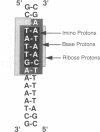
Abstract
The high mobility group (HMG) protein HMG-D from Drosophila melanogaster is a highly abundant chromosomal protein that is closely related to the vertebrate HMG domain proteins HMG1 and HMG2. In general, chromosomal HMG domain proteins lack sequence specificity. However, using both NMR spectroscopy and standard biochemical techniques we show that binding of HMG-D to a single DNA site is sequence selective. The preferred duplex DNA binding site comprises at least 5 bp and contains the deformable dinucleotide TG embedded in A/T-rich sequences. The TG motif constitutes a common core element in the binding sites of the well-characterized sequence-specific HMG domain proteins. We show that a conserved aromatic residue in helix 1 of the HMG domain may be involved in recognition of this core sequence. In common with other HMG domain proteins HMG-D binds preferentially to DNA sites that are stably bent and underwound, therefore HMG-D can be considered an architecture-specific protein. Finally, we show that HMG-D bends DNA and may confer a superhelical DNA conformation at a natural DNA binding site in the Drosophila fushi tarazu scaffold-associated region.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexander-Bridges M., Ercolani L., Kong X. F., Nasrin N. Identification of a core motif that is recognized by three members of the HMG class of transcriptional regulators: IRE-ABP, SRY, and TCF-1 alpha. J Cell Biochem. 1992 Feb;48(2):129–135. doi: 10.1002/jcb.240480204. [DOI] [PubMed] [Google Scholar]
- Amati B., Pick L., Laroche T., Gasser S. M. Nuclear scaffold attachment stimulates, but is not essential for ARS activity in Saccharomyces cerevisiae: analysis of the Drosophila ftz SAR. EMBO J. 1990 Dec;9(12):4007–4016. doi: 10.1002/j.1460-2075.1990.tb07622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazett-Jones D. P., Leblanc B., Herfort M., Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994 May 20;264(5162):1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- Bellon S. F., Coleman J. H., Lippard S. J. DNA unwinding produced by site-specific intrastrand cross-links of the antitumor drug cis-diamminedichloroplatinum(II). Biochemistry. 1991 Aug 13;30(32):8026–8035. doi: 10.1021/bi00246a021. [DOI] [PubMed] [Google Scholar]
- Bellon S. F., Lippard S. J. Bending studies of DNA site-specifically modified by cisplatin, trans-diamminedichloroplatinum(II) and cis-[Pt(NH3)2(N3-cytosine)Cl]+. Biophys Chem. 1990 Apr;35(2-3):179–188. doi: 10.1016/0301-4622(90)80007-t. [DOI] [PubMed] [Google Scholar]
- Berta P., Hawkins J. R., Sinclair A. H., Taylor A., Griffiths B. L., Goodfellow P. N., Fellous M. Genetic evidence equating SRY and the testis-determining factor. Nature. 1990 Nov 29;348(6300):448–450. doi: 10.1038/348448A0. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Falciola L., Ferrari S., Lilley D. M. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992 Mar;11(3):1055–1063. doi: 10.1002/j.1460-2075.1992.tb05144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy E., Rouvière-Yaniv J. HU, the major histone-like protein of E. coli, modulates the binding of IHF to oriC. EMBO J. 1992 Dec;11(12):4489–4496. doi: 10.1002/j.1460-2075.1992.tb05550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles S. S., Pettijohn D. E. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J Mol Biol. 1986 Jan 5;187(1):47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- Bruhn S. L., Housman D. E., Lippard S. J. Isolation and characterization of cDNA clones encoding the Drosophila homolog of the HMG-box SSRP family that recognizes specific DNA structures. Nucleic Acids Res. 1993 Apr 11;21(7):1643–1646. doi: 10.1093/nar/21.7.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn S. L., Pil P. M., Essigmann J. M., Housman D. E., Lippard S. J. Isolation and characterization of human cDNA clones encoding a high mobility group box protein that recognizes structural distortions to DNA caused by binding of the anticancer agent cisplatin. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2307–2311. doi: 10.1073/pnas.89.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A. P., Mardian J. K., Olins D. E. Nonhistone chromosomal protein HMG 1 interactions with DNA. Fluorescence and thermal denaturation studies. J Biol Chem. 1985 Sep 5;260(19):10613–10620. [PubMed] [Google Scholar]
- Cheung S., Arndt K., Lu P. Correlation of lac operator DNA imino proton exchange kinetics with its function. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3665–3669. doi: 10.1073/pnas.81.12.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E., Suzuki M. 'SPKK' motifs prefer to bind to DNA at A/T-rich sites. EMBO J. 1989 Dec 20;8(13):4189–4195. doi: 10.1002/j.1460-2075.1989.tb08604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E., Travers A. A. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991 Mar;16(3):92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Kallenbach N. R., Seeman N. C. A Holliday recombination intermediate is twofold symmetric. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4653–4656. doi: 10.1073/pnas.85.13.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Klug A. Mode of interaction of the zinc finger protein TFIIIA with a 5S RNA gene of Xenopus. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5528–5532. doi: 10.1073/pnas.87.14.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver G. P., Putnam C. D., Denton M. L., Pikaard C. S. The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids. Nucleic Acids Res. 1994 Jul 11;22(13):2651–2657. doi: 10.1093/nar/22.13.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M. Architectural elements in nucleoprotein complexes. Curr Biol. 1993 Oct 1;3(10):675–676. doi: 10.1016/0960-9822(93)90065-v. [DOI] [PubMed] [Google Scholar]
- Dooijes D., van de Wetering M., Knippels L., Clevers H. The Schizosaccharomyces pombe mating-type gene mat-Mc encodes a sequence-specific DNA-binding high mobility group box protein. J Biol Chem. 1993 Nov 25;268(33):24813–24817. [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Harley V. R., Pontiggia A., Goodfellow P. N., Lovell-Badge R., Bianchi M. E. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992 Dec;11(12):4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta-Stogniew E., Russu I. M. Sequence dependence of base-pair opening in a DNA dodecamer containing the CACA/GTGT sequence motif. Biochemistry. 1994 Sep 13;33(36):11016–11024. doi: 10.1021/bi00202a022. [DOI] [PubMed] [Google Scholar]
- Gerchman S. E., Graziano V., Ramakrishnan V. Expression of chicken linker histones in E. coli: sources of problems and methods for overcoming some of the difficulties. Protein Expr Purif. 1994 Jun;5(3):242–251. doi: 10.1006/prep.1994.1037. [DOI] [PubMed] [Google Scholar]
- Giese K., Amsterdam A., Grosschedl R. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 1991 Dec;5(12B):2567–2578. doi: 10.1101/gad.5.12b.2567. [DOI] [PubMed] [Google Scholar]
- Giese K., Cox J., Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992 Apr 3;69(1):185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Giese K., Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994 Mar;10(3):94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Haqq C. M., King C. Y., Ukiyama E., Falsafi S., Haqq T. N., Donahoe P. K., Weiss M. A. Molecular basis of mammalian sexual determination: activation of Müllerian inhibiting substance gene expression by SRY. Science. 1994 Dec 2;266(5190):1494–1500. doi: 10.1126/science.7985018. [DOI] [PubMed] [Google Scholar]
- Harrison S. D., Travers A. A. Identification of the binding sites for potential regulatory proteins in the upstream enhancer element of the Drosophila fushi tarazu gene. Nucleic Acids Res. 1988 Dec 23;16(24):11403–11416. doi: 10.1093/nar/16.24.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haykinson M. J., Johnson R. C. DNA looping and the helical repeat in vitro and in vivo: effect of HU protein and enhancer location on Hin invertasome assembly. EMBO J. 1993 Jun;12(6):2503–2512. doi: 10.1002/j.1460-2075.1993.tb05905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen H. M., Admon A., Bell S. P., Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990 Apr 26;344(6269):830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- Jones D. N., Searles M. A., Shaw G. L., Churchill M. E., Ner S. S., Keeler J., Travers A. A., Neuhaus D. The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster. Structure. 1994 Jul 15;2(7):609–627. doi: 10.1016/s0969-2126(00)00063-0. [DOI] [PubMed] [Google Scholar]
- Kim J. L., Nikolov D. B., Burley S. K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993 Oct 7;365(6446):520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger J. H., Hahn S., Sigler P. B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993 Oct 7;365(6446):512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- King C. Y., Weiss M. A. The SRY high-mobility-group box recognizes DNA by partial intercalation in the minor groove: a topological mechanism of sequence specificity. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11990–11994. doi: 10.1073/pnas.90.24.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlarz D., Fritsch A., Buc H. Variations of intramolecular ligation rates allow the detection of protein-induced bends in DNA. EMBO J. 1986 Apr;5(4):799–803. doi: 10.1002/j.1460-2075.1986.tb04284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm A., Suck D. DNase I-induced DNA conformation. 2 A structure of a DNase I-octamer complex. J Mol Biol. 1991 Dec 5;222(3):645–667. doi: 10.1016/0022-2836(91)90502-w. [DOI] [PubMed] [Google Scholar]
- Landsman D., Bustin M. A signature for the HMG-1 box DNA-binding proteins. Bioessays. 1993 Aug;15(8):539–546. doi: 10.1002/bies.950150807. [DOI] [PubMed] [Google Scholar]
- Landsman D., Bustin M. Assessment of the transcriptional activation potential of the HMG chromosomal proteins. Mol Cell Biol. 1991 Sep;11(9):4483–4489. doi: 10.1128/mcb.11.9.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus L. R., Travers A. A. The Escherichia coli FIS protein is not required for the activation of tyrT transcription on entry into exponential growth. EMBO J. 1993 Jun;12(6):2483–2494. doi: 10.1002/j.1460-2075.1993.tb05903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc B., Read C., Moss T. Recognition of the Xenopus ribosomal core promoter by the transcription factor xUBF involves multiple HMG box domains and leads to an xUBF interdomain interaction. EMBO J. 1993 Feb;12(2):513–525. doi: 10.1002/j.1460-2075.1993.tb05683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy J. L., Kochoyan M., Huynh-Dinh T., Guéron M. Characterization of base-pair opening in deoxynucleotide duplexes using catalyzed exchange of the imino proton. J Mol Biol. 1988 Mar 20;200(2):223–238. doi: 10.1016/0022-2836(88)90236-7. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem Biophys Res Commun. 1983 Jun 29;113(3):967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Megraw T. L., Chae C. B. Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. J Biol Chem. 1993 Jun 15;268(17):12758–12763. [PubMed] [Google Scholar]
- Nakaseko Y., Neuhaus D., Klug A., Rhodes D. Adjacent zinc-finger motifs in multiple zinc-finger peptides from SWI5 form structurally independent, flexibly linked domains. J Mol Biol. 1992 Nov 20;228(2):619–636. doi: 10.1016/0022-2836(92)90845-b. [DOI] [PubMed] [Google Scholar]
- Nasrin N., Buggs C., Kong X. F., Carnazza J., Goebl M., Alexander-Bridges M. DNA-binding properties of the product of the testis-determining gene and a related protein. Nature. 1991 Nov 28;354(6351):317–320. doi: 10.1038/354317a0. [DOI] [PubMed] [Google Scholar]
- Ner S. S., Churchill M. E., Searles M. A., Travers A. A. dHMG-Z, a second HMG-1-related protein in Drosophila melanogaster. Nucleic Acids Res. 1993 Sep 11;21(18):4369–4371. doi: 10.1093/nar/21.18.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner S. S. HMGs everywhere. Curr Biol. 1992 Apr;2(4):208–210. doi: 10.1016/0960-9822(92)90541-h. [DOI] [PubMed] [Google Scholar]
- Ner S. S., Travers A. A., Churchill M. E. Harnessing the writhe: a role for DNA chaperones in nucleoprotein-complex formation. Trends Biochem Sci. 1994 May;19(5):185–187. doi: 10.1016/0968-0004(94)90017-5. [DOI] [PubMed] [Google Scholar]
- Ner S. S., Travers A. A. HMG-D, the Drosophila melanogaster homologue of HMG 1 protein, is associated with early embryonic chromatin in the absence of histone H1. EMBO J. 1994 Apr 15;13(8):1817–1822. doi: 10.1002/j.1460-2075.1994.tb06450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T. T., Haykinson M. J., Johnson R. C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993 Aug;7(8):1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- Pil P. M., Chow C. S., Lippard S. J. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9465–9469. doi: 10.1073/pnas.90.20.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pil P. M., Lippard S. J. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992 Apr 10;256(5054):234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- Pollock R., Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990 Nov 11;18(21):6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontiggia A., Negri A., Beltrame M., Bianchi M. E. Protein HU binds specifically to kinked DNA. Mol Microbiol. 1993 Feb;7(3):343–350. doi: 10.1111/j.1365-2958.1993.tb01126.x. [DOI] [PubMed] [Google Scholar]
- Read C. M., Cary P. D., Crane-Robinson C., Driscoll P. C., Norman D. G. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 1993 Jul 25;21(15):3427–3436. doi: 10.1093/nar/21.15.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read C. M., Cary P. D., Preston N. S., Lnenicek-Allen M., Crane-Robinson C. The DNA sequence specificity of HMG boxes lies in the minor wing of the structure. EMBO J. 1994 Dec 1;13(23):5639–5646. doi: 10.1002/j.1460-2075.1994.tb06902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell S. C., Drew H. R., Travers A. A. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986 Oct 20;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Sheflin L. G., Fucile N. W., Spaulding S. W. The specific interactions of HMG 1 and 2 with negatively supercoiled DNA are modulated by their acidic C-terminal domains and involve cysteine residues in their HMG 1/2 boxes. Biochemistry. 1993 Apr 6;32(13):3238–3248. doi: 10.1021/bi00064a005. [DOI] [PubMed] [Google Scholar]
- Sheflin L. G., Spaulding S. W. High mobility group protein 1 preferentially conserves torsion in negatively supercoiled DNA. Biochemistry. 1989 Jun 27;28(13):5658–5664. doi: 10.1021/bi00439a048. [DOI] [PubMed] [Google Scholar]
- Smith J. M., Thomas D. J. Quantitative analysis of one-dimensional gel electrophoresis profiles. Comput Appl Biosci. 1990 Apr;6(2):93–99. doi: 10.1093/bioinformatics/6.2.93. [DOI] [PubMed] [Google Scholar]
- Stros M., Stokrová J., Thomas J. O. DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res. 1994 Mar 25;22(6):1044–1051. doi: 10.1093/nar/22.6.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Travers A. A., Klug A. The bending of DNA in nucleosomes and its wider implications. Philos Trans R Soc Lond B Biol Sci. 1987 Dec 15;317(1187):537–561. doi: 10.1098/rstb.1987.0080. [DOI] [PubMed] [Google Scholar]
- Travers A. A., Ner S. S., Churchill M. E. DNA chaperones: a solution to a persistence problem? Cell. 1994 Apr 22;77(2):167–169. doi: 10.1016/0092-8674(94)90306-9. [DOI] [PubMed] [Google Scholar]
- Vipond I. B., Halford S. E. Structure-function correlation for the EcoRV restriction enzyme: from non-specific binding to specific DNA cleavage. Mol Microbiol. 1993 Jul;9(2):225–231. doi: 10.1111/j.1365-2958.1993.tb01685.x. [DOI] [PubMed] [Google Scholar]
- Wagner C. R., Hamana K., Elgin S. C. A high-mobility-group protein and its cDNAs from Drosophila melanogaster. Mol Cell Biol. 1992 May;12(5):1915–1923. doi: 10.1128/mcb.12.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir H. M., Kraulis P. J., Hill C. S., Raine A. R., Laue E. D., Thomas J. O. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993 Apr;12(4):1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski J. R., Schulze E. High affinity interaction of dipteran high mobility group (HMG) proteins 1 with DNA is modulated by COOH-terminal regions flanking the HMG box domain. J Biol Chem. 1994 Apr 8;269(14):10713–10719. [PubMed] [Google Scholar]
- van de Wetering M., Clevers H. Sequence-specific interaction of the HMG box proteins TCF-1 and SRY occurs within the minor groove of a Watson-Crick double helix. EMBO J. 1992 Aug;11(8):3039–3044. doi: 10.1002/j.1460-2075.1992.tb05374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M., Oosterwegel M., van Norren K., Clevers H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J. 1993 Oct;12(10):3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Berg O. G. On the specificity of DNA-protein interactions. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1608–1612. doi: 10.1073/pnas.83.6.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H. Protein-DNA recognition: new perspectives and underlying themes. Science. 1994 Feb 11;263(5148):769–770. doi: 10.1126/science.8303292. [DOI] [PubMed] [Google Scholar]