Abstract
Quinolones are one of the most commonly prescribed classes of antibacterials in the world and are used to treat a variety of bacterial infections in humans. Because of the wide use (and overuse) of these drugs, the number of quinolone-resistant bacterial strains has been growing steadily since the 1990s. As is the case with other antibacterial agents, the rise in quinolone resistance threatens the clinical utility of this important drug class. Quinolones act by converting their targets, gyrase and topoisomerase IV, into toxic enzymes that fragment the bacterial chromosome. This review describes the development of the quinolones as antibacterials, the structure and function of gyrase and topoisomerase IV, and the mechanistic basis for quinolone action against their enzyme targets. It will then discuss the following three mechanisms that decrease the sensitivity of bacterial cells to quinolones. Target-mediated resistance is the most common and clinically significant form of resistance. It is caused by specific mutations in gyrase and topoisomerase IV that weaken interactions between quinolones and these enzymes. Plasmid-mediated resistance results from extrachromosomal elements that encode proteins that disrupt quinolone–enzyme interactions, alter drug metabolism, or increase quinolone efflux. Chromosome-mediated resistance results from the underexpression of porins or the overexpression of cellular efflux pumps, both of which decrease cellular concentrations of quinolones. Finally, this review will discuss recent advancements in our understanding of how quinolones interact with gyrase and topoisomerase IV and how mutations in these enzymes cause resistance. These last findings suggest approaches to designing new drugs that display improved activity against resistant strains.
Over a period of a few decades, quinolones have transformed from a small and unimportant class of drugs used primarily to treat urinary tract infections to some of the most commonly prescribed antibacterials in the world.1−3 Today, they are used to treat a wide variety of Gram-negative and Gram-positive bacterial infections. Unfortunately, quinolone usage is threatened by the rising occurrence of resistance, which has been observed in every species that is treated by this drug class.4−6
The cellular targets for quinolones are the bacterial type II topoisomerases, gyrase and topoisomerase IV.5,7−10 Recent work has helped to define how quinolones interact with these enzymes and how mutations in gyrase or topoisomerase IV can lead to resistance.11−13 Furthermore, additional resistance mechanisms caused by altered protein interactions, drug metabolism, and uptake and/or efflux have been described.1,2,5,14,15 This review will discuss our current knowledge of quinolone mechanism and resistance and how that information may be used to design drugs that are capable of overcoming the most common forms of resistance.
Quinolones
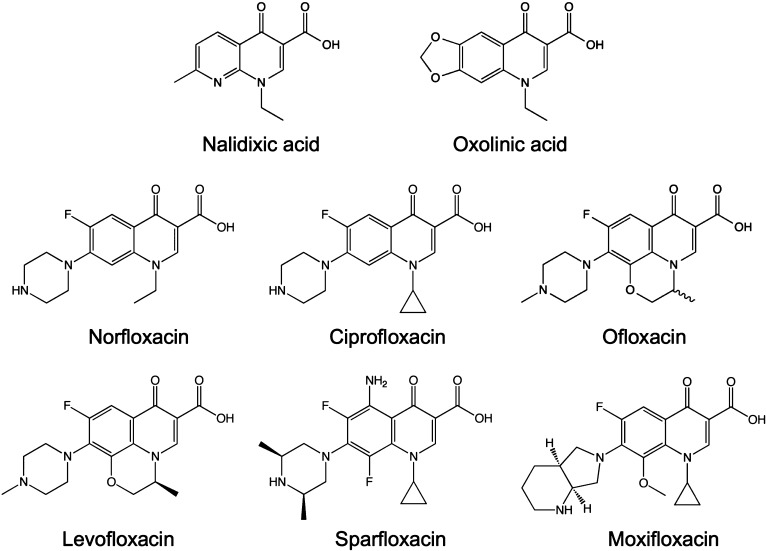
The founding member of the quinolone drug class, nalidixic acid, is a naphthyridine that was first isolated by George Lesher and colleagues in 1962 as a byproduct of chloroquine synthesis (Figure 1).16 Nalidixic acid was introduced into the clinic in the 1960s for the treatment of uncomplicated urinary tract infections caused by enteric bacteria.1 By the 1970s, several first-generation quinolones, oxolinic acid being the most notable, had been synthesized and introduced into the clinic (Figure 1).1,2,4,17
Figure 1.
Quinolone structures. Nalidixic acid and oxolinic acid were the first-generation quinolones that were used most often in the clinic. Norfloxacin, ciprofloxacin, and ofloxacin are the most relevant second-generation quinolones. Levofloxacin (the levorotary isomer of ofloxacin), sparfloxacin, and moxifloxacin are newer-generation quinolones.
The quinolones were a little-used drug class until the early 1980s, when a second generation of compounds was developed (Figure 1).1,2,4,17 These newer drugs, highlighted by norfloxacin, ciprofloxacin, and ofloxacin, displayed considerably improved activity against gyrase, greater penetration into Gram-positive organisms, and enhanced pharmacokinetics and pharmacodynamics. The most critical changes to the quinolone skeleton were the introduction of a fluorine at position C6 and a major ring substituent (piperazine or methyl-piperazine) at C7.1,2,4,17 Because of the inclusion of the fluorine, quinolones are often termed “fluoroquinolones”.
Norfloxacin is considered to be the first broad-spectrum quinolone and was utilized to a far greater extent than nalidixic acid.1,2,4,17 Unfortunately, because of low serum levels and poor tissue penetration, norfloxacin was still confined to use for the treatment of urinary tract infections and sexually transmitted diseases. Ciprofloxacin was the first quinolone that displayed significant activity outside of the urinary tract.1,2,4,17 After more than 20 years in clinical use, ciprofloxacin remains one of the most commonly prescribed antibacterial drugs and is used to treat a variety of Gram-negative and, to a lesser extent, Gram-positive infections.1,2,4
The clinical success of ciprofloxacin spawned an array of newer-generation quinolones that displayed an even broader spectrum of activity, especially against Gram-positive species.1,2,4,17 Levofloxacin, moxifloxacin, and sparfloxacin (Figure 1) have enjoyed the most success and display good activity against Gram-positive respiratory tract infections. Furthermore, the pharmacokinetics of levofloxacin are advantageous compared to those of other members of the drug class, and treatment requires only a single pill per day.18,19
A number of diseases currently are treated with quinolones, including urinary tract infections and pyelonephritis, sexually transmitted diseases, prostatitis, skin and tissue infections, chronic bronchitis, community-acquired and nosocomial pneumonia, and intra-abdominal and pelvic infections.6 Quinolones also are used to treat tuberculosis, the deadliest infectious disease on the planet, which has an annual death toll that exceeds 1 million.20
Bacterial Type II Topoisomerases
Most bacterial species encode two distinct, but homologous, type II topoisomerases, gyrase and topoisomerase IV.9,21−26 These enzymes play essential roles in most nucleic acid processes, help control levels of DNA under- and overwinding, and remove knots and tangles from the bacterial chromosome. Gyrase and topoisomerase IV modulate the topological state of DNA by passing an intact double helix through a transient double-stranded break that they generate in a separate segment of DNA. Progression through the catalytic cycle is driven by ATP binding and hydrolysis.9,21−26
The DNA cleavage and ligation reactions, which constitute the core of enzyme function, utilize a noncanonical two-metal ion mechanism.27−29 Gyrase and topoisomerase IV generate staggered cuts in the DNA backbone that are 4 bp apart and on opposite strands (leaving a 5′-overhang). To maintain genomic integrity during this process, the enzymes form covalent bonds between active site tyrosine residues and the newly generated 5′-DNA termini.9,21,22,25 These covalent enzyme-cleaved DNA complexes are known as “cleavage complexes”.
Despite their mechanistic and structural similarities, gyrase and topoisomerase IV have separate physiological functions.21,22,25,26 Gyrase is the only type II topoisomerase that can actively introduce negative supercoils into DNA. The enzyme works in conjunction with the ω protein (a type I topoisomerase) to set the superhelical density of the bacterial chromosome. In addition, gyrase is primarily responsible for removing the torsional stress that accumulates in front of replication forks and transcription complexes.21,22,25,26
Topoisomerase IV appears to play a lesser role than gyrase in maintaining chromosomal superhelical density and alleviating torsional stress. Its major function is removing knots that accumulate in the bacterial chromosome as a result of fundamental cellular processes and decatenating daughter chromosomes following replication.21,22,25,30,31
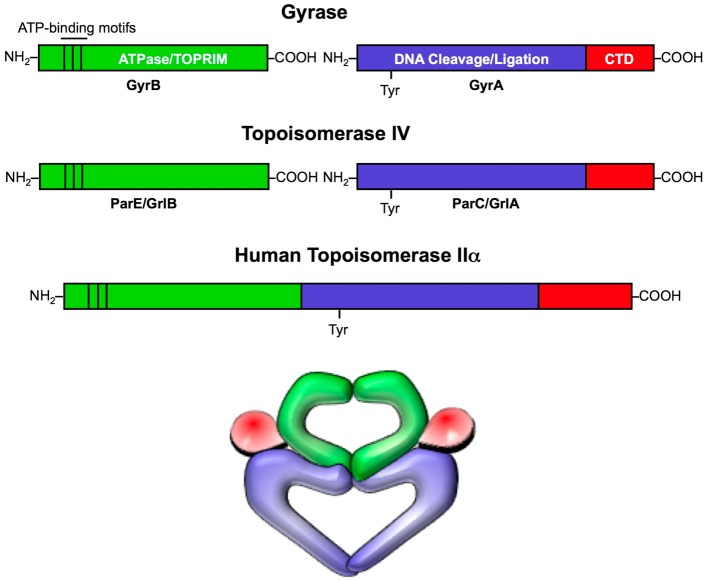
Gyrase and topoisomerase IV both are comprised of two distinct functional subunits and function as A2B2 heterotetramers (Figure 2).21,22,25,26 The subunits in gyrase are GyrA and GyrB. The homologous subunits in topoisomerase IV are ParC and ParE in Gram-negative species and GrlA and GrlB in Gram-positive species. GyrA (and the equivalent topoisomerase IV subunit) contains the active site tyrosine residue. GyrB (and the equivalent topoisomerase IV subunit) contains the ATPase domain as well as the TOPRIM domain, which binds the divalent metal ions involved in DNA cleavage and ligation. Despite the strong sequence similarity between gyrase and topoisomerase IV, the C-termini of the A subunits are not well conserved. This portion of the protein is involved in topology recognition and allows gyrase, but not topoisomerase IV, to generate supercoils in DNA.21,22,25,32−34
Figure 2.
Domain structures of type II topoisomerases. Gyrase and topoisomerase IV are heterotetrameric enzymes consisting of two A subunits and two B subunits. The A subunits (blue and red; GyrA in gyrase and ParC and GrlA in Gram-negative and Gram-positive topoisomerase IV, respectively) contain the active site tyrosine residue that covalently attaches to the newly generated 5′-termini of DNA during the cleavage reaction. The C-terminal domains (CTDs; red) of the A subunits are variable and allow gyrase to introduce negative supercoils into DNA. The B subunits (green; GyrB in gyrase and ParE and GrlB in Gram-negative and Gram-positive topoisomerase IV, respectively) contain the ATPase and TOPRIM domains, the latter of which binds the catalytic divalent metal ions essential for enzyme activity. Human topoisomerase IIα is homologous to the bacterial type II enzymes. However, during the course of evolution, the A and B subunits fused into a single polypeptide chain. Therefore, eukaryotic type II topoisomerases function as homodimers. A representation of the three-dimensional structure of type II topoisomerases is shown at the bottom.
It is notable that humans also express two type II enzymes, topoisomerase IIα and topoisomerase IIβ.22,25,26,35,36 Both share significant amino acid sequence similarity with the bacterial enzymes. However, during the course of evolution, the genes encoding the A and B subunits have fused, resulting in a single polypeptide chain (Figure 2). Consequently, the human type II enzymes function as homodimers.22,25,36 As discussed later, specific amino acid differences provide the basis for clinically relevant quinolones to discriminate between the human and bacterial type II topoisomerases. However, the similarity of these enzymes presents a confounding issue for designing quinolone-like drugs that overcome bacterial drug resistance.
Quinolone Action
In order to carry out their critical physiological functions, gyrase and topoisomerase IV generate double-stranded breaks in the bacterial chromosome. Thus, while essential for cell survival, these enzymes have the potential to fragment the genome.9,21,22,25,26,35 Quinolones take advantage of this latter, and potentially lethal, characteristic and kill cells by increasing the concentration of enzyme–DNA cleavage complexes.2,5,7−10,37 Thus, these drugs are termed “topoisomerase poisons” because they convert gyrase and topoisomerase IV into cellular toxins.37 In contrast, “catalytic inhibitors” block the overall catalytic functions of these enzymes without increasing levels of DNA strand breaks.
Quinolones bind in a noncovalent manner at the enzyme–DNA interface in the cleavage–ligation active site.11,38−40 Drugs interact with the protein and intercalate into the DNA at both cleaved scissile bonds (Figure 3). Because the scissile bonds on each strand are staggered, two drug molecules are required to increase levels of double-stranded DNA breaks. As a result of their intercalation, quinolones increase the steady-state concentration of cleavage complexes by acting as physical blocks to ligation.2,5,7−10
Figure 3.
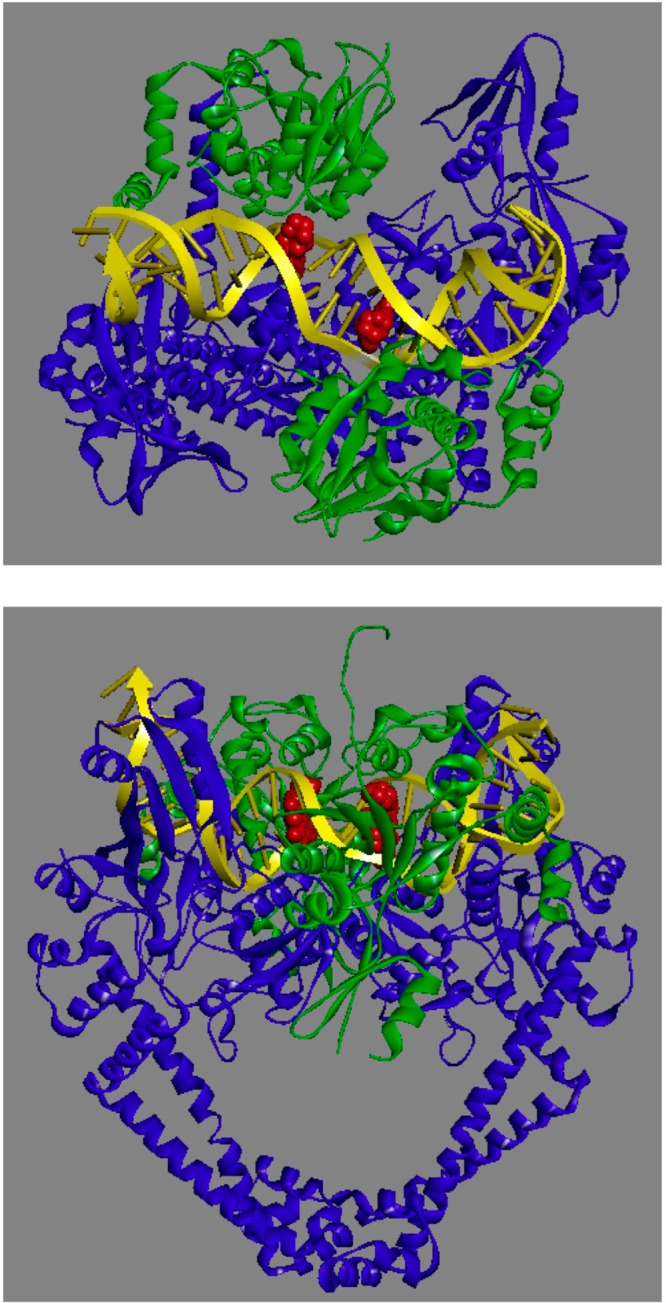
Crystal structure of a moxifloxacin-stabilized Acinetobacter baumannii topoisomerase IV–DNA cleavage complex. The catalytic core of the enzyme is shown. Moxifloxacin is colored red; the topoisomerase IV A and B subunits are colored blue and green, respectively, and DNA is colored yellow. The top panel is a top view of the cleavage complex showing two quinolone molecules intercalating 4 bp apart at the sites of DNA cleavage. The bottom panel is a front view (rotated by 90° from the top view) of the cleavage complex. Protein Data Bank accession 2XKK was visualized using Discovery Studio 3.5 Visualizer (Accelrys Software Inc.). Adapted from ref (11).
When replication forks, transcription complexes, or other DNA tracking systems collide with drug-stabilized gyrase– or topoisomerase IV–DNA cleavage complexes, these complexes are converted to permanent chromosomal breaks. In turn, the generation of these DNA breaks triggers the SOS response and other DNA repair pathways. If the strand breaks overwhelm these processes, they can lead to cell death. This is the primary mechanism that quinolones use to kill bacterial cells (Figure 4).2,5,7−10
Figure 4.
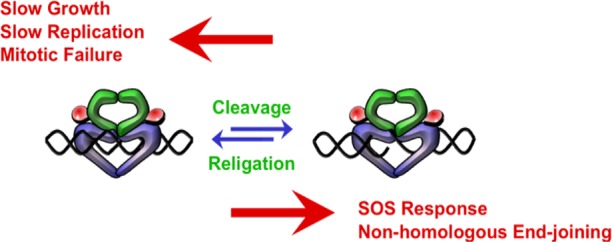
Bacterial type II topoisomerases are essential but potentially toxic enzymes. The balance between enzyme-mediated DNA cleavage and religation is critical for cell survival. If the level of gyrase-mediated DNA cleavage decreases, rates of DNA replication slow and impair cell growth (left). If the level of topoisomerase IV-mediated DNA cleavage decreases, cells are not able to untangle daughter chromosomes and ultimately die of mitotic failure (left). If the level of gyrase- or topoisomerase IV-mediated DNA cleavage becomes too high (right), the actions of DNA tracking systems can convert these transient complexes to permanent double-stranded breaks. The resulting DNA breaks initiate the SOS response and other DNA repair pathways and can lead to cell death.
Because quinolones stabilize cleavage complexes by inhibiting DNA ligation, they also impair the overall catalytic functions of gyrase and topoisomerase IV. Thus, in addition to acting as poisons, quinolones act as catalytic inhibitors. This accompanying loss of enzyme activity affects a number of nucleic acid processes and likely contributes to the overall toxicity of these drugs (Figure 4).2,5,7−10
Quinolone Targeting
Gyrase was first identified as the cellular target for quinolones in 1977.41,42 The later discovery of topoisomerase IV43 raised the question of whether this enzyme also was a target for quinolones. Based on an analysis of Escherichia coli strains carrying drug resistance mutations in one, the other, or both enzymes, it was concluded that gyrase is the primary toxic target for quinolones and that topoisomerase IV is a secondary drug target.44 Consistent with this conclusion, quinolones are more potent against E. coli gyrase than topoisomerase IV44 and induce higher levels of gyrase–DNA cleavage complexes in cells.45
Surprisingly, genetic studies in Streptococcus pneumoniae found that topoisomerase IV, rather than gyrase, was the primary cellular target for ciprofloxacin.46 This led to the concept that gyrase was the primary target for quinolones in Gram-negative bacteria but that the opposite was true in Gram-positive species. However, subsequent studies found that this paradigm did not hold in many cases. There are examples of Gram-positive bacteria in which gyrase is the primary target for quinolones. Furthermore, in a given bacterial species, different quinolones have been shown to have different primary targets.47−49 Ultimately, the issue of quinolone targeting is still a matter of debate, and the relative contributions of gyrase versus topoisomerase IV to quinolone action need to be evaluated on a species-by-species and drug-by-drug basis.
Quinolone–Topoisomerase Interactions
The fact that specific mutations in gyrase or topoisomerase IV cause quinolone resistance strongly suggests that drug–protein interactions play an important role in stabilizing cleavage complexes. As discussed below, the amino acids that most frequently are associated with quinolone resistance are Ser83 (based on E. coli GyrA numbering) and an acidic residue four amino acids downstream (Figure 5).5,7−9,47,50,51 Thus, it has been assumed that these two amino acid residues play an integral role in mediating quinolone–enzyme interactions.
Figure 5.
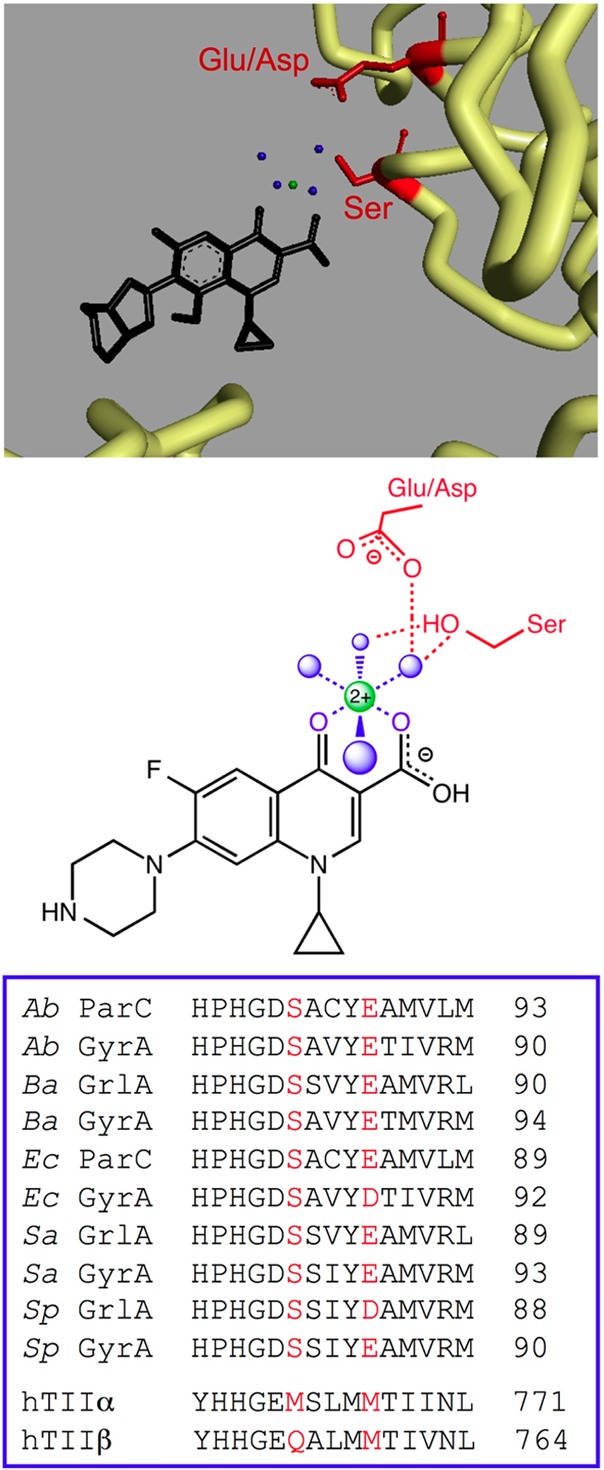
Quinolone–topoisomerase binding is facilitated through a water–metal ion bridge. The top panel shows the crystal structure of a moxifloxacin-stabilized A. baumannii topoisomerase IV–DNA cleavage complex. Moxifloxacin is colored black, and the noncatalytic Mg2+ ion that is chelated by the C3/C4 keto acid of the quinolone and participates in the bridge interaction is colored green. The four water molecules that fill out the coordination sphere of the Mg2+ ion are colored blue. The backbone of selected portions of the protein amino acid chain is colored yellow. The side chains of the serine and acidic residues that form hydrogen bonds with the water molecules in the water–metal ion bridge are colored red. In A. baumannii topoisomerase IV, these residues are Ser84 and Glu88, respectively. For clarity, DNA has been omitted from the picture. Protein Data Bank accession 2XKK was visualized using Discovery Studio 3.5 Visualizer (Accelrys Software Inc.). Adapted from ref (11). The middle panel shows a simplified diagram of the water–metal ion bridge. Only interactions with the protein (and not DNA) are shown. Ciprofloxacin (a representative quinolone) is colored black, and the noncatalytic Mg2+, water molecules, and coordinating serine and acidic residues are colored as described above. Blue dashed lines indicate the octahedral coordination sphere of the divalent metal ion interacting with four water molecules and the C3/C4 keto acid of the quinolone. The red dashed lines represent hydrogen bonds between the serine side chain hydroxyl group or the acidic residue side chain carboxyl group and the water molecules. Adapted from ref (13). The bottom panel shows a sequence alignment of the A subunits highlighting the conserved serine and acidic residues (red) that coordinate the water–metal ion bridge. Sequences of A. baumannii (Ab), Bacillus anthracis (Ba), E. coli (Ec), Staphylococcus aureus (Sa), and S. pneumoniae (Sp) gyrase (GyrA) and topoisomerase IV (ParC/GrlA) are shown. The homologous regions of human topoisomerase IIα (hTIIα) and IIβ (hTIIβ), which lack the residues necessary to coordinate the water–metal ion bridge interaction, are shown for comparison.
A series of crystallographic studies with gyrase and topoisomerase IV have shed considerable light on quinolone–enzyme interactions. Initial structures placed the drug near the serine and acidic resides, but the amino acids were not sufficiently close in space to mediate drug binding directly.38−40 In contrast, a later structure captured a quinolone complex that contained a noncatalytic Mg2+ ion that was chelated by the C3/C4 keto acid of the drug (Figure 5).11 The metal ion was coordinated to four water molecules, and two of these water molecules were situated close enough to the serine and acidic residues to form hydrogen bonds. On the basis of this structure and an earlier study that suggested a role for metal ions in quinolone action,52 the authors suggested that this water–metal ion interaction “bridged” the drug to the enzyme.11
Recent functional studies have characterized the role of the proposed water–metal ion bridge in mediating quinolone–enzyme interactions.12,13 Findings indicate that mutation of either the serine or acidic residue restricts the variety of metal ions that can be used to support drug activity and decreases the affinity of the quinolone–enzyme complex for the noncatalytic Mg2+ ion. Furthermore, mutation of either residue significantly decreases the affinity of gyrase or topoisomerase IV for quinolones, and mutation of both residues abolishes the ability of clinically relevant quinolones to stabilize cleavage complexes.12,13 Taken together, these results confirm the existence of the water–metal ion bridge and provide evidence that the serine and acidic residues act as the anchor points that coordinate the bridge to the enzyme. Moreover, they demonstrate that the water–metal ion bridge is the primary interaction between quinolones and the bacterial type II enzymes. A significant ramification of the above is that the most important interactions between clinically relevant quinolones and their enzyme targets are mediated through the C3/C4 keto acid of the drug skeleton (Figure 1). This may explain the tolerance for the structural diversity of substituents at positions N1, C7, and C8 of this drug class.53
It is notable that the human type II topoisomerases lack the serine and acidic residues that anchor the water–metal ion bridge (Figure 5) and are unable to utilize this critical mechanism to interact with quinolones.53 This difference provides the basis by which quinolones discriminate between the bacterial and human enzymes and accounts, in part, for the therapeutic window of this drug class.
Quinolone Resistance
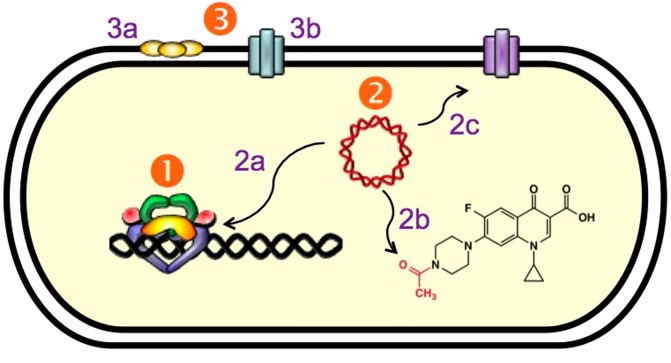
As discussed earlier, quinolone resistance is becoming a prevalent clinical issue that is threatening the use of these drugs. Resistance mechanisms are grouped into three distinct categories that are discussed below (Figure 6). The cellular alterations associated with each mechanism are not mutually exclusive and can accumulate to create strains that exhibit very high levels of quinolone resistance.
Figure 6.
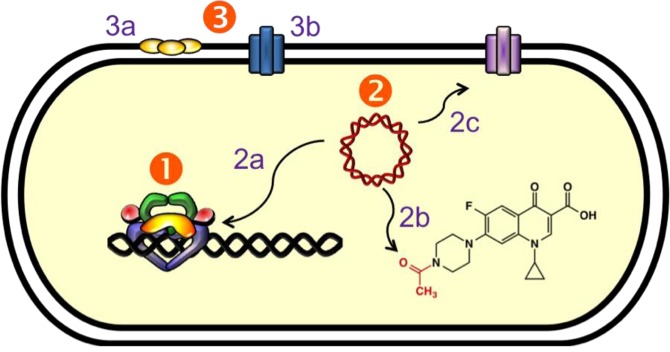
Mechanisms of quinolone resistance. (1) Target-mediated resistance. Mutations in gyrase and topoisomerase IV weaken quinolone–enzyme interactions. (2) Plasmid-mediated resistance. (2a) Qnr proteins (yellow) decrease topoisomerase–DNA binding and protect enzyme–DNA complexes from quinolones. (2b) Aac(6′)-Ib-cr is an aminoglycoside acetyltransferase that acetylates the free nitrogen on the C7 ring of ciprofloxacin and norfloxacin, decreasing their effectiveness. (2c) Plasmid-encoded efflux pumps decrease the concentration of quinolones in the cell. (3) Chromosome-mediated resistance. (3a) Underexpression of porins in Gram-negative species decreases drug uptake. (3b) Overexpression of chromosome-encoded efflux pumps decreases drug retention in the cell.
Target-Mediated Quinolone Resistance
Quinolone resistance is most often associated with specific mutations in gyrase and/or topoisomerase IV (Figure 6). Generally, mutation of one type II enzyme confers ≤10-fold drug resistance. Selection for higher levels of resistance (∼10–100-fold) usually yields strains with mutations in both enzymes.5,7−9,47,50
Although mutations have been mapped throughout the A and B subunits of gyrase and topoisomerase IV in quinolone-resistant strains, the most commonly mutated amino acids are the serine and acidic residues that anchor the water–metal ion bridge (Figure 5).5,51,54,55 Presumably, disruption of the water–metal ion bridge causes the observed quinolone resistance.
Typically, in both laboratory and clinical isolates, alterations at the serine comprise >90% of the mutant pool, with changes at the acidic residue comprising the bulk of the other mutations.50,51 In general, mutant gyrase and topoisomerase IV maintain wild-type DNA cleavage activity in the absence of drugs.12,13,56 However, quinolones display little ability to increase levels of enzyme-mediated DNA cleavage at clinically relevant concentrations.12,13,56−61 Furthermore, drug–enzyme binding is significantly reduced, and quinolones lose much of their ability to inhibit DNA ligation or to form stable ternary enzyme–DNA–drug complexes.12,13,57,62−65
Resistance mutations at the serine residue in gyrase and topoisomerase IV do not appear to adversely affect catalytic activity in the absence of drug.12,56,65 In contrast, mutations at the acidic residue decrease overall catalytic activity ∼5–10-fold.13,66 This may explain, at least in part, why the serine mutation is found much more frequently.
The serine residue is highly conserved across bacterial species (Figure 5). This raises the question of why an amino acid residue in gyrase and topoisomerase IV that has no apparent function other than to provide sensitivity to a class of synthetic antibacterials is maintained so consistently throughout the bacterial kingdom. An intriguing possibility comes from a study on nybomycin, an antibiotic produced by Streptomyces spp. This compound displays little activity against S. aureus strains that express wild-type gyrase. However, nybomycin is active against strains that express a Ser → Leu quinolone-resistant GyrA.67 Thus, the conserved serine residue may represent a “resistance mutation” that provides protection against naturally occurring antibiotics.
Plasmid-Mediated Quinolone Resistance
Recently, plasmids that carry quinolone resistance genes have been identified as an emerging clinical problem that generally cause low-level (≤10-fold) resistance (Figure 6).5,14,68−75 However, resistance as high as ∼250-fold has been reported.5,71,73 Unlike target-mediated resistance, which is transmitted vertically from generation to generation, plasmid-mediated quinolone resistance can be transmitted horizontally (through bacterial conjugation) as well as vertically. Plasmids that confer quinolone resistance typically carry additional genes that cause resistance to other drug classes.68,70,71,73 However, this review will focus on those that affect quinolone sensitivity.
Three families of genes are associated with plasmid-mediated quinolone resistance. The first are the Qnr genes, which encode proteins (∼200 amino acids in length) that are part of the pentapeptide repeat protein family.71,73,76,77 Approximately 100 Qnr variants have been identified to date and have been classified into at least five distinct subfamilies.73,74,78,79 These proteins share homology with McbG and MfpA, which are DNA mimics.71,73,76 The Qnr proteins appear to confer quinolone resistance by two different mechanisms. Like McbG and MfpA, they decrease the binding of gyrase and topoisomerase IV to DNA. Thus, they protect cells from quinolones by lowering the number of available enzyme targets on the chromosome. They also bind to gyrase and topoisomerase IV and inhibit quinolones from entering cleavage complexes formed by the enzymes.76,77,80,81
The second plasmid-encoded protein associated with quinolone resistance is aac(6′)-Ib-cr.82,83 This protein is a variant of an aminoglycoside acetyltransferase that contains two specific point mutations, W102R and D179Y. The enzyme acetylates the unsubstituted nitrogen of the C7 piperazine ring that is found in norfloxacin and ciprofloxacin, which decreases drug activity. Although the wild-type and mutant aminoglycoside acetyltransferases are capable of acetylating other drugs, only the mutant enzyme is active against quinolones.82,83
The third group of plasmid-encoded quinolone resistance proteins is comprised of efflux pumps. Thus far, three have been identified: OqxAB, QepA1, and QepA2.73,84,85 Whereas the latter two proteins have been found in human bacterial infections, OqxAB is seen almost exclusively in animal infections.73,86,87
Chromosome-Mediated Quinolone Resistance
The cellular concentration of quinolones is regulated by the opposing actions of diffusion-mediated drug uptake and pump-mediated efflux. In contrast to Gram-positive species, the outer membrane of Gram-negative bacteria poses an additional barrier that drugs must cross to enter the cell. Therefore, drug influx in Gram-negative species is facilitated by protein channels called porins. If the expression of porins is downregulated, it can lead to low-level resistance to quinolones (Figure 6).2,71,73,88,89
In addition to the introduction of plasmid-encoded efflux pumps, enhanced expression of chromosome-encoded efflux pumps also can lead to quinolone resistance (Figure 6). Most commonly, the upregulation of these pumps is caused by mutations in regulatory proteins.73,89,90 In general, changes in quinolone uptake and retention cause low-level resistance and, in the absence of additional resistance mechanisms, do not appear to be a major clinical issue.90 However, lowering the cellular concentration of quinolones creates a favorable background for other forms of resistance to develop and propagate.68,72,91,92
Overcoming Quinolone Resistance
As discussed above, mutations in gyrase and topoisomerase IV are the most common cause of high-level quinolone resistance. Therefore, the development of novel quinolones that retain activity against these mutated enzymes has the potential to greatly extend the clinical efficacy of this drug class. Unfortunately, no such drugs have appeared in the clinic.
The initial hurdle to the development of drugs against resistant gyrase and topoisomerase IV was the concept that it would be impractical to design quinolones against every common mutation. This concept was based on the assumption that different mutations caused resistance by different mechanisms. Recent studies have demonstrated that this is not the case. As discussed above, the vast majority of clinically relevant mutations cause quinolone resistance through a common mechanism, namely by disrupting the water–metal ion bridge.11−13 Therefore, at least in theory, it should be possible to overcome most instances of target-mediated resistance by designing a quinolone-like drug that no longer depends on the water–metal ion bridge for its primary interaction with gyrase or topoisomerase IV.
Recent studies that examined quinazolinediones have provided insight into how this might be possible.12,13,53,56,61,93−95 Quinazolinediones are structurally similar to quinolones (Figure 7); however, they lack the keto acid that chelates the bridging metal ion. In contrast to clinically relevant quinolones, some quinazolinediones retain (or actually display higher) activity against gyrase and topoisomerase IV that carry mutations in the anchoring serine or acidic residue. This observation has been reported using purified enzymes as well as cultured resistant bacterial strains.12,13,53,56,61,93−95 Structural studies indicate that quinolones and quinazolinediones interact with topoisomerase IV in the same drug-binding pocket.39 Thus, it was believed that the quinazolinedione skeleton was able to mediate drug–enzyme binding directly and did not require a divalent metal ion. To this point, the C2 carbonyl of quinazolinediones likely forms a hydrogen bond with a conserved arginine residue (Arg121 based on E. coli GyrA numbering).39,95 However, this interaction is expected to be weaker than the water–metal ion bridge interaction utilized by quinolones. Indeed, in the absence of additional substituents that form strong interactions in the cleavage complex, quinazolinediones have poorer activity than their structurally cognate quinolones.53,94,95 Recent studies demonstrated that interactions between highly active quinazolinediones and bacterial type II topoisomerases are mediated primarily through novel contacts made by the 3′-(aminomethyl)pyrrolidinyl (and related) C7 substituent.53,96 These C7 groups are not represented in any clinically relevant quinolone.
Figure 7.
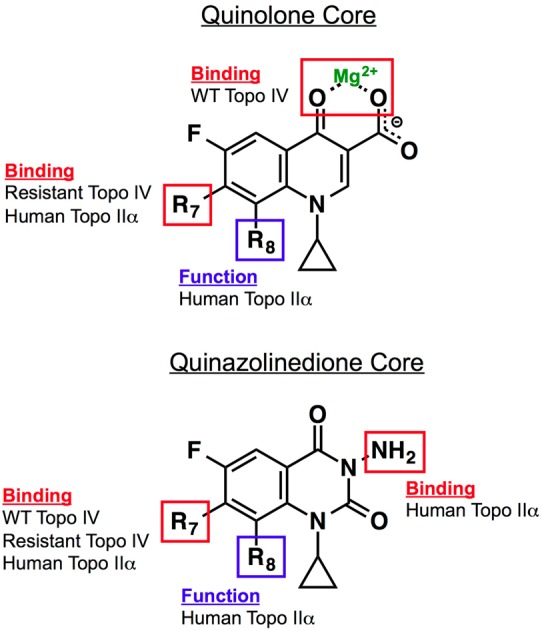
Roles of substituents and core elements of quinolones and quinazolinediones that mediate drug activity against bacterial and human type II topoisomerases. Results are based on studies with B. anthracis topoisomerase IV and human topoisomerase IIα.53 For quinolones, the binding of clinically relevant drugs to topoisomerase IV is mediated primarily through the water–metal ion bridge. The binding of quinolones that overcome resistance is mediated primarily by the substituent at C7. Binding of quinolones to human topoisomerase IIα also is mediated by the C7 substituent. The group at C8 affects the ability of quinolones to act against the human type II enzyme but is not required for drug binding. For quinazolinediones, interactions between drugs and topoisomerase IV (wild-type and resistant) are mediated through the C7 substituent. The effects of the C7 and C8 substituents on quinazolinedione activity against topoisomerase IIα are the same as described for the quinolones. The N3 amino group plays a role in the binding of quinazolinediones to the human enzyme. Adapted from ref (53).
Unfortunately, the 3′-(aminomethyl)pyrrolidinyl group also appears to mediate novel contacts with topoisomerase IIα and results in a drug that poisons the human enzyme.53 Thus, one of the major challenges in overcoming quinolone resistance will be the identification of C7 (and potentially other) substituents that can differentiate between the bacterial and human type II enzymes. Initial studies have dissected the roles of quinolone and quinazolinedione substituents in mediating drug binding and function against topoisomerase IIα (Figure 7).53 They indicate that it is possible to generate drugs that retain activity against quinolone-resistant bacterial enzymes without cross-reacting with the human enzyme.53 Thus, a “mechanistic” approach to drug discovery holds potential.
Summary
Quinolones are one of the most important classes of antibacterials available for the treatment of infectious diseases in humans. However, the clinical utility of these drugs is being impacted by the growing number of resistant bacterial strains. Although several resistance mechanisms have been described, the most common and significant form of resistance is caused by specific mutations in gyrase and topoisomerase IV that disrupt the water–metal ion bridge interaction.
Quinolones have been in the clinics since the 1960s, but the molecular details of how these drugs interact with their topoisomerase targets and how mutations cause resistance have only recently been described. Hopefully, this new information can be used to direct the discovery of a new generation of quinolones with improved activity against wild-type and mutant gyrase and topoisomerase IV to extend the clinical use of these drugs well into the future.
Acknowledgments
We are grateful to MaryJean Pendleton, Rachel E. Ashley, and Kendra R. Vann for critical reading of the manuscript.
K.J.A. was a trainee under Grant T32 CA09582 from the National Institutes of Health. This work was supported by National Institutes of Health Grants AI87671 (to R.J.K.) and GM033944 (to N.O.).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Emmerson A. M.; Jones A. M. (2003) The quinolones: Decades of development and use. J. Antimicrob. Chemother. 51(Suppl. 1), 13–20. [DOI] [PubMed] [Google Scholar]
- Mitscher L. A. (2005) Bacterial topoisomerase inhibitors: Quinolone and pyridone antibacterial agents. Chem. Rev. 105, 559–592. [DOI] [PubMed] [Google Scholar]
- Linder J. A.; Huang E. S.; Steinman M. A.; Gonzales R.; Stafford R. S. (2005) Fluoroquinolone prescribing in the United States: 1995 to 2002. Am. J. Med. 118, 259–268. [DOI] [PubMed] [Google Scholar]
- Andriole V. T. (2005) The quinolones: Past, present, and future. Clin. Infect. Dis. 41(Suppl. 2), S113–S119. [DOI] [PubMed] [Google Scholar]
- Drlica K.; Hiasa H.; Kerns R.; Malik M.; Mustaev A.; Zhao X. (2009) Quinolones: Action and resistance updated. Curr. Top. Med. Chem. 9, 981–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhoff A. (2012) Resistance surveillance studies: A multifaceted problem—the fluoroquinolone example. Infection 40, 239–262. [DOI] [PubMed] [Google Scholar]
- Hooper D. C. (1999) Mode of action of fluoroquinolones. Drugs 58(Suppl. 2), 6–10. [DOI] [PubMed] [Google Scholar]
- Hooper D. C. (2001) Mechanisms of action of antimicrobials: Focus on fluoroquinolones. Clin. Infect. Dis. 32(Suppl. 1), S9–S15. [DOI] [PubMed] [Google Scholar]
- Anderson V. E.; Osheroff N. (2001) Type II topoisomerases as targets for quinolone antibacterials: Turning Dr. Jekyll into Mr. Hyde. Curr. Pharm. Des. 7, 337–353. [DOI] [PubMed] [Google Scholar]
- Drlica K.; Malik M.; Kerns R. J.; Zhao X. (2008) Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 52, 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlkonig A.; Chan P. F.; Fosberry A. P.; Homes P.; Huang J.; Kranz M.; Leydon V. R.; Miles T. J.; Pearson N. D.; Perera R. L.; Shillings A. J.; Gwynn M. N.; Bax B. D. (2010) Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struct. Mol. Biol. 17, 1152–1153. [DOI] [PubMed] [Google Scholar]
- Aldred K. J.; McPherson S. A.; Wang P.; Kerns R. J.; Graves D. E.; Turnbough C. L. Jr.; Osheroff N. (2012) Drug interactions with Bacillus anthracis topoisomerase IV: Biochemical basis for quinolone action and resistance. Biochemistry 51, 370–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldred K. J.; McPherson S. A.; Turnbough C. L. Jr.; Kerns R. J.; Osheroff N. (2013) Topoisomerase IV-quinolone interactions are mediated through a water-metal ion bridge: Mechanistic basis of quinolone resistance. Nucleic Acids Res. 41, 4628–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper D. C. (2001) Emerging mechanisms of fluoroquinolone resistance. Emerging Infect. Dis. 7, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X.; Xue X.; Liu Y.; Wang J.; Wang Y.; Wang K.; Jiang H.; Zhang L.; Yang B.; Wang N.; Pan L. (2013) Plasmid-mediated quinolone resistance-current knowledge and future perspectives. J. Int. Med. Res. 41, 20–30. [DOI] [PubMed] [Google Scholar]
- Lesher G. Y.; Froelich E. J.; Gruett M. D.; Bailey J. H.; Brundage R. P. (1962) 1,8-Naphthyridine derivatives. A new class of chemotherapeutic agents. J. Med. Pharm. Chem. 91, 1063–1065. [DOI] [PubMed] [Google Scholar]
- Stein G. E. (1988) The 4-quinolone antibiotics: Past, present, and future. Pharmacotherapy 8, 301–314. [DOI] [PubMed] [Google Scholar]
- Anderson V. R.; Perry C. M. (2008) Levofloxacin: A review of its use as a high-dose, short-course treatment for bacterial infection. Drugs 68, 535–565. [DOI] [PubMed] [Google Scholar]
- Noel G. J. (2009) A review of levofloxacin for the treatment of bacterial infections. Clin. Med.: Ther. 1, 433–458. [Google Scholar]
- World Health Organization (2013) Global Tuberculosis Control http://www.who.int/tb/publications/global_report/en/(accessed February 25, 2014).
- Levine C.; Hiasa H.; Marians K. J. (1998) DNA gyrase and topoisomerase IV: Biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta 1400, 29–43. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. (2001) DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413. [DOI] [PubMed] [Google Scholar]
- Forterre P.; Gribaldo S.; Gadelle D.; Serre M. C. (2007) Origin and evolution of DNA topoisomerases. Biochimie 89, 427–446. [DOI] [PubMed] [Google Scholar]
- Forterre P.; Gadelle D. (2009) Phylogenomics of DNA topoisomerases: Their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 37, 679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y.; Leo E.; Zhang H.; Marchand C. (2010) DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry A. C., and Osheroff N. (2013) DNA topoisomerases: Type II. In Encyclopedia of Biological Chemistry, pp 163–168, Elsevier Inc., Amsterdam. [Google Scholar]
- Schmidt B. H.; Burgin A. B.; Deweese J. E.; Osheroff N.; Berger J. M. (2010) A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature 465, 641–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deweese J. E.; Osheroff N. (2010) The use of divalent metal ions by type II topoisomerases. Metallomics 2, 450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts S. L.; Liou G. F.; Mitchenall L. A.; Burgin A. B.; Maxwell A.; Neuman K. C.; Osheroff N. (2011) Use of divalent metal ions in the DNA cleavage reaction of topoisomerase IV. Nucleic Acids Res. 39, 4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechiedrich E. L.; Khodursky A. B.; Bachellier S.; Schneider R.; Chen D.; Lilley D. M.; Cozzarelli N. R. (2000) Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 275, 8103–8113. [DOI] [PubMed] [Google Scholar]
- Deibler R. W.; Rahmati S.; Zechiedrich E. L. (2001) Topoisomerase IV, alone, unknots DNA in E. coli. Genes Dev. 15, 748–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K. D.; Shultzaberger R. K.; Berger J. M. (2004) The C-terminal domain of DNA gyrase A adopts a DNA-bending β-pinwheel fold. Proc. Natl. Acad. Sci. U.S.A. 101, 7293–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter E. M.; Berger J. M. (2012) Mechanisms for defining supercoiling set point of DNA gyrase orthologs: I. A nonconserved acidic C-terminal tail modulates Escherichia coli gyrase activity. J. Biol. Chem. 287, 18636–18644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter E. M.; Berger J. M. (2012) Mechanisms for defining supercoiling set point of DNA gyrase orthologs: II. The shape of the GyrA subunit C-terminal domain (CTD) is not a sole determinant for controlling supercoiling efficiency. J. Biol. Chem. 287, 18645–18654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deweese J. E.; Osheroff N. (2009) The DNA cleavage reaction of topoisomerase II: Wolf in sheep’s clothing. Nucleic Acids Res. 37, 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deweese J. E.; Osheroff M. A.; Osheroff N. (2009) DNA topology and topoisomerases: Teaching a “knotty” subject. Biochem. Mol. Biol. Educ. 37, 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer K. N.; Cozzarelli N. R. (1979) Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: Effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J. Bacteriol. 140, 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laponogov I.; Sohi M. K.; Veselkov D. A.; Pan X. S.; Sawhney R.; Thompson A. W.; McAuley K. E.; Fisher L. M.; Sanderson M. R. (2009) Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 16, 667–669. [DOI] [PubMed] [Google Scholar]
- Laponogov I.; Pan X. S.; Veselkov D. A.; McAuley K. E.; Fisher L. M.; Sanderson M. R. (2010) Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One 5, e11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax B. D.; Chan P. F.; Eggleston D. S.; Fosberry A.; Gentry D. R.; Gorrec F.; Giordano I.; Hann M. M.; Hennessy A.; Hibbs M.; Huang J.; Jones E.; Jones J.; Brown K. K.; Lewis C. J.; May E. W.; Saunders M. R.; Singh O.; Spitzfaden C. E.; Shen C.; Shillings A.; Theobald A. J.; Wohlkonig A.; Pearson N. D.; Gwynn M. N. (2010) Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 466, 935–940. [DOI] [PubMed] [Google Scholar]
- Sugino A.; Peebles C. L.; Kreuzer K. N.; Cozzarelli N. R. (1977) Mechanism of action of nalidixic acid: Purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. U.S.A. 74, 4767–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M.; Mizuuchi K.; O’Dea M. H.; Itoh T.; Tomizawa J. (1977) Nalidixic acid resistance: A second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. U.S.A. 74, 4772–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J.; Nishimura Y.; Imamura R.; Niki H.; Hiraga S.; Suzuki H. (1990) New topoisomerase essential for chromosome segregation in E. coli. Cell 63, 393–404. [DOI] [PubMed] [Google Scholar]
- Khodursky A. B.; Zechiedrich E. L.; Cozzarelli N. R. (1995) Topoisomerase IV is a target of quinolones in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 92, 11801–11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aedo S.; Tse-Dinh Y. C. (2012) Isolation and quantitation of topoisomerase complexes accumulated on Escherichia coli chromosomal DNA. Antimicrob. Agents Chemother. 56, 5458–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X.-S.; Ambler J.; Mehtar S.; Fisher L. M. (1996) Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40, 2321–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier B.; Zhao X.; Lu T.; Drlica K.; Hooper D. C. (2000) Selective targeting of topoisomerase IV and DNA gyrase in Staphylococcus aureus: Different patterns of quinolone-induced inhibition of DNA synthesis. Antimicrob. Agents Chemother. 44, 2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X.-S.; Fisher L. M. (1997) Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: Selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob. Agents Chemother. 41, 471–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X.-S.; Fisher L. M. (1998) DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42, 2810–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price L. B.; Vogler A.; Pearson T.; Busch J. D.; Schupp J. M.; Keim P. (2003) In vitro selection and characterization of Bacillus anthracis mutants with high-level resistance to ciprofloxacin. Antimicrob. Agents Chemother. 47, 2362–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan-Linnell S. K.; Becnel Boyd L.; Steffen D.; Zechiedrich L. (2009) Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob. Agents Chemother. 53, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sissi C.; Perdona E.; Domenici E.; Feriani A.; Howells A. J.; Maxwell A.; Palumbo M. (2001) Ciprofloxacin affects conformational equilibria of DNA gyrase A in the presence of magnesium ions. J. Mol. Biol. 311, 195–203. [DOI] [PubMed] [Google Scholar]
- Aldred K. J.; Schwanz H. A.; Li G.; McPherson S. A.; Turnbough C. L. Jr.; Kerns R. J.; Osheroff N. (2013) Overcoming target-mediated quinolone resistance in topoisomerase IV by introducing metal-ion-independent drug-enzyme interactions. ACS Chem. Biol. 8, 2660–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K.; Zhao X. (1997) DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61, 377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Deguchi T.; Yasuda M.; Kawamura T.; Kanematsu E.; Nishino Y.; Ishihara S.; Kawada Y. (1998) Alteration in the GyrA subunit of DNA gyrase and the ParC subunit of DNA topoisomerase IV in quinolone-resistant clinical isolates of Staphylococcus epidermidis. Antimicrob. Agents Chemother. 42, 3293–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X. S.; Gould K. A.; Fisher L. M. (2009) Probing the differential interactions of quinazolinedione PD 0305970 and quinolones with gyrase and topoisomerase IV. Antimicrob. Agents Chemother. 53, 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson V. E.; Zaniewski R. P.; Kaczmarek F. S.; Gootz T. D.; Osheroff N. (2000) Action of quinolones against Staphylococcus aureus topoisomerase IV: Basis for DNA cleavage enhancement. Biochemistry 39, 2726–2732. [DOI] [PubMed] [Google Scholar]
- Pan X. S.; Yague G.; Fisher L. M. (2001) Quinolone resistance mutations in Streptococcus pneumoniae GyrA and ParC proteins: Mechanistic insights into quinolone action from enzymatic analysis, intracellular levels, and phenotypes of wild-type and mutant proteins. Antimicrob. Agents Chemother. 45, 3140–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yague G.; Morris J. E.; Pan X. S.; Gould K. A.; Fisher L. M. (2002) Cleavable-complex formation by wild-type and quinolone-resistant Streptococcus pneumoniae type II topoisomerases mediated by gemifloxacin and other fluoroquinolones. Antimicrob. Agents Chemother. 46, 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer E. S.; Hiasa H. (2007) Determination of the primary target of a quinolone drug and the effect of quinolone resistance-conferring mutations by measuring quinolone sensitivity based on its mode of action. Antimicrob. Agents Chemother. 51, 3410–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppegard L. M.; Streck K. R.; Rosen J. D.; Schwanz H. A.; Drlica K.; Kerns R. J.; Hiasa H. (2010) Comparison of in vitro activities of fluoroquinolone-like 2,4- and 1,3-diones. Antimicrob. Agents Chemother. 54, 3011–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmott C. J.; Maxwell A. (1993) A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 37, 126–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson V. E.; Gootz T. D.; Osheroff N. (1998) Topoisomerase IV catalysis and the mechanism of quinolone action. J. Biol. Chem. 273, 17879–17885. [DOI] [PubMed] [Google Scholar]
- Anderson V. E.; Zaniewski R. P.; Kaczmarek F. S.; Gootz T. D.; Osheroff N. (1999) Quinolones inhibit DNA religation mediated by Staphylococcus aureus topoisomerase IV: Changes in drug mechanism across evolutionary boundaries. J. Biol. Chem. 274, 35927–35932. [DOI] [PubMed] [Google Scholar]
- Barnard F. M.; Maxwell A. (2001) Interaction between DNA gyrase and quinolones: Effects of alanine mutations at GyrA subunit residues Ser(83) and Asp(87). Antimicrob. Agents Chemother. 45, 1994–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiasa H. (2002) The Glu-84 of the ParC subunit plays critical roles in both topoisomerase IV-quinolone and topoisomerase IV-DNA interactions. Biochemistry 41, 11779–11785. [DOI] [PubMed] [Google Scholar]
- Hiramatsu K.; Igarashi M.; Morimoto Y.; Baba T.; Umekita M.; Akamatsu Y. (2012) Curing bacteria of antibiotic resistance: Reverse antibiotics, a novel class of antibiotics in nature. Int. J. Antimicrob. Agents 39, 478–485. [DOI] [PubMed] [Google Scholar]
- Martinez-Martinez L.; Pascual A.; Jacoby G. A. (1998) Quinolone resistance from a transferable plasmid. Lancet 351, 797–799. [DOI] [PubMed] [Google Scholar]
- Martinez-Freijo P.; Fluit A. C.; Schmitz F. J.; Grek V. S.; Verhoef J.; Jones M. E. (1998) Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J. Antimicrob. Chemother. 42, 689–696. [DOI] [PubMed] [Google Scholar]
- Wang M.; Tran J. H.; Jacoby G. A.; Zhang Y.; Wang F.; Hooper D. C. (2003) Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47, 2242–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek A.; Jacoby G. A.; Hooper D. C. (2006) The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6, 629–640. [DOI] [PubMed] [Google Scholar]
- Poirel L.; Cattoir V.; Nordmann P. (2008) Is plasmid-mediated quinolone resistance a clinically significant problem?. Clin. Microbiol. Infect. 14, 295–297. [DOI] [PubMed] [Google Scholar]
- Strahilevitz J.; Jacoby G. A.; Hooper D. C.; Robicsek A. (2009) Plasmid-mediated quinolone resistance: A multifaceted threat. Clin. Microbiol. Rev. 22, 664–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Martinez J. M.; Cano M. E.; Velasco C.; Martinez-Martinez L.; Pascual A. (2011) Plasmid-mediated quinolone resistance: An update. J. Infect. Chemother. 17, 149–182. [DOI] [PubMed] [Google Scholar]
- Carattoli A. (2013) Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304. [DOI] [PubMed] [Google Scholar]
- Tran J. H.; Jacoby G. A. (2002) Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. U.S.A. 99, 5638–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X.; Bromley E. H.; Oelschlaeger P.; Woolfson D. N.; Spencer J. (2011) Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: Conserved surface loops direct the activity of a Qnr protein from a Gram-negative bacterium. Nucleic Acids Res. 39, 3917–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. I.; Jeong da U.; Lee J. H.; Wu X.; Park K. S.; Lee J. J.; Jeong B. C.; Lee S. H. (2010) A novel family (QnrAS) of plasmid-mediated quinolone resistance determinant. Int. J. Antimicrob. Agents 36, 578–579. [DOI] [PubMed] [Google Scholar]
- Lahey Clinic. qnr Numbering and Sequence (Jacoby G. A., Ed.) http://www.lahey.org/qnrstudies/ (accessed January 9, 2014).
- Tran J. H.; Jacoby G. A.; Hooper D. C. (2005) Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49, 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran J. H.; Jacoby G. A.; Hooper D. C. (2005) Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob. Agents Chemother. 49, 3050–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek A.; Strahilevitz J.; Jacoby G. A.; Macielag M.; Abbanat D.; Park C. H.; Bush K.; Hooper D. C. (2006) Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12, 83–88. [DOI] [PubMed] [Google Scholar]
- Guillard T.; Cambau E.; Chau F.; Massias L.; de Champs C.; Fantin B. (2013) Ciprofloxacin treatment failure in a murine model of pyelonephritis due to an AAC(6′)-Ib-cr-producing Escherichia coli strain susceptible to ciprofloxacin in vitro. Antimicrob. Agents Chemother. 57, 5830–5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K.; Wachino J.; Suzuki S.; Kimura K.; Shibata N.; Kato H.; Shibayama K.; Konda T.; Arakawa Y. (2007) New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 51, 3354–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoir V.; Poirel L.; Nordmann P. (2008) Plasmid-mediated quinolone resistance pump QepA2 in an Escherichia coli isolate from France. Antimicrob. Agents Chemother. 52, 3801–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L. H.; Sorensen S. J.; Jorgensen H. S.; Jensen L. B. (2005) The prevalence of the OqxAB multidrug efflux pump amongst olaquindox-resistant Escherichia coli in pigs. Microb. Drug Resist. 11, 378–382. [DOI] [PubMed] [Google Scholar]
- Kim H. B.; Wang M.; Park C. H.; Kim E. C.; Jacoby G. A.; Hooper D. C. (2009) oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 3582–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Martinez L.; Pascual A.; Garcia I.; Tran J.; Jacoby G. A. (2003) Interaction of plasmid and host quinolone resistance. J. Antimicrob. Chemother. 51, 1037–1039. [DOI] [PubMed] [Google Scholar]
- Jacoby G. A. (2005) Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41(Suppl. 2), S120–S126. [DOI] [PubMed] [Google Scholar]
- Poole K. (2007) Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 39, 162–176. [DOI] [PubMed] [Google Scholar]
- Goldman J. D.; White D. G.; Levy S. B. (1996) Multiple antibiotic resistance (mar) locus protects Escherichia coli from rapid cell killing by fluoroquinolones. Antimicrob. Agents Chemother. 40, 1266–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.; Swick M. C.; Ledesma K. R.; Yang Z.; Hu M.; Zechiedrich L.; Tam V. H. (2012) Temporal interplay between efflux pumps and target mutations in development of antibiotic resistance in Escherichia coli. Antimicrob. Agents Chemother. 56, 1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T. P.; Ellsworth E. L.; Sanchez J. P.; Watson B. M.; Stier M. A.; Showalter H. D.; Domagala J. M.; Shapiro M. A.; Joannides E. T.; Gracheck S. J.; Nguyen D. Q.; Bird P.; Yip J.; Sharadendu A.; Ha C.; Ramezani S.; Wu X.; Singh R. (2007) Structure-activity relationships of 3-aminoquinazolinediones, a new class of bacterial type-2 topoisomerase (DNA gyrase and topo IV) inhibitors. Bioorg. Med. Chem. Lett. 17, 1312–1320. [DOI] [PubMed] [Google Scholar]
- German N.; Malik M.; Rosen J. D.; Drlica K.; Kerns R. J. (2008) Use of gyrase resistance mutants to guide selection of 8-methoxy-quinazoline-2,4-diones. Antimicrob. Agents Chemother. 52, 3915–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik M.; Marks K. R.; Mustaev A.; Zhao X.; Chavda K.; Kerns R. J.; Drlica K. (2011) Fluoroquinolone and quinazolinedione activities against wild-type and gyrase mutant strains of Mycobacterium smegmatis. Antimicrob. Agents Chemother. 55, 2335–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustaev A.; Malik M.; Zhao X.; Kurepina N.; Luan G.; Oppegard L. M.; Hiasa H.; Marks K. R.; Kerns R. J.; Berger J. M.; Drlica K. (2014) Fluoroquinolone-gyrase-DNA complexes: Two modes of drug binding. J. Biol. Chem. DOI: 10.1074/jbc.M113.529164. [DOI] [PMC free article] [PubMed] [Google Scholar]