Abstract
The extrinsic apoptosis pathway is initiated by binding of death ligands to death receptors resulting in the formation of the death-inducing signaling complex (DISC). Activation of procaspase-8 within the DISC and its release from the signaling complex is required for processing executor caspases and commiting cell death. Here, we report that the atypical cadherin FAT1 interacts with caspase-8 preventing the association of caspase-8 with the DISC. We identified FAT1 in a genome-wide siRNA screen for synthetic lethal interactions with death receptor-mediated apoptosis. Knockdown of FAT1 sensitized established and patient-derived glioblastoma cell lines for apoptosis transduced by cell death ligands. Depletion of FAT1 resulted in enhanced procaspase-8 recruitment to the DISC and increased formation of caspase-8 containing secondary signaling complexes. In addition, FAT1 knockout cell lines generated by CRISPR/Cas9-mediated genome engineering were more susceptible for death receptor-mediated apoptosis. Our findings provide evidence for a mechanism to control caspase-8-dependent cell death by the atypical cadherin FAT1. These results contribute towards the understanding of effector caspase regulation in physiological conditions.
Keywords: apoptosis, FAT1, glioblastoma, TRAIL
See also: M Herold and A Strasser (January 2014)
Introduction
Apoptosis or ‘programmed cell death’ is a controlled mechanism to remove old, unhealthy or damaged cells in multicellular organisms to maintain tissue homeostasis. Deregulation of apoptosis contributes to neurodegenerative or autoimmune diseases and evasion of apoptosis is a hallmark of cancerogenesis (Hanahan & Weinberg, 2011). The apoptosis pathway is divided into two major branches: The intrinsic pathway is induced intracellularly by stress responses and/or DNA damage. The extrinsic pathway is activated extracellularly by binding of death ligands to a subclass of the tumor necrosis factor (TNF) receptor family.
TNF-related apoptosis-inducing ligand (TRAIL/Apo2L) is a member of death receptor ligands (Wiley et al, 1995; Pitti et al, 1996). TRAIL can selectively induce apoptosis in tumor cells without severly affecting normal tissue (Roth et al, 1999; Walczak et al, 1999; Ashkenazi et al, 2008). Despite caspase activation, TRAIL can induce NF-κB, c-Jun N-terminal kinase (JNK) and p38 MAPK pathways, albeit much less compared to other death ligands CD95L and TNFα (Gonzalvez & Ashkenazi, 2010).
Receptor-mediated apoptosis is initiated by binding of the death ligand to its corresponding receptor, triggering the formation of the death-inducing signaling complex (DISC) (Gonzalvez & Ashkenazi, 2010). After binding of the adaptor protein FADD (Fas-associated death domain protein) to the death receptors via the death-domains (DD), the death effector domain (DED) of FADD recruits procaspase-8/procaspase-10 or cFLIP (cellular FLICE-inhibitory protein). Within the DISC, procaspase-8/-10 is activated by proteolytic cleavage (Muzio et al, 1996). Active caspase-8/-10 in turn activates executor caspases like procaspase-3/procaspase-7. In ‘type I’ cells, there is sufficient caspase-8 activation to induce effector caspases. In ‘type II’ cells, lower quantities of the DISC are formed resulting in less caspase-8 activation (Scaffidi et al, 1998; Krammer et al, 2007). They require an amplification loop via caspase-8-mediated cleavage of the proapoptotic Bcl2 family member BID (BH3-interacting-domain death agonist) and subsequent caspase activation via the mitochondrial pathway.
The first proposed mechanism for initiator caspase activation was based on the induced proximity model. According to this model, the dimerization of caspase-8 monomers (p55/p53) results in a conformational change of caspase-8 and the exposure of its active caspase site (Muzio et al, 1998). The dimerization seems to be sufficient to activate caspase-8 (Martin et al, 1998; Boatright et al, 2003; Chang et al, 2003). However, additional mutational analysis of caspase-8 showed that its full activation requires both dimerization and subsequent cleavage of procaspase-8 (Hughes et al, 2009). Posttranslational modifications provide an additional mechanism to regulate caspase-8 activation. Cullin-3-mediated polyubiquitination of caspase-8 promoted caspase activation and apoptosis induction (Jin et al, 2009). TRAF2 (TNF-receptor-associated factor 2)-mediated ubiquitination of activated caspase-8 set a threshold for extrinsic apoptosis (Gonzalvez et al, 2012).
Previous studies suggested that one ligand trimer initiates receptor trimerization, each recruiting one FADD molecule, so that each FADD in turn interacts with one DED-protein (Krammer et al, 2007). However, two recent studies challenge this ‘one-to-one’ model. Quantitative mass spectrometry analysis of immunoprecipitated TRAIL-DISC and CD95-DISC revealed an abundance of DED-proteins (procaspase-8 and cFLIP) several folds higher than FADD, proposing a model in which DED-chain formation triggers caspase activation and apoptosis (Dickens et al, 2012; Schleich et al, 2012).
The composition and activation of the DISC has been extensively studied, emphasizing the essential role of procaspase-8 in the extrinsic apoptosis pathway. However, little is known about the regulation of caspase-8. The importance of secondary signaling complexes, downstream of the DISC, is just emerging and not well defined. The formation, the timing of assembly and the composition of secondary multiprotein complexes remains an open question.
Here, we searched for proteins that sensitize cells for death receptor-induced apoptosis and identified FAT1 which upon depletion resulted in increased apoptosis upon stimulation with death-inducing ligands. FAT1 knockdown and TRAIL stimulation promoted procaspase-8 recruitment to the DISC and the formation of native high molecular weight complexes containing caspase-8. Co-immunoprecipitation experiments revealed a previously unknown interaction between FAT1 and caspase-8. These findings identify a mechanism controlling caspase activation and thus death receptor-mediated apoptosis under physiological conditions.
Results
A genome-wide RNAi screen identifies FAT1 as a negative regulator of TRAIL-induced apoptosis
To identify novel regulators of death-receptor pathways, we performed a genome-wide siRNA screen in the human glioblastoma cell line U251MG using SMART-pool siRNA library (Dharmacon). Forty-eight hours after siRNA transfection, cells were treated with a sublethal dose of 10 ng/ml TRAIL (Ganten et al, 2006) or medium as a control. Cell viability was measured 2 days later using the CellTiterGlo-assay. We were interested in siRNAs that were synthetic lethal with TRAIL treatment but had no effect on viability themselves. To focus on siRNAs with a wide difference between TRAIL-treated and control-treated cells, we determined a differential score (for further details see Materials and Methods). As shown in Fig 1A, the majority of siRNAs followed a normal Gaussian distribution without effecting TRAIL sensitivity. By contrast, all siRNAs in the quantile-quantile-plot deviating from the straight line (normal distribution) indicate a TRAIL-sensitizing phenotype (Fig 1B, Supplementary Table S1).
Figure 1.
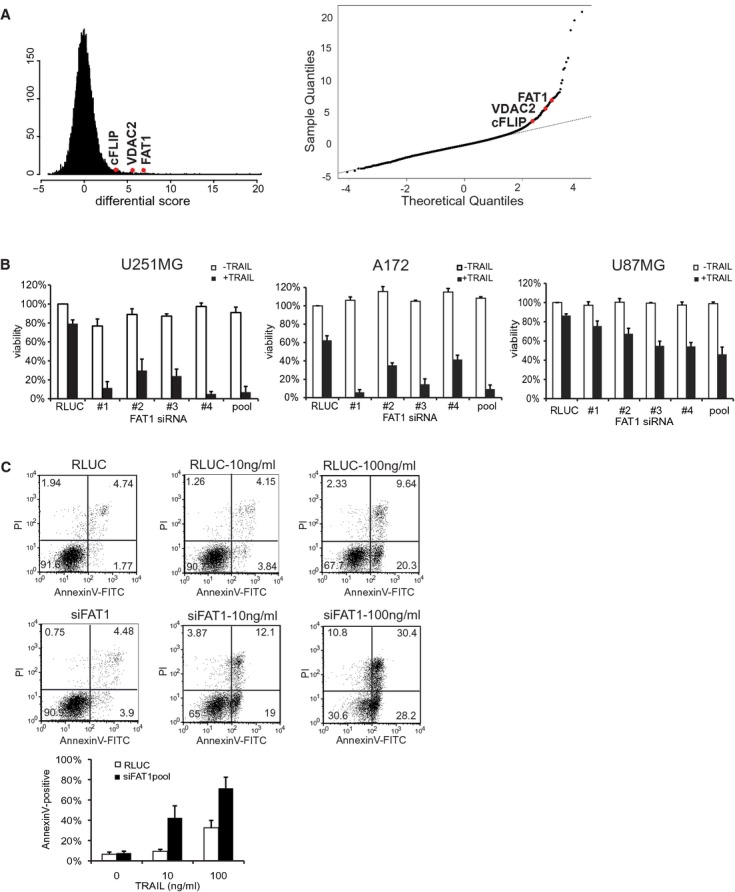
- Left: Histogram of the calculated differential score indicates Gaussian (normal) distribution. Right: Q-Q-plot of genome-wide siRNA screen for the identification of TRAIL sensitizing siRNAs. All siRNAs deviating from the straight line indicate TRAIL sensitizers. Known anti-apoptotic regulators (cFLIP, VDAC2) and the candidate FAT1 are depicted in red.
- Glioma cell lines U251MG, A172 or U87MG were transfected with RLUC (control) or siRNA-FAT no. 1–4, pool. Cells were stimulated with 10 ng/ml TRAIL or left untreated. Viability was determined by luminescence-based CellTiterGlo-Assay 48 h after treatment. Results are shown as percentage of RLUC-untreated cells. The mean ± s.d. of at least three independent experiments is shown.
- U251MG cells were transfected with RLUC (control) or FAT1pool and treated with 10 or 100 ng/ml TRAIL. After 16 h, cells were double stained with AnnexinV-FITC and propidium iodide and analysed by FACS. The graph depicts the amount of AnnexinV-positive cells and the mean ± s.d. of four independent experiments is shown.
Among these candidates were the siRNAs targeting cFLIP/CFLAR and the voltage-dependent anion channel 2 (VDAC2), both previously described as inhibitors of apoptosis. cFLIP is recruited into the DISC, resulting in the inhibition of caspase-8 activation and subsequently suppression of apoptosis (Krueger et al, 2001). VDAC2-deficient cells exhibited increased oligomerization of the pro-apoptotic mitochondrial membrane protein BAK, thus enhancing apoptosis induction after treatment with different chemotherapeutic agents (Cheng et al, 2003). Identification of known apoptotic regulators confirmed the reliability of the screen.
For further analysis we selected the candidate FAT1 because of the following reasons: First, four single siRNAs as well as the pool resulted in increased sensitivity towards TRAIL. Second, siRNA no. 1, siRNA no. 2, siRNA no. 3 and the siRNApool decreased FAT1 mRNA levels to 90% (Supplementary Fig S1A), suggesting that knockdown of FAT1 is responsible for the increased TRAIL sensitivity. Of note, siRNA no. 4 only showed a moderate knockdown of FAT1 but still increased sensitivity towards TRAIL, thus suggesting an off-target effect. Third, the sensitivity towards TRAIL was observed in different glioblastoma cell lines U251MG, A172 and U87MG, excluding a cell type-specific effect (Fig 1B). Moreover, this sensitisation effect was not restricted to glioma cell lines, since the cervical carcinoma cell line HeLa, the osteosarcoma cell line U2OS and the hepatocellular carcinoma cell line HepG2 were also sensitized towards TRAIL by FAT1 depletion (Supplementary Fig S1B). Most importantly, FAT1 has not been connected to death receptor-mediated apoptosis yet. The atypical cadherin FAT1 has been associated with cell adhesion and cell-cell signaling (Tanoue & Takeichi, 2004; Hou & Sibinga, 2009).
To confirm that the decreased viability corresponds to an increase in apoptosis induction, we measured AnnexinV-propidium iodide(PI)-positive cells by flow cytometry analysis. Knockdown of FAT1 increased both fractions, single AnnexinV-positive and AnnexinV-PI-double-positive cells (Fig 1C), indicating that cells lacking FAT1 were dying via apoptosis more rapidly than control cells.
Taken together, our genome-wide screen identified FAT1 as a novel negative regulator of TRAIL-induced apoptosis.
FAT1 depletion increases caspase activation upon TRAIL treatment
The first fat gene was identified in Drosophila melanogaster and following studies indicated the conservation of the Fat family from flies to mammals (Mahoney et al, 1991). Two members (fat and fat-like) exist in Drosophila and four family members (FAT1-4) in mammalians (Tanoue & Takeichi, 2004). FAT4 shows the highest homology to the Drosophila fat, whereas FAT1-3 resemble the Drosophila fat-like. Accordingly, previous studies indicate functional differences between FAT1-3 and FAT4 (Tanoue & Takeichi, 2005). To test the role of the different family members in TRAIL-induced apoptosis, we determined caspase activity upon depletion of each FAT member in combination with TRAIL stimulation. Knockdown of FAT1 but not of any other family member increased caspase activity upon TRAIL treatment (Fig 2A). Specificity of the different siRNAs was confirmed by qPCR indicating no cross regulation of the FAT family members (Supplementary Fig S1C). This result indicates a unique/specific role of FAT1 in TRAIL-induced apoptosis differing from its family members.
Figure 2.
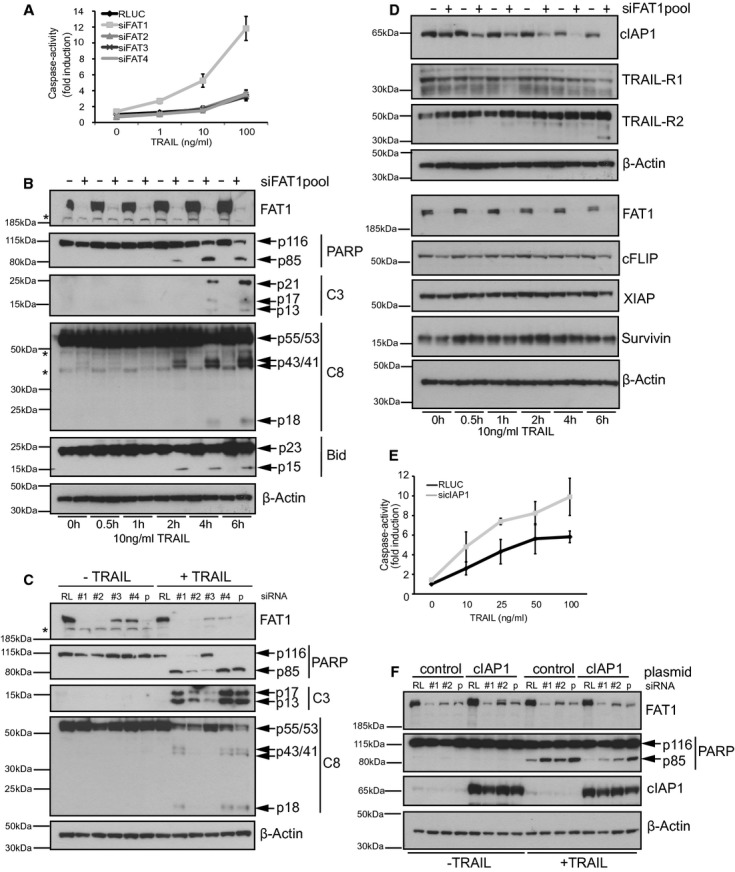
- U251MG cells were transfected with RLUC or siRNApools targeting FAT1,FAT2,FAT3 or FAT4. 72 h later, cells were treated with indicated concentrations of TRAIL. After 6 h, caspase-activity was determined by fluorescence-based ApoOne-assay (Promega). Data is shown as normalized to RLUC untreated cells and as mean ± s.e.m of five independent experiments.
- U251MG cells were transfected with RLUC (control) or siRNA-FAT1pool. Cells were treated with 10 ng/ml TRAIL and harvested at the indicated time points. Cell lysates were analyzed by FAT1, PARP, cleaved caspase-3 (C3) and caspase-8 (C8) immunoblot. β-actin serves as a loading control. Asterisk indicates non-specific band.
- U251MG cells were transfected with RL (control) or different single FAT1 siRNAs. Cells were stimulated with 10 ng/ml TRAIL for 6 h. Cell lysates were analyzed by western blot. β-actin serves as a loading control. Asterisk indicates non-specific band.
- U251MG cells were transfected with RLUC (control) or siRNA-FAT1pool. Cells were treated with 10 ng/ml TRAIL and harvested at the indicated time points for western blot analysis. β-actin serves as a loading control.
- U251MG cells were transfected with RLUC or siRNA-cIAP1pool. Seventy-two hours later, cells were treated with indicated concentrations of TRAIL. After 6 h, caspase-activity was determined by fluorescence-based ApoOne-assay (Promega). Data is shown as normalized to RLUC untreated cells and as mean ± s.d. of three independent experiments.
- U251MG were transduced with control or cIAP1 lentiviral vector, transfected with RL (control) or siRNA-FAT1 no.1, no.2 or pool and induced with hydroxytamoxifen to express cIAP1. Cells were stimulated with 100 ng/ml TRAIL and lysed for immunoblot analysis 6 h later. β-actin serves as a loading control.
Source data are available online for this figure.
In line with the augmented caspase activity, we observed an increase in caspase cleavage products over time in TRAIL-stimulated FAT1 knockdown cells. Immunoblot analysis indicated more cleaved Poly(ADP-ribose)-Polymerase (PARP-p85), more cleaved caspase-3 fragments (p13/p17/p21) and more cleaved tBID (p15) in FAT1-depleted cells starting already 2 h after TRAIL addition (Fig 2B). Interestingly, we detected less procaspase-8 (p55/p53) but likewise more cleavage fragments (p43/p41 and p18) under these conditions. The increase in caspase cleavage products was confirmed using different single siRNAs targeting FAT1 (Fig 2C).
Next, we investigated the involvement of other components of the TRAIL pathway in this sensitization effect. FAT1 knockdown had no effect on the protein levels of the TRAIL-receptor 2 (TRAIL-R2), whereas TRAIL-receptor 1 (TRAIL-R1) staining revealed multiple bands suggesting a very low expression in these cells (Fig 2D). Combinatorial siRNA treatment targeting TRAIL-R2 in combination with FAT1 reversed the increase in caspase cleavage compared to FAT1 knockdown alone, but the combinatorial treatment of siRNA-TRAIL-R1 and siRNA-FAT1 failed to do so (Supplementary Fig S2A and S2B) suggesting that TRAIL-R2 is the main death-receptor required for TRAIL-induced apoposis in these cells. Of note, FAT1 knockdown did not affect surface expression of TRAIL-R2 (Supplementary Fig S2C).
Previous studies indicated the important regulatory role of inhibitor of apoptosis proteins (IAPs) for death receptor-mediated apoptosis (Vaux & Silke, 2005; Varfolomeev et al, 2007). We asked whether anti-apoptotic proteins were affected by the increased TRAIL sensitivity upon FAT1 depletion. Immunoblot analysis showed no change of protein levels, neither for the BIR domain-containing proteins XIAP and Survivin nor for the cellular FLICE-inhibitory protein cFLIP (Fig 2D). However, we observed time-dependent decrease of the anti-apoptotic protein cellular inhibitor of apoptosis-1 (cIAP1) after TRAIL treatment and FAT1 knockdown (Fig 2D) that inversely correlated to the increase of caspase activity (Fig 2B). Previous reports described lysosomal, proteasomal or caspase-dependent cIAP1 degradation (Varfolomeev et al, 2007; Vince et al, 2008; Guicciardi et al, 2010). Only prior addition of pan-caspase inhibitor Z-VAD-FMK and caspase-8 inhibitor Z-IETD-FMK reduced TRAIL-dependent cIAP1 degradation but not treatment with the lysosomal inhibitor bafilomycin A1 or the proteasomal inhibitor MG132 (Supplementary Fig S2D). Accordingly, depleting cIAP1 by siRNA treatment increased caspase activity after TRAIL stimulation (Fig 2E), presumably due to the formation of the previously described ‘ripoptosome’ complex (Geserick et al, 2009; Tenev et al, 2011). To test the involvement of cIAP1 in our sensitivity phenotype, we induced overexpression of cIAP1 after FAT1 depletion and TRAIL stimulation. Independent of FAT1 knockdown we observed reduced PARP cleavage (Fig 2F). Accordingly, it was previously reported that overexpression of cIAP1 itself confers resistance to apoptosis (Gyrd-Hansen & Meier, 2010). However, cells depleted of FAT1 using different siRNAs still showed increased PARP cleavage compared to control cells, indicating that these cells were more sensitive towards TRAIL despite increased cIAP1 protein levels. This result suggests that depletion of FAT1 results in rapid caspase-8 activation leading to caspase-mediated degradation of cIAP1, further facilitating apoptosis induction.
Loss of FAT1 sensitizes for death receptor-induced apoptosis
The best-characterized members of death ligands comprise TRAIL, CD95L/FASL and TNFα. Each of them bind to their respective receptors, but all result in the formation of DISC (Gonzalvez & Ashkenazi, 2010). Therefore, we investigated whether FAT1 knockdown affects sensitivity towards other death-inducing ligands. In line with our previous data, we observed reduced viability upon stimulation with death-inducing ligands TRAIL and CD95L in FAT1 knockdown cells (Fig 3A). Cells depleted of FAT1 were also more susceptible for TNFα treatment (Fig 3B).
Figure 3.
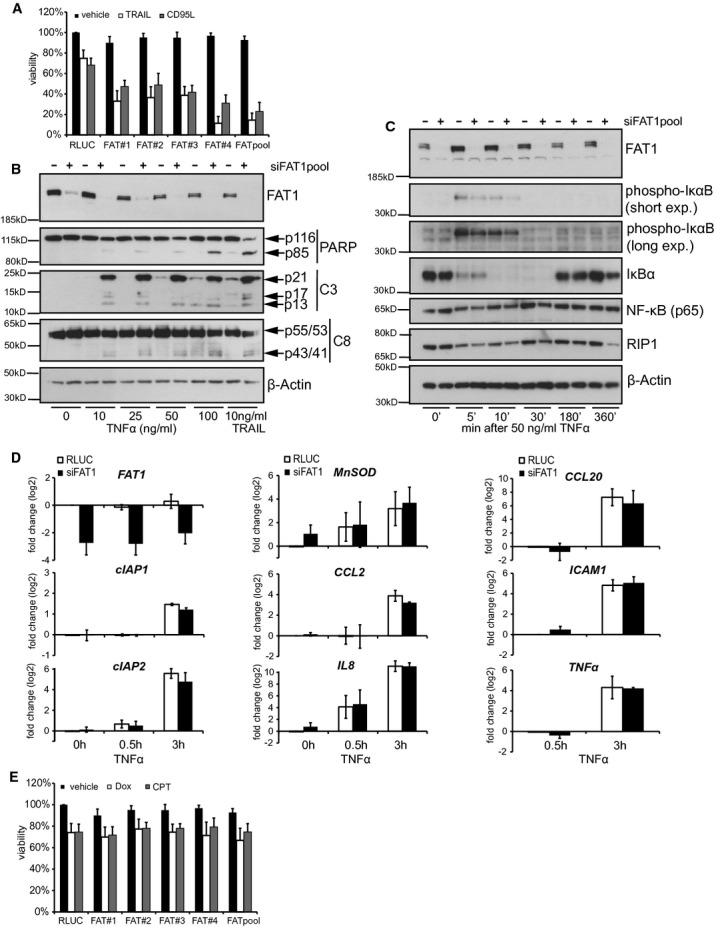
- Cells were transfected with indicated siRNAs, stimulated with 10 ng/ml TRAIL and 50 ng/ml CD95L or left untreated (vehicle). Viability was determined by CellTiterGlo-Assay 48 h after treatment. Results are shown as percentage of RLUC-untreated cells (mean ± s.d., n = 4).
- U251MG cells were transfected with control siRNA (RLUC) or FAT1pool-siRNA. Cells were treated with 10 ng/ml TRAIL or different concentrations of TNFα. 24 h later, cell lysates were analyzed by immunoblot. β-actin serves as a loading control.
- U251MG cells were transfected with control siRNA (RLUC) or FAT1pool-siRNA. Seventy-two hours later, cells were stimulated with 50 ng/ml TNFα. At the indicated time points, cells were lysed for western blot analysis. β-actin serves as a loading control.
- U251MG cells were transfected with control siRNA (RLUC) or FAT1pool-siRNA. Seventy-two hours later, cells were stimulated with 50 ng/ml TNFα. After 0.5 and 3 h, cells were lysed and the expression of the indicated mRNAs was analysed by qPCR. Data is presented as log2-fold change after calibration to the house keeping gene GAPDH and normalization to RLUC-transfected cells. The mean ± s.d. of three independent experiments is shown.
- Cells were transfected with indicated siRNAs and treated with 10 μM doxorubicin (Dox), 100 μM camptothecin (CPT) or DMSO as a control. Forty-eight hours later, viability was determined by CellTiterGlo-Assay. Results are shown as percentage of RLUC-untreated cells (mean ± s.d., n = 4).
Source data are available online for this figure.
Besides mediating apoptosis, TNFα induces the canonical NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) pathway triggering the expression of genes involved in inflammation, immunity, cell migration and cell survival (Gyrd-Hansen & Meier, 2010). TNFα stimulation results in the phosphorylation of IκB (inhibitor of NF-κB) and its subsequent proteasomal degration, releasing the transcription factor NF-κB to enter the nucleus. We observed phosphorylation of IκBα 5 and 10 min after TNFα stimulation and the levels were comparable between control and FAT1 knockdown (Fig 3C). Degradation of IκBα was already detected after 5 min of TNFα stimulation, but the levels were reestablished 180 min after treatment. Again, FAT1 knockdown cells did not differ from the control knockdown cells. Next, we investigated the induction of NF-κB target gene expression by quantitative real-time PCR. We observed that TNFα stimulation resulted in increased expression of genes encoding for anti-apoptotic proteins (cIAP1, cIAP2), chemokines (CCL2, CCL20, IL8, TNFα), the Intercellular adhesion molecule 1 (ICAM1) and Manganese Superoxide Dismutase (MnSOD) but without any difference between control and siRNA-FAT1-transfected cells (Fig 3D).
In order to determine whether FAT1 depletion affects apoptosis induction in general, we measured cell viability after treatment with chemotherapeutic drugs doxorubicin (Dox) and camptothecin (CPT). Our results showed that the reduction in cell viability was indistinguishable, comparing FAT1-depleted and control cells (Fig 3E).
Thus, knockdown of FAT1 sensitizes for death receptor-mediated apoptosis but it does not affect general apoptosis induction. FAT1 depletion did not interfere with activation of the classical NF-κB pathway despite sensitizing for TNFα-mediated apoptosis suggesting an apoptosis-specific role of FAT1.
Depletion of FAT1 enhances caspase-8 recruitment to the DISC
So far our data suggest an essential role of caspase-8 in the sensitivity towards death ligands upon loss of FAT1. Thus, we combined knockdown of FAT1 with depletion of caspase-8. Knockdown of caspase-8 completely rescued the siRNA-FAT1 mediated phenotype as the combinatorial knockdown of both genes resulted in loss of caspase cleavage (Fig 4A).
Figure 4.
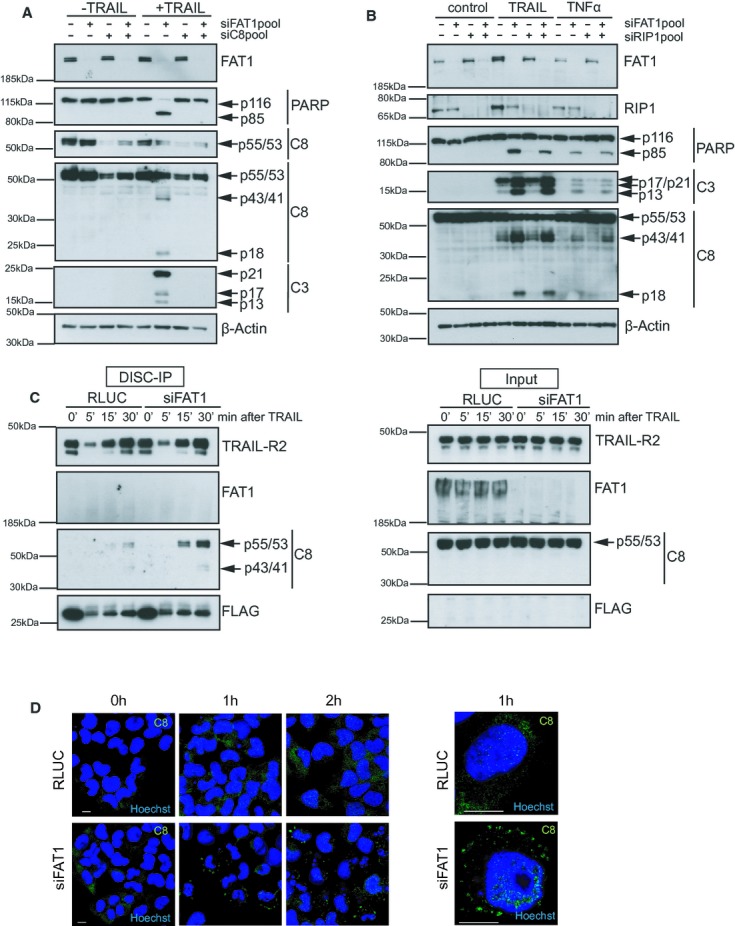
- U251MG cells were transfected with siRNAs targeting FAT1 (siFAT1) or caspase-8 (siC8). Cells were treated with 10 ng/ml TRAIL for 6 h. Cell lysates were analyzed by western blot. β-actin serves as a loading control.
- U251MG cells were transfected with indicated siRNAs. Cells were treated with 10 ng/ml TRAIL or 50 ng/ml TNFα for 6 h. Cell lysates were analyzed by western blot. β-actin serves as a loading control.
- U251MG cells were transfected with control siRNA (RLUC) or FAT1pool-siRNA. Seventy-two hours later, cells were stimulated with precomplexed FLAG-tagged TRAIL (500 ng/ml) and anti-FLAG-antibody (3μg/ml). After 5, 15 and 30 min, cells were lysed and TRAIL DISC was immunopurified with protein G beads. Total cell lysates (Input) and immunoprecipitated DISC (DISC-IP) were analysed by immunoblot.
- U251MG cells were transfected with control siRNA (RLUC) or FAT1pool-siRNA. 72 h later, cells were stimulated with 100 ng/ml TRAIL for 0, 1 and 2 h. Then cells were fixed, stained with C8-antibody (green) and Hoechst (blue), and analyzed by confocal microscopy. Panel on the right represents a higher magnification of cells after 1 h TRAIL treatment. Scale bar, 10 μm.
Source data are available online for this figure.
Previous studies showed that caspase-8 participates in other secondary signaling complexes (Micheau & Tschopp, 2003; Varfolomeev et al, 2005). Recently, the ‘ripoptosome’ complex consisting of protein kinase RIP1, FADD and procaspase-8 was described to assemble upon loss of anti-apoptotic proteins and genotoxic stress (Geserick et al, 2009; Feoktistova et al, 2011; Tenev et al, 2011). To test whether RIP1 affects the observed sensitivity phenotype, we performed combinatorial knockdowns depleting FAT1 and RIP1 either alone or together (Fig 4B). Our experiments revealed that RIP1 depletion alone had no effect on caspase cleavage neither after TRAIL nor after TNFα stimulation. Cells depleted of both RIP1 and FAT1 showed similar caspase cleavage as FAT1-only knockdown cells, indicating that the increased sensitivity mediated by siRNA FAT1 is independent of RIP1.
Based on our results that the observed sensitivity phenotype completely depended on caspase-8, we reasoned that FAT1 works upstream or at the level of procaspase-8 activation. Formation of the death-inducing signaling complex (DISC) is the key event in the extrinsic pathway to activate procaspase-8 and subsequently to induce apoptosis.
Therefore, we immunopurified the DISC and analysed DISC-bound proteins by western blotting as described previously (Walczak & Haas, 2008). As expected, TRAIL-R2 but not procaspase-8 was detected in RLUC-transfected and siRNA-FAT1-transfected cells under unstimulated conditions (Fig 4C). But in TRAIL-stimulated cells knockdown of FAT1 increased binding of caspase-8 to the DISC and also its processing leading to more caspase-8 cleavage products p43/p41. However, FAT1 itself was not associated with the DISC.
Recent studies indicate that caspase-8 chains are formed at the DISC as a prerequisite for caspase activation (Dickens et al, 2012; Schleich et al, 2012). Immunofluorescence microscopy showed that after ligand addition control RLUC cells showed similar cytoplasmic staining of caspase-8 as unstimulated cells (Fig 4D). Depletion of FAT1 resulted in a ligand-dependent increase of caspase-8 intensity and the formation of spot-like structures at the membrane presumably representing caspase-8 activating chains. However, it should be noted that these structures indicated accumulation of caspase-8, but the images did not allow a clear distinction between filaments.
Thus, we concluded that the increased sensitivity upon loss of FAT1 is mediated by enhanced caspase-8 recruitment to the DISC and the formation of caspase-8 activating chains.
FAT1 associates with caspase-8 and its loss promotes the formation of caspase-8-containing HMW complexes
Given that FAT1 depletion enhanced caspase-8 recruitment to the DISC in the membrane, we intended to investigate the subcellular localization of TRAIL pathway components in more detail. We separated cell lysates into different subcellular fractions and confirmed the purity of the subcellular fractions by different cellular markers: N-cadherin for the membrane fraction (F2), histone H3 for the nuclear fraction (F3) and intermediate filament glial fibrillary acidic protein (GFAP) for the cytokeleton fraction (F4) (Fig 5A). FAT1 as an atypical cadherin and class I-transmembrane protein was detected in the membrane and cytoskeleton fraction, similar to N-cadherin. Knockdown of FAT1 neither affected localization of the TRAIL-receptor (TRAIL-R2) nor the adaptor protein FADD (Fig 5A). FADD was only found in the membrane fraction upon TRAIL addition, since it is recruited to the DISC upon ligand stimulation (Kischkel et al, 1995; Sprick et al, 2000). In line with the increased recruitment of caspase-8 to the DISC (Fig 4C), more caspase-8 cleavage fragments (p43/41) were detected in the absence of FAT1 and upon TRAIL stimulation in the membrane fraction (Fig 5A). We also observed increased amount of p18 fragments of caspase-8 in the cytoplasmic fraction (F1) of FAT1-depleted cells, according to previous results describing the rapid release of the subunit p18 from the DISC after CD95L stimulation (Hughes et al, 2009).
Figure 5.
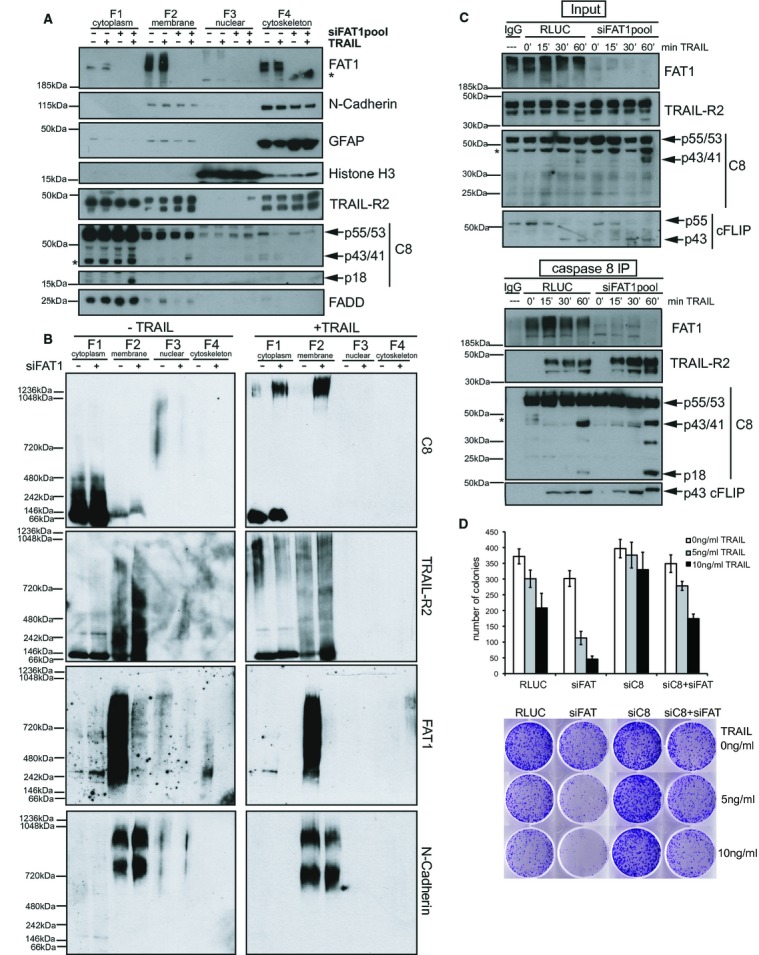
- U251MG cells were transfected with control siRNA (RLUC) or FAT1pool-siRNA. Cells were treated with 100 ng/ml TRAIL for 1 h and subjected to subcellular fractionation protocol. Fractions were analysed by immunoblots. Asterisk indicates non-specific band.
- Subcellular fractions from (A) were separated by blue native gelelectrophoresis and analysed by immunoblots.
- U251MG cells were transfected with indicated siRNAs and treated with 500 ng/ml TRAIL. After 15, 30 and 60 min, cells were lysed and subjected to immunoprecipitations with caspase-8 antibody (1 μg/ml) and protein G beads. Total cell lysates (Input) and immunoprecipitations (C8-IP) were analysed by immunoblot. Asterisk indicates non-specific band.
- U251MG cells were transfected with indicated siRNA, then re-plated into six-well dishes with 1000 cells/well and treated for 24 h with TRAIL. After 7–9 days colonies were stained with crystal violet. Graph depicts number of colonies of five biological replicates (mean ± s.e.m., n = 5). Below one representative picture per well for each condition is shown.
Source data are available online for this figure.
Stimulation with death ligands results in the formation of high molecular weight (HMW) DISC, composed of ligand, the TRAIL-receptor, FADD, procaspase-8/10 and c-FLIP, that can be detected by size-exclusion chromatography or sucrose gradient fractionation (Wagner et al, 2007; Dickens et al, 2012). Recent quantitative mass spectrometry analysis of HMW DISC suggests the formation of procaspase-8 activating chains within the DISC (Dickens et al, 2012). Furthermore, TRAIL stimulation can induce the assembly of other secondary signaling complexes (Varfolomeev et al, 2005). Extending these previous approaches, we wanted to investigate whether FAT1 affects formation of other HMW complexes than DISC by blue native polyacrylamide gel electrophoresis (BN-PAGE). BN-PAGE has been previously used to identify and analyze multiprotein complexes (Wittig & Schagger, 2009). We observed FAT1-containing HMW complexes in the membrane-and cytoskeleton-enriched fraction indicating that FAT1 is part of an even larger complex (Fig 5B). Upon depletion of FAT1, caspase-8 containing HMW complexes (>1236 kDa) were formed in the membrane-and the cytoplasmic-enriched fraction after TRAIL stimulation. However, TRAIL-R2-containing HMW complexes were absent in the cytoplasmic fraction and showed a distinct pattern differing from the caspase-8 native complexes in the membrane fraction.
Since our data indicated that the sensitivity phenotype completely depended on caspase-8, we hypothesized that FAT1 interacts with caspase-8. Immunoprecipitation of caspase-8 after TRAIL stimulation revealed that FAT1 associated with procaspase-8 (Fig 5C). Supporting our results from DISC immunoprecipitations, we observed increased association between procaspase-8 and the TRAIL-R2 in the immunopurified fraction of FAT1 knockdown cells. FAT1 depletion did not affect the binding of cFLIP to caspase-8 or its processing.
In order to investigate the long-term survival effect of FAT1 and caspase-8 depletion, we performed colony forming assays after TRAIL stimulation (Fig 5D). As expected, knockdown of caspase-8 alone protected cells from death ligand-induced apoptosis. In support of the previously observed decreased cell viability, knockdown of FAT1 drastically reduced the number of surviving colonies after TRAIL treatment. Importantly, the simultaneous depletion of caspase-8 and FAT1 rescued the effect, resulting in equal colony numbers as the control knockdown.
Taken together, these findings indicate that FAT1 interacts with caspase-8 independently of the DISC preventing induction of extrinsic apoptosis. Depletion of FAT1 facilitates caspase-8 recruitment to the DISC and the formation of caspase-8-containing secondary HMW complexes (>1236 kDa). Such secondary complexes might facilitate activation of effector caspases and subsequent apoptosis induction resulting in decreased clonogenic survival.
FAT1 depletion sensitizes non-adherent and primary glioblastoma cells for extrinsic apoptosis
Similar to its family members, FAT1 has a huge extracellular region comprising 34 cadherin-like repeats, one laminin A-G domain and five EGF-like domains, but the cytoplasmic domain of each FAT family member seems to be unique. FAT1 was previously linked to cell adhesion and migration (Moeller et al, 2004; Tanoue & Takeichi, 2004; Hou & Sibinga, 2009). To address the question whether adherence status contributes to the increased TRAIL sensitivity, we compared adherent cells with cells growing in suspension. After TRAIL stimulation, we observed that low-adherent cells were less sensitive to TRAIL despite declining FAT1 protein levels (Fig 6A). Accordingly, it has been demonstrated that glioblastoma cells are capable of switching from cell-substrate tethering to cell-cell adhesion and both forms are required for apoptosis resistance (Westhoff et al, 2008). However, FAT1-depleted cells were still more sensitive to TRAIL regardless of their adherence status (Fig 6A). This result suggests that the increased sensitivity is presumably not attributed to the adherent functions of FAT1.
Figure 6.
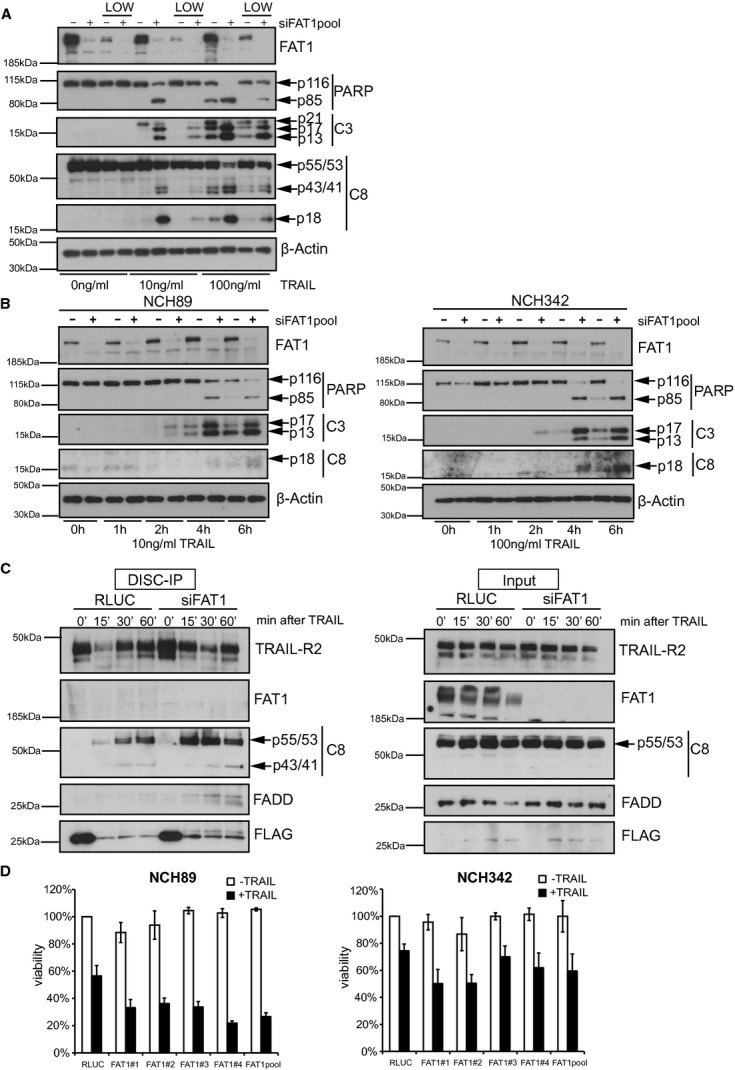
- U251MG cells were transfected with indicated siRNAs and seeded on adherent plates and ultra-low attachment plates (LOW). Cells were treated with 10 ng/ml TRAIL for 6 h. Cell lysates were analyzed by western blot. β-actin serves as a loading control.
- NCH89 (left) and NCH342 (right) were transfected with control siRNA (RLUC) or FAT1pool-siRNA. Cells were treated with 10 ng/ml TRAIL (NCH89) or 100 ng/ml TRAIL (NCH342) and harvested at the indicated time points for western blot analysis. β-actin serves as a loading control.
- NCH89 cells were transfected with control siRNA (RLUC) or FAT1pool-siRNA. Seventy-two hours later, cells were stimulated with precomplexed FLAG-tagged TRAIL (500 ng/ml) and anti-FLAG-M2-antibody (3 μg/ml). After 15, 30 and 60 min, cells were lysed and TRAIL DISC was immunopurified with protein G beads. Total cell lysates (Input) and immunprecipitated DISC (DISC-IP) were analysed by immunoblot.
- NCH89 and NCH342 were transfected with indicated siRNAs, stimulated with 2.5 ng/ml TRAIL or 25 ng/ml TRAIL, respectively. Viability was determined by CellTiterGlo-Assay 24 h after treatment. Results are shown as percentage of RLUC-untreated cells (mean ± s.d., n = 3).
Source data are available online for this figure.
Glioblastoma multiforme is the most common and most malignant brain tumor with poor prognosis regardless of treatment. Glioma cell lines vary in their sensitivity towards TRAIL, presumably as a consequence of transcriptional and epigenetic regulation of TRAIL receptors and anti-apoptotic proteins (Hawkins, 2004). To assess the impact of FAT1 depletion on death receptor-mediated apoptosis in primary cell lines, we obtained previously characterized cell lines from patients with glioblastoma multiforme, namely NCH89 and NCH342 (Karcher et al, 2006). Knockdown of FAT1 increased cleavage of procaspase-8, caspase-3 and PARP in both cell lines after TRAIL stimulation (Fig 6B). In agreement with our previous data, we observed increased binding of procaspase-8 to the DISC in FAT1-depleted cells that led to more processed cleavage products (Fig 6C). Furthermore, we found some FADD bound to the DISC in these cells indicating increased recruitment of adaptor protein FADD to the DISC.
Accordingly, stimulation of TRAIL during FAT1 depletion resulted in decreased cell viability in both cell lines, NCH89 and NCH342 (Fig 6D).
These results demonstrate that patient-derived glioma cell lines are sensitized towards TRAIL-induced apoptosis by releasing caspase-8 from its association with FAT1.
Deficiency of FAT1 causes higher susceptibility towards TRAIL-induced apoptosis
Recently, new genome engineering technologies such as zinc finger nucleases, transcription activator-like effector nucleases (TALEN) and CRISPR (clustered regulatory interspaced short palindromic repeat)/Cas9 evolved as powerful tools for precise genome editing (Gaj et al, 2013). Previously, it has been demonstrated that human codon-optimized Cas9 nucleases can be directed to target sites in the human genome to induce precise cleavage at genomic loci in human cells (Cong et al, 2013; Mali et al, 2013). To induce targeted cleavage in U251MG cells, we transfected cells with the bicistronic expression vector encoding for human codon-optimized Cas9 and a single-guide RNA (sgRNA) targeting endogenous sites within the FAT1 locus and the caspase-8 locus (Cong et al, 2013; Mali et al, 2013). Since caspase-8 knockout cells are resistant to death receptor-mediated apoptosis in vitro and in vivo, we used caspase-8 as a control to show the feasibility of this approach (Juo et al, 1998; Varfolomeev et al, 1998; Kang et al, 2004).
Sequence alignment of two FAT1 clones and two caspase-8 clones indicated indels of up to 14 bp and insertions of up to 82 bp (Fig 7A), events described previously for genome editing technologies (Reyon et al, 2012; Cong et al, 2013; Mali et al, 2013). To verify the knockout on the protein level and to investigate the phenotype of the different clones after death ligand addition, we performed immunoblot analysis with increasing concentrations of TRAIL (Fig 7B). Both caspase-8 mutant clones neither showed any detectable caspase-8 nor any induction of caspase cleavage after TRAIL stimulation (Fig 7B). According to the sequence analysis, both FAT1 mutant clones lacked FAT1 protein. Furthermore, mutant clone F1 showed strong enhancement of caspase cleavage, whereas mutant clone H3 indicated weaker induction of caspase cleavage after TRAIL stimulation (Fig 7B).
Figure 7.
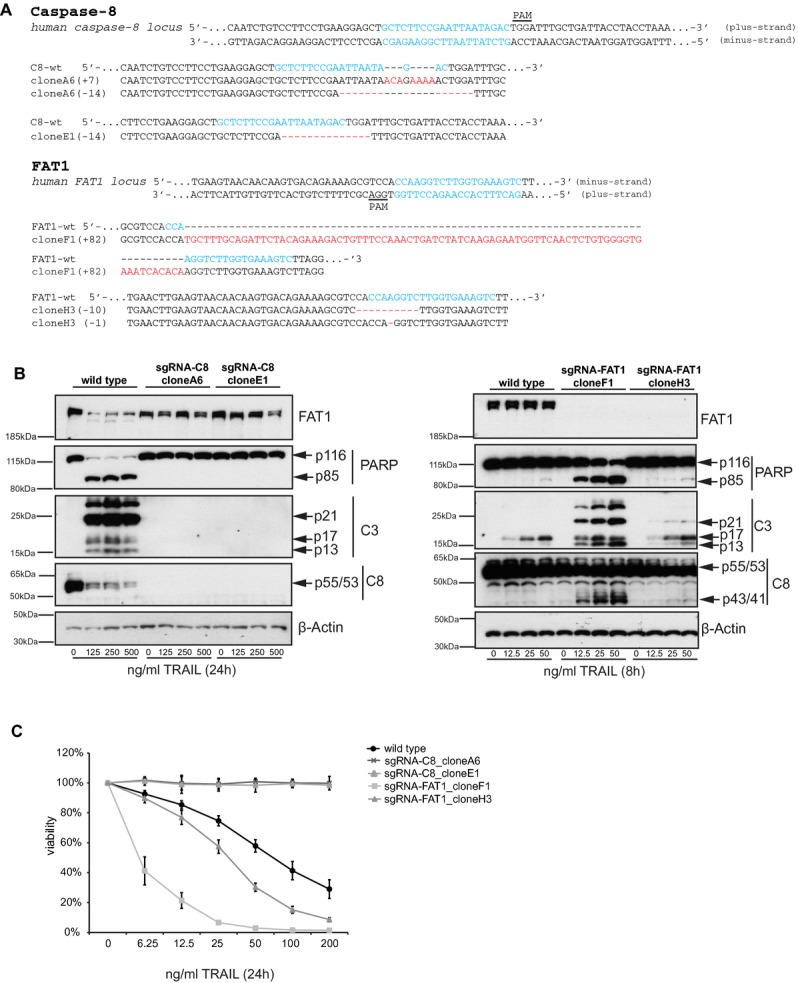
- Schematic of the human caspase-8 and FAT1 loci. 20nt-targeting sequence of the sgRNA is labeled in blue and protospacer-adjacent motif (PAM) is underlined. Cas9/sgRNA target sites for the minus strand of human FAT1 locus and for the plus strand of human caspase-8 locus were chosen as described previously (Mali et al, 2013). Sequence alignment of wild type (wt) and depicted mutant clones surrounding the Cas9/sgRNA targeting sites (in blue) are shown. Red dashes indicates deleted bases and red bases represents insertions. Number of deleted/inserted base pairs is indicated on the left side.
- Wild type and mutant cells were stimulated with increasing concentrations of TRAIL and harvested 8 or 24 h after treatment, respectively. Cell lysates were analyzed by immunoblots. β-actin serves as a loading control.
- Wild type and mutant cells were treated with increasing concentrations of TRAIL. Viability was determined by CellTiterGlo-Assay 24 h after treatment. Values were normalized to untreated sample of each clone (mean ± s.d., n = 4).
Source data are available online for this figure.
Finally, we wanted to determine the effect of TRAIL on the survival of these mutant cell lines. In agreement with the immunoblot analysis, deficiency of caspase-8 rendered the cells completely insensitive towards death receptor-mediated apoptosis, whereas FAT1 mutant cells were more susceptible towards TRAIL-induced apoptosis (Fig 7C).
Together, these data support the observed RNAi-mediated phenotype that, indeed, cells lacking FAT1 are more susceptible for death receptor-mediated apoptosis.
Discussion
RNA interference screening is a valuable tool to identify novel components of signaling pathways (Boutros & Ahringer, 2008). In this study, we identified the atypical cadherin FAT1 in a synthetic lethal RNAi screen for death receptor-mediated apoptosis. For the first time, we provide evidence that the atypical cadherin FAT1 is linked to extrinsic apoptosis. Previous data indicated that activation of procaspase-8 is regulated by cullin-3-mediated polyubiquitination (Jin et al, 2009). Here, we found an ubiquitin-independent mechanism controlling caspase-8 activation, wherein FAT1 impedes caspase-8 recruitment to the DISC and/or its activation in non-apoptotic conditions (Fig 8). This could be mediated by an interaction between FAT1 and caspase-8 or via the adaptor protein FADD, whereas FAT1 might interfere with the incorporation of both proteins into the DISC.
Figure 8.
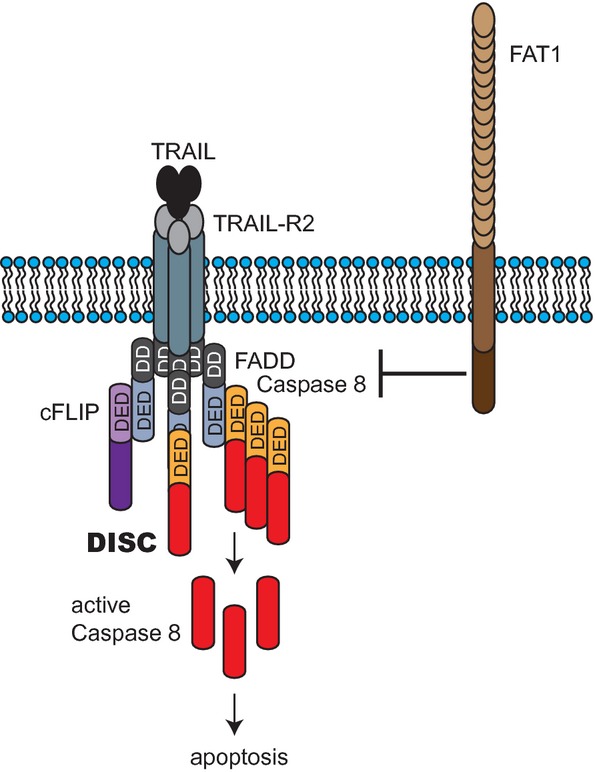
Model of death receptor signaling.
FAT1 inhibits the binding of caspase-8 either by interacting with caspase-8 or via FADD to the DISC upon TRAIL stimulation.
We confirmed the RNAi phenotype by generating FAT1 mutant cell lines by CRISPR/Cas9-mediated genome engineering. CRISPR-mediated mutagenesis might be an approach to independently confirm knockdown phenotypes and it could be used as a general strategy for candidate selection of RNAi screens in future. However, it should be noted that in our experiments different CRISPR-generated FAT1 mutant clones increased sensitivity towards TRAIL, but they differed in the magnitude of susceptibility. Thus, it seems equally important to test several clones for each targeting site or/and testing different target sites to exclude potential off-targets or adaptations of the system.
FAT1 fulfills tasks differing from the other FAT family members, supporting the idea of distinct functions for each member. We observed increased sensitivity towards TRAIL after knockdown of FAT1 but not of the other family members. Comparative sequence analysis shows the highest homology between FAT4 and the Drosophila fat, whereas FAT1-3 resemble the Drosophila fat-like (Castillejo-Lopez et al, 2004). Lack of Fat4 in mice resulted in severe planar polarity phenotypes including misorientation of hair cells (Saburi et al, 2012). Accordingly, Drosophila fat mutant flies showed hyperplastic, tumor-like overgrowth of larval imaginal discs, defects in differentiation and morphogenesis (Mahoney et al, 1991). In contrast to mice lacking Fat4,Fat1−/− mice have no planar polarity phenotype, but they are perinatal lethal with severe defects in brain, kidney and eye development (Ciani et al, 2003; Saburi et al, 2012).
Our data indicate that loss of FAT1 promoted the formation of HMW caspase-8-containing complexes (>1,200 kDa). Recent studies suggest that pre-formation of DED-chains of procaspase-8 are required for death receptor-induced apoptosis (Dickens et al, 2012; Schleich et al, 2012). Polyubiquitinated caspase-8 was found at HMW DISC in lipid raft/cytoskeleton fractions (Jin et al, 2009). However, we detected caspase-8 containing complexes in the cytoplasmic fraction that were distinct from TRAIL-R2 complexes, presumably not representing ligand-receptor subcomplexes. We surmise that our HMW complexes represent other previously described secondary cytoplasmic signaling complexes. TRAIL induces the formation of a secondary complex consisting of FADD, caspase-8, RIP1, TRAF2, and NEMO that leads to kinase pathway activation (Varfolomeev et al, 2005). It is conceivable that different secondary complexes are formed containing overlapping and/or distinct components. In such a scenario, FAT1 could impede the formation of such secondary signaling complexes.
Comparing cells growing in suspension with adherent cells, we found that non-adherent cells were less sensitive to TRAIL, but still capable of undergoing apoptosis. Cancer cells can acquire resistance to anoikis, enabling them to grow in non-adherent conditions (Westhoff & Fulda, 2009). Depletion of FAT1 could still sensitize non-adherent cells towards death receptor-mediated apoptosis suggesting the sensitivity effect is independent from cell-extracellular matrix interactions. However, we cannot exclude that FAT1 is still required for cell-cell interactions. Previous data indicate the alternation between cell-extracellular matrix and cell-cell adhesion in glioblastoma cells and only inhibiting both forms sensitized for apoptosis (Westhoff et al, 2008). Previously, it was shown that FAT1 interacts with Ena/Vasp to control actin dynamics modulating cell contacts and polarity (Moeller et al, 2004; Tanoue & Takeichi, 2004). FAT1 also regulates migration of vascular smooth muscle cells via binding to atrophin proteins (Hou & Sibinga, 2009). Accordingly, FAT1 is implicated in invasion and migration of gliomas and oral squamous cell carcinomas (Nishikawa et al, 2011; Dikshit et al, 2012). In support of these findings, FAT1 was overexpressed in leukemias and invasive breast cancer (Kwaepila et al, 2006; de Bock et al, 2012). Recently, FAT1 mutations were found across multiple cancer types including gliomas, colon carcinomas and head and neck squamous cell carcinomas implying that FAT1 regulates aberrant WNT pathway activation in these tumors (Morris et al, 2013). These studies suggest that FAT1 might contribute to epithelial-mesenchymal transition during tumorigenesis and the generation of invasive and metastatic cancer cells. The interaction between FAT1 and caspase-8 in these cells may represent a mechanism to evade apoptosis.
Taken together, our data suggest a mechanism to regulate death receptor-mediated apoptosis via an interaction between FAT1 and procaspase-8. Since caspase-8 has to be tightly regulated in physiological conditions, this might represent a previously unrecognized mechanism to control the induction of cell death.
Materials and Methods
Cell lines, transfection and treatment
All cell lines were cultured in DMEM (supplemented with 10% FBS, 1% penicillin/streptamycin) and maintained in 5% CO2 at 37°C. Primary derived human glioblastoma cell lines NCH89 and NCH342 were kindly provided by C. Herold-Mende and maintained as previously described (Karcher et al, 2006). Cells were reverse transfected with siGenome Upgrade siRNAs (Dharmacon, Thermo Scientific) and Dharmafect1 (Thermo Scientific) according to the manufacturer's instruction with a final concentration of 20 nM siRNAs. Treatments were performed 48 or 72 h after transfection according to the assay. siRNAs used in this study are listed in Supplementary Table S2.
Stable overexpression in U251MG cells was performed by lentiviral transduction of the inducible lentiviral vectors pF-cIAP1 vector or pF-control (kindly provided by J. Silke) and addition of 100 nM 4-hydroxytamoxifen (Sigma).
In all experiments (except the DISC-IPs) cells were stimulated with IZ-TRAIL. IZ-TRAIL and CD95L were kindly provided by H. Walzcak. Doxorubicin-hydrochloride was purchased from AppliChem, Irinotecan hydrochloride (Camptothecin) from Sigma, TNFα from Life Technologies, Z-VAD-FMK from Promega, Z-IETD-FMK from Enzo Life Sciences, MG132 from Merck Biosciences and bafilomycin A1 from LC Laboratories. Ultra-low attachment plates were purchased from Greiner.
Genome-wide siRNA screen
RNAi screen was performed in 384-well plates using a pooled siRNA library (Dharmacon SMART pool library; Thermo Scientific), where each gene was targeted by a pool of four single siRNAs. The transfection mix for a single well contained 5 μl siRNApool, 0.05 μl of Dharmafect1 and 14.95 μl RPMI. 30 μl of U251MG cells (1,500 cells) were seeded on top of this. The final siRNA concentration was 25 nM. Plates were incubated for 48 h. Then, plates were treated with 20 μl medium as a control or 20 μl medium containing IZ-TRAIL achieving a final concentration of 10 ng/ml TRAIL. After 48 h, CellTiterGlo assay (Promega) was performed to measure cell viability. Medium was removed with a 24 channel comb and 20 μl of 1:4 diluted CellTiterGlo reagent was added for 20 min. Luminescence was measured with Mithras LB940 plate reader (Berthold Technologies).
Data were analyzed with web-cellHTS2 (Pelz et al, 2010) to obtain plate-based median normalized z-scores for control and TRAIL-treated plates separately. The mean of the two biological replicates was determined. Strong viability hits were removed from the list (viability z-score > 4). After quantile normalization, the difference between the z-scores was calculated (Supplementary Table S1). The complete data set is available in the GenomeRNAi-database (http://www.genomernai.org) (Schmidt et al, 2013).
Viability and apoptosis assays
Cell viability was measured by CellTiterGlo assay (Promega). Caspase-3/7 activity was determined by fluorescence-based ApoOne kit (Promega). Both assays were performed in 384-well plates according to the manufacturer's instruction. Luminescence and fluorescence were measured with Mithras LB940 plate reader (Berthold Technologies). For measurement of AnnexinV-positive cells, cells were transfected with indicated siRNAs and treated with TRAIL for 16 h. Then, cells were stained with AnnexinV-FITC and propidium iodide (Annexin V FLUOS Staining kit from Roche) and analyzed by flow cytometry (FACSCantoII; BD Biosciences).
Long-term survival was determined by colony-forming assays. Cells were reverse transfected with indicated siRNAs. After 72 h, cells were counted and plated in 6-well plates in triplicates (1,000 cells per well). The next day, cells were treated with TRAIL for 24 h. Then, plates were washed with PBS and drug-free medium was added. Plates were maintained for 7–9 days to allow formation of colonies. Colonies were fixed and stained for 30 min at room temperature with crystal violet solution (0.1% crystal violet, 10% formaldehyde). Plates were washed with water, air-dried and colonies were scanned. Colonies were counted using ImageJ.
Protein assays
Cells were lysed in urea lysis buffer [8M Urea, PBS pH 7.5 and protease inhibitor tablets (Roche)]. Protein concentration was measured with BCA-protein assay kit (Pierce, Thermo Scientific). Ten to twenty microgram sample were supplemented with 5× Laemmli buffer and boiled for 5 min at 96°C. Cell lysates were separated on 4–12% NuPAGE Bis-TRIS gels (Life Technologies) and transfered to Immobilon PVDF membrane (Millipore, Merck Biosciences).
Subcellular fractions were obtained using the ProteoExtract Subcellular Proteome Extraction Kit according to the manufacturer's instruction (Merck Biosciences).
For blue native gel electrophoresis, subcellular fractions were separated on NativePAGE Novex 3–12% Bis–Tris gels (Invitrogen, Life Technologies) according to the manufacturer's instruction and transfered to Immobilon PVDF membrane (Millipore, Merck Biosciences).
Commercial antibodies used in this study are listed in Supplementary Table S5. Anti-cIAP1 antibody was kindly provided by J. Silke.
Immunoprecipitation
DISC immunoprecipitations were performed as described previously (Walczak & Haas, 2008). In brief, 1.5 × 106 cells were reverse transfected in 10-cm dishes with indicated siRNAs and Dharmafect1 according to the manufacturer's instruction for 72 h. Then, 500 ng/ml of FLAG-tagged TRAIL (Enzo Life Sciences) was incubated with 3 μg/ml FLAG-M2 antibody (Sigma) for 30 min. Cells were harvested by centrifugation and incubated with 1 ml precomplexed TRAIL-M2-antibody stimulation mix. After the indicated time points, cells were washed with ice-cold PBS and lysed in 1 ml DISC-IP buffer (30 mM TRIS-HCl pH 7.5, 150 mM NaCl, 10% glycerole, 1% Triton X-100 and protease inhibitor tablets). The resulting cleared lysate was subjected to immunopecipitations with protein G at 4°C overnight. As a control for unstimulated conditions, FLAG-TRAIL and FLAG-M2-antibody were added after cell lysis. After several washes with ice-cold DISC-buffer, TRAIL-DISC was eluted with 2× Laemmli buffer at 95°C.
Similarily, caspase-8 immunoprecipitations were performed overnight in DISC-IP buffer using 1 μg/ml caspase-8 IgG2b antibody and protein G beads (GE Healthcare).
Quantitative real-time PCR (qPCR)
Total RNA was isolated with the RNeasy Mini kit (Qiagen) and 1 μg RNA was used as a template for cDNA synthesis using oligo-dT primer and SuperscriptIII cDNA synthesis kit (Invitrogen, Life Technologies). qPCR was performed using the Universal Probe Library (Roche) on a Light Cycler 480 (Roche). Expression levels were quantified using ΔΔCt method using GAPDH as calibrator. Primers used in this study are listed in Supplementary Table S3A.
Immunofluorescence
U251MG cells were reverse transfected with siRNAs and seeded on coverslips. Seventy-two hours later, cells were treated with 100 ng/ml TRAIL for 0, 1 and 2 h. Then, cells were fixed for 20 min with 4% paraformaldehyde, permeabilized for 25 min with 0.2% Triton X-100 and blocked for 30 min with 5% BSA. Primary caspase-8 antibody was incubated for 2 h at room temperature, followed by 1 h incubation with secondary anti-mouse-Alexa 488 antibody (Molecular Probes, Invitrogen). Nuclei were stained with Hoechst 33342 (Molecular Probes, Invitrogen). Coverslips were mounted onto slides. Multiple fields were imaged at room temperature using a confocal laser scanning microscope (model TCS SP5, Leica; operated with the LAS AF acquisition software) equipped with a Plan Apochromat 40×/1.25 oil immersion objective lens for imaging. An Argon laser was used for the excitation of the Alexa 488 nm and a 405-nm diode laser for Hoechst. Pictures were processed using Adobe Photoshop or ImageJ.
Procedures for generating sgRNA vectors and CRISPR/Cas9-mediated genome engineering in human cells
Bi-cistronic expression vector pX330 [Addgene no. 42230, (Cong et al, 2013] containing hCas9 and single-guide RNA (sgRNA) was digested with BbsI, and a pair of annealed oligonucleotides [Supplementary Table S4, (Mali et al, 2013)] for each targeting site was cloned into the guide RNA according to the protocol from Cong et al (Cong et al, 2013). Designs were chosen to target the gene in one of the first exons and were tested for obvious potential off-targets by bioinformatics analysis (http://www.e-crisp.org).
U251MG cells were transfected with TransIT (Mirus Bio LLC) and pX330-Cas9-sgRNA along with pEGFP-puro [Addgene no. 45561, (Abbate et al, 2001)] (ratio of 10:1) according to the manufacturer's instruction. Cells were selected for 7 days with 1 μg/ml puromycin. Then, cells were seeded as single colonies (10 cells/well) in 96-well plates. After 2–3 weeks, clones were selected based on the TRAIL sensitivity phenotype in the previously described CellTiterGlo assay.
Sequence verification of endogenous human genes
Genomic DNA was extracted from cell lines arising from single clones with the DNeasy Blood & Tissue kit (Qiagen). PCR reactions to amplify targeted loci were performed using the primers presented in Supplementary Table S4. Standard PCR conditions were used with Phusion High-Fidelity DNA Polymerase (Fermentas) and 250 ng of genomic DNA according to the manufacturer's instruction for 35 cycles (98°C, 10 s denaturation; 60°C, 30 s annealing; 72°C, 30 s elongation). Correct size of PCR products was checked by agarose gel electrophoresis, which were then excised from the gel, purified using the QIAquick Gel Extraction kit (Qiagen) according to the manufacturer's instruction and sent for DNA sequencing. Additionally, PCR products were cloned into the pCR2.1-TOPO vector using the TOPO-TA subcloning kit (Life Technologies) according to the manufacturer's instruction and transformed into One Shot TOP10 chemically competent cells. Plasmid DNA was isolated from multiple colonies of each transformation and sequenced.
Acknowledgments
We thank Thomas Sandmann for help with data analysis, Marco Breinig and Julia Christina Gross for helpful discussions and Matthias Dobbelstein for comments on the manuscript. We are grateful to H. Walzcak, J. Silke and C. Herold-Mende for reagents. This work was in part supported by a Marie Curie Excellence Grant (‘Cellular Signaling’) from the European Commission and the ApoNet project.
Author contributions
DK designed and performed the experiments. DK and MB wrote the manuscript. MB supervised the study.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Abbate J, Lacayo JC, Prichard M, Pari G, McVoy MA. Bifunctional protein conferring enhanced green fluorescence and puromycin resistance. Biotechniques. 2001;31:336–340. doi: 10.2144/01312st05. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Holland P, Eckhardt SG. Ligand-based targeting of apoptosis in cancer: the potential of recombinant human apoptosis ligand 2/Tumor necrosis factor-related apoptosis-inducing ligand (rhApo2L/TRAIL) J Clin Oncol. 2008;26:3621–3630. doi: 10.1200/JCO.2007.15.7198. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- de Bock CE, Ardjmand A, Molloy TJ, Bone SM, Johnstone D, Campbell DM, Shipman KL, Yeadon TM, Holst J, Spanevello MD, Nelmes G, Catchpoole DR, Lincz LF, Boyd AW, Burns GF, Thorne RF. The Fat1 cadherin is overexpressed and an independent prognostic factor for survival in paired diagnosis-relapse samples of precursor B-cell acute lymphoblastic leukemia. Leukemia. 2012;26:918–926. doi: 10.1038/leu.2011.319. [DOI] [PubMed] [Google Scholar]
- Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- Castillejo-Lopez C, Arias WM, Baumgartner S. The fat-like gene of Drosophila is the true orthologue of vertebrate fat cadherins and is involved in the formation of tubular organs. J Biol Chem. 2004;279:24034–24043. doi: 10.1074/jbc.M313878200. [DOI] [PubMed] [Google Scholar]
- Chang DW, Xing Z, Capacio VL, Peter ME, Yang X. Interdimer processing mechanism of procaspase-8 activation. EMBO J. 2003;22:4132–4142. doi: 10.1093/emboj/cdg414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng EH, Sheiko TV, Fisher JK, Craigen WJ, Korsmeyer SJ. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science. 2003;301:513–517. doi: 10.1126/science.1083995. [DOI] [PubMed] [Google Scholar]
- Ciani L, Patel A, Allen ND, ffrench-Constant C. Mice lacking the giant protocadherin mFAT1 exhibit renal slit junction abnormalities and a partially penetrant cyclopia and anophthalmia phenotype. Mol Cell Biol. 2003;23:3575–3582. doi: 10.1128/MCB.23.10.3575-3582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens LS, Boyd RS, Jukes-Jones R, Hughes MA, Robinson GL, Fairall L, Schwabe JW, Cain K, Macfarlane M. A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Mol Cell. 2012;47:291–305. doi: 10.1016/j.molcel.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikshit B, Irshad K, Madan E, Aggarwal N, Sarkar C, Chandra PS, Gupta DK, Chattopadhyay P, Sinha S, Chosdol K. FAT1 acts as an upstream regulator of oncogenic and inflammatory pathways, via PDCD4, in glioma cells. Oncogene. 2012;32:3798–3808. doi: 10.1038/onc.2012.393. [DOI] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Cain K, MacFarlane M, Hacker G, Leverkus M. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell. 2011;43:449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganten TM, Koschny R, Sykora J, Schulze-Bergkamen H, Buchler P, Haas TL, Schader MB, Untergasser A, Stremmel W, Walczak H. Preclinical differentiation between apparently safe and potentially hepatotoxic applications of TRAIL either alone or in combination with chemotherapeutic drugs. Clin Cancer Res. 2006;12:2640–2646. doi: 10.1158/1078-0432.CCR-05-2635. [DOI] [PubMed] [Google Scholar]
- Geserick P, Hupe M, Moulin M, Wong WW, Feoktistova M, Kellert B, Gollnick H, Silke J, Leverkus M. Cellular IAPs inhibit a cryptic CD95-induced cell death by limiting RIP1 kinase recruitment. J Cell Biol. 2009;187:1037–1054. doi: 10.1083/jcb.200904158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalvez F, Ashkenazi A. New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene. 2010;29:4752–4765. doi: 10.1038/onc.2010.221. [DOI] [PubMed] [Google Scholar]
- Gonzalvez F, Lawrence D, Yang B, Yee S, Pitti R, Marsters S, Pham VC, Stephan JP, Lill J, Ashkenazi A. TRAF2 sets a threshold for extrinsic apoptosis by tagging caspase-8 with a ubiquitin shutoff timer. Mol Cell. 2012;48:888–899. doi: 10.1016/j.molcel.2012.09.031. [DOI] [PubMed] [Google Scholar]
- Guicciardi ME, Mott JL, Bronk SF, Kurita S, Fingas CD, Gores GJ. Cellular inhibitor of apoptosis 1 (cIAP-1) degradation by caspase 8 during TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. Exp Cell Res. 2010;317:107–116. doi: 10.1016/j.yexcr.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer. 2010;10:561–574. doi: 10.1038/nrc2889. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hawkins CJ. TRAIL and malignant glioma. Vitam Horm. 2004;67:427–452. doi: 10.1016/S0083-6729(04)67022-1. [DOI] [PubMed] [Google Scholar]
- Hou R, Sibinga NE. Atrophin proteins interact with the Fat1 cadherin and regulate migration and orientation in vascular smooth muscle cells. J Biol Chem. 2009;284:6955–6965. doi: 10.1074/jbc.M809333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA, Harper N, Butterworth M, Cain K, Cohen GM, MacFarlane M. Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Mol Cell. 2009;35:265–279. doi: 10.1016/j.molcel.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Juo P, Kuo CJ, Yuan J, Blenis J. Essential requirement for caspase-8/FLICE in the initiation of the Fas-induced apoptotic cascade. Curr Biol. 1998;8:1001–1008. doi: 10.1016/s0960-9822(07)00420-4. [DOI] [PubMed] [Google Scholar]
- Kang TB, Ben-Moshe T, Varfolomeev EE, Pewzner-Jung Y, Yogev N, Jurewicz A, Waisman A, Brenner O, Haffner R, Gustafsson E, Ramakrishnan P, Lapidot T, Wallach D. Caspase-8 serves both apoptotic and nonapoptotic roles. J Immunol. 2004;173:2976–2984. doi: 10.4049/jimmunol.173.5.2976. [DOI] [PubMed] [Google Scholar]
- Karcher S, Steiner HH, Ahmadi R, Zoubaa S, Vasvari G, Bauer H, Unterberg A, Herold-Mende C. Different angiogenic phenotypes in primary and secondary glioblastomas. Int J Cancer. 2006;118:2182–2189. doi: 10.1002/ijc.21648. [DOI] [PubMed] [Google Scholar]
- Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer PH, Arnold R, Lavrik IN. Life and death in peripheral T cells. Nat Rev Immunol. 2007;7:532–542. doi: 10.1038/nri2115. [DOI] [PubMed] [Google Scholar]
- Krueger A, Baumann S, Krammer PH, Kirchhoff S. FLICE-inhibitory proteins: regulators of death receptor-mediated apoptosis. Mol Cell Biol. 2001;21:8247–8254. doi: 10.1128/MCB.21.24.8247-8254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaepila N, Burns G, Leong AS. Immunohistological localisation of human FAT1 (hFAT) protein in 326 breast cancers. Does this adhesion molecule have a role in pathogenesis? Pathology. 2006;38:125–131. doi: 10.1080/00313020600559975. [DOI] [PubMed] [Google Scholar]
- Mahoney PA, Weber U, Onofrechuk P, Biessmann H, Bryant PJ, Goodman CS. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell. 1991;67:853–868. doi: 10.1016/0092-8674(91)90359-7. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DA, Siegel RM, Zheng L, Lenardo MJ. Membrane oligomerization and cleavage activates the caspase-8 (FLICE/MACHalpha1) death signal. J Biol Chem. 1998;273:4345–4349. doi: 10.1074/jbc.273.8.4345. [DOI] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Moeller MJ, Soofi A, Braun GS, Li X, Watzl C, Kriz W, Holzman LB. Protocadherin FAT1 binds Ena/VASP proteins and is necessary for actin dynamics and cell polarization. EMBO J. 2004;23:3769–3779. doi: 10.1038/sj.emboj.7600380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan S, Eng S, Kannan K, Zou Y, Peng L, Banuchi VE, Paty P, Zeng Z, Vakiani E, Solit D, Singh B, Ganly I, Liau L, Cloughesy TC, Mischel PS, et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45:253–261. doi: 10.1038/ng.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M, Chinnaiyan AM, Kischkel FC, O'Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME, Dixit VM. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death–inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. An induced proximity model for caspase-8 activation. J Biol Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- Nishikawa Y, Miyazaki T, Nakashiro K, Yamagata H, Isokane M, Goda H, Tanaka H, Oka R, Hamakawa H. Human FAT1 cadherin controls cell migration and invasion of oral squamous cell carcinoma through the localization of beta-catenin. Oncol Rep. 2011;26:587–592. doi: 10.3892/or.2011.1324. [DOI] [PubMed] [Google Scholar]
- Pelz O, Gilsdorf M, Boutros M. web cellHTS2: a web-application for the analysis of high-throughput screening data. BMC Bioinformatics. 2010;11:185. doi: 10.1186/1471-2105-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth W, Isenmann S, Naumann U, Kugler S, Bahr M, Dichgans J, Ashkenazi A, Weller M. Locoregional Apo2L/TRAIL eradicates intracranial human malignant glioma xenografts in athymic mice in the absence of neurotoxicity. Biochem Biophys Res Commun. 1999;265:479–483. doi: 10.1006/bbrc.1999.1693. [DOI] [PubMed] [Google Scholar]
- Saburi S, Hester I, Goodrich L, McNeill H. Functional interactions between Fat family cadherins in tissue morphogenesis and planar polarity. Development. 2012;139:1806–1820. doi: 10.1242/dev.077461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ, Debatin KM, Krammer PH, Peter ME. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich K, Warnken U, Fricker N, Ozturk S, Richter P, Kammerer K, Schnolzer M, Krammer PH, Lavrik IN. Stoichiometry of the CD95 death-inducing signaling complex: experimental and modeling evidence for a death effector domain chain model. Mol Cell. 2012;47:306–319. doi: 10.1016/j.molcel.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Pelz O, Buhlmann S, Kerr G, Horn T, Boutros M. GenomeRNAi: a database for cell-based and in vivo RNAi phenotypes, 2013 update. Nucleic Acids Res. 2013;41:D1021–D1026. doi: 10.1093/nar/gks1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprick MR, Weigand MA, Rieser E, Rauch CT, Juo P, Blenis J, Krammer PH, Walczak H. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity. 2000;12:599–609. doi: 10.1016/s1074-7613(00)80211-3. [DOI] [PubMed] [Google Scholar]
- Tanoue T, Takeichi M. Mammalian Fat1 cadherin regulates actin dynamics and cell-cell contact. J Cell Biol. 2004;165:517–528. doi: 10.1083/jcb.200403006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T, Takeichi M. New insights into Fat cadherins. J Cell Sci. 2005;118:2347–2353. doi: 10.1242/jcs.02398. [DOI] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K, Meier P. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell. 2011;43:432–448. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ, Flygare JA, Fairbrother WJ, Deshayes K, Dixit VM, Vucic D. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Maecker H, Sharp D, Lawrence D, Renz M, Vucic D, Ashkenazi A. Molecular determinants of kinase pathway activation by Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand. J Biol Chem. 2005;280:40599–40608. doi: 10.1074/jbc.M509560200. [DOI] [PubMed] [Google Scholar]
- Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL, Rebrikov D, Brodianski VM, Kemper OC, Kollet O, Lapidot T, Soffer D, Sobe T, Avraham KB, Goncharov T, Holtmann H, Lonai P, Wallach D. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity. 1998;9:267–276. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- Vince JE, Chau D, Callus B, Wong WW, Hawkins CJ, Schneider P, McKinlay M, Benetatos CA, Condon SM, Chunduru SK, Yeoh G, Brink R, Vaux DL, Silke J. TWEAK-FN14 signaling induces lysosomal degradation of a cIAP1-TRAF2 complex to sensitize tumor cells to TNFalpha. J Cell Biol. 2008;182:171–184. doi: 10.1083/jcb.200801010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, Lee D, von GoetzM, Yee SF, Totpal K, Huw L, Katta V, Cavet G, Hymowitz SG, Amler L, Ashkenazi A. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. 2007;13:1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- Walczak H, Haas TL. Biochemical analysis of the native TRAIL death-inducing signaling complex. Methods Mol Biol. 2008;414:221–239. doi: 10.1007/978-1-59745-339-4_16. [DOI] [PubMed] [Google Scholar]
- Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C, Smolak P, Goodwin RG, Rauch CT, Schuh JC, Lynch DH. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- Westhoff MA, Fulda S. Adhesion-mediated apoptosis resistance in cancer. Drug Resist Update. 2009;12:127–136. doi: 10.1016/j.drup.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Westhoff MA, Zhou S, Bachem MG, Debatin KM, Fulda S. Identification of a novel switch in the dominant forms of cell adhesion-mediated drug resistance in glioblastoma cells. Oncogene. 2008;27:5169–5181. doi: 10.1038/onc.2008.148. [DOI] [PubMed] [Google Scholar]
- Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, Goodwin RG. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- Wittig I, Schagger H. Native electrophoretic techniques to identify protein-protein interactions. Proteomics. 2009;9:5214–5223. doi: 10.1002/pmic.200900151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


