Abstract
The genetic basis of hybrid male sterility in house mice is complex, highly polygenic, and strongly X linked. Previous work suggested that there might be interactions between the Mus musculus musculus X and the M. m. domesticus Y with a large negative effect on sperm head morphology in hybrid males with an F1 autosomal background. To test this, we introgressed the M. m. domesticus Y onto a M. m. musculus background and measured the change in sperm morphology, testis weight, and sperm count across early backcross generations and in 11th generation backcross males in which the opportunity for X–autosome incompatibilities is effectively eliminated. We found that abnormality in sperm morphology persists in M. m. domesticus Y introgression males, and that this phenotype is rescued by M. m. domesticus introgressions on the X chromosome. In contrast, the severe reductions in testis weight and sperm count that characterize F1 males were eliminated after one generation of backcrossing. These results indicate that X–Y incompatibilities contribute specifically to sperm morphology. In contrast, X–autosome incompatibilities contribute to low testis weight, low sperm count, and sperm morphology. Restoration of normal testis weight and sperm count in first generation backcross males suggests that a small number of complex incompatibilities between loci on the M. m. musculus X and the M. m. domesticus autosomes underlie F1 male sterility. Together, these results provide insight into the genetic architecture of F1 male sterility and help to explain genome-wide patterns of introgression across the house mouse hybrid zone.
Keywords: Dobzhansky–Muller incompatibilities, negative epistasis, sex chromosomes, speciation, spermiogenesis
ACROSS sexually reproducing animals, intrinsic barriers to gene flow between species are caused primarily by deleterious interactions between loci that function normally within species (Bateson 1909; Dobzhansky 1937; Muller 1942). These reproductive incompatibilities manifest first in heterogametic hybrids and often involve the sex chromosomes (Haldane 1922; Laurie 1997; Presgraves 2002; Price and Bouvier 2002). In taxa with heterogametic males (e.g., Drosophila and mammals), X-linked hybrid male sterility is a prominent feature of the earliest stages of speciation (Forejt 1996; Presgraves 2008). Nonetheless, the genetic architecture of hybrid male sterility is typically complex, both in terms of the number of loci involved in any one incompatibility and the total number of incompatibilities (True et al. 1996; Tao et al. 2003; Masly and Presgraves 2007; Reed et al. 2008; Phadnis 2011; White et al. 2011; Dzur-Gejdosova et al. 2012).
Theory predicts that complex incompatibilities should accrue more readily than simple ones (Cabot et al. 1994; Orr 1995). In fact, although the probability that any two substitutions will result in an incompatibility is small, the genetic basis of intrinsic isolation grows “very complex very quickly” (Orr and Turelli 2001). This is largely a consequence of the increased potential for negative epistasis as multiple loci accumulate substitutions in diverging lineages. An additional explanation for the genetic complexity of hybrid sterility is simply that sterility is a composite phenotype with a correspondingly polygenic basis. If hybrid male sterility is the product of multiple incompatibilities that act at different time points in spermatogenesis, then a smaller number of interactions might underlie any one sterility phenotype. Thus, decomposing hybrid male sterility into distinct phenotypes may help us understand the genetic complexity of intrinsic reproductive isolation. Here, we demonstrate that reproductive deficits in hybrid male house mice are genetically separable. Incompatibilities between the X and Y chromosomes contribute to sperm abnormality, but we find no evidence that X–Y interactions contribute to low testis weight and sperm count. In contrast, X–autosome interactions explain low testis weight and sperm count and also contribute to sperm abnormality in F1 males.
House mice in the Mus musculus species complex are a classic mammalian model for the genetics of postzygotic reproductive isolation. Three lineages, M. musculus musculus, M. musculus domesticus, and M. musculus castaneus, split from a common ancestor ∼350,000 years ago (Geraldes et al. 2011) and hybridize where their ancestral ranges come into secondary contact (Tucker et al. 1992; Boursot et al. 1993; Spiridonova et al. 2011; Janoušek et al. 2012). Despite recent common ancestry and incomplete reproductive isolation, barriers to gene flow between M. m. domesticus and M. m. musculus are strong and highly polygenic. These subspecies form a stable hybrid zone across Central Europe in which males are subfertile (Hunt and Selander 1973; Turner et al. 2012). Patterns of gene flow across the hybrid zone suggest that both sex chromosomes contain loci that reduce hybrid fitness (Tucker et al. 1992; Dod et al. 1993). Although a few autosomal markers show reduced introgression across different transects of this hybrid zone, X-linked markers consistently show little introgression (Payseur et al. 2004; Macholán et al. 2007; Teeter et al. 2010; Janoušek et al. 2012). Similarly, Y-linked markers typically do not introgress across the hybrid zone (Vanlerberghe et al. 1986; Tucker et al. 1992; Prager et al. 1997). In the only exceptions to this pattern, gene flow is strictly unidirectional, with introgression of the M. m. musculus Y into M. m. domesticus territory (Macholán et al. 2008; Jones et al. 2010; Albrechtová et al. 2012).
Consistent with the large role of the X in reproductive isolation in nature, hybrid male sterility in lab crosses is strongly X-linked and is often asymmetric. Males with all or part of a M. m. musculus X chromosome are sterile or subfertile, whereas males with a M. m. domesticus X are usually reproductively normal (Forejt and Ivanyi 1974; Storchová et al. 2004; Britton-Davidian et al. 2005; Good et al. 2008a,b). In mapping studies, associations between sterility phenotypes and M. m. musculus genotype are significant for most or all of the X chromosome (Storchová et al. 2004; Good et al. 2008b; White et al. 2011), whereas the estimated number and individual effect size of autosomal incompatibilities varies among crosses. For example, Prdm9 is an autosomal gene of large effect that segregates “sterile” and “fertile” alleles in M. m. domesticus (Forejt and Iványi 1974; Forejt 1996). Heterozygosity for the sterile allele, in combination with the M. m. musculus X and an undefined number of unmapped autosomal loci, causes complete meiotic arrest in F1 males (Mihola et al. 2009; Dzur-Gejdosova et al. 2012). In contrast, QTL mapping in a cross that does not involve the Prdm9 sterile allele suggests that sterility-associated autosomal loci with individually small effect sizes are distributed throughout the genome (White et al. 2011).
In lab crosses between M. m. musculus and M. m. domesticus, the Y chromosome does not appear to be required for hybrid male sterility (White et al. 2011; Dzur-Gejdosova et al. 2012). This is surprising since, like the X, the Y chromosome typically does not introgress across the hybrid zone. Like others, we recently found that sterility does not require interactions involving the Y chromosome (Campbell et al. 2012). However, our crosses also suggested that incompatibilities between the M. m. musculus X and the M. m. domesticus Y might explain a large proportion of the phenotypic variance in F1 male sperm abnormality (Campbell et al. 2012). We used the wild-derived inbred strains, PWK/PiJ (musculusPWK) and LEWES/EiJ (domesticusLEWES). In this cross, F1 males with a musculusPWK X have severe reproductive deficits, whereas F1 males with a domesticusLEWES X do not (Good et al. 2008a; Campbell et al. 2012). We tested for a contribution of the domesticusLEWES Y to hybrid sterility phenotypes using low resolution QTL mapping on the X in F1 males with either domesticusLEWES or musculusPWK Y chromosomes. We identified an interval between ∼38 and 91 Mb (∼32% of the X) for which there was a large negative effect on sperm morphology of a musculusPWK genotype in males with a domesticusLEWES Y (Campbell et al. 2012). This experimental design could not, however, control for the possible contribution of X–autosome incompatibilities in F1 males. Here, we explicitly test the hypothesis that X–Y incompatibilities underlie sperm abnormality by isolating the domesticusLEWES Y on a musculusPWK background in which the opportunity for X–autosome incompatibilities is eliminated.
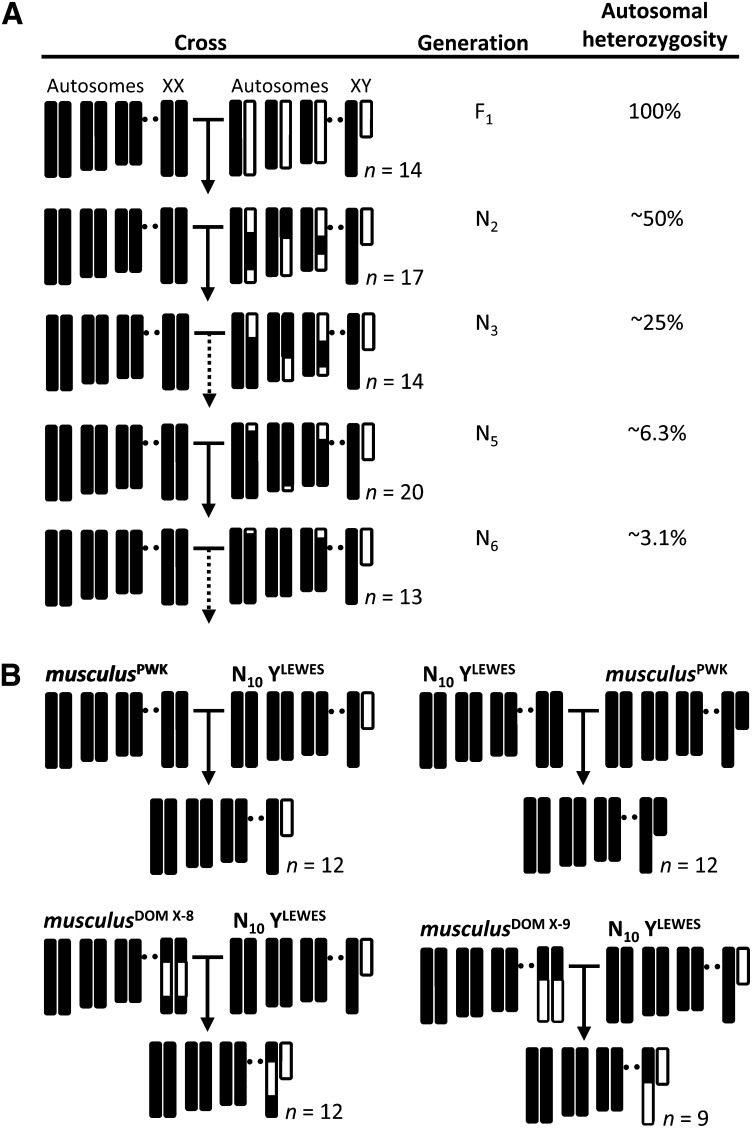
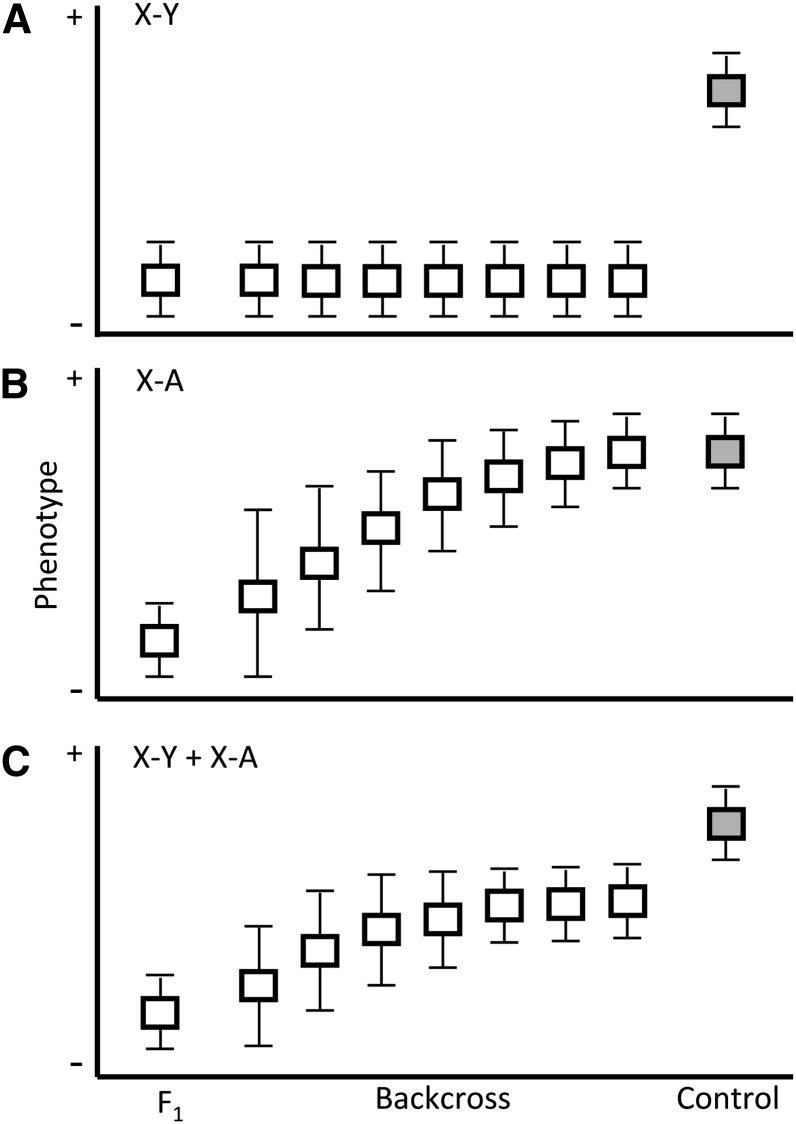
We introgressed the domesticusLEWES Y onto a musculusPWK background and measured the change in male reproductive phenotypes across early to mid (N2–N6) backcross generations (Figure 1A). This design allowed us to test competing predictions about the contribution of the Y chromosome to hybrid sterility (Figure 2). In particular, the predicted pattern of phenotypic change depends on whether X-linked incompatibilities interact with Y-linked loci, with autosomal loci, or with both. If sperm abnormality is primarily due to X–Y interactions, then the progressive reduction in the proportion of the autosomal genome derived from domesticusLEWES should have little effect on this phenotype. In contrast, if deficits in testis mass and sperm count are caused exclusively by X–autosome interactions, phenotypic means should improve with each generation. We then measured reproductive phenotypes in males from the 11th backcross generation (N11) with either a complete musculusPWK X or with two different domesticusLEWES X introgressions (Figure 1B). If X–Y interactions contribute specifically to sperm abnormality, domesticusLEWES Y introgression males should have excess abnormal sperm but normal testis weight and sperm count, and a domesticusLEWES introgression on the X between 38 and 91 Mb should rescue sperm abnormality.
Figure 1.
Crossing design and genotypes of experimental males. (A) The domesticusLEWES Y chromosome (white) was introgressed onto a musculusPWK background (black) by backcrossing hybrid males to musculusPWK females for 11 generations. Only generations for which male reproductive phenotypes were measured are shown. Expected autosomal heterozygosity is reduced by 50% each generation. (B) Tenth generation backcross males (N10 YLEWES) were crossed to either pure musculusPWK females or females with domesticusLEWES introgressions on the central (musculusDOM X-8) or distal (musculusDOM X-9) part of the X chromosome. Control males were generated by crossing N10 females to musculusPWK males. Sample sizes (n) are the number of males in each generation for which reproductive phenotypes were measured.
Figure 2.
Expected distributions of reproductive phenotypes in backcross males relative to infertile F1’s and fertile controls (gray) depend on whether negative epistasis between X-linked loci and (A) Y-linked loci, (B) autosomal loci, or (C) a combination of both, is the primary cause of sterile (−) phenotypes. Note that the change in phenotypic variance in B and C approximates the expectation for multiple X–A incompatibilities of small effect, one of several plausible scenarios for an autosomal contribution to hybrid male infertility.
Materials and Methods
Animals
The wild-derived inbred strains used in this study, PWK/PhJ and LEWES/EiJ, were originally purchased from The Jackson Laboratory and were maintained at the University of Arizona (UA) Central Animal Care Facility under standard conditions in accordance with the UA Animal Care and Use Committee regulations. Males used for reproductive assays were separated from same-sex siblings for at least 15 days prior to kill at 70 days. Males used in crosses were paired with nulliparous musculusPWK females at 55–60 days, or 76 days (F1 males only).
Crossing design and data collection
We introgressed the domesticusLEWES Y chromosome onto the musculusPWK background by backcrossing male progeny to musculusPWK females for 11 generations. Males in all generations have the same sex chromosome and mitochondrial genotypes, whereas autosomal heterozygosity is reduced by half each generation (Figure 1A). By N11, expected heterozygosity for domesticusLEWES autosomal alleles is <0.1%. For each backcross generation we set up an average of four crosses (range 2–7) and recorded litter size at birth and weaning, and sex ratio. Litters were checked regularly during the first week postpartum. Dead neonates were removed and sexed by PCR assay based on amplification of the Y-linked gene, Sry, together with male and female controls. Pups could be sexed with confidence by visual examination after the first week. Each male contributed a maximum of one litter to the next generation. Whenever possible, males from different litters were used in crosses. To generate N11 experimental males, N10 males were crossed to either pure musculusPWK females or to females from two X introgression lines that are homozygous for domesticusLEWES introgressions from ∼37–126 Mb (musculusDOM X-8; Campbell et al. 2012) or ∼106–164 Mb (musculusDOM X-9) on an otherwise musculusPWK background (Figure 1B). N11 control males with a musculusPWK Y were produced by backcrossing N10 females to musculusPWK males.
We assayed testis mass, sperm count, and sperm head morphology in N11 males (n = 9–12/genotype, Figure 1B), and in the progeny of F1, N2, N4, N5, and N6 males (n = 13–20/generation, Figure 1A). Reproductive measures for musculusPWK × domesticusLEWES F1 males (n = 14) are from Campbell et al. (2012). Detailed methods for reproductive assays are provided in Good et al. (2008a,b). Briefly, males were weighed to the nearest 0.01 g and freshly dissected testes were weighed to the nearest 0.1 mg. Mature spermatozoa were obtained from the cauda epididymis. Sperm count was estimated as millions/milliliter using a Makler counting chamber. Heat-shocked sperm suspension was spread on slides and stained with 1% eosin yellow. Sperm head morphology was scored on a phase contrast microscope, blind to genotype. A minimum of 100 sperms/male were evaluated and assigned to one of four categories: (1) normal, characterized by a rounded head and a strongly curved apical hook (Russell et al. 1990); (2) moderately abnormal, characterized by a flattened head and shortened hook; (3) abnormal, characterized by a shortened head and a hook reduced to a short point; and (4) severely abnormal, characterized by a small, asymmetrical head lacking a hook. Because category 4 sperm were not observed in 37% of backcross males (28/76), we combined categories 3 and 4 for analysis (severely abnormal, hereafter).
Data analysis
We corrected for the correlation between testis and body weight by using relative testis weight (RTW = milligrams of testis/gram of body weight) in all analyses. With the exception of RTW and litter sex ratio, none of the reproductive variables were normally distributed and transformations did not improve the normal fit. Significant differences between genotypes were tested with ANOVA followed by parametric (RTW, sex ratio) or nonparametric (all other variables) post hoc tests with corrections for multiple comparisons. All statistical analyses were carried out in JMP v10.01.
Results
Fertility in F1 and backcross males
F1 males with a musculusPWK X and a domesticusLEWES Y have severe reproductive deficits, including small testes, sperm counts up to an order of magnitude below controls, and <5% of sperm with normal head morphology (Good et al. 2008a; Campbell et al. 2012). Nonetheless, these males are not completely sterile. Whereas crosses between musculusPWK females and 55-day-old musculusPWK × domesticusLEWES F1 males (n = 4) produced no progeny, 100% of 76-day-old F1 males (n = 6) sired litters. Given that musculusPWK males are reproductively mature by 6 weeks (48 days), this pattern suggests that F1 males with a musculusPWK X experience a moderate reproductive delay.
Mean litter size, percentage of preweaning mortality, and percentage of male progeny for F1, backcross, and control males are shown in Table 1. For all generations in which at least three crosses were attempted (F1, N2–N5, N8–N10), we compared litter size and sex ratio at birth, and preweaning pup mortality to that in control crosses between pure musculusPWK males and nulliparous musculusPWK females. Although there was a suggestive trend toward male-biased litters sired by F1 and early backcross males (N2–N5; Table 1), litter sex ratio did not differ statistically from controls in any generation.
Table 1. Litter size, survivorship, and sex ratio for F1, backcross, and control males.
| Experimental crosses (n) | Litters | Litter sizea (SD) | % pup mortality (SD) | % male progenyb (SD) |
|---|---|---|---|---|
| ♀ musculusPWK × ♂ (musculusPWK/ domesticusLEWES) F1 (6) | 6 | 5.7 (1.5) | 12.9 (15.5) | 66.6 (27.5) |
| ♀ musculusPWK × ♂ N2 YLEWES (3) | 3 | 8.0 (0.0) | 12.5 (21.7) | 66.7 (19.1) |
| ♀ musculusPWK × ♂ N3 YLEWES (6) | 5 | 2.6 (2.3) | 75.0 (43.3) | 58.3 (11.8) |
| ♀ musculusPWK × ♂ N4 YLEWES (4) | 4 | 7.3 (1.5) | 0 | 68.1 (13.0) |
| ♀ musculusPWK × ♂ N5 YLEWES (3) | 3 | 7.7 (0.6) | 0 | 64.3 (22.4) |
| ♀ musculusPWK × ♂ N6 YLEWES (2) | 2 | 5 (1.4) | 0 | 37.5 (18.7) |
| ♀ musculusPWK × ♂ N7 YLEWES (2) | 2 | 5 (0.0) | 0 | 40.0 (0.0) |
| ♀ musculusPWK × ♂ N8 YLEWES (4) | 4 | 6.5 (1.7) | 0 | 46.7 (23.7) |
| ♀ musculusPWK × ♂ N9 YLEWES (6) | 6 | 7.5 (1.9) | 0 | 50.1 (15.9) |
| ♀ musculusPWK × ♂ N10 YLEWES (4) | 4 | 8.3 (2.1) | 0 | 53.7 (16.2) |
| Control crosses (n) | ||||
| ♀ N10 × ♂ musculusPWK (3) | 3 | 7.7 (1.5) | 0 | 61.6 (5.6) |
| ♀ musculusPWK × ♂ musculusPWK (9) | 9 | 6.3 (0.9) | 14.9 (32.8) | 53.1 (0.13) |
Mean litter size at birth.
Mean percent male progeny at birth.
Severe fertility deficits were only apparent in the N3 generation (Table 1). N3 males sired significantly smaller litters (Wilcoxon P = 0.004; Bonferroni-corrected α = 0.007) and only two of six males sired surviving offspring, resulting in higher pup mortality (P = 0.04) relative to controls. However, sperm count and relative testis weight in N3 males were not different from controls, and N3 males had significantly more normal sperm than N2 males (Figure 3; see below). The genetic basis of the fertility defects seen in the N3 generation is not readily apparent. In this experiment, any X–Y or Y–mitochondrial incompatibilities were exposed in all generations; the opportunity for dominant-acting X–autosome incompatibilities was higher in F1 and N2 relative to N3 males, whereas the opportunity for Y–autosomal recessive incompatibilities increased with each backcross generation. However, in early backcross generations, autosomal recessive–autosomal dominant incompatibilities are exposed, and these are masked in later backcross generations as the genome becomes dominated by musculusPWK. We speculate that such incompatibilities may be responsible for the reduced litter size seen in the N3 generation (Table 1), possibly mediated by phenotypes not measured in this study. For example, excess DNA fragmentation in sperm is a phenotype strongly associated with zygotic, embryonic, and postnatal mortality in mammals (Cho et al. 2003; Ruiz-López et al. 2010; Robinson et al. 2012).
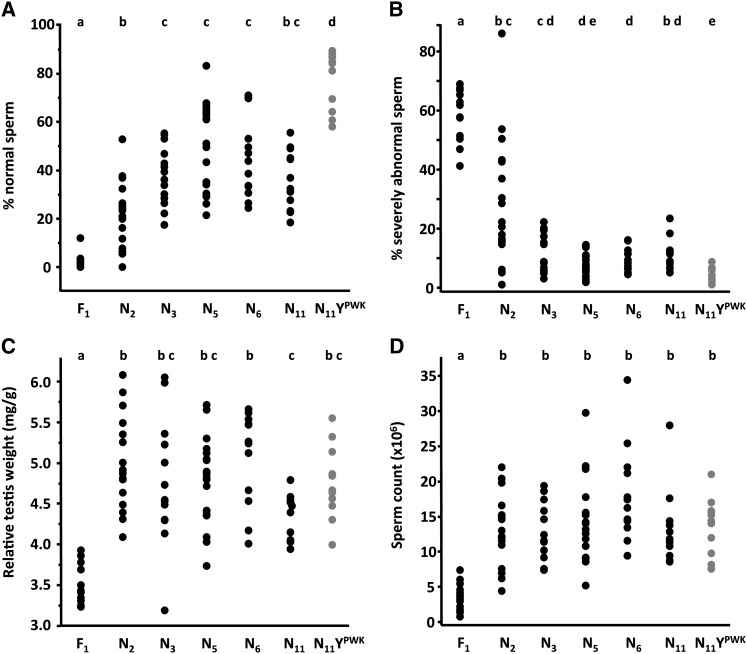
Figure 3.
Reproductive phenotypes in infertile F1’s, backcross, and control (N11 YPWK, gray) males (A-D). Pairwise differences were tested with ANOVA followed by post hoc tests (Steel–Dwass, sperm phenotypes, and sperm count; Tukey HSD, relative testis weight). Sample sizes are shown in Figure 1A. Genotypes not connected by the same letter are significantly different at experiment-wise α = 0.05.
Change in male reproductive parameters across backcross generations
There was a significant improvement from the F1 to the first backcross generation (N2) for the three descriptors of male reproductive phenotype: sperm head morphology (Steel–Dwass P = 0.001, Figure 3A; P = 0.003, Figure 3B), RTW [Turkey honest significant difference (HSD) P < 0.0001, Figure 3C], and sperm count (Steel–Dwass P = 0.0002, Figure 3D). This indicates that X–autosomal dominant incompatibilities contribute significantly to reproductive defects in F1 males. However, the pattern of recovery in backcross males relative to controls differed between the three phenotypes. Consistent with a large negative effect of X–Y interactions on sperm morphology, all backcross generations had significantly fewer normal sperm relative to controls (Figure 3A). Likewise, there was a moderate but significant excess of severely abnormal sperm in all backcross generations except N5 (Figure 3B). These patterns are most consistent with the prediction shown in Figure 2C, in which both X–Y and X–autosome incompatibilities contribute to F1 sperm abnormality. Notably, only X–Y effects can explain the persistence of abnormalities in later backcross generations.
In contrast, RTW and sperm count were statistically equivalent in all backcross generations relative to controls (Figure 3, C and D). This indicates that X–Y incompatibilities do not influence these phenotypes. In the N2 generation, all males had testis weight in the normal range and 82% (14/17) had normal sperm counts (Table 2). These patterns suggest that the probability of recovering the combination of X–autosome incompatibilities required for infertile phenotypes is greatly reduced when autosomal heterozygosity for domesticusLEWES alleles is decreased to ∼50%. To better understand the architecture of X–autosome incompatibilities responsible for the severe deficits in F1 males, we compared the observed numbers of early backcross males with phenotypic values in the F1 range to those expected if F1 phenotypes were due to incompatibilities between the X and one (X–A), two (X–2A), or three (X–3A) autosomal dominant loci (Table 2). While our power to discriminate between these simple models was very low, for the N2 sample, X–A was rejected for both phenotypes (RTW, chi-square = 17.0, P < 0.001; sperm count, chi-square = 7.1, P = 0.0008), and X–2A was rejected for RTW (chi-square = 5.7, P = 0.02). Chi-square values for all comparisons are provided in Supporting Information, Table S1.
Table 2. Observed percentages of males with phenotypic values in infertile F1 range vs. expected percentages under three alternative hypotheses for the minimum number of X–autosome incompatibilities required for an infertile phenotype.
| % (n) males with phenotypic values in infertile F1 range | ||||||
|---|---|---|---|---|---|---|
| Observed | Expected | |||||
| Generation (n) | RTWa | Sperm count | X–Ab | X–2Ac | X–3Ad | |
| N2 (17) | 0 (0) | 17.6 (3) | 50 (8.5) | 25 (4.3) | 6.3 (1.1) | |
| N3 (14) | 7.1 (1) | 14.3 (2) | 25 (3.5) | 6.3 (0.9) | 0.4 (0.6) | |
RTW, relative testis weight.
Infertility requires incompatibility between X and one autosomal dominant locus.
Infertility requires incompatibilities between X and two unlinked autosomal dominant loci with additive effects.
Infertility requires incompatibilities between X and three unlinked autosomal dominant loci with additive effects.
Unexpectedly, RTW was significantly lower in N11 relative to N2 and N6 males, with a trend in the same direction relative to controls (Figure 3C). Reduced RTW relative to earlier backcross generations could be explained by the effects of inbreeding in N11 males, whereas reduction relative to controls cannot. Therefore, we compared RTW in the three inbred genotypes with a domesticusLEWES Y (N11, musculusDOM X-8, musculusDOM X-9) to inbred controls. With fewer comparisons to correct for, there was a modest but significant reduction in RTW in all genotypes with a domesticusLEWES Y (Figure S1). Given the rapid recovery of RTW in N2–N3 males, a reasonable interpretation is that this mild deficit is caused by Y–autosomal recessive incompatibilities that are missing in F1 and early backcross generations.
The contribution of X–Y interactions to hybrid male sperm abnormality
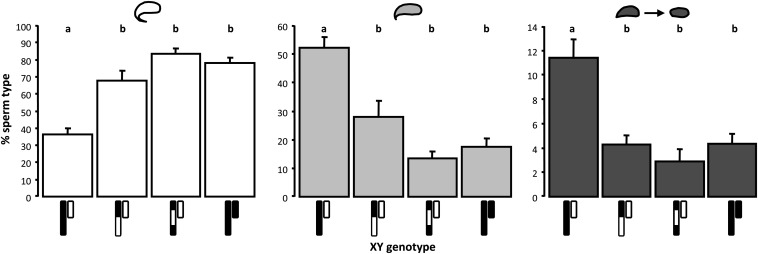
To verify that X–Y interactions were the cause of abnormal sperm morphology in N11 males, we used X chromosome introgression lines to see if we could rescue the phenotype. N11 males with a domesticusLEWES Y on an otherwise musculusPWK background had significantly more abnormal sperm than males with the same autosomal and Y chromosome genotypes paired with domesticusLEWES introgressions on either the central (musculusDOM X-8, Steel–Dwass P = 0.0008) or distal (musculusDOM X-9, P = 0.005) part of the X chromosome (Figure 4). In contrast, neither X introgression genotype had excess abnormal sperm relative to controls (P > 0.6).
Figure 4.
Sperm phenotypes in domesticusLEWES Y introgression males. Males with a domesticusLEWES Y (white) and a complete musculusPWK X (black) have significantly fewer normal sperm (white bars) and significantly more moderately abnormal (light gray bars) and severely abnormal sperm (dark gray bars) than males with domesticusLEWES introgressions on the musculusPWK X, or control males with a musculusPWK Y; domesticusLEWES X introgressions eliminate excess sperm abnormality. Bars represent genotypic means; error bars are +1 SE; sample sizes are shown in Figure 1B. Pairwise differences were tested with ANOVA followed by Steel–Dwass post hoc tests. Genotypes not connected by the same letter are significantly different at experiment-wise α = 0.05.
The interval on the musculusPWK X for which we previously found a strong negative effect on sperm morphology when combined with a domesticusLEWES Y is replaced with a domesticusLEWES introgression in musculusDOM X-8, but not in musculusDOM X-9 (Campbell et al. 2012). We did not, therefore, expect the musculusDOM X-9 introgression to rescue sperm phenotypes. This result is likely explained by limited power to detect and resolve the location of X-linked QTL in our earlier study. One interpretation of the current data is that the causative X-linked locus (or loci) is in the region between ∼106 and 126 Mb that is domesticusLEWES derived in both introgression genotypes and therefore at least 10 Mb distal to the interval implicated in Campbell et al. (2012). Alternatively, loci in several regions of the musculusPWK X might interact negatively with the domesticusLEWES Y, and replacement of one or more of these with a domesticusLEWES genotype is sufficient to rescue sperm abnormality. Importantly, while resolution of these issues awaits fine-scale mapping on the X, rescue of sperm phenotypes in genotypes with reduced mismatch between the X and Y provides strong support for the proposition that incompatibilities between the domesticusLEWES Y and musculusPWK X are important for hybrid sperm abnormality.
Discussion
The genetic basis of hybrid male sterility in house mice is complex, polygenic, and strongly X linked (Storchová et al. 2004; Good et al. 2008b; White et al. 2011, 2012; Dzur-Gejdosova et al. 2012). Whereas a consistently large role of the X, but not the Y, is found in lab crosses between M. m. musculus and M. m. domesticus, large effects of both sex chromosomes are inferred from hybrid zone studies in nature (Vanlerberghe et al. 1986; Tucker et al. 1992; Payseur et al. 2004; Teeter et al. 2010; Janoušek et al. 2012). We dissected the relative contributions of X–Y and X–autosomal dominant incompatibilities to three reproductive phenotypes in an 11-generation backcross experiment in which the M. m. domesticus Y chromosome was introgressed onto a M. m. musculus background. We found a significant negative effect of X–Y interactions that was specific to sperm morphology: males with a M. m. domesticus Y and a M. m. musculus X have excess abnormal sperm, regardless of autosomal background, and M. m. domesticus introgressions on the X rescue this phenotype. In contrast, the severe reductions in testis weight and sperm count that characterize F1 males were explained by incompatibilities between the M. m. musculus X and loci in the M. m. domesticus autosomal genome. Strikingly, these deficits were largely eliminated after just one generation of backcrossing. These results provide insight into the genetic architecture of F1 male sterility, and help to explain genome-wide patterns of introgression across the hybrid zone.
The genetic architecture of sperm abnormality in hybrid males
We previously suggested that incompatibilities between the M. m. musculus X and M. m. domesticus Y chromosomes might have a negative effect on sperm head morphology in males with an F1 autosomal background (Campbell et al. 2012). Here, we tested this hypothesis and demonstrate that sperm abnormality persists on a genetic background in which potential X–autosome incompatibilities are progressively removed. Thus, the genetic architecture of this sterility phenotype is distinct from that underlying reduced testis weight and sperm count. Although the overall contribution of X–Y interactions to hybrid male sterility is small relative to that of X–autosome incompatibilities, this result is important for two main reasons.
First, the specificity of X–Y incompatibilities to sperm abnormality delimits the search for candidate loci to a specific spermatogenic time point and cell type, thereby reducing the genetic complexity of hybrid male sterility. Whereas relative testis weight is a general index of male reproductive fitness and sperm count provides a cumulative measure of the successful progression of germ cells through spermatogenesis, sperm morphology is largely dependent on processes acting in postmeiotic germ cells. During this final stage of spermatogenesis, chromatin is progressively remodeled and condensed, and nuclear morphology undergoes a dramatic transformation, culminating in the highly differentiated structure of mature spermatozoa (reviewed in Oliva and Castillo 2011). Incomplete chromatin compaction is a common cause of abnormal sperm head morphology in mammals (Balhorn 2007; Revay et al. 2009). While autosomal genes play the major roles in chromatin repackaging and condensation (e.g., transition nuclear proteins and protamines; reviewed in Sassone-Corsi 2002), a small subset of X- and Y-linked genes are highly transcribed in postmeiotic spermatids (Namekawa et al. 2006; Mueller et al. 2008), and several are required for normal sperm differentiation (e.g., Cocquet et al. 2009, 2012; Vernet et al. 2012). Thus, candidate gene-targeted fine-scale mapping on the X could accelerate identification of the X-linked component of this X–Y incompatibility.
Second, although excess sperm abnormality does not reduce the fecundity of M. m. domesticus Y introgression males under noncompetitive lab conditions, this phenotype should have significant fitness consequences in nature where multiple mating in females (Dean et al. 2006) promotes sperm competition. In mice, sperm head morphology is highly correlated with competitive ability (Immler et al. 2007) and fertilization success (Kawai et al. 2006), with lower fertilization rate associated with abnormal head shape and particularly with reduction or absence of the apical hook (Krzanowska and Lorenc 1983; Krzanowska et al. 1995; Oka et al. 2007). This suggests that even moderate levels of sperm abnormality could have large negative effects on male fitness in natural populations. Thus, the effect of X–Y incompatibilities on sperm morphology may explain the complete absence of M. m. domesticus Y chromosome introgression across the hybrid zone, a hypothesis that could be tested by evaluating the contribution of Y genotype to sperm abnormality in hybrid zone males (Albrechtová et al. 2012).
The pattern of recovery from F1 to N2 males indicates that X–Y and X–autosome effects on sperm abnormality are compounded in F1 males. We recently discovered that widespread overexpression of the musculusPWK X chromosome on an F1 autosomal background is explained by partial failure of meiotic sex chromosome inactivation (MSCI) in primary spermatocytes (Good et al. 2010; Campbell et al. 2013). X overexpression persists in postmeiotic round spermatids, and the negative correlation between whole testis X expression and reproductive parameters is strongest for sperm morphology. Importantly, there is no association between Y chromosome genotype and disrupted MSCI (Campbell et al. 2013). Thus, the X–autosome incompatibilities that underlie disrupted MSCI may be a major cause of the severe sterility phenotypes that, in this study, were unique to the F1 generation.
The genetic architecture of X–autosome incompatibilities in F1 males
This and previous studies demonstrate that X–autosome incompatibilities are essential for sterility and subfertility in F1 hybrid male house mice. Above, we suggest that disrupted MSCI may explain the severe reproductive deficits in F1 males from the musculusPWK × domesticusLEWES cross. But what genetic architecture is consistent with the restoration of normal sperm count and testis weight when the opportunity for X–autosomal dominant incompatibilities is reduced from 100 to 50%?
If many X–autosome incompatibilities act additively to produce sterile values for testis weight and sperm count, a larger sample of genotypes might be required to observe the full F1 phenotype in early backcross males. However, we would still expect a more gradual recovery in these phenotypes as deleterious M. m. domesticus alleles are progressively removed. In contrast, if simple incompatibilities between the X and one or two autosomal loci are sufficient for F1 sterility we would expect a bimodal distribution of phenotypes in early backcross generations, with males that retain the incompatible M. m. domesticus alleles having phenotypic values in the F1 range. The sharp transition between F1 and backcross phenotypes is also inconsistent with this simple architecture. We confirmed this quantitatively by comparing the observed percentages of N2 and N3 males with testis weight or sperm count in the F1 range to those expected if F1 phenotypes were due to an incompatibility between the X and one, two, or three autosomal loci. Even with limited power, the one autosomal locus model was rejected for both phenotypes in N2 males, and the two autosomal loci model was rejected for testis weight.
Together, these observations suggest that, while a relatively small number of individual X–autosome incompatibilities may underlie F1 sterility, each incompatibility is complex. This is in agreement with the theoretical expectation that complex incompatibilities evolve more readily than simple ones (Orr 1995), and with empirical work in house mice, Drosophila, and other taxa, demonstrating that complex negative epistasis is a common feature of the genetic architecture of sterility in hybrids between incipient or recently diverged species (Kao et al. 2010; Dzur-Gejdosova et al. 2012; and reviewed in Coyne and Orr 2004). For example, in crosses between subspecies of Drosophila pseudoobscura, at least seven interacting genes underlie a single incompatibility that causes hybrid male sterility (Phadnis 2011).
Conclusions
This study provides direct evidence that the Y chromosome contributes to hybrid male sterility in house mice. Lack of introgression of the M. m. domesticus Y chromosome across the European hybrid zone suggests that the moderate negative effects of X–Y interactions on sperm phenotypes in the lab may be amplified by sperm competition in nature. In contrast, significant recovery of testis weight and sperm count after one generation of backcrossing suggests that male reproductive fitness is robust to substantial autosomal heterozygosity for M. m. domesticus alleles on a M. m. musculus background. This inference is consistent with asymmetric introgression of M. m. domesticus autosomal alleles into M. m. musculus populations in nature (Vanlerberghe et al. 1988; Raufaste et al. 2005; Teeter et al. 2008, 2010).
Supplementary Material
Acknowledgments
Comments from Associate Editor B. Payseur and two anonymous reviewers significantly improved an earlier version of this article. C. W. Birky provided equipment used during the course of the experiment. P.C. was supported by a G. G. Simpson Postdoctoral Fellowship from the University of Arizona. This work was funded by National Science Foundation and National Institutes of Health grants to M.W.N.
Footnotes
Communicating editor: B. Payseur
Literature Cited
- Albrechtová J., Albrecht T., Baird S. J. E., Macholán M., Rudolfsen G., et al. , 2012. Sperm-related phenotypes implicated in both maitenance and breakdown of a natural species barrier in the house mouse. Proc. Biol. Sci. 279: 4803–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R., 2007. The protamine family of sperm nuclear proteins. Genome Biol. 8: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W., 1909. Heredity and variation in modern lights, pp. 85–101 in Darwin and Modern Science, edited by Seward A. C., Cambridge University Press, Cambridge, UK. [Google Scholar]
- Britton-Davidian J., Fel-Clair F., Lopez J., Alibert P., Boursot P., 2005. Postzygotic isolation between the two European subspecies of the house mouse: estimates from fertility patterns in wild and laboratory-bred hybrids. Biol. J. Linn. Soc. Lond. 84: 379–393. [Google Scholar]
- Cabot E. L., Davis A. W., Johnson N. A., Wu C.-I., 1994. Genetics of reproductive isolation in the Drosophila simulans clade: complex epistasis underlying hybrid male sterility. Genetics 137: 175–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P., Good J. M., Dean M. D., Tucker P. K., Nachman M. W., 2012. The contribution of the Y chromosome to hybrid male sterility in house mice. Genetics 191: 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P., Good J. M., Nachman M. W., 2013. Meiotic sex chromosome inactivation is disrupted in sterile hybrid male house mice. Genetics 193: 819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C., Jung-Ha H., Willis W. D., Goulding E. H., Stein P., et al. , 2003. Protamine-2 deficiency leads to sperm DNA damage and embryo death in mice. Biol. Reprod. 69: 211–217. [DOI] [PubMed] [Google Scholar]
- Cocquet J., Ellis P. J. I., Yamauchi Y., Mahadevaiah S. K., Affara N. A., et al. , 2009. The multicopy gene Sly repressed the sex chromosomes in the male mouse germline after meiosis. PLoS Biol. 7: e1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J., Ellis P. J. I., Mahadevaiah S. K., Affara N. A., Vaiman D. et al, 2012. A genetic basis for a postmeiotic X vs. Y chromosome intragenomic conflict in the mouse. PLoS Genet. 8: e1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Dean M. D., Ardlie K. G., Nachman M. W., 2006. The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus). Mol. Ecol. 15: 4141–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1937. Genetics and the Origin of Species, Columbia University Press, New York. [Google Scholar]
- Dod B., Jermin L. S., Boursot P., Chapman V. M., Nielsen J. T., et al. , 1993. Counterselection on sex chromosomes in the Mus musculus European hybrid zone. J. Evol. Biol. 6: 529–546. [Google Scholar]
- Dzur-Gejdosova M., Simecek P., Gregorova S., Bhattacharyya T., Forejt J., 2012. Dissecting the genetic architecture of F1 hybrid sterility in house mice. Evolution 66: 3321–3335. [DOI] [PubMed] [Google Scholar]
- Forejt J., 1996. Hybrid sterility in the mouse. Trends Genet. 12: 412–417. [DOI] [PubMed] [Google Scholar]
- Forejt J., Iványi P., 1974. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.). Genet. Res. 24: 189–206. [DOI] [PubMed] [Google Scholar]
- Geraldes A., Basset P., Smith K. L., Nachman M. W., 2011. Higher differentiation among subspecies of the house mouse (Mus musculus) in genomic regions with low recombination. Mol. Ecol. 20: 4722–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Handel M. A., Nachman M. W., 2008a Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution 62: 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Dean M. D., Nachman M. W., 2008b A complex genetic basis to X- linked hybrid male sterility between two species of house mice. Genetics 179: 2213–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Giger T., Dean M. D., Nachman M. W., 2010. Widespread over- expression of the X chromosome in sterile F1 hybrid mice. PLoS Genet. 6: e1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S., 1922. Sex ratio and unisexual sterility in animal hybrids. J. Genet. 12: 101–109. [Google Scholar]
- Hunt W. G., Selander R. K., 1973. Biochemical genetics of hybridization in European house mice. Heredity 31: 11–33. [DOI] [PubMed] [Google Scholar]
- Immler S., Moore H. D., Breed W. G., and T. R. Birkhead, 2007. By hook or by crook? Morphometry, competition and cooperation in rodent sperm. PLoS ONE 2: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoušek V., Wang L., Luzynski K., Dufková P., Vyskočilová M. M., et al. , 2012. Genome-wide architecture of reproductive isolation in a naturally occurring hybrid zone between Mus musculus musculus and M. m. domesticus. Mol. Ecol. 21: 3032–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. P., Van Der Kooij J., Solheim R., Searle J. B., 2010. Norwegian house mice (Mus musculus musculus/domesticus): distributions, routes of colonization and patterns of hybridization. Mol. Ecol. 19: 5252–5264. [DOI] [PubMed] [Google Scholar]
- Kao K. C., Schwartz K., Sherlock G., 2010. A genome-wide analysis reveals no nuclear Dobzhansky-Muller pairs of determinants of speciation between S. cervisiae and S. paradox, but suggests more complex incompatibilities. PLoS Genet. 6: e1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y., Hata T., Suzuki O., Matsuda J., 2006. The relationship between sperm morphology and in vitro fertilization ability in mice. J. Reprod. Dev. 52: 561–568. [DOI] [PubMed] [Google Scholar]
- Krzanowska H., Lorenc E., 1983. Influence of egg investments on in-vitro penetration of mouse eggs by misshapen spermatozoa. J. Reprod. Fertil. 68: 57–62. [DOI] [PubMed] [Google Scholar]
- Krzanowska H., Styrna J., Wabiksliz B., 1995. Analysis of sperm quality in recombinant inbred mouse strains: correlation of sperm head shape with sperm abnormalities and with the incidence of supplementary spermatozoa in the perivitelline space. J. Reprod. Fertil. 104: 347–354. [DOI] [PubMed] [Google Scholar]
- Laurie C. C., 1997. The weaker sex is heterogametic: 75 years of Haldane’s rule. Genetics 147: 937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholán M., Munclinger P., Šugerková M., Dufková P., Bímová B., et al. , 2007. Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution 61: 746–771. [DOI] [PubMed] [Google Scholar]
- Macholán M., Baird S. J. E., Munclinger P., Dufková P., Bímová B., et al. , 2008. Genetic conflict outweighs heterogametic incompatibility in the mouse hybrid zone? BMC Evol. Biol. 8: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly J. P., Presgraves D. C., 2007. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 5: 1890–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O., Trachtulec Z., Vlcek C., Schimenti J. C., Forejt J., 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323: 373–375. [DOI] [PubMed] [Google Scholar]
- Mueller J. L., Mahadevaiah S. K., Park P. J., Warburton P. E., Page D. C., et al. , 2008. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 40: 794–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1942. Isolating mechanisms, evolution and temperature. Biol. Symposia 6: 71–125. [Google Scholar]
- Namekawa S. H., Park P. J., Zhang L. F., Shima J. E., McCarrey J. R., et al. , 2006. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 16: 660–667. [DOI] [PubMed] [Google Scholar]
- Oka A., Aoto T., Totsuka Y., Takahashi R., Ueda M., et al. , 2007. Disruption of genetic interaction between two autosomal regions and the X chromosome causes reproductive isolation between mouse strains derived from different subspecies. Genetics 175: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R., Castillo J., 2011. Proteomics and the genetics of sperm chromatin condensation. Asian J. Androl. 13: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139: 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., Turelli M., 2001. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution 55: 1085–1094. [DOI] [PubMed] [Google Scholar]
- Payseur B. A., Krenz J. G., Nachman M. W., 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58: 2064–2078. [DOI] [PubMed] [Google Scholar]
- Phadnis N., 2011. Genetic architecture of male sterility and segregation distortion in Drosophila pseudoobscura Bogota-USA hybrids. Genetics 189: 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager E. M., Boursot P., Sage R. D., 1997. New assays for Y chromosome and p53 pseudogene clines among East Holstein mice. Mamm. Genome 8: 279–281. [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2002. Patterns of postzygotic isolation in Lepidoptera. Evolution 56: 1168–1183. [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24: 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price T. D., Bouvier M. M., 2002. The evolution of postzygotic incompatibilities in birds. Evolution 56: 2083–2089. [PubMed] [Google Scholar]
- Raufaste N., Orth A., Belkhir K., Senet D., Smadja C., et al. , 2005. Inferences of selection and migration in the Danish house mouse hybrid zone. Biol. J. Linn. Soc. Lond. 84: 593–616. [Google Scholar]
- Reed L. K., LaFlamme B. A., Markow T. A., 2008. Genetic architecture of hybrid male sterility in Drosophila: analysis of intraspecies variation for interspecies isolation. PLoS ONE 3: e3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revay T., Nagy S., Kopp C., Flyckt A., Rens W., et al. , 2009. Macrocephaly in bull spermatozoa is associated with nuclear vacuoles, diploidy and alteration of chromatin condensation. Cytogenet. Genome Res. 126: 202–209. [DOI] [PubMed] [Google Scholar]
- Robinson L., Gallos I. D., Conner S. J., Rajkhowa M., Miller D., et al. , 2012. The effect of sperm DNA fragmentation on miscarriage rates: a systematic review and meta- analysis. Hum. Reprod. 27: 2908–2917. [DOI] [PubMed] [Google Scholar]
- Ruiz-López M. J., Espeso G., Evenson D. P., Roldan E. R. S., Gomendio M., 2010. Paternal levels of DNA damage in spermatozoa and maternal parity influence offspring mortality in an endangered ungulate. Proc. Biol. Sci. 277: 2541–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L. D., Ettlin R. A., Sinha Hikin A. P., Clegg E. D., 1990. Histological and Histopathological Evaluation of the Testis, Cache River Press, Clearwater, FL. [Google Scholar]
- Sassone-Corsi P., 2002. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296: 2176–2178. [DOI] [PubMed] [Google Scholar]
- Spiridonova L. N., Kiselev K. V., Korobitsyna K. V., 2011. Discordance in the distribution of markers of different inheritance systems (nDNA, mtDNA, and chromosomes) in the superspecies complex Mus musculus as a result of extensive hybridization in Primorye. Russ. J. Genet. 47: 100–109. [PubMed] [Google Scholar]
- Storchová R., Gregorová S., Buckiová D., Kyselová V., Divina P., et al. , 2004. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm. Genome 15: 515–524. [DOI] [PubMed] [Google Scholar]
- Tao Y., Zeng Z.-B., Li J., Hartl D. L., Laurie C. C., 2003. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. II. Mapping hybrid male sterility loci on the third chromosome. Genetics 164: 1399–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter K. C., Payseur B. A., Harris L. W., Bakewell M. A., Thibodeau L. M., et al. , 2008. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Res. 18: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter K. C., Thibodeau L. M., Gompert Z., Buerkle C. A., Nachman M. W., et al. , 2010. The variable genomic architecture of isolation between hybridizing species of house mice. Evolution 64: 472–485. [DOI] [PubMed] [Google Scholar]
- True J. R., Weir B. S., Laurie C. C., 1996. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics 142: 819–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker P. K., Sage R. D., Warner J., Wilson A. C., Eicher E. M., 1992. Abrupt cline for sex chromosomes in a hybrid zone between two species of mice. Evolution 46: 1146–1163. [DOI] [PubMed] [Google Scholar]
- Turner L. M., Schwahn D. J., Harr B., 2012. Reduced male fertility is common but highly variable in form and severity in a natural house mouse hybrid zone. Evolution 66: 443–458. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe F., Dod B., Boursot P., Bellis M., Bonhomme F., 1986. Absence of Y chromosome introgression across the hybrid zone between Mus musculus domesticus and Mus musculus musculus. Genet. Res. 48: 191–197. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe F., Boursot P., Catalan J., Gerasimov S., Bonhomme F., 1988. Genetic analysis of the hybrid zone between two murine subspecies, Mus musculus musculus and Mus musculus domesticus in Bulgaria. Genome 30: 427–437. [PubMed] [Google Scholar]
- Vernet N., Mahadevaiah S. K., Ellis P. J. I., de Rooij D. G., Burgoyne P. S., 2012. Spermatid development in X0 male mice with varying Y chromosome short- arm gene content: evidence for a Y gene controlling the initiation of sperm morphogenesis. Reproduction 144: 433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Steffy B., Wiltshire T., Payseur B. A., 2011. Genetic dissection of a key reproductive barrier between nascent species of house mice. Genetics 189: 289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Stubbings M., Dumont B. L., Payseur B. A., 2012. Genetics and evolution of hybrid male sterility in house mice. Genetics 191: 917–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.