressing GFP were treated with vehicle or nilotinib. Fluorescent surface lung metastases were quantitated on Day 34. vehicle, n=10; nilotinib, n=9. *p<0.05 by t-test. (e,g) Cells expressing non-targeting vector (PLK01) or NM23-H1 shRNA were serum-starved and treated with imatinib for 8h (435s; 10μM) or 16h (BT-549; 5μM), and utilized in a matrigel invasion assay. (f) PLK01/NM23 shRNA-expressing 435s cells, treated with imatinib (24h), were used in single-cell 3D-collagen invasion assays (2h). Lysates from a representative experiment were probed with antibodies. Brackets indicate comparisons among groups (***p<0.001), whereas dotted lines indicate post-hoc pairwise comparisons (*p<0.05, **p<0.01, or n.s. non-significant). (h) Lung sections from mice, injected with 435s cells expressing GFP/luciferase and treated with nilotinib (n=7) (14), were stained with NM23-H1 or pCrk/CrkL antibodies. Representative lungs from responding (low IVIS luminescence; mouse #1) and non-responding (high IVIS luminescence; mouse #2) mice (left) are shown. IVIS flux values for all mice (right) (14). Arrows indicate mice utilized for photographs. (i) NM23-H1 IHC scores were plotted against IVIS flux (left) or pCrk/CrkL scores (right). NM23-H1/Flux Pearson's correlation coefficient = 0.92, 95% confidence interval −0.988 to −0.544, p=0.0033. NM23-H1/pCrk/CrkL Pearson's correlation coefficient =0.78, 95% confidence interval −0.965 to −0.068, p=0.038. Rabbit IgG control is in Supplementary Figure S7b.
Previously, we showed that c-Abl/Arg kinase activity (pCrk/CrkL immunohistochemical (IHC) staining) correlated with metastatic burden (IVIS luminescence) in lung nodules from nilotinib-treated mice injected (i.v.) with 435s-GFP/luciferase cells (14). Responding mice had a few small nodules that had low pCrk/CrkL staining whereas a non-responding mouse had large nodules that stained intensely with pCrk/CrkL antibody, indicating that the anti-metastatic capability of nilotinib is linked to inhibition of c-Abl/Arg kinase activity (14). To determine whether nilotinib-mediated c-Abl/Arg inhibition is associated with upregulation of NM23-H1 in vivo, we stained lungs from nilotinib-treated mice with NM23-H1 antibody. Significantly, NM23-H1 expression was intense in small lesions from mice that responded to nilotinib (mouse #1), whereas metastases from a non-responding mouse (high flux, high pCrk/CrkL staining; mouse #2) expressed low levels of NM23-H1, and NM23-H1 expression was inversely correlated with c-Abl/Arg activity and metastatic burden in all nilotinib-treated mice (Figure 6h,i and Supplementary Figure 7b). Thus, c-Abl/Arg inhibition induces NM23-H1 stabilization, in vitro and in vivo.
c-Abl/Arg activity and NM23-H1 expression are inversely correlated in primary breast cancers and melanomas
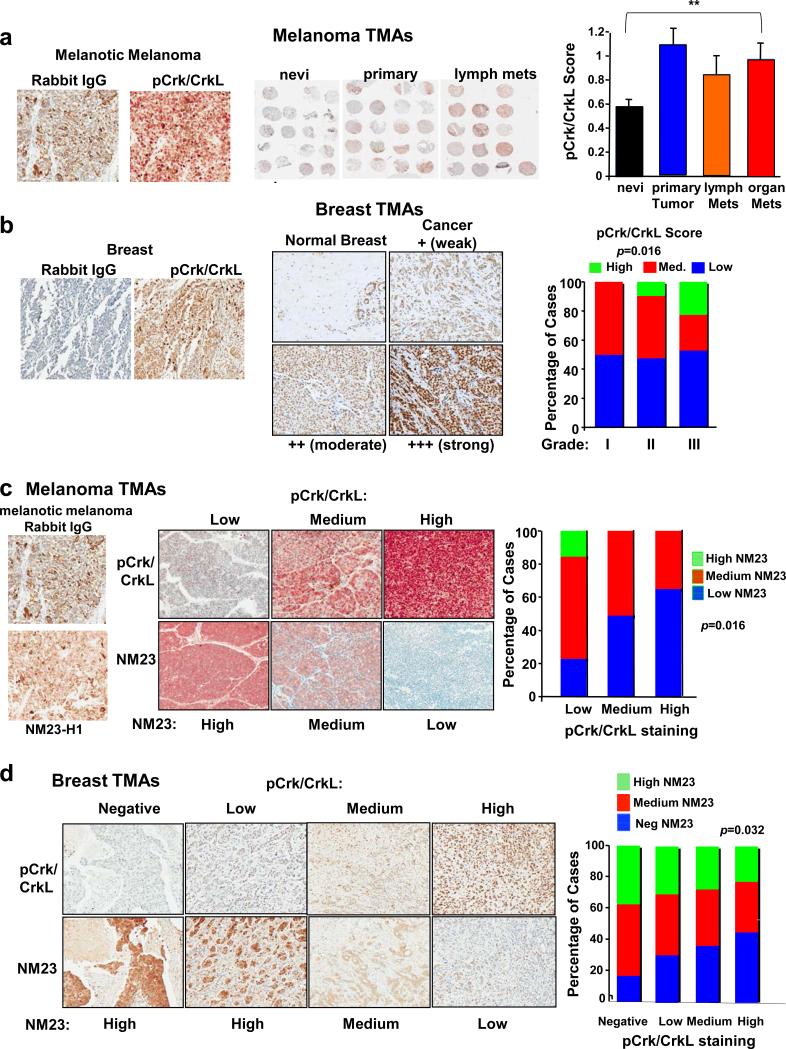
Consistent with our previous data using a small TMA (14), phosphorylation of Crk/CrkL on c-Abl/Arg phosphorylation sites (c-Abl/Arg activity), was elevated in melanomas relative to benign nevi in a second large TMA (Figure 7a). c-Abl/Arg expression levels also were increased in melanoma lines relative to primary melanocytes (14). In addition, c-Abl (Abl1) and Arg (Abl2) mRNAs were increased in primary melanomas versus normal skin in two Oncomine microarray datasets, and Arg mRNA was elevated in metastatic versus primary melanomas (Supplementary Figure S8a-c). Thus, c-Abl/Arg expression and activity are correlated with melanoma disease progression.
Figure 7. c-Abl/Arg activation is inversely correlated with NM23-H1 expression in primary melanomas and breast cancers.
(a) A large melanoma TMA (NCI) containing nevi (n=81), primary melanomas (n=60), and metastases (lymph, n=37; organ, n=67) was stained with pCrk/CrkL or IgG antibody and DakoRed-conjugated secondary antibody. Cores of bad quality were not scored. Score=Intensity (1-3+)*Proportion of positively staining cells. Representative regions of slides (middle), and pCrk/CrkL scores (Mean±SEM, right) are shown. Bracket indicates overall difference among groups (**p<0.01). (b) Breast TMAs (BCO8118, BR1502, BR10010a) were stained with pCrk/CrkL or IgG antibody and DAB-conjugated secondary antibody. pCrk/CrkL staining of normal breast (normal ducts and lymphocytes stain positive) and breast cancers representing weak, moderate and strong intensity staining are shown (TMA BR08188; middle). Distribution of pCrk/CrkL scores among breast cancer grades (grade I, n=26; grade II, n=63; grade III, n=53; right). Scores were grouped into high, medium, and low scores. p=0.016 based on a Fisher's Exact Test (3X3). (c) Melanoma TMAs containing benign nevi (n=24), primary melanomas (n=56), and melanoma metastases (n=20) (ME1003) were stained with pCrk/CrkL, NM23-H1 (D98), or IgG antibodies; representative cores are on left. Cores were grouped into low, medium and high NM23 and pCrk/CrkL staining, and the percentage of cases containing negative, low, or high NM23-H1 staining in each pCrk/CrkL group was graphed (right). An inverse correlation was observed: p=0.016 with a Fisher's Exact test-3X3 and Kendall's tau-b=−0.2637, 95% CI −0.4540 to −0.0734, p=0.009. (d) Breast cancer TMA serial sections (BR10010a; n=100), stained with pCrk/CrkL and NM23-H1 antibodies. IgG control is in Supplementary Figure S8G. Representative cores (left), and IHC score distributions (right) are shown. An inverse correlation between pCrk/CrkL and NM23-H1 was observed (p=0.032; Kendall's tau-b = −0.177 with 95% confidence interval −0.339 to −0.015).
Unlike melanoma, c-Abl/Arg expression does not correlate with activity in breast cancer cell lines (11), and c-Abl/Arg activation has not been assessed in primary breast cancers (15). We found that many breast cancers as well as normal lymphocytes and ducts stained positively with pCrk/CrkL antibody (Figure 7b). Significantly, in contrast to reports suggesting that c-Abl may prevent breast cancer progression (36, 37), we found that c-Abl/Arg activities (pCrk/CrkL scores: intensity * proportion of positively staining tumor cells) increased with increasing breast cancer grade, indicating that c-Abl/Arg activation is associated with disease aggressiveness (Figure 7b-right). While activation of c-Abl/Arg was observed in tumors from all subtypes (estrogen/progesterone receptor-positive, Her-2+, triple-negative, basal-like), there was a trend towards higher activation in triple-negative tumors (Supplementary Figure S8d,e).
In contrast to c-Abl/Arg, NM23-H1 expression is induced in primary breast cancers and melanomas and suppressed at later stages during progression (4). Thus, we examined whether there was a relationship between c-Abl/Arg activity (pCrk/CrkL staining) and NM23-H1 expression in primary tumors. IgG controls lacked staining and NM23-H1 antibody specificity was confirmed by silencing NM23-H1 (Figure 7c-left, Supplementary Figure 8f,g). Significantly, high pCrk/CrkL staining (c-Abl/Arg activity) was associated with low NM23-H1 expression and vice-versa in primary melanomas (Figure 7c-middle, right) and primary breast cancers (Figure 7d). These important data indicate that the Abl/Arg-NM23 axis we identified in cell lines also occurs in the human diseases.
DISCUSSION
Here, we show that cathepsins, proteases with known roles in invasion and metastasis, directly cleave and degrade NM23-H1, which not only explains how NM23-H1 expression is lost, but also provides new insight into how intracellular cathepsins contribute to invasion and metastasis (degradation of metastasis suppressors). We also show that c-Abl/Arg are upstream regulators of NM23-H1 during invasion and metastasis, which uncovers novel crosstalk between metastasis suppressor and oncogenic signaling pathways. Furthermore, c-Abl/Arg induction of cathepsin mRNA demonstrates a new mechanism to explain cathepsin upregulation in cancer cells.
Interestingly, c-Abl also activates cathepsins by promoting endosome maturation, which is important since abnormal vesicular trafficking plays a key role in cancer progression (38). The effects of silencing/inhibiting c-Abl on endosome distribution are reminiscent of the effects observed following overexpression of wild-type Rab5 or dominant-negative Rab7, indicating that depletion of c-Abl may prevent Rab5/Rab7 switching (32, 39). Alternatively, since endosome maturation requires actin polymerization, which is driven by the Arp2/3 complex (40), and c-Abl interacts with Arp2/3 regulators (41), c-Abl may promote trafficking by inducing actin polymerization. Interestingly, silencing Arg had no affect on cathepsin activation in 435s cells, but it did decrease procathepsin expression, and prevent NM23-H1 degradation. Thus, cathepsin proforms, which are transcriptionally upregulated by both c-Abl and Arg, also may be involved in NM23-H1 degradation. Alternatively, Arg may affect a different step in vesicular trafficking, or may promote activation of another cathepsin, not yet identified. Interestingly, NM23-H1 represses aspartyl cathepsin D mRNA, indicating that there may be bidirectional regulation between cathepsins and NM23-H1 (42).
Lysosomal proteases degrade plasma membrane-bound proteins (e.g. RTKs, E-cadherin) via clathrin-mediated endocytosis followed by trafficking to early endosomes (43). Interestingly, NM23-H1, which is degraded by lysosomal proteases, also functions in endocytosis. Drosophila NM23 (awd) promotes synaptic vesicle internalization, and endocytosis of adherens junction components (44, 45). Mammalian NM23 promotes endocytosis of E-cadherin and fibroblast growth factor receptor and decreases phagocytosis of Dictyostelium (46-48). Like NM23, c-Abl/Arg also regulate endocytosis, preventing endocytosis of EGFR and H. pylori, and promoting endocytosis of Notch and Leishmania (49-54). Thus, the balance between c-Abl/Arg activation and NM23-H1 expression may be critical for endocytosis of receptors critical for cancer progression.
Here, we demonstrate that c-Abl/Arg-dependent degradation of NM23-H1 occurs in two different cancer types, in cell lines and primary tumors, in vitro and in vivo, and degradation of NM23-H1 is necessary for c-Abl/Arg to promote cancer progression. Agents that induce upregulation of NM23-H1 are currently being pursued in the clinic (7). Thus, these data have important clinical implications as they demonstrate that FDA-approved c-Abl/Arg inhibitors increase NM23-H1 expression, and thus, could potentially be utilized to prevent metastatic progression.
MATERIALS AND METHODS
Cell Lines and Reagents
MDA-MB-435s cells were obtained from University of North Carolina Tissue Culture Facility (Chapel Hill, NC), BT-549 cells were from Rolf Craven (University of Kentucky), and A375 were from Dr. Suyan Huang (M.D. Anderson Cancer Center, Houston, TX) (14, 55). All sources obtained the lines from ATCC (Manassas, VA). 435s is genetically identical to melanoma M14 (14, 25), and BT-549 cells are identical to ATCC BT-549 (56, 57). WM melanoma cells were obtained from Dr. Meenhard Herlyn (Wistar Institute, Philadephia, PA) (14, 58). Other breast cancer lines were previously described (11).
Stable expression of c-Abl/Arg-PP
The stop codons of c-Abl-PP and Arg-PP (29) were mutated (Quikchange; Clontech, Mountain View, CA), and cDNAs cloned into XhoI/EcoRI sites of Migr1 (c-Abl) or PK1 (Arg) (59). DD tags, which target the proteins for degradation thereby preventing toxic high-level expression, were liberated (NheI fragment; Proteotuner, Clontech) blunted, and cloned in-frame onto the C-termini (blunted EcoRI sites). Transfected cells were selected with puromycin (1μg/ml), and GFP-sorted. Leakiness was detected in untreated cells; Shield1 treatment further increased expression.
The following antibodies were purchased commercially: c-Abl (K12; kinase assay), Arg (9H5-western blot), α-tubulin, NM23-H1 (sc-465; immunofluorescence), LAMP1, cathepsin L1, HRP-conjugated secondary antibodies (Santa Cruz Biotechnology; Santa Cruz, CA); pCrk/CrkL (Y221/Y207), c-Abl (8E9; western blotting), GAPDH, (BD Biosciences; Chicago, IL); EEA1, Rab7, p27, NM23-H1 (D98-immunohistochemistry, D14H1-western blotting), fluorescent secondary antibodies (Cell Signaling; Danvers, MA); cathepsin B (R&D; Minneapolis, MN), cyclin D1 (Millipore; Billerica, MA); Rab5 (Abcam, Cambridge, England) and β-actin (Sigma Aldrich, St. Louis, MO). The Arg kinase assay antibody was previously described (59). Imatinib and nilotinib were provided by Novartis (Basel, Switzerland); IGF-1 was from Upstate Biotechnology (Charlottesville, VA), matrigel invasion chambers and collagen I were from BD Biosciences; chloroquine and ammonium chloride were from Sigma; MG132 and proteasome inhibitor I (PS1) were from Millipore; and E64d was from Tocris (Minneapolis, MN).
Single-Cell 3D-Invasion Assays (33)
One milliliter of collagen I/media solution (pH>9) was allowed to solidify (37°C, 1h). Treated (24h) or siRNA-transfected cells (72h) were plated on collagen, incubated at 37°C/10% CO2, and invasive (containing invasive extensions) and non-invasive (refractile) cells scored from 20 random fields.
Immunofluorescence and Confocal Microscopy
Cells were plated on coverslips, fixed in paraformaldehyde (4%), blocked in goat serum (5%), incubated with primary antibodies (EEA1-1:100, Rab7-1:25, Rab5-1:400, NM23-H1 sc465-1:400, LAMP1-1:10 in PBS/1%BSA/0.3% Triton-X-100), secondary antibodies (488-green or CY3-red; 1:1000), and mounted in ProlongGold Antifade (Invitrogen). Cells were photographed on an Olympus FV1000 laser scanning confocal microscope, 63X objective, magnified 3X. Staining was quantitated on Leica AOBS TCS SP5 inverted Laser Scanning Confocal Microscope with Leica DMI 6000 microscope using LAS AF 2.6.0.7266 build 5194 software (Buffalo Grove, IL).
Percoll-Gradient Fractionation (32)
Cells were suspended in homogenization buffer, dounced, postnuclear supernatants (equal protein) layered on 20% Percoll, spun (20,000 × g; 50’), 12-1ml fractions were collected from the bottom, and CHAPS (10mM final) was added. SDS-PAGE buffer was added to equivalent aliquots, boiled, spun, and equivalent volumes were loaded.
In Vitro Proteolysis (60)
Recombinant NM23-H1 (1μl; 875ng)(61) was mixed with 3μl Buffer L (25mM HEPES pH 7.4, 5mM MgCl2, 1mM EGTA, 0.5% Triton X-100, 5mM DTT), 5μl of recombinant cathepsins, 5μl 3X reaction buffer (150mM sodium acetate pH 6.0, 12mM EDTA, 24mM DTT) and 1μl water at 37°C, and terminated with SDS-PAGE buffer or with glacial acetic acid (15% final, mass spectrometry) (62). Mass analysis was performed on reactions (1:10 dilution) or mock (enzyme or substrate) with an AB Sciex 4800 MALDI TOF/TOF mass spectrometer (Framingham, MA) in linear, mid-mass mode (2-20 kDa) using the Chait thin layer method (63). C-terminal fragments were sequenced by Edman degradation.
Matrigel Boyden Chamber Invasion Assays (11)
Cells were serum-starved and treated with c-Abl/Arg inhibitors or transfected with siRNAs and serum-starved overnight 48h after transfection. Cells were allowed to migrate through matrigel (BD Biosciences, Chicago, IL) towards IGF-1 (10nM, 48h) (11).
Transfection and RNAi (11)
Cells were transfected with the following siRNAs (Applied Biosystems; Carlsbad, CA) using Lipofectamine 2000 (Invitrogen). Abl #1,2 (1336, s886, respectively; 20nM), Arg #1,2 (1478, s872; 20nM), NM23-H1 (s9588; 1nM); cathepsin L1 (s3753; 5nM); cathepsin B (s3740; 5nM), scrambled control #1 (control for non-silencer select), silencer select scrambled #1 (for silencer select). siRNAs were transfected on two consecutive days (except Abl#2 and Arg #2) to increase silencing efficiency. Cells were infected with PLK01-NM23-H1 shRNA (182s1c1) or PLK01 non-targeted shRNA Mission lentiviruses (MOI6; Sigma;12h; 8μg/ml polybrene) and selected with puromycin (1μg/ml).
Kinase Assays and Western Blots
Immunoblotting using fractions or cell lysates (in RIPA buffer) (57), was performed using antibody manufacturers’ protocols. Bands were quantified with ImageQuant (GE Healthcare, Piscataway, NJ) or ImageJ64 (freeware). Kinase assays were performed as previously described (64).
Soft agar Colony Formation
Cells expressing PLK01 or NM23-H1 shRNA (4 ×103 were plated in soft agar −/+ imatinib (12). Colonies (≥ 100 microns) were counted after 3 weeks.
RT-PCR (11)
Semi-quantitative
DNase-treated cDNA was subjected to PCR using specific primers (below) together with β-actin control primers. Aliquots were taken at cycles 27-35 to check for linearity. Bands were quantified with ImageQuant (GE Healthcare) and normalized to β-actin. c-Abl, Arg and β-actin primers were previously described (11).
cathepsin B forward primer, 5’TGGACAAGAAAAGGCCTGGTT3’
cathepsin B reverse primer, 5’CCCAGTCAGTGTTCCAGGA3’
cathepsin L1 forward primer, 5’CTGTGAAGAATCAGGGTCAGTG3’
cathepsin L1 reverse primer, 5’CTGGCTGCTGAGGCAATTC3’.
Real-Time RT-qPCR (Bio-Rad; Hercules, CA)
Primers were designed and blasted with Primer Blast (NCBI website) to rule out homology to other genes. Five reference genes were tested for stability among cell lines (β-actin, HSPCB, RPS13, RPS11, RPII) (27, 65). RPS13 was the most stable (Supplementary Fig. S1). DNase-treated cDNA (50ng; iScript; Bio-Rad) was amplified using SYBER green and NM23-H1 or RPS13 primers (500nM; 40 cycles, 62°C annealing temperature). Results were analyzed with CFX Manager (Bio-Rad).
NM23-H1 forward primer, 5’CAGTGTTACCATCCCCGACC3’
NM23-H1 reverse primer, 5’CAACAATATGAAGTAACCAACTCA3’
RPS13 forward primer, 5’CGAAAGCATCTTGAGAGGAACA3’
RPS13 reverse primer, 5’GCACCACGTCCAATGACAT3'
Immunohistochemistry (IHC) (14)
Tissue microarrays (TMAs) (Biomax ME1003, BR1502, BR10010a, BCO8118, NCI melanoma progression array) or mouse lungs were stained with phospho-Crk/CrkL-Y221/Y207 (1:10), NM23-H1 (1:75) or normal rabbit serum as described (14). Dako Red AEC+ High Sensitivity Chromagen RTU was used for melanoma, and DAB (3,3'-diaminobenzidine) for breast cancer and mouse lungs. Slides were scanned on an AperioScope (20X objective, 20X zoom; Vista, CA). Pathologists scored TMAs blindly on a microscope, except the melanoma array, which they scored at the computer.
Lung Colonization/Metastasis Assays (14)
WM3248 cells, stably transfected with GFP (pcDNA-EGFP-N1), were injected (2×106 cells/100μl Hanks Balanced Salt Solution; Invitrogen) into the tail vein of 7-8 week-old nude (nu/nu) mice (Harlan, Indianapolis, IN). Mice were treated with vehicle (0.5% hydroxymethylcellulose/0.05% Tween-80) or nilotinib (33mg/kg in vehicle; b.i.d.) by oral gavage. On day 34, lungs were fixed in formalin (100%), metastatic nodules counted and photographed on an Olympus MVX-10 stereomicroscope with fluorescence illumination (0.63X objective, 2X-Zoom). SCID-beige mice injected with 435s-GFP/luciferase cells and treated with vehicle/nilotinib were previously described (14). Lungs from nilotinib-treated mice were stained with pCrk/CrkL or NM23-H1 antibody, and scanned on the Aperio (above). The University IACUC Committee approved all studies.
Statistics
Statistical analyses were performed with SAS or the Vassar Website. Some data were normalized to vehicle or scrambled control, and analyzed with one-sample t-tests. Two-tailed values are shown. 0.01≤*p<0.05; 0.001≤**p<0.01; ***p<0.001.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the following individuals for their assistance. Garretson Epperly and James Begley (confocal), Spear/Peterson Labs (qPCR), Jonathan Sims and Divya Srinivasan (cell lysates), Vivek Rangnekar, Suzanne Ridges, Aditi Jain (manuscript review), and Woodrow Friend (Fig. 1a, 6d). Mass spectrometry was performed at the University of Kentucky Proteomics Core Facility, which is supported in part by the Office of the Vice President for Research. Edman Degradation was performed at the University of Nebraska Protein Structure Core Facility. This work was supported by NIH/R01 grants to R.P. (CA116784, CA166499).
Financial Support: This work was supported by NIH grants R01 CA116784 and R01 CA166499 to R.P.
Footnotes
Author contributions: L.S.F. performed experiments for Figs. 2,5,7 and S.S.G. performed experiments in Figs. 3,4,6. L.S.F. and S.S.G. equally contributed data in Fig. 1.
J.S. assisted with experiments performed in Figure 2.
Pathologists, M.L.S., D.L.R., and J.M.N. blindly scored primary tumors.
C.W. performed statistics.
C.B. performed mass spectrometry.
J.R.M. made NM23-H1 constructs and lentiviral stocks.
D.M.K. was involved in the experimental planning and design.
R.P. directed the project and wrote the manuscript.
CONFLICT OF INTEREST
Dr. Plattner is funded by NIH/NCI. D.M.K. also is funded by NIH/NCI, but this work was not supported by his grants. All other authors declare no potential conflicts of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
REFERENCES
- 1.Novak M, Jarrett SG, McCorkle JR, Mellon I, Kaetzel DM. Multiple mechanisms underlie metastasis suppressor function of NM23-H1 in melanoma. Naunyn Schmiedebergs Arch Pharmacol. 2011;384(4-5):433–8. doi: 10.1007/s00210-011-0621-2. Epub 2011/03/31. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS, Horak CE, Miller KD. Clinical-translational approaches to the Nm23-H1 metastasis suppressor. Clin Cancer Res. 2008;14(16):5006–12. doi: 10.1158/1078-0432.CCR-08-0238. Epub 2008/08/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith SC, Theodorescu D. Learning therapeutic lessons from metastasis suppressor proteins. Nat Rev Cancer. 2009;9(4):253–64. doi: 10.1038/nrc2594. Epub 2009/02/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saha A, Robertson ES. Functional modulation of the metastatic suppressor Nm23-H1 by oncogenic viruses. FEBS Lett. 2011;585(20):3174–84. doi: 10.1016/j.febslet.2011.08.007. Epub 2011/08/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horak CE, Lee JH, Marshall JC, Shreeve SM, Steeg PS. The role of metastasis suppressor genes in metastatic dormancy. APMIS. 2008;116(7-8):586–601. doi: 10.1111/j.1600-0463.2008.01213.x. Epub 2008/10/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boissan M, De Wever O, Lizarraga F, Wendum D, Poincloux R, Chignard N, et al. Implication of metastasis suppressor NM23-H1 in maintaining adherens junctions and limiting the invasive potential of human cancer cells. Cancer Res. 2010;70(19):7710–22. doi: 10.1158/0008-5472.CAN-10-1887. Epub 2010/09/16. [DOI] [PubMed] [Google Scholar]
- 7.Marshall JC, Collins J, Marino N, Steeg P. The Nm23-H1 metastasis suppressor as a translational target. Eur J Cancer. 2010;46(7):1278–82. doi: 10.1016/j.ejca.2010.02.042. Epub 2010/03/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, et al. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012;1824(1):68–88. doi: 10.1016/j.bbapap.2011.10.002. Epub 2011/10/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiser J, Adair B, Reinheckel T. Specialized roles for cysteine cathepsins in health and disease. J Clin Invest. 2010;120(10):3421–31. doi: 10.1172/JCI42918. Epub 2010/10/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Braekeleer E, Douet-Guilbert N, Rowe D, Bown N, Morel F, Berthou C, et al. ABL1 fusion genes in hematological malignancies: a review. Eur J Haematol. 2011;86(5):361–71. doi: 10.1111/j.1600-0609.2011.01586.x. Epub 2011/03/26. [DOI] [PubMed] [Google Scholar]
- 11.Srinivasan D, Plattner R. Activation of Abl tyrosine kinases promotes invasion of aggressive breast cancer cells. Cancer Res. 2006;66(11):5648–55. doi: 10.1158/0008-5472.CAN-06-0734. [DOI] [PubMed] [Google Scholar]
- 12.Srinivasan D, Sims JT, Plattner R. Aggressive breast cancer cells are dependent on activated Abl kinases for proliferation, anchorage-independent growth and survival. Oncogene. 2008;27(8):1095–105. doi: 10.1038/sj.onc.1210714. [DOI] [PubMed] [Google Scholar]
- 13.Srinivasan D, Kaetzel DM, Plattner R. Reciprocal regulation of Abl and receptor tyrosine kinases. Cell Signal. 2009;21(7):1143–50. doi: 10.1016/j.cellsig.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganguly SS, Fiore LS, Sims JT, Friend JW, Srinivasan D, Thacker MA, et al. c-Abl and Arg are activated in human primary melanomas, promote melanoma cell invasion via distinct pathways, and drive metastatic progression. Oncogene. 2012;31(14):1804–16. doi: 10.1038/onc.2011.361. Epub 2011/09/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganguly SS, Plattner R. Activation of Abl family kinases in solid tumors. Genes Cancer. 2012;3(5-6):414–25. doi: 10.1177/1947601912458586. Epub 2012/12/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iizuka N, Oka M, Noma T, Nakazawa A, Hirose K, Suzuki T. NM23-H1 and NM23-H2 messenger RNA abundance in human hepatocellular carcinoma. Cancer Res. 1995;55(3):652–7. Epub 1995/02/01. [PubMed] [Google Scholar]
- 17.Ma D, Luyten GP, Luider TM, Jager MJ, Niederkorn JY. Association between NM23-H1 gene expression and metastasis of human uveal melanoma in an animal model. Invest Ophthalmol Vis Sci. 1996;37(11):2293–301. Epub 1996/10/01. [PubMed] [Google Scholar]
- 18.Easty DJ, Maung K, Lascu I, Veron M, Fallowfield ME, Hart IR, et al. Expression of NM23 in human melanoma progression and metastasis. Br J Cancer. 1996;74(1):109–14. doi: 10.1038/bjc.1996.323. Epub 1996/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodall RJ, Dawkins HJ, Robbins PD, Hahnel E, Sarna M, Hahnel R, et al. Evaluation of the expression levels of nm23-H1 mRNA in primary breast cancer, benign breast disease, axillary lymph nodes and normal breast tissue. Pathology. 1994;26(4):423–8. doi: 10.1080/00313029400169122. Epub 1994/10/01. [DOI] [PubMed] [Google Scholar]
- 20.Hwang BG, Park IC, Park MJ, Moon NM, Choi DW, Hong WS, et al. Role of the nm23-H1 gene in the metastasis of gastric cancer. J Korean Med Sci. 1997;12(6):514–8. doi: 10.3346/jkms.1997.12.6.514. Epub 1998/01/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myeroff LL, Markowitz SD. Increased nm23-H1 and nm23-H2 messenger RNA expression and absence of mutations in colon carcinomas of low and high metastatic potential. J Natl Cancer Inst. 1993;85(2):147–52. doi: 10.1093/jnci/85.2.147. Epub 1993/01/20. [DOI] [PubMed] [Google Scholar]
- 22.Sgouros J, Galani E, Gonos E, Moutsatsou P, Belechri M, Skarlos D, et al. Correlation of nm23-H1 gene expression with clinical outcome in patients with advanced breast cancer. In Vivo. 2007;21(3):519–22. Epub 2007/06/27. [PubMed] [Google Scholar]
- 23.Lin LI, Lee PH, Wu CM, Lin JK. Significance of nm23 mRNA expression in human hepatocellular carcinoma. Anticancer Res. 1998;18(1B):541–6. Epub 1998/05/06. [PubMed] [Google Scholar]
- 24.Engel M, Theisinger B, Seib T, Seitz G, Huwer H, Zang KD, et al. High levels of nm23-H1 and nm23-H2 messenger RNA in human squamous-cell lung carcinoma are associated with poor differentiation and advanced tumor stages. Int J Cancer. 1993;55(3):375–9. doi: 10.1002/ijc.2910550306. Epub 1993/09/30. [DOI] [PubMed] [Google Scholar]
- 25.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 melanoma cells--a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2007;104(1):13–9. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 26.Chambers AF. MDA-MB-435 and M14 cell lines: identical but not M14 melanoma? Cancer Res. 2009;69(13):5292–3. doi: 10.1158/0008-5472.CAN-09-1528. Epub 2009/06/25. [DOI] [PubMed] [Google Scholar]
- 27.Jacob F, Guertler R, Naim S, Nixdorf S, Fedier A, Hacker NF, et al. Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLoS One. 2013;8(3):e59180. doi: 10.1371/journal.pone.0059180. Epub 2013/04/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yogalingam G, Pendergast AM. Abl kinases regulate autophagy by promoting the trafficking and function of lysosomal components. J Biol Chem. 2008;283(51):35941–53. doi: 10.1074/jbc.M804543200. Epub 2008/10/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barila D, Superti-Furga G. An intramolecular SH3-domain interaction regulates c-Abl activity. Nature Genet. 1998;18:280–2. doi: 10.1038/ng0398-280. [DOI] [PubMed] [Google Scholar]
- 30.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30(17):3481–500. doi: 10.1038/emboj.2011.286. Epub 2011/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mesaki K, Tanabe K, Obayashi M, Oe N, Takei K. Fission of tubular endosomes triggers endosomal acidification and movement. PLoS One. 2011;6(5):e19764. doi: 10.1371/journal.pone.0019764. Epub 2011/05/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Press B, Feng Y, Hoflack B, Wandinger-Ness A. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J Cell Biol. 1998;140(5):1075–89. doi: 10.1083/jcb.140.5.1075. Epub 1998/04/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Wever O, Hendrix A, De Boeck A, Westbroek W, Braems G, Emami S, et al. Modeling and quantification of cancer cell invasion through collagen type I matrices. Int J Dev Biol. 2010;54(5):887–96. doi: 10.1387/ijdb.092948ow. Epub 2009/09/17. [DOI] [PubMed] [Google Scholar]
- 34.McDermott WG, Boissan M, Lacombe ML, Steeg PS, Horak CE. Nm23-H1 homologs suppress tumor cell motility and anchorage independent growth. Clin Exp Metastasis. 2008;25(2):131–8. doi: 10.1007/s10585-007-9128-0. Epub 2007/12/07. [DOI] [PubMed] [Google Scholar]
- 35.Horak CE, Lee JH, Elkahloun AG, Boissan M, Dumont S, Maga TK, et al. Nm23-H1 suppresses tumor cell motility by down-regulating the lysophosphatidic acid receptor EDG2. Cancer Res. 2007;67(15):7238–46. doi: 10.1158/0008-5472.CAN-07-0962. Epub 2007/08/03. [DOI] [PubMed] [Google Scholar]
- 36.Allington TM, Galliher-Beckley AJ, Schiemann WP. Activated Abl kinase inhibits oncogenic transforming growth factor-beta signaling and tumorigenesis in mammary tumors. FASEB J. 2009;23(12):4231–43. doi: 10.1096/fj.09-138412. Epub 2009/08/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allington TM, Schiemann WP. The Cain and Abl of Epithelial-Mesenchymal Transition and Transforming Growth Factor-beta in Mammary Epithelial Cells. Cells Tissues Organs. 2011;193(1-2):98–113. doi: 10.1159/000320163. Epub 2010/11/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright PK. Targeting vesicle trafficking: an important approach to cancer chemotherapy. Recent Pat Anticancer Drug Discov. 2008;3(2):137–47. doi: 10.2174/157489208784638730. Epub 2008/06/10. [DOI] [PubMed] [Google Scholar]
- 39.Rosenfeld JL, Moore RH, Zimmer KP, Alpizar-Foster E, Dai W, Zarka MN, et al. Lysosome proteins are redistributed during expression of a GTP-hydrolysis-defective rab5a. J Cell Sci. 2001;114(Pt 24):4499–508. doi: 10.1242/jcs.114.24.4499. Epub 2002/01/17. [DOI] [PubMed] [Google Scholar]
- 40.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14(1):7–12. doi: 10.1038/nrm3492. Epub 2012/12/06. [DOI] [PubMed] [Google Scholar]
- 41.Sossey-Alaoui K, Li X, Cowell JK. c-Abl-mediated phosphorylation of WAVE3 is required for lamellipodia formation and cell migration. J Biol Chem. 2007;282(36):26257–65. doi: 10.1074/jbc.M701484200. Epub 2007/07/12. [DOI] [PubMed] [Google Scholar]
- 42.Curtis CD, Likhite VS, McLeod IX, Yates JR, Nardulli AM. Interaction of the tumor metastasis suppressor nonmetastatic protein 23 homologue H1 and estrogen receptor alpha alters estrogen-responsive gene expression. Cancer Res. 2007;67(21):10600–7. doi: 10.1158/0008-5472.CAN-07-0055. Epub 2007/11/03. [DOI] [PubMed] [Google Scholar]
- 43.Platta HW, Stenmark H. Endocytosis and signaling. Curr Opin Cell Biol. 2011;23(4):393–403. doi: 10.1016/j.ceb.2011.03.008. Epub 2011/04/09. [DOI] [PubMed] [Google Scholar]
- 44.Woolworth JA, Nallamothu G, Hsu T. The Drosophila metastasis suppressor gene Nm23 homolog, awd, regulates epithelial integrity during oogenesis. Mol Cell Biol. 2009;29(17):4679–90. doi: 10.1128/MCB.00297-09. Epub 2009/07/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishnan KS, Rikhy R, Rao S, Shivalkar M, Mosko M, Narayanan R, et al. Nucleoside diphosphate kinase, a source of GTP, is required for dynamin-dependent synaptic vesicle recycling. Neuron. 2001;30(1):197–210. doi: 10.1016/s0896-6273(01)00273-2. Epub 2001/05/10. [DOI] [PubMed] [Google Scholar]
- 46.Annesley SJ, Bago R, Bosnar MH, Filic V, Marinovic M, Weber I, et al. Dictyostelium discoideum nucleoside diphosphate kinase C plays a negative regulatory role in phagocytosis, macropinocytosis and exocytosis. PLoS One. 2011;6(10):e26024. doi: 10.1371/journal.pone.0026024. Epub 2011/10/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palacios F, Schweitzer JK, Boshans RL, D'Souza-Schorey C. ARF6-GTP recruits Nm23-H1 to facilitate dynamin-mediated endocytosis during adherens junctions disassembly. Nat Cell Biol. 2002;4(12):929–36. doi: 10.1038/ncb881. Epub 2002/11/26. [DOI] [PubMed] [Google Scholar]
- 48.Hsu T, Adereth Y, Kose N, Dammai V. Endocytic function of von Hippel-Lindau tumor suppressor protein regulates surface localization of fibroblast growth factor receptor 1 and cell motility. J Biol Chem. 2006;281(17):12069–80. doi: 10.1074/jbc.M511621200. Epub 2006/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacob M, Todd LA, Majumdar RS, Li Y, Yamamoto K, Pure E. Endogenous cAbl regulates receptor endocytosis. Cell Signal. 2009;21(8):1308–16. doi: 10.1016/j.cellsig.2009.03.016. Epub 2009/04/07. [DOI] [PubMed] [Google Scholar]
- 50.Wetzel DM, McMahon-Pratt D, Koleske AJ. The Abl and Arg kinases mediate distinct modes of phagocytosis and are required for maximal Leishmania infection. Mol Cell Biol. 2012;32(15):3176–86. doi: 10.1128/MCB.00086-12. Epub 2012/06/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong W, Morillo SA, Rebay I. The Abelson tyrosine kinase regulates Notch endocytosis and signaling to maintain neuronal cell fate in Drosophila photoreceptors. Development. 2012;140(1):176–86. doi: 10.1242/dev.088799. Epub 2012/11/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanos B, Pendergast AM. Abl tyrosine kinase regulates endocytosis of the epidermal growth factor receptor. J Biol Chem. 2006;281(43):32714–23. doi: 10.1074/jbc.M603126200. [DOI] [PubMed] [Google Scholar]
- 53.Bauer B, Bartfeld S, Meyer TF. H. pylori selectively blocks EGFR endocytosis via the non-receptor kinase c-Abl and CagA. Cell Microbiol. 2009;11(1):156–69. doi: 10.1111/j.1462-5822.2008.01246.x. Epub 2008/11/20. [DOI] [PubMed] [Google Scholar]
- 54.Balaji K, Mooser C, Janson CM, Bliss JM, Hojjat H, Colicelli J. RIN1 Orchestrates the Activation of RAB5 GTPases and ABL Tyrosine Kinases to Determine EGFR Fate. J Cell Sci. 2012 doi: 10.1242/jcs.113688. Epub 2012/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang S, DeGuzman A, Bucana CD, Fidler IJ. Level of interleukin-8 expression by metastatic human melanoma cells directly correlates with constitutive NF-kappaB activity. Cytokines Cell Mol Ther. 2000;6(1):9–17. doi: 10.1080/13684730050515868. Epub 2000/09/08. [DOI] [PubMed] [Google Scholar]
- 56.Lorenzi PL, Reinhold WC, Varma S, Hutchinson AA, Pommier Y, Chanock SJ, et al. DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther. 2009;8(4):713–24. doi: 10.1158/1535-7163.MCT-08-0921. Epub 2009/04/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sims JT, Ganguly SS, Bennett H, Friend WJ, J. T, Plattner R. Imatinib reverses doxorubicin resistance by affecting activation of STAT3-dependent NF-κΒ and HSP27/p38/AKT pathways and by inhibiting ABCB1. PLoS One. 2013;8(1):e55509. doi: 10.1371/journal.pone.0055509. Epub 2013 Jan 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, et al. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J Clin Invest. 2005;115(11):3166–76. doi: 10.1172/JCI25001. Epub 2005/10/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plattner R, Kadlec L, DeMali KA, Kazlauskas A, Pendergast AM. c-Abl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes Dev. 1999;13(18):2400–11. doi: 10.1101/gad.13.18.2400. Epub 1999/09/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taha TA, El-Alwani M, Hannun YA, Obeid LM. Sphingosine kinase-1 is cleaved by cathepsin B in vitro: identification of the initial cleavage sites for the protease. FEBS Lett. 2006;580(26):6047–54. doi: 10.1016/j.febslet.2006.09.070. Epub 2006/10/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma D, McCorkle JR, Kaetzel DM. The metastasis suppressor NM23-H1 possesses 3′-5′ exonuclease activity. J Biol Chem. 2004;279(17):18073–84. doi: 10.1074/jbc.M400185200. Epub 2004/02/13. [DOI] [PubMed] [Google Scholar]
- 62.Authier F, Metioui M, Bell AW, Mort JS. Negative regulation of epidermal growth factor signaling by selective proteolytic mechanisms in the endosome mediated by cathepsin B. J Biol Chem. 1999;274(47):33723–31. doi: 10.1074/jbc.274.47.33723. Epub 1999/11/24. [DOI] [PubMed] [Google Scholar]
- 63.Fenyo D, Wang Q, DeGrasse JA, Padovan JC, Cadene M, Chait BT. MALDI sample preparation: the ultra thin layer method. J Vis Exp. 2007;(3):192. doi: 10.3791/192. Epub 2008/11/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitra S, Beach C, Feng GS, Plattner R. SHP-2 is a novel target of Abl kinases during cell proliferation. J Cell Sci. 2008;121(Pt 20):3335–46. doi: 10.1242/jcs.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313(4):856–62. doi: 10.1016/j.bbrc.2003.11.177. Epub 2004/01/07. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.