Abstract
Polymerases evolved in nature to synthesize DNA and RNA, and they underlie the storage and flow of genetic information in all cells. The availability of these enzymes for use at the bench has driven a revolution in biotechnology and medicinal research; however, polymerases did not evolve to function efficiently under the conditions required for some applications and their high substrate fidelity precludes their use for most applications that involve modified substrates. To circumvent these limitations, researchers have turned to directed evolution to tailor the properties and/or substrate repertoire of polymerases for different applications, and several systems have been developed for this purpose. These systems draw on different methods of creating a pool of randomly mutated polymerases and are differentiated by the process used to isolate the most fit members. A variety of polymerases have been evolved, providing new or improved functionality, as well as interesting new insight into the factors governing activity.
Keywords: Directed evolution, polymerase, modified nucleotide, protein engineering
1. Introduction
In addition to mediating the storage, retrieval, and transfer of information in all cells, polymerases are the cornerstone of a variety of technologies, ranging from the now indispensible polymerase chain reaction (PCR) [1] to DNA sequencing, and their availability has revolutionized virtually all areas of the biological and medical sciences. Other applications include cloning [2], next generation sequencing by synthesis [3], the diagnosis of genetic diseases [4], the detection of single-nucleotide polymorphisms [5, 6], personal identification [7], and systematic evolution of ligands by exponential enrichment (SELEX) [8]. In all of these applications, the polymerase is used to copy a deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) template into its complement strand of DNA or RNA using monomeric deoxynucleotide triphosphate (dNTP) or ribonucleotide triphosphate (rNTP) precursors. Under physiological conditions, polymerases are remarkably efficient at utilizing their natural substrates, with polymerization rates as high as 103 nucleotides per second and typical fidelities of less than one error for every 107 nucleotides replicated (polymerases with proofreading ability can reach a fidelity of less than one error for every 109 nucleotides replicated) [9]. However, polymerases are poorly active under some conditions of interest due to instability or the presence of inhibitors, and their optimization for better performance under these conditions would enable or facilitate a variety of additional applications. Also, the ability to control transcription from multiple promoters not used by natural RNA polymerases would increase our ability to manipulate cellular pathways for synthetic biology applications [10].
The four nucleotide monomers of DNA and RNA are composed of a nucleobase (guanine, cytosine, adenine, and thymine or uracil), a ribotyl or deoxyribotyl sugar, and a phosphate that links the nucleotide to the preceding nucleotide. Possibly because nucleic acids evolved to be stable, their functionality is actually quite limited from a physiochemical perspective, and this precludes or limits many of the possible applications of polymerases as well as the biopolymers they synthesize. The ability to recognize modified triphosphates would enable synthesis of the corresponding polymers, and the ability to recognize these polymers as templates would enable their amplification or conversion into DNA or RNA.
A related and particularly interesting class of nucleotide modification is replacement of the entire natural nucleobase with an unnatural one, because two such unnatural nucleobases that are efficiently and selectively paired during enzymatic synthesis could form the foundation of an expanded genetic alphabet [11, 12]. In addition to providing unnatural codons that might be used to direct the incorporation of unnatural amino acids into proteins, the modification of unnatural base pairs with linkers could enable the attachment of different functionalities of interest for materials, diagnostics, or SELEX applications [13-16]. Poor recognition of the modified nucleotides by natural polymerases currently limits the development of such expanded genetic systems and thus is a major obstacle toward achieving these ambitious goals.
It is clear that despite the remarkable ability of polymerases to efficiently synthesize natural DNA or RNA, and the revolutionary applications that this has enabled, even more applications would be possible if the properties and activities of polymerases could be appropriately tailored. In nature, this tailoring works by adaptive evolution, an iterative process of selective amplification of the “most fit” members of a population of mutants and their further optimization by mutation and recombination. This process may be recapitulated and accelerated in the laboratory setting, a process commonly known as directed evolution (directed in the sense that a selection pressure is specified by the experimentalist). Over the last several decades, directed evolution has produced a myriad of proteins with optimized properties and reactivities. In addition, unlike with natural evolution, it is straightforward to identify the mutations responsible for the selected phenotype, making it possible to gain unique insights into mechanism.
Herein, we review the directed evolution of polymerases. After a brief discussion of the structures, properties, and substrate repertoires of the natural polymerases, which serve as starting points for directed evolution efforts, we describe the approaches that have been used to generate diversity. We then review in more detail the strategies that have been employed to selectively amplify the most fit mutants. Lastly, we review the polymerase variants that have been evolved from a perspective of the applications that they may eventually enable and the insight that they provide into polymerase function.
2. Polymerases in nature
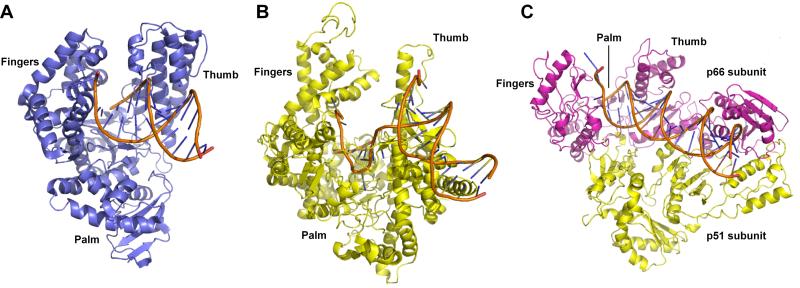
In general, evolution in nature is thought to proceed from progenitor proteins where some level of the desired activity is already present [17, 18]. While it is obvious that directed polymerase evolution should begin with a natural polymerase, the natural polymerases constitute an ancient class of proteins that diverged long ago into different families with subtly different properties and activities, which might make one more suitable than another as starting points. Most generally, polymerases can be classified into three categories based on their substrates and products: DNA polymerases (DNAPs), RNA polymerases (RNAPs), and Reverse Transcriptases (RTs). DNAPs synthesize strands of DNA complementary to DNA templates (following Watson-Crick base pairing), and are responsible for all DNA replication during cell division as well as the DNA replication involved in DNA recombination and repair, and lateral gene transfer in prokaryotes. Generally, polymerases fold into a common overall structure that resembles a right hand with a palm, fingers, and thumb subdomain (Fig. 1). DNAP-catalyzed synthesis of DNA generally includes five steps: (1) binary complex formation between the DNAP and a DNA template; (2) ternary “open” complex formation upon binding a dNTP; (3) a conformational change from the open to a closed state when the bound dNTP forms a correct Watson-Crick base pair with the template nucleotide in the active site; (4) catalysis of phosphodiester bond formation between the dNTP and the 3’OH of the primer terminus; (5) a conformational change from the closed to the open state and release of an inorganic pyrophosphate [19-22]. During processive synthesis, the DNAP then translocates the newly elongated primer/template to position the next templating nucleotide in the active site.
Fig. 1.
Common polymerase fold, with palm, thumb, and fingers domains, illustrated with SF DNAP (PDB ID: 1QSY) (A), T7 RNAP (PDB ID: 1QLN) (B), and RT from HIV-1 (PDB ID: 1RTD) (C). Primer/template is shown in orange.
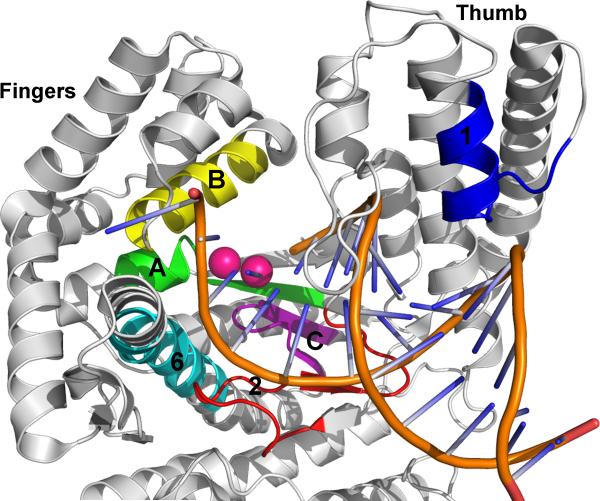
Based on sequence homology, DNAPs are classified into six families: A, B, C, D, X, and Y [23]. Classification within families A, B, and C is based on homology with E. coli polymerase genes, polA, polB, and polC, respectively, which correspondingly encode Pol I, Pol II, and the α subunit of Pol III [24]. Extensive effort has been focused on rationally optimizing family A DNAPs, especially E. coli Pol I, or its truncated variant Klenow fragment (Kf); Taq DNAP from Thermus aquaticus (Taq), or one of its truncated variants Stoffel fragment (Sf) or KlenTaq; and the DNAP from T7 bacteriophage, due to their already widespread use and because they are functional in the absence of accessory proteins (with the exception of T7 DNAP, which requires E. coli thioredoxin as a cofactor). Family A DNAPs have six common structural motifs: A, B, C, 1, 2, and 6 that surround and form the active site and interact with substrates (Fig. 1A and 2) [25]. Motifs A, C, and 2 are located in the palm domain; Motifs B and 6 are located in the fingers domain; and motif 1 is located in thumb domain. Motifs A, B, and C are more conserved than motifs 1, 2, and 6, especially at the amino acid level, consistent with their functional importance.
Fig. 2.
Structure of SF DNAP with A (green), B (yellow), C (purple), 1 (blue), 2 (red), and 6 (cyan) structural motifs illustrated (orange: primer/template DNA; pink spheres: catalytic magnesium ions).
RNAPs synthesize RNA molecules complementary to DNA templates, and are responsible in all cells for the initiation of DNA synthesis and all RNA synthesis, and in RNA viruses for information storage. RNAPs recognize specific sequences of DNA, referred to as promoters and terminators, to initiate and halt transcription, respectively. Relatively simple, single subunit RNAPs include those from many viruses, such as T3, T7, SP6, and K11 phages, and from the mitochondria of eukaryotic cells [26], which like their DNAP counterparts, assume a right-hand-like overall structure (Fig. 1B). However, core bacterial RNAPs, such as that from E. coli, include five subunits (β’, β, αI, αII, ω), as well as another small subunit, σ, which is responsible for promoter recognition [27, 28]. Eukaryotic genomes encode five multiple subunit RNAPs, RNAP I–V, which are responsible for the synthesis of different RNAs [29-33]. The RNAPs of bacteria and archaea are most structurally and mechanically similar to eukaryotic RNAP II [34-36]. Regardless of their subunit architecture, RNAPs function similarly to DNAPs, except that full length transcription is preceded by an abortive phase, during which short RNAs are synthesized [37].
RTs synthesize DNA from an RNA template, and are employed by RNA viruses to synthesize complementary DNAs (cDNAs) that guide subsequent transcription and replication processes. Although most well-studied RTs are from retroviruses, they are also found in prokaryotes and eukaryotes. For example, telomerases, which are responsible for maintaining the ends of chromosomes of eukaryotic cells and which carry their own RNA templates, belong to the RT family of polymerases [38]. Retroviral RTs commonly have three distinct activities: an RNA-dependent DNAP activity (responsible for synthesis of single-strand DNA from RNA templates), ribonuclease H activity (responsible for degradation of RNA), and a DNA-dependent DNAP activity (responsible for synthesizing another strand of DNA to form double-strand DNA) [39]. The polymerase domains of RTs, such as the p66 polymerase domain of HIV-1 RT, are generally structurally and mechanistically similar to their distant DNAP and RNAP cousins (Fig. 1C).
3. Diversification Strategies
Directed evolution methodologies simulate the processes of adaptive evolution in nature, in which species are diversified via the mutagenesis and recombination of their DNA, and then subjected to the natural selection provided by environmental pressures. Similarly, directed evolution typically includes: 1) construction of a pool (or “library”) of variants of the gene of interest differentiated from each other by an at least roughly controllable number of mutations; 2) enriching the pool in members with a desired activity (i.e. selection of the most fit mutants); and 3) analysis of individual members of the enriched pool or subjecting the pool to further rounds of diversification and enrichment.
During directed evolution, random mutagenesis is used to simulate natural spontaneous mutation and may be performed using error-prone PCR (epPCR) in which the mutation rate can be controlled by adding different amounts of manganese to reduce polymerase fidelity [40]. Random mutations may also be introduced via gene shuffling, in which the gene is first digested with DNase I, and then reassembled by the inherently mutagenic process of PCR fragment assembly and amplification [41]. EpPCR and gene shuffling subject the entire gene, or at least the PCR amplified portion, to mutagenesis, and if more focused libraries are desired, for example, focused to an active site or specific secondary structural elements, then synthetic oligonucleotides may be used, either in a simple PCR assembly reaction [42] or in a variant of shuffling, known as synthetic shuffling, where synthetic oligonucleotides are added to the reassembly reaction [43].
Adaptive evolution in nature is thought to proceed in part via beneficial epistatic (non-additive) effects of otherwise neutral or nearly neutral mutations [44]. Practitioners of directed evolution may draw upon these effects via a variant of gene shuffling, known as family shuffling. In family shuffling, the DNase I fragments used in the reassembly step are generated from a set of homologs, and thus the diversity present in nature is sampled in novel combinations. Family shuffling has proven to be an effective way to construct high-quality libraries; however, reassembly is relatively inefficient when the method is applied to regions with low sequence homology [45]. To improve reassembly, RACHITT (random chimeragenesis on transient templates) was developed as a substitute for DNA shuffling. RACHITT is similar to traditional shuffling in that parent sequences are first fragmented, but before the reassembly step, the resulting fragments are hybridized to a scaffold template and subjected to overhanging tail digestion and gap filling [46]. In addition to sampling novel reassortments of natural diversity, the mutations introduced are less likely to cause misfolding, which otherwise reduces the size of the library. As with the original implementation of DNA shuffling, family shuffling has the additional advantage of providing a mechanism to separate (or “unlink”) beneficial and deleterious mutations [45].
Recently, various semi-rational approaches have been developed that combine various randomization methods with sequence, structure, or structure-activity data to more efficiently engineer proteins, including iterative saturation mutagenesis (ISM), combinatorial active-site saturation testing (CASTing), and statistical analysis of protein sequence activity relationships (ProSAR). In ISM, a limited number of residues within focused regions of the protein of interest are randomized in separated libraries and then subjected to selection or screening [47]. The most fit mutants are then used as the templates for the next round of mutation and selection. The process is repeated iteratively, possibly including exploration of the same site in nonsequential rounds of evolution. An advantage of this method is that sequence space at the selected sites may be fully explored; however, its focus on the sequential examination of different sites may preclude the identification of epistatic mutations. In contrast, epistatic effects between physically interacting residues are sought in the CASTing approach by focusing mutations to sets of spatially localized residues, for example, n and n+1, 2, 3, or 4 residues in a loop, β sheet, 310 helix, or α helix, respectively [48]. Lastly, the most guided of these newer approaches, statistical analysis of protein sequence activity relationships (ProSAR), assumes that the contributions of different mutations are additive (non-epistatic), and generates linear equations based on sequence-activity data of mutants from previous rounds of evolution [49]. Regression of these linear equations yields output parameters that indicate the contribution of different residues when at different positions, which are then used to guide further engineering of the protein.
4. Screening and selection strategies
In order to enrich a library for members with a desired property or activity, an efficient selection or screening method is required. Screening and selection differ in that the former samples every member individually while the latter provides a method to selectively isolate or amplify the most fit members directly from the library.
4.1. Screening
Screens are conceptually straightforward to implement, but they are only capable of sorting through a relatively small number of mutants, typically on the order of 1000 (or higher if automated liquid handling systems are used). Traditionally, screens for polymerase activity have employed gel electrophoresis to detect radio- or fluorophore-labeled primer extension or have directly detected the amplification products of a PCR reaction. Recently, ELISA-like screening methods have also been developed [50]. In a typical ELISA-like screen, the primer/template complex is attached to the surface of individual wells of a 96 or 384-well plate, and a probe is incorporated into the primer during extension, or annealed onto an extended primer after denaturing the primer/template complex. The probe may be a fluorophore [51], allowing for direct spectrophotometric detection, or an affinity tag (i.e. biotin or an antigen), allowing for a traditional sandwich assay with a streptavidin- or antibody-enzyme conjugate and detection with enzyme-catalyzed chromogenic reaction [50]. A high-throughput ELISA screen for polymerases using oligonucleotide arrays instead of multiwell plates has also been reported [52].
4.2. Selections
Selections are capable of sorting through much larger libraries, with as many as 1011 to 1012 members, depending on the specific methodology utilized, but they are complicated by the need to maintain a link between phenotype and genotype. Because no selection is capable of enriching such large libraries to a single member, screens, as described above, are often run to sort through a selection-enriched library. For directed polymerase evolution, three selection methodologies have been developed.
4.2.1. Genetic complementation and autogene selection
Loeb and co-workers developed a genetic complementation system for the in vivo selection of DNAP mutants in a strain of E. coli in which Pol I is inactive at 37 °C [53]. After transformation with a library of mutant polymerase genes, growth at the non-permissive temperature selects for mutants that retain sufficient natural activity to support genome replication. This approach may be used with Pol I, Taq DNAP, or HIV RT, which are each able to complement deficient host Pol I activity [53-55]. Because Pol I is required for the replication of DNA immediately downstream of a ColE1 origin of replication, genes included within this region may be used to provide specific selection pressures. For example, Brackmann and co-workers evolved a mutagenic variant of T7 DNAP by selecting for mutational reversion of a β–lactamase gene and growth in the presence of ampicillin [56].
An approach referred to as autogene selection has been developed to evolve RNAPs that recognize unique promoter sequences, and has been run in two formats. In the original format, transcription of a mutant RNAP is driven from the target promoter, and RNAP recognition of the promoter results in RNAP over-expression and reduced growth [57]. To select instead for increased growth, a second format has been developed in which an essential gene, such as one whose protein product confers antibiotic resistance, is placed behind the altered promoter [58]. Recently, Liu and co-workers developed an autogene selection for altered T7 RNAP promoter recognition that they have termed phage-assisted continuous evolution [59]. In this system, hypermutagenic E. coli cells harbor a plasmid carrying the altered promoter that controls transcription of the M13 phage pIII gene. When these cells are infected with M13 phage bereft of the gene encoding their own pIII and harboring a mutant RNAP, growth in a “lagoon” with constant inflow and outflow of cell culture allows for RNAP evolution without the laborious steps of gene isolation, mutagenesis, and re-transformation between rounds of selection.
4.2.2. Activity-based phage display selection
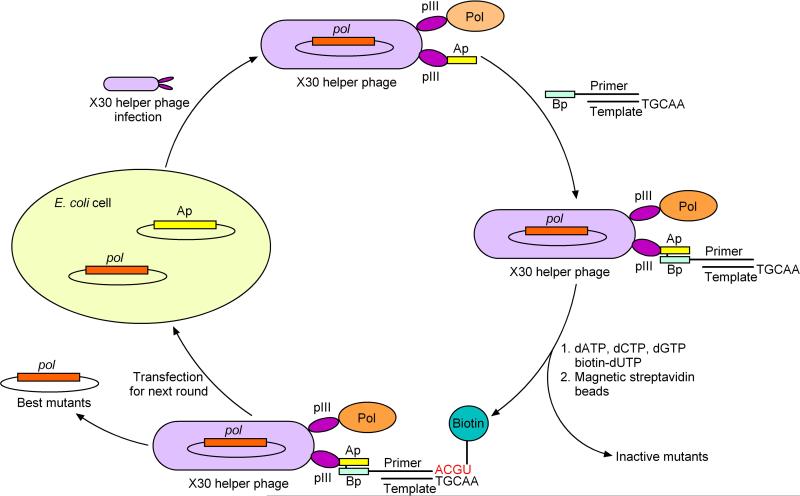
A general disadvantage of both the complementation and autogene approaches for directed polymerase evolution is that they may not be used to select for novel activities involving unnatural substrates. Phage display has proven to be a robust methodology for the selection of proteins from a library based on affinity for a target, and our laboratory has developed a modified version that selects for polymerase mutants based in their having desired activities and that may be straightforwardly applied to selections with unnatural substrates (Fig. 3) [60]. In this method, the polymerase mutant is expressed as an N-terminal fusion to the minor M13 phage coat protein pIII, and on average one fusion protein is packaged within the coat of each phage particle. The remainder of the five pIII proteins are fused to a short acid peptide that contains a cysteine residue and that forms a coiled-coil with a basic peptide. The basic peptide is chemically synthesized and covalently coupled to an oligonucleotide that acts as a primer for oligonucleotide synthesis. After the basic peptide-primer conjugate is annealed to a template oligonucleotide, it is added to the phage particles, resulting in coiled-coil and disulfide bond formation and the localization of a polymerase mutant, the gene that encoded it, and a substrate on the same phage particle. Active polymerases extend their attached primers, which leads to the incorporation of a biotinylated nucleotide. When incorporation of the biotinylated nucleotide occurs after the desired activity (i.e. after incorporation of a natural or modified nucleotide triphosphate opposite a natural or modified nucleotide in the template), phage particles whose mutant polymerases have an increased ability to catalyze the desired reaction will be preferentially biotinylated and recovered with magnetic beads coated with streptavidin. Selection proceeds through multiple rounds until fit mutants become sufficiently enriched in the population that intramolecular reactivity does not dominate intermolecular activity. At this point the library may be screened to identify individual mutants. An advantage of this approach is that the incorporation of unnatural substrates is possible in the template, primer, or triphosphate, and that selection does not require amplification.
Fig. 3.
Phage display-based selection technique (Ap: acidic peptide; Bp: basic peptide; Pol: polymerase protein; pol: polymerase gene). Only two copies of the pIII protein are shown for clarity.
4.2.3. Compartmentalized Self-Replication (CSR) and its derivatives
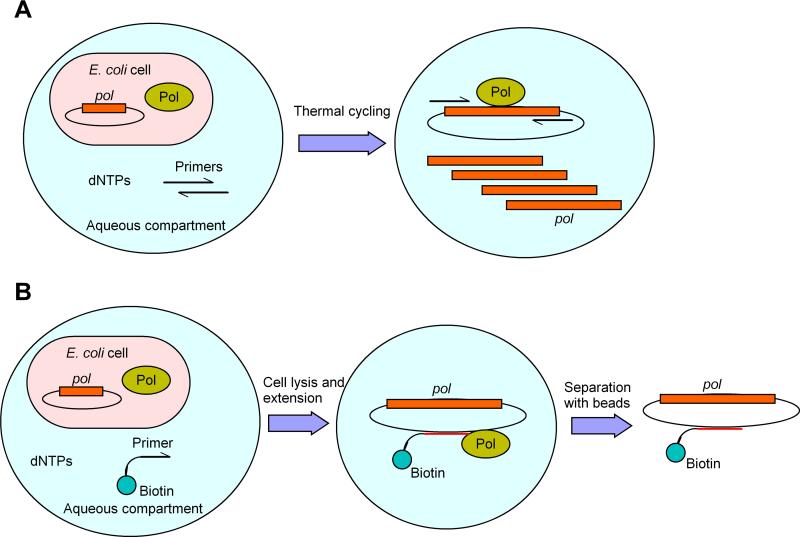
Water-in-oil emulsions may be used to provide transiently stable microcompartments and Holliger and co-workers have used this methodology for polymerase evolution (Fig. 4) [61]. In this system, E. coli cells harboring a mutant polymerase gene are encapsulated in water-in-oil emulsion bubbles such that each bubble contains only a single E. coli cell. Triphosphates and primers suitable for amplification of the polymerase gene are also encapsulated, the polymerase gene is expressed, and the bubbles are subjected to thermal cycling. During thermal cycling, the E. coli cells are lysed, releasing the polymerase mutant and the gene that encoded it. If a particular mutant is active with the provided substrates, then its corresponding gene is amplified and thus the total population of DNA will be enriched for those that encode active mutants. The enriched library may be subjected to further rounds of selection or screened for active members. A modified CSR method, spCSR (short-patched CSR), focuses amplification to a target region of the polymerase gene, which after amplification is assembled into a full length gene and assayed or further evolved [50]. Another variant of CSR, termed compartmentalized self-tagging (CST), has been developed wherein a biotinylated primer is used that hybridizes to the polymerase gene, allowing plasmid recovery even after only limited extension. Recently, Ellington and co-workers adapted the approach for in vitro use, employing water-in-oil emulsion to link genotype and phenotype [62]. In their method, the transcription and translation of T7 RNA polymerase proceeded within emulsified E. coli cell lysate, instead of the whole cells used in the traditional autogene selection, and SP6 RNA polymerase was employed to trigger the initial in vitro transcription via an SP6 promoter embedded downstream of the T7 promoter, which is responsible for subsequent positive-feedback transcription as in autogene selection.
Fig. 4.
Water-in-oil emulsion-based polymerase selection technique (Pol: polymerase protein; pol: polymerase gene). (A) CSR; (B) CST.
5. Evolved polymerase mutants with various properties and applications
Using a combination of methods for library construction and selection and/or screening for fit mutants, a variety of different polymerases with improved or novel properties or activities have been evolved for various practical applications, and these are reviewed here.
5.1. Polymerase mutants with altered fidelity
Although natural DNAPs typically have extremely high fidelity, even higher fidelities would facilitate several applications, such as next generation sequencing by synthesis and single nucleotide polymorphism detection. Towards this goal, Summerer et al. evolved exonuclease deficient Kf for higher fidelity against mismatch extension [63]. A library was constructed by randomizing residues of motif C (879-881), which is involved in mismatch recognition in family A and B polymerases [64], and screened using a primer extension assay in a 384-well plate format. The most promising three mutants, PLQ, LVG, and LVL (referring to the residues present at positions 879-881), showed significantly decreased mismatch extension, and when the latter was moved into Taq (replacing QVH), the resulting DNAP exhibited discrimination that translated into improved performance in allele specific PCR. This work demonstrates that similar mutations can confer similar activities in related DNAPs. Strerath et al. created a Taq library with mutations focused to residues Q782, V783 and H784 within motif C that they screened for increased fidelity against mismatched pair extension using a 384-well plate PCR assay with SYBRgreen I–mediated detection [65]. The best mutants they obtained, ILL, IVF, CLV and SFN (referring to the residues at positions 782-784), showed significantly increased discrimination against mismatch extension.
At the other extreme of fidelity, DNAPs with specific mutations that reduce fidelity are useful for understanding the mechanism by which fidelity is achieved and could possibly also find practical applications for epPCR. Using genetic complementation followed by gel-based screening, Loeb and co-workers isolated a series of active Taq DNAPs with high activity but significantly reduced fidelity. Interestingly, such mutations were preferentially found on the O helix, which is conserved in family A DNAPs and forms the wall of the active site that is packed against the end of the primer/template duplex [53]. Specifically, they found that the A661E or T664R mutation results in at least a 7- and 25-fold increase in mutation frequency. With the same method, Taq mutant I614K, which is located in Motif A (amino acids 605-617) of the dNTP binding site, was identified and shown to have a greater than 20-fold higher mutation frequency [66].
Ghadessy et al. used CSR to select for mutants of Taq DNAP that can extend mismatched primer termini [67]. The two best mutants isolated from the selection, M1 (G84A, D144G, K314R, E520G, F598L, A608V, E742G) and M4 (D58G, R74P, A109T, L245R, R343G, G370D, E520G, N583S, E694K, A743P), not only had an increased ability to extend mismatches, but also acquired the ability to incorporate various noncanonical substrates, including 7-deaza-dGTP, αS-dNTPs, FITC-12-dATP, and biotin-16-dUTP. The fidelity of M1 for incorporating dye-modified bases was estimated based on sequencing to be one or fewer errors in 500 nucleotides synthesized.
Brakmman et al. evolved an error prone T7 RNAP variant (F11L, C515Y, T613A) by virtue of increased reversion rate of a mutated resistance gene [68]. This T7 RNA polymerase was also shown to be able to reduce the replication efficiency of T7 phage during infection. Based on in vitro transcription, reverse transcription, and sequencing, the substitution error rate of this mutant is at least 20-fold greater than that of wild-type.
5.2. Polymerase mutants with improved thermal stability
High DNAP thermostability is important for PCR applications and identifying the mutations that confer increased thermal stability is also useful for understanding the biophysical origins of protein stability and thermostability, in general. By using CSR with increased time at elevated temperatures, Ghadessy et al. selected a Taq DNAP mutant, T8 (F73S, R205K, K219E, M236T, E434D, A608V), with an 11-fold increased half-life at 97.5 °C [61]. Most of the mutations are located in the 5’-3’ exonuclease domain, and truncation of this domain also leads to greater thermostability [69, 70], suggesting that this domain is relatively less stable than the rest of the protein and that its presence in a denatured or non-native state causes loss of DNAP activity.
5.3. Polymerase mutants with enhanced inhibitor tolerance
Many potentially important diagnostic and medical applications of DNAPs, for example, disease diagnosis, personal identification, pathogen detection, and microbiome characterization, currently require the time and labor intensive purification of DNA from complex sources, such as biological fluids or soil extracts to remove inhibitors such as heparin, hemoglobin, lactoferrin, serum IgG, and humic acid. Kermekchiev et al. created a library of KlenTaq 1 (a truncated Taq DNAP lacking the N-terminal 278 amino acids) focused to codons 626, 706, 707 and 708, which they screened using a PCR assay in the presence of blood or soil extract [71]. One mutant, KT 7 (E626K, I707L, E708W) was further diversified by saturation mutagenesis at codon 708 and selection yielded KT 10 (E626K, I707L, E708K) and KT 12 (E626K, I707L, E708L). By introducing the KT 10 and KT 12 mutations into full-length Taq and another round saturation mutagenesis at codon 708, they isolated Taq 10 (E626K, I707L, E708K) and Taq 22 (E626K, I707L, E708Q), which showed significantly enhanced PCR performance in the presence of human blood, soil extracts, or high concentration of SYBR Green I. By applying CSR in the presence of heparin, Ghadessy et al. evolved H15 Taq DNAP (K225E, E388V, K540R, D578G, N583S, M747R), which increased resistance 130-fold compared to the wild type parent enzyme [61]. Baar et al. also evolved a DNAP for enhanced resistance to inhibitors by constructing a library via family shuffling of Pol I genes and CSR selection in the presence of inhibitors [72]. A chimera of Thermus aquaticus, Thermus oshimai, Thermus thermophilus, and Thermus brockianus DNAPs showed significantly enhanced resistance toward bone dust, humic acid, peat extract, coprolite, clay-rich soil, cave sediment and tar.
5.4. Polymerase mutants with altered substrate repertoires
5.4.1. Nucleotide triphosphates with modified triphosphate moieties
Polymerases are usually specific for triphosphate monomers, however, some applications would be facilitated by the synthesis of DNA or RNA from monomers with modified triphosphate moieties. Hansen et al. evolved the family B DNAP from Pyrococcus furiosus (Pfu) for improved incorporation of nucleotides with modified γ-phosphates upon library construction by epPCR and CSR selection [73]. A variant bearing the Q484R mutation and an internal split (resulting from the addition of a premature stop codon and the re-initiation of transcription downstream, and thus the production of two fragments that associate to form the active DNAP) was identified that gained the ability to incorporate γ-phosphate-O-linker-dabcyl derivatives.
5.4.2. Nucleotide triphosphates with modified sugars
One of the earliest reports of directed polymerase evolution used the phage display-based system to evolve Sf DNAP variants that efficiently synthesize RNA [60]. RNAP activity was targeted for these early efforts because understanding the mechanism by which dNTPs are selected in the presence of higher cellular concentrations rNTP is of fundamental importance, and many rational-based efforts had been directed toward the goal with only moderate success [74, 75]. A polymerase library was constructed by primer-directed random mutation of the substrate binding site and O-helix. The most efficient RNA polymerase evolved, SFR3 (A597T, W604R, L605Q, I614T, E615G), was able to incorporate rNTPs with rates increased 103-104-fold relative to the wild type enzyme. This demonstrates that a relatively small number of mutations interconvert DNAP and RNAP activity.
Ong et al. employed spCSR to select for variants of Taq DNAP that can incorporate both dNTPs and rNTPs [50]. Two libraries were constructed by randomizing codons 611-617, or 597-609, and after one round of selection, the enriched libraries were combined and subjected to a second round of selection. The best mutant obtained, AA40 (E602V, A608V, I614M, E615G), retained DNAP activity and gained RNAP and RT activity. This mutant can also incorporate various 2’-substituted triphosphates, such as 2’-N3-dATP and 2’-F-dUTP, further revealing an insensitivity to the nature of the substituent at the 2’ position. AA40 was able to amplify C2’-F modified templates, suggesting that the modifications were tolerated in both the template and triphosphate, however, primer extension processivity was poor with C2’ -NH2 or -OMe substituents, with only 2 modified nucleotides incorporated. Unfortunately, fidelity was not characterized.
Using the phage selection system, Fa et al. expanded the substrate repertoire of Sf DNAP to include 2’-OMe modified triphosphates [76]. 2’-OMe modified DNA is resistant to nucleases, and thus an enzyme that efficiently amplifies it could be used to evolve aptamers by SELEX that are stable in biological solutions [77]. For these selections, libraries were constructed by focusing mutations to residues proximal to the triphosphate binding site (Ile64, Glu615, Phe667, Tyr671, Asn750, and Gln754 or Arg573, Met 747, Gln754, Val783, His784, and Glu786). The most efficient mutant identified, SFM19, possessed two mutations, I614E and E615G, and showed increased rates for the incorporation of each 2’-OMe modified triphosphate of at least 104-fold relative to the parental enzyme, with no significant loss in fidelity.
Using CST, TgoT DNAP (a mutant of the family B DNAP from Thermococcus gorgonarius bearing four mutations: V93Q, which decreases uracil stalling; D141A and E143A, which disable exonulease proofreading; and A485L, which enhances the recognition of unnatural substrates [62]) has been evolved to interconvert DNA and polymers of peptide nucleic acid (PNA), 2’-O,4’-C-methylene-β-D-ribonucleic acid or locked nucleic acid (LNA), α-L-threofuranosyl nucleic acid (TNA), glycol nucleic acid (GNA), anhydrohexitol nucleic acid (HNA), cyclohexenyl nucleic acid (CeNA), arabinonucleic acid (ANA), or 2’-fluoro-arabinonucleic acid (FANA), collectively referred to as xeno-nucleic acids (XNAs). CST selection for HNA synthesis proceeded with a library constructed by random or homology-based diversification focused to 22 motifs within 10 Å of the nascent strand yielded Pol6G12 (TgoT: V589A, E609K, I610M, K659Q, E664Q, Q665P, R668K, D669Q, K671H, K674R, T676R, A681S, L704P, E730G). Similarly, polymerase for CeNA or LNA synthesis, PolC7 (TgoT: E654Q, E658Q, K659Q, V661A, E664Q, Q665P, D669A, K671Q, T676K, R709K), and ANA or FANA synthesis, PolD4K (TgoT: L403P, P657T, E658Q, K659H, Y663H, E664K, D669A, K671N, T676I) were also evolved. By combining saturation mutagenesis based on statistical correlation analysis (SCA) [78] and ELISA-like screening (CST may not be used for this activity as non-natural nucleic acids cannot be included in the template without selecting for amplification), they also evolved TgoT mutant that can reverse transcribe some XNAs back to DNA. Mutant RT521 (TgoT: E429G, I521L, K726R) was shown to have moderate RT activity for HNA, ANA, and FANA, and RT521K (RT521: A385V, F445L, E664K) for CeNA and LNA. Using these polymerases and a modification of SELEX similar to that originally used by Klussmann [79] and later by Eaton and co-workers [80], pools of DNA oligomers were first transcribed into XNAs, subjected to selection for binding to various targets, and then reverse transcribed back in to DNA for amplification. This work demonstrates that family B DNAPs may be evolved to have altered substrate repertoires, and that modified oligonucleotides may be used for the selection phase of evolution (amplification was still performed with DNA).
RNAPs are also obvious starting points for the evolution of enzymes capable of synthesizing DNA with 2’-modified nucleotides. Chelliserrykattil et al. evolved T7 RNAP for better incorporation of 2’-O-methyl nucleotide triphosphates using a library constructed by mutagenizing residues R425, G542, Y639 and H784, and autogene selection with a chloramphenicol resistance gene [58]. Members of the enriched library were then screened using plate and gel assays. The best mutants, Y639V/H784G/E593G/V685A and Y639L/A255T, can incorporate multiple triphosphates with C’2-modified sugars into a transcript with good processivities.
Clearly, both the substrate repertoires of DNAPs and RNAPs can be evolved to include C2’-modified substrates, but different consideration appear to be required. For DNAPs, it is likely that initial mutations must “open” the active site to make room for the 2’-substituents, while subsequent mutations are then required to further refine and optimize activity. Because RNAPs already accommodate C2’ substituents, they may represent more useful starting points, as the evolution of efficient activity appears to require only mutations that allow for nature or size of the substituent to be changed. However, if thermal stability is desired, then thermostable DNAPs may represent attractive starting points due to the absence of well characterized thermostable RNAPs.
5.4.3. Nucleotide triphosphates with modified nucleobases
Nucleotide triphosphates with modifications to their nucleobases are widely used in sequencing and for the labeling of nucleic acids. For example, the incorporation of dNTPs with removable fluorophores attached to their nucleobases forms the foundation of several next generation approaches currently under development. However, after fluorophore removal, a portion of the linker remains attached to the nucleobase and polymerase recognition of the modified triphosphates in the context of an increasingly modified primer terminus can significantly limit sequence read length. Toward the goal of optimizing a polymerase for these applications, Leconte et al. generated libraries of Sf by synthetic shuffling, focusing mutations to 21 positions that are within 14 Å of the incoming dNTP and that show high levels of variation among only similar amino acids in related DNAPs [81]. After four rounds of selection with the phage-based system, Sf197 was identified by screening the enriched library with a gel extension assay. The mutant possessed 14 mutations (V618I, L619M, V631A, I638V, T640K, M646V, M658L, A661I, T664A, I665V, L670M, F700Y, T756S, A757G) that were all clustered in the O- and N-helices. The resulting DNAP after moving the mutations into a full length Taq background, Taq197, incorporated each modified dNTP onto a modified primer terminus 50- to 400-fold more efficiently than wild-type Taq, and showed potential for significantly increased read length in sequencing applications without any apparent loss in fidelity.
Ramsay et al. evolved a variant of Pfu DNAP with mutations that disable both exonuclease activity and uracil-stalling (V93Q, D141A, E143A; referred to as Pfuexo-) for enhanced PCR amplification of DNA with Cy3- or Cy5-labeled dCTP [82]. Residues 399-415 of motif A were diversified via a combination of random mutagenesis and synthetic shuffling (leaving the catalytically critical residue D405 intact), and the resulting library was subjected to one round of spCSR selection, with dCTP replaced by Cy5-dCTP, and ELISA-based screening. Residues 537-546 of motif C of the four most promising leads were again diversified (avoiding catalytically residues D541 and D543) and after a second round of selection, followed by ELISA-and PCR-based screening, mutant E10 (V337I, E399D, N400D, R407I, Y546H) was identified which can amplify DNA with Cy5- or Cy3-dC. Sequencing of the amplification products revealed no loss in fidelity relative to Pfuexo-, although the fidelity of the mutant was more sensitive to additives such as formamide, glycerol, RNase, and DTT.
Chen et al. evolved Taq DNAP to accept 3’-ONH2 nucelotides (dNTP-ONH2), which because the 3’OH may be deprotected with sodium nitrite (pH 5.5) treatment, serves as a reversible terminator for next generation sequencing methodologies [83]. A library of 93 Taq variants was constructed based on structural analysis and a sequence-based approach termed “Reconstructed Evolutionary Adaptive Path” (REAP) [84], and screened for the ability to incorporate dTTP-ONH2. Mutants REAP-42 (A597T, L616A, F667Y, E745H) and REAP-58 (E520G, K540I, L616A), were isolated, with the later shown to efficiently incorporate dNTP-ONH2 and ddNTPs with good fidelity. Interestingly, L616A is present in both mutants, and analysis of the individual mutations indicated that it is sufficient for the selected activity.
Efforts to expand the genetic alphabet to include a third, unnatural base pair are limited by poor recognition of the unnatural nucleotides by DNAPs, which provides an obvious goal for directed polymerase evolution. Towards this goal, Leconte et al. reported the evolution of a variant of Sf DNAP that more efficiently synthesizes DNA containing an unnatural base pair formed between two nucleotides bearing isocarbostyryl nucleobase analogs [85]. A library was constructed by randomization of five regions of the polymerase that mediate substrate recognition [86], and enriched for mutants better able to synthesize DNA containing the unnatural base pair via four rounds of selection with the activity-based phage system and a gel-based extension screen. One mutant, P2 (F598I, I614F, Q489H), was found to synthesize the unnatural base pair (by incorporation of the unnatural triphosphate opposite the unnatural nucleotide in a template) and then extend it (by incorporation of the next correct natural triphosphate) with efficiencies that were >30- and >300-fold increased relative to the parental enzyme, respectively. Importantly, it was shown that this activity did not come at the expense of decreased fidelity.
Loakes et al. also evolved thermophilic DNAPs for the more efficient extension of primers terminating with a variety of pairs formed between nucleotides with modified nucleobases [87]. They shuffled family A DNAPs from the genus Thermus, including Taq from T. aquaticus, Tth from T. thermophilus, and Tfl from T. flavus, and subjected the library to five rounds of CSR selection, in which flanking primers containing hydrophobic nucleobase analogues, including 5-nitroindole and its 3-carboxamide. The best mutant isolated, 5D4, which is a chimera of Tth and Taq DNAPs with 14 additional mutations (V62I, Y78H, T88S, P114Q, P264S, E303V, G389V, E424G, E432G, E602G, A608V, I614M, M761T, M775T), was able to generate and extend several of the pairs, as well as the pair formed between analogs with isocarbostyryl and 7-azaindole. However, mispairs between the unnatural nucleotides and natural nucleotides were also synthesized and extended, suggesting that the activity may have come at the expense of decreased fidelity.
Laos et al. generated an exonuclease deficient Taq DNAP library by random mutagenesis and then subjected it to CSR with primers containing the unnatural nucleotide 2-amino-8-(1’-β-d-2’-deoxyribofuranosyl)imidazo[1,2-a]-1,3,5-triazin-4(8H)-one (abbreviated as P) and 6-amino-5-nitro-3-(1’-β-d-2’-deoxyribofuranosyl)-2(1H)-pyridone (abbreviated as Z), which form an unnatural base pair via a nonstandard pattern hydrogen-bonding [88]. The most promising mutants ((M444V/P527A/D551E/E832V) and (N580S/L628V/E832V)) showed reduced pausing when dZTP was incorporated opposite dP in a template. The fidelity of unnatural triphosphate insertion was examined with N580S/L628V/E832V, which appeared good for the incorporation of dZTP opposite dP, but poor for the incorporation dPTP opposite dZ.
Clearly, DNAPs have the potential for synthesizing DNA with unnatural base pairs, and directed evolution is likely to play an increasing role in the effort to expand the genetic alphabet. Similar experiments with RNAPs may facilitate the transcription of the expanded genetic alphabet.
5.4.4. Modified templates
RT-PCR is an important method for detection of RNAs, or generation and amplification of complementary DNAs (cDNAs) in transcriptome analysis, cloning of eukaryotic genes into prokaryotes, genetic disease diagnosis, and pathogen detection. The development of thermophilic DNAPs with RT activity could simplify RT-PCR and also allow for reverse transcription at higher temperatures, which would be useful with RNA that forms secondary structure. Towards this goal, Sauter et al. evolved thermostable KlenTaq DNAP for RT activity via a library constructed using epPCR, a PCR-based plate screen to ensure the retention of DNAP activity, followed by an RT-PCR-based plate screen [89]. Two mutants, M1 (L322M, L459M, S515R, I638F, S739G, E773G) and M2 (L322M, L459M, S515R, I638F, S739G, E773G, L789F), displayed significantly increased RT activity, and can be used directly in one-step RT-PCR of RNA at different concentrations. Guerre et al. also obtained a series of evolved Taq DNAPs with thermostable RT activity by creating multiple focused libraries and subsequent selection using a phage display method similar to that described above, but where the substrates are non-specifically attached via 5’-maleimidyl linkers [90]. Mutants 5 (A608T, E520G, W827R), 14 (M747K, E742K), and 21 (M761T, D547G, I584V), were identified and shown to be capable of synthesizing long cDNAs using mRNA as a template. The ability to amplify modified oligonucleotides, which by definition include the recognition of modified templates, has received somewhat less attention.
5.5. Polymerase mutants with other properties
In some PCR applications, such as in paleontological and forensic investigations, template DNA is highly damaged. For these applications, Gloeckner et al. evolved KlenTaq DNAP to efficiently amplify from damaged DNA by epPCR and a plate-based primer extension assay using only three of the four dNTPs (to mimic lesion bypass) and which was monitored by SYBRgreen I fluorescence [91]. One mutant, M747K, was 100-fold more tolerant of UV irradiation than the wild type DNAP. Obeid et al. combined the M747K mutation with the I614K mutation (see above), and showed that the resulting double mutant had even higher lesion bypass activity, although it was also more error prone [92].
d'Abbadie et al. shuffled A-family DNA polymerases from Thermus aquaticus (Taq), Thermus thermophilus (Tth), and Thermus flavus (Tfl), and applied CSR selection to this library for mismatch extension [93]. The best mutants 3D1 (a Tth-Taq chimera with six additional mutations: L33P, E76K, D145G, P552S, E775G, and M777T) and 3A10 (a Tth-Taq chimera with eight additional mutations: E76K, E91Q, D145G, R336Q, A448T, I616M, V739M, and E744G) were able to extend primers terminating with up to four mismatches, and also able to amplify 47,000–60,000-year-old cave bear DNA at concentrations which wild type Taq is not. Based on the amplification of undamaged DNA, the fidelity of 3D1 and 3A10 was estimated to be decreased 2- to 4- and 7-fold, respectively.
The availability of RNAPs that recognize alternate promoters would enable simultaneous modular control over multiple cellular pathways [10]. Toward this goal, Chelliserrykattil et al. successfully evolved T7 RNAP variants that recognize a T3-like promoter sequence by randomizing codons 746, 747, and 748, and then performing autogene selection [57]. After 192 hours of continuous evolution, Esvelt et al. also evolved a T7 RNAP with 89-fold higher transcription from a T3 promoter [59].
6. Conclusion and perspectives
The effort to evolve enzymes with novel DNAP, RNAP, and RT activity has had considerable success. While the effort is still at a very early stage, it is perhaps not too early to offer some possible trends and generalizations. Surprisingly, despite the development and implementation of sophisticated strategies for introducing mutations in a better than random manner, random approaches, such as epPCR appear to have had no less success. This may be due to epistatic interactions throughout the protein and/or other long-distance effects that are difficult to predict or include in library design. The two selections capable of sorting through the largest libraries of mutants, the phage-based approach and CSR and its modified variants, have several important differences. The CSR-based techniques have the advantage that they strictly enforce the link between genotype-phenotype via physical separation and also that they may be used to select for amplification. The phage display approach offers the advantage of finer control over selection pressure, for example allowing for enrichment based on single nucleotide turnover and for varying triphosphate concentration to select for increased binding or increased catalytic turnover. In addition, the fully in vitro phage method permits single turnover selections with templates containing modified nucleotides, which is difficult with the CSR-based techniques. The phage display method has the disadvantage that with sufficient levels of enrichment of an active mutant in a population, cross-reactivity between phage can sever the link between phenotype and genotype, placing a higher demand on a post-selection screening step.
Interestingly, there are common mutations among several of the evolved mutants, suggesting that only a limited number of pathways to the selected phenotype exist. For example, in each of the Sf or Taq DNAPs evolved to incorporate 2’ modified triphosphates, residues I614 and E615 are mutated. This is consistent with the hypothesis that in the wild type enzyme E615 plays a key role as a steric gate to exclude incorporation of rNTPs [94]. Interestingly, residue I614 is also mutated both in polymerase that incorporate nucleotides with nucleobase modifications (5D4 [87] and P2 [85]), possibly suggesting a more general role, such as in facilitating the required conformational changes in the DNAP.
Several other common mutations in Taq also appear to contribute to disparate activities. A608 is located in motif A, and its mutation to Val or Thr has been linked to a broad range of activities, including increased extension of distorting 3’ mismatches [67], thermostability [61], RNAP activity [50], RT activity [90], and substrate discrimination [67, 87]. E520 is positioned at the interface of the thumb and fingers domains within the thumb domain and its mutation to Gly has been linked to 3’ mismatch extension [67], RT activity [90] and substrate discrimination [67]. M747 is located within the Q helix of the fingers domain and its mutation has been linked to inhibitor resistance [[61], RT activity [90], and replication from damaged templates [91, 92]. Interestingly, the structure of KlenTaq indicates that M747 interacts with the 2’-deoxyribose of the first (n+1) nucleotide in the template [86, 95]. Many of these mutations are located away form the active site, and would likely not have been included in rationally or semi-rationally designed libraries.
While polymerases with diverse functions have been evolved, future efforts should include increased use of both recombination during selection, the addition of selections for stability, and the addition of counter selections for fidelity. Recombination is necessary to combine beneficial mutations from different clones and also to unlink beneficial and detrimental mutations. Mutations that are neutral, or even adaptive, with regard to the selected phenotype commonly destabilize the protein [96-98], and this destabilization may limit evolution [99-101]. Thus, the inclusion of rounds of selection for stability may introduce compensatory stabilizing mutations that allow for the acquisition of additional adaptive mutations. Finally, it is clear that the easiest way to increase the substrate repertoire of a polymerase (or perhaps any enzyme) is to acquire mutations that reduce fidelity. All too often the characterization of evolved mutants focuses on the selected activity with little or only characterization of fidelity. Variants with significantly reduced fidelity are unlikely to be of any practical value. The inclusion of additional rounds of selection for fidelity, for example, with large concentrations of dideoxy nucleotide triphosphates that are incorporated only upon mispairing and whose incorporation prevents recovery, should be explored. With these and other advances in protein engineering, polymerases with a broad range of activities should be available to enable an equally broad range of novel applications.
Table 1.
Comparison of selection strategies for polymerase evolution
| Strategy | Advantages | Disadvantages |
|---|---|---|
| Phage Display | •Straightforward to select for modifications in primer, template and/or nucleotide triphsophate, including modifications in both strands simultaneously; •Control of selection pressure; •Challenging conditions easy to apply; •Selection possible without amplification and even with the incorporation of a single nucleotide. |
•Cross reactivity with sufficient enrichment, mandating the use of post-selection screen; •Complicated. |
| CSR and derivatives | •No cross reaction between mutants; •Challenging conditions easy to apply. |
•Requires amplification to select for modifications in both strands of DNA; •Less control over selection pressure. •Complicated. |
| Genetic complementation and derivatives | •No cross reaction between mutants; •Sufficient activity to replicate genome ensured •Simple. |
• Only natural substrates possible; •Selection pressure difficult to control. |
| Autogene selection and derivatives | •No cross reaction between mutants •Simple. |
•Only natural substrates possible; •Selection pressure difficult to control. |
Acknowledgements
We thank the National Institutes of Health (GM097489) for supporting our work on directed evolution of DNA polymerases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 2.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 4.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Syvanen AC. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- 6.Orita M, Suzuki Y, Sekiya T, Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989;5:874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- 7.Hammond HA, Jin L, Zhong Y, Caskey CT, Chakraborty R. Evaluation of 13 short tandem repeat loci for use in personal identification applications. Am. J. Hum. Genet. 1994;55:175–189. [PMC free article] [PubMed] [Google Scholar]
- 8.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 9.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Temme K, Hill R, Segall-Shapiro TH, Moser F, Voigt CA. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 2012;40:8773–8781. doi: 10.1093/nar/gks597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry AA, Romesberg FE. Beyond A, C, G and T: augmenting nature's alphabet. Curr. Opin. Chem. Biol. 2003;7:727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Hirao I, Kimoto M. Unnatural base pair systems toward the expansion of the genetic alphabet in the central dogma. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2012;88:345–367. doi: 10.2183/pjab.88.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirao I, Ohtsuki T, Fujiwara T, Mitsui T, Yokogawa T, Okuni T, Nakayama H, Takio K, Yabuki T, Kigawa T, Kodama K, Yokogawa T, Nishikawa K, Yokoyama S. An unnatural base pair for incorporating amino acid analogs into proteins. Nat. Biotechnol. 2002;20:177–182. doi: 10.1038/nbt0202-177. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Lavergne T, Malyshev DA, Zimmermann J, Adhikary R, Dhami K, Ordoukhanian P, Sun Z, Xiang J, Romesberg FE. Site-Specifically Arraying Small Molecules or Proteins on DNA Using An Expanded Genetic Alphabet. Chem. Eur. J. 2013;19:14205–14209. doi: 10.1002/chem.201302496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo YJ, Malyshev DA, Lavergne T, Ordoukhanian P, Romesberg FE. Site-specific labeling of DNA and RNA using an efficiently replicated and transcribed class of unnatural base pairs. J. Am. Chem. Soc. 2011;133:19878–19888. doi: 10.1021/ja207907d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimoto M, Yamashige R, Matsunaga K, Yokoyama S, Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 17.Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 18.Zimmermann J, Oakman EL, Thorpe IF, Shi X, Abbyad P, Brooks CL, 3rd, Boxer SG, Romesberg FE. Antibody evolution constrains conformational heterogeneity by tailoring protein dynamics. Proc. Natl. Acad. Sci. USA. 2006;103:13722–13727. doi: 10.1073/pnas.0603282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel PH, Suzuki M, Adman E, Shinkai A, Loeb LA. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. J. Mol. Biol. 2001;308:823–837. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- 20.Eger BT, Benkovic SJ. Minimal kinetic mechanism for misincorporation by DNA polymerase I (Klenow fragment) Biochemistry. 1992;31:9227–9236. doi: 10.1021/bi00153a016. [DOI] [PubMed] [Google Scholar]
- 21.Dahlberg ME, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 22.Kuchta RD, Benkovic P, Benkovic SJ. Kinetic mechanism whereby DNA polymerase I (Klenow) replicates DNA with high fidelity. Biochemistry. 1988;27:6716–6725. doi: 10.1021/bi00418a012. [DOI] [PubMed] [Google Scholar]
- 23.Filee J, Forterre P, Sen-Lin T, Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- 24.Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loh E, Loeb LA. Mutability of DNA polymerase I: implications for the creation of mutant DNA polymerases. DNA Repair (Amst) 2005;4:1390–1398. doi: 10.1016/j.dnarep.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Cermakian N, Ikeda TM, Cedergren R, Gray MW. Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucleic Acids Res. 1996;24:648–654. doi: 10.1093/nar/24.4.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cramer P. Common structural features of nucleic acid polymerases. Bioessays. 2002;24:724–729. doi: 10.1002/bies.10127. [DOI] [PubMed] [Google Scholar]
- 28.Cramer P. RNA polymerase II structure: from core to functional complexes. Curr. Opin. Genet. Dev. 2004;14:218–226. doi: 10.1016/j.gde.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- 30.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willis IM. RNA polymerase III. Genes, factors and transcriptional specificity. Eur. J. Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 32.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 33.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebright RH. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 35.Korkhin Y, Unligil UM, Littlefield O, Nelson PJ, Stuart DI, Sigler PB, Bell SD, Abrescia NG. Evolution of complex RNA polymerases: the complete archaeal RNA polymerase structure. PLoS Biol. 2009;7:e1000102. doi: 10.1371/journal.pbio.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Werner F. Structure and function of archaeal RNA polymerases. Mol. Microbiol. 2007;65:1395–1404. doi: 10.1111/j.1365-2958.2007.05876.x. [DOI] [PubMed] [Google Scholar]
- 37.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 39.Pata JD, Stirtan WG, Goldstein SW, Steitz TA. Structure of HIV-1 reverse transcriptase bound to an inhibitor active against mutant reverse transcriptases resistant to other nonnucleoside inhibitors. Proc. Natl. Acad. Sci. USA. 2004;101:10548–10553. doi: 10.1073/pnas.0404151101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cirino PC, Mayer KM, Umeno D. Generating mutant libraries using error-prone PCR. Methods Mol. Biol. 2003;231:3–9. doi: 10.1385/1-59259-395-X:3. [DOI] [PubMed] [Google Scholar]
- 41.Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 42.Sneeden JL, Loeb LA. Random oligonucleotide mutagenesis. Methods Mol. Biol. 2003;231:65–73. doi: 10.1385/1-59259-395-X:65. [DOI] [PubMed] [Google Scholar]
- 43.Ness JE, Kim S, Gottman A, Pak R, Krebber A, Borchert TV, Govindarajan S, Mundorff EC, Minshull J. Synthetic shuffling expands functional protein diversity by allowing amino acids to recombine independently. Nat. Biotechnol. 2002;20:1251–1255. doi: 10.1038/nbt754. [DOI] [PubMed] [Google Scholar]
- 44.DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat. Rev. Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 45.Crameri A, Raillard SA, Bermudez E, Stemmer WP. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391:288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 46.Coco WM, Levinson WE, Crist MJ, Hektor HJ, Darzins A, Pienkos PT, Squires CH, Monticello DJ. DNA shuffling method for generating highly recombined genes and evolved enzymes. Nat. Biotechnol. 2001;19:354–359. doi: 10.1038/86744. [DOI] [PubMed] [Google Scholar]
- 47.Reetz MT, Carballeira JD. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat. Protoc. 2007;2:891–903. doi: 10.1038/nprot.2007.72. [DOI] [PubMed] [Google Scholar]
- 48.Reetz MT, Bocola M, Carballeira JD, Zha D, Vogel A. Expanding the range of substrate acceptance of enzymes: combinatorial active-site saturation test. Angew. Chem. Int. Ed. Engl. 2005;44:4192–4196. doi: 10.1002/anie.200500767. [DOI] [PubMed] [Google Scholar]
- 49.Fox RJ, Davis SC, Mundorff EC, Newman LM, Gavrilovic V, Ma SK, Chung LM, Ching C, Tam S, Muley S, Grate J, Gruber J, Whitman JC, Sheldon RA, Huisman GW. Improving catalytic function by ProSAR-driven enzyme evolution. Nat. Biotechnol. 2007;25:338–344. doi: 10.1038/nbt1286. [DOI] [PubMed] [Google Scholar]
- 50.Ong JL, Loakes D, Jaroslawski S, Too K, Holliger P. Directed evolution of DNA polymerase, RNA polymerase and reverse transcriptase activity in a single polypeptide. J. Mol. Biol. 2006;361:537–550. doi: 10.1016/j.jmb.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 51.Staiger N, Marx A. A DNA polymerase with increased reactivity for ribonucleotides and C5-modified deoxyribonucleotides. Chembiochem. 2010;11:1963–1966. doi: 10.1002/cbic.201000384. [DOI] [PubMed] [Google Scholar]
- 52.Kranaster R, Marx A. Taking fingerprints of DNA polymerases: multiplex enzyme profiling on DNA arrays. Angew. Chem. Int. Ed. Engl. 2009;48:4625–4628. doi: 10.1002/anie.200900953. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki M, Baskin D, Hood L, Loeb LA. Random mutagenesis of Thermus aquaticus DNA polymerase I: concordance of immutable sites in vivo with the crystal structure. Proc. Natl. Acad. Sci. USA. 1996;93:9670–9675. doi: 10.1073/pnas.93.18.9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki M, Avicola AK, Hood L, Loeb LA. Low fidelity mutants in the O-helix of Thermus aquaticus DNA polymerase I. J. Biol. Chem. 1997;272:11228–11235. doi: 10.1074/jbc.272.17.11228. [DOI] [PubMed] [Google Scholar]
- 55.Kim B. Genetic selection in Escherichia coli for active human immunodeficiency virus reverse transcriptase mutants. Methods. 1997;12:318–324. doi: 10.1006/meth.1997.0485. [DOI] [PubMed] [Google Scholar]
- 56.Söte S, Kleine S, Schlicke M, Brakmann S. Directed evolution of an error-prone T7 DNA polymerase that attenuates viral replication. Chembiochem. 2011;12:1551–1558. doi: 10.1002/cbic.201000799. [DOI] [PubMed] [Google Scholar]
- 57.Chelliserrykattil J, Cai G, Ellington AD. A combined in vitro/in vivo selection for polymerases with novel promoter specificities. BMC Biotechnol. 2001;1:13. doi: 10.1186/1472-6750-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chelliserrykattil J, Ellington AD. Evolution of a T7 RNA polymerase variant that transcribes 2′-O-methyl RNA. Nat. Biotechnol. 2004;22:1155–1160. doi: 10.1038/nbt1001. [DOI] [PubMed] [Google Scholar]
- 59.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia G, Chen L, Sera T, Fa M, Schultz PG, Romesberg FE. Directed evolution of novel polymerase activities: mutation of a DNA polymerase into an efficient RNA polymerase. Proc. Natl. Acad Sci. USA. 2002;99:6597–6602. doi: 10.1073/pnas.102577799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghadessy FJ, Ong JL, Holliger P. Directed evolution of polymerase function by compartmentalized self-replication. Proc. Natl. Acad Sci. USA. 2001;98:4552–4557. doi: 10.1073/pnas.071052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, Chaput JC, Wengel J, Peak-Chew SY, McLaughlin SH, Herdewijn P, Holliger P. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336:341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Summerer D, Rudinger NZ, Detmer I, Marx A. Enhanced fidelity in mismatch extension by DNA polymerase through directed combinatorial enzyme design. Angew. Chem. Int. Ed. Engl. 2005;44:4712–4715. doi: 10.1002/anie.200500047. [DOI] [PubMed] [Google Scholar]
- 64.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 65.Strerath M, Gloeckner C, Liu D, Schnur A, Marx A. Directed DNA polymerase evolution: effects of mutations in motif C on the mismatch-extension selectivity of thermus aquaticus DNA polymerase. Chembiochem. 2007;8:395–401. doi: 10.1002/cbic.200600337. [DOI] [PubMed] [Google Scholar]
- 66.Patel PH, Kawate H, Adman E, Ashbach M, Loeb LA. A single highly mutable catalytic site amino acid is critical for DNA polymerase fidelity. J. Biol. Chem. 2001;276:5044–5051. doi: 10.1074/jbc.M008701200. [DOI] [PubMed] [Google Scholar]
- 67.Ghadessy FJ, Ramsay N, Boudsocq F, Loakes D, Brown A, Iwai S, Vaisman A, Woodgate R, Holliger P. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nat. Biotechnol. 2004;22:755–759. doi: 10.1038/nbt974. [DOI] [PubMed] [Google Scholar]
- 68.Brakmann S, Grzeszik S. An error-prone T7 RNA polymerase mutant generated by directed evolution. Chembiochem. 2001;2:212–219. doi: 10.1002/1439-7633(20010302)2:3<212::AID-CBIC212>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 69.Barnes WM. The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene. 1992;112:29–35. doi: 10.1016/0378-1119(92)90299-5. [DOI] [PubMed] [Google Scholar]
- 70.Lawyer FC, Stoffel S, Saiki RK, Chang SY, Landre PA, Abramson RD, Gelfand DH. High-level expression, purification, and enzymatic characterization of full-length Thermus aquaticus DNA polymerase and a truncated form deficient in 5′ to 3′ exonuclease activity. PCR Methods Appl. 1993;2:275–287. doi: 10.1101/gr.2.4.275. [DOI] [PubMed] [Google Scholar]
- 71.Kermekchiev MB, Kirilova LI, Vail EE, Barnes WM. Mutants of Taq DNA polymerase resistant to PCR inhibitors allow DNA amplification from whole blood and crude soil samples. Nucleic Acids Res. 2009;37:e40. doi: 10.1093/nar/gkn1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baar C, d'Abbadie M, Vaisman A, Arana ME, Hofreiter M, Woodgate R, Kunkel TA, Holliger P. Molecular breeding of polymerases for resistance to environmental inhibitors. Nucleic Acids Res. 2011;39:e51. doi: 10.1093/nar/gkq1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hansen CJ, Wu L, Fox JD, Arezi B, Hogrefe HH. Engineered split in Pfu DNA polymerase fingers domain improves incorporation of nucleotide gamma-phosphate derivative. Nucleic Acids Res. 2011;39:1801–1810. doi: 10.1093/nar/gkq1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Astatke M, Ng K, Grindley ND, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl. Acad Sci. USA. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Joyce CM. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc. Natl. Acad Sci. USA. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fa M, Radeghieri A, Henry AA, Romesberg FE. Expanding the substrate repertoire of a DNA polymerase by directed evolution. J. Am. Chem. Soc. 2004;126:1748–1754. doi: 10.1021/ja038525p. [DOI] [PubMed] [Google Scholar]
- 77.Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, McCauley TG, Kurz JC, Epstein DM, Wilson C, Keefe AD. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 2005;12:25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 78.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 79.Schoetzau T, Langner J, Moyroud E, Roehl I, Vonhoff S, Klussmann S. Aminomodified nucleobases: functionalized nucleoside triphosphates applicable for SELEX. Bioconjug. Chem. 2003;14:919–926. doi: 10.1021/bc0256547. [DOI] [PubMed] [Google Scholar]
- 80.Vaught JD, Bock C, Carter J, Fitzwater T, Otis M, Schneider D, Rolando J, Waugh S, Wilcox SK, Eaton BE. Expanding the chemistry of DNA for in vitro selection. J. Am. Chem. Soc. 2010;132:4141–4151. doi: 10.1021/ja908035g. [DOI] [PubMed] [Google Scholar]
- 81.Leconte AM, Patel MP, Sass LE, McInerney P, Jarosz M, Kung L, Bowers JL, Buzby PR, Efcavitch JW, Romesberg FE. Directed evolution of DNA polymerases for next-generation sequencing. Angew. Chem. Int. Ed. Engl. 2010;49:5921–5924. doi: 10.1002/anie.201001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramsay N, Jemth AS, Brown A, Crampton N, Dear P, Holliger P. CyDNA: synthesis and replication of highly Cy-dye substituted DNA by an evolved polymerase. J. Am. Chem. Soc. 2010;132:5096–5104. doi: 10.1021/ja909180c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen F, Gaucher EA, Leal NA, Hutter D, Havemann SA, Govindarajan S, Ortlund EA, Benner SA. Reconstructed evolutionary adaptive paths give polymerases accepting reversible terminators for sequencing and SNP detection. Proc. Natl. Acad Sci. USA. 2010;107:1948–1953. doi: 10.1073/pnas.0908463107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gaucher EA. In: Ancestral sequence reconstruction as a tool to understand natural history and guide synthetic biology: realizing and extending the vision of zuckerkandl and pauling in Ancestral Sequence Reconstruction. Liberles DA, editor. Oxford University Press; New York: 2007. pp. 20–33. [Google Scholar]
- 85.Leconte AM, Chen L, Romesberg FE. Polymerase evolution: efforts toward expansion of the genetic code. J. Am. Chem. Soc. 2005;127:12470–12471. doi: 10.1021/ja053322h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Loakes D, Gallego J, Pinheiro VB, Kool ET, Holliger P. Evolving a polymerase for hydrophobic base analogues. J. Am. Chem. Soc. 2009;131:14827–14837. doi: 10.1021/ja9039696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Laos R, Shaw R, Leal NA, Gaucher E, Benner S. Directed Evolution of Polymerases To Accept Nucleotides with Nonstandard Hydrogen Bond Patterns. Biochemistry. 2013;52:5288–5294. doi: 10.1021/bi400558c. [DOI] [PubMed] [Google Scholar]
- 89.Sauter KB, Marx A. Evolving thermostable reverse transcriptase activity in a DNA polymerase scaffold. Angew. Chem. Int. Ed. Engl. 2006;45:7633–7635. doi: 10.1002/anie.200602772. [DOI] [PubMed] [Google Scholar]
- 90.Vichier-Guerre S, Ferris S, Auberger N, Mahiddine K, Jestin JL. A population of thermostable reverse transcriptases evolved from Thermus aquaticus DNA polymerase I by phage display. Angew. Chem. Int. Ed. Engl. 2006;45:6133–6137. doi: 10.1002/anie.200601217. [DOI] [PubMed] [Google Scholar]
- 91.Gloeckner C, Sauter KB, Marx A. Evolving a thermostable DNA polymerase that amplifies from highly damaged templates. Angew. Chem. Int. Ed. Engl. 2007;46:3115–3117. doi: 10.1002/anie.200603987. [DOI] [PubMed] [Google Scholar]
- 92.Obeid S, Schnur A, Gloeckner C, Blatter N, Welte W, Diederichs K, Marx A. Learning from directed evolution: Thermus aquaticus DNA polymerase mutants with translesion synthesis activity. Chembiochem. 2011;12:1574–1580. doi: 10.1002/cbic.201000783. [DOI] [PubMed] [Google Scholar]
- 93.d'Abbadie M, Hofreiter M, Vaisman A, Loakes D, Gasparutto D, Cadet J, Woodgate R, Paabo S, Holliger P. Molecular breeding of polymerases for amplification of ancient DNA. Nat. Biotechnol. 2007;25:939–943. doi: 10.1038/nbt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Y, Mitaxov V, Waksman G. Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation. Proc. Natl. Acad Sci. USA. 1999;96:9491–9496. doi: 10.1073/pnas.96.17.9491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bloom JD, Raval A, Wilke CO. Thermodynamics of neutral protein evolution. Genetics. 2007;175:255–266. doi: 10.1534/genetics.106.061754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am. J. Hum. Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tokuriki N, Stricher F, Serrano L, Tawfik DS. How protein stability and new functions trade off. PLoS Comput. Biol. 2008;4:e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol. 2002;320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 100.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc. Natl. Acad Sci. USA. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell. Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]