Abstract
Genome analysis of the yeast Saccharomyces cerevisiae identified 68 genes encoding flavin-dependent proteins (1.1% of protein encoding genes) to which 47 distinct biochemical functions were assigned. The majority of flavoproteins operate in mitochondria where they participate in redox processes revolving around the transfer of electrons to the electron transport chain. In addition, we found that flavoenzymes play a central role in various aspects of iron metabolism, such as iron uptake, the biogenesis of iron–sulfur clusters and insertion of the heme cofactor into apocytochromes. Another important group of flavoenzymes is directly (Dus1-4p and Mto1p) or indirectly (Tyw1p) involved in reactions leading to tRNA-modifications. Despite the wealth of genetic information available for S. cerevisiae, we were surprised that many flavoproteins are poorly characterized biochemically. For example, the role of the yeast flavodoxins Pst2p, Rfs1p and Ycp4p with regard to their electron donor and acceptor is presently unknown. Similarly, the function of the heterodimeric Aim45p/Cir1p, which is homologous to the electron-transferring flavoproteins of higher eukaryotes, in electron transfer processes occurring in the mitochondrial matrix remains to be elucidated. This lack of information extends to the five membrane proteins involved in riboflavin or FAD transport as well as FMN and FAD homeostasis within the yeast cell. Nevertheless, several yeast flavoproteins, were identified as convenient model systems both in terms of their mechanism of action as well as structurally to improve our understanding of diseases caused by dysfunctional human flavoprotein orthologs.
Abbreviations: DHAP, dihydroxy acetone phosphate; DHBP, 3,4-dihydroxy-2-butanone-4-phosphate; DRAP, 2,5-diamino-6-(ribosylamino)-4-(3H)-pyrimidinone 5′-phosphate; ER, endoplasmic reticulum; ETC, electron transport chain; Gly3p, glycerol 3-phosphate; gluSA, γ-glutamic acid semialdehyde; Mia(40), mitochondrial intermembrane space import and assay/oxidoreductase 40; ORF, open reading frame; Q, ubiquionone
Keywords: Iron metabolism, Mitochondrion, Redox balance, tRNA-modifications, Membrane transporters
Highlights
-
•
Overview of flavin-dependent proteins in S. cerevisiae.
-
•
The role of yeast flavoproteins in iron metabolism.
-
•
Biosynthesis and transport of flavins.
-
•
Yeast as a model organism for investigating human diseases linked to flavoproteins.
1. Introduction to the history of Flavoprotein discovery
The yeast Saccharomyces cerevisiae played a central role in the discovery of flavoproteins as Otto Warburg and his collaborators were the first to isolate a “yellow ferment” from yeast cells [1]. Further studies by Theorell led to the concept of reversible association of co-enzyme and apo-enzyme to form the active holo-enzyme [2]. The isolation of other (new) yellow ferments from yeast prompted the renaming of the original ferment in to “old yellow ferment” (old yellow enzyme = OYE) [3]. Although it was demonstrated that OYE is reduced by NADPH [1], [4] the physiological electron accepting substrate(s) remains uncertain despite its reported role in the maintenance of the cytoskeleton [5]. This may have contributed to the persistent use of OYE instead of the official classification as NAD(P)H dehydrogenase (EC 1.6.99.1).
Despite the elusive nature of the physiological substrate(s), OYE rapidly developed in to an important model flavoenzyme culminating in the determination of the nucleotide sequence of oye2 and oye3 as well as the elucidation of its three-dimensional structure by X-ray crystallography [6], [7], [8]. The detailed biochemical characterisation of OYE also led to the identification of a number of artificial substrates, such as N-ethylmaleimide, cyclohex-2-enone and nitroolefins [8], [9], [10]. All of these substrates share a common structural motif consisting of an electron-withdrawing group (e.g. a carbonyl or nitro group) in α-position of a carbon–carbon double bond. The remarkably broad range of accepted substrates rendered OYE an ideal tool for biocatalytic applications exploited in numerous studies [11], [12], [13], [14], [15]. These efforts were further stimulated by the discovery of OYE homologs in many eubacteria as well as plant species in the 1990s [16], [17], [18], [19], [20], [21], [22], [23]. Plant OYE homologs were of particular interest because of their well-defined role in the biosynthesis of the plant hormone jasmonate, which plays a crucial role in the plant's defense response to pathogens [24], [25]. In all reported cases, the natural substrates exhibited the structural motif discovered in previous studies with yeast OYE. Hence, the yeast enzyme also became the paradigm for the class of “ene-reductases” now widely used for the synthesis of a variety of useful chemicals. Curiously, the broad range of activated “enes” accepted by OYEs as substrates is in stark contrast to its invoked physiological role as a reductase of disulphide bonds in oxidatively damaged proteins of the cytoskeleton, such as actin [5].
2. General aspects of the yeast flavoproteome
The yeast genome contains 68 genes encoding for a flavin-dependent protein and thus 1.1% of all yeast proteins (5885 protein-encoding genes [26]) have a requirement for either FMN or FAD. Owing to the presence of several flavoprotein families, which will be discussed further below, these 68 genes give rise to 47 defined biochemical roles. Thirty-five flavoproteins require FAD (74%) and fifteen require FMN (26%). Yeast also possesses three diflavin enzymes, which harbor both FMN and FAD (Table 1). The utilization of FMN and FAD in yeast flavoproteins is very similar to the distribution found in a global analysis across all kingdoms of life [27] and does not have the bias towards FAD as found for the human flavoproteome [28]. Covalent flavinylation, which is statistically found in ca. 10% of flavoproteins, is underrepresented in the yeast flavoproteome with only two enzymes, succinate dehydrogenase (Sdh1p) and l-arabinono-1,4-lactone oxidase (Alo1p) featuring a covalent bond between the N(3)-nitrogen of a histidine residue and the 8-methyl group of the isoalloxazine ring system (Table 1). Both of these enzymes operate in yeast mitochondria and are located in the inner (Sdh1p) and outer (Alo1p) membrane. The scarcity of covalent flavoproteins is linked to the relative absence of the structural clan FAD_PCMH (with the exception of Alo1p), which features many examples of mono- and even bi-covalent flavinylation [27], [28].
Table 1.
Yeast flavoproteins and genes.
| No. | E.C. | Enzyme | Cofactor | Structure clan (family)a | Localization | Abbrev. | Syst. name |
|---|---|---|---|---|---|---|---|
| 1 | 1.1.2.3 | l-Lactate:cytochrome c oxidoreductase (flavocytochrome b2) | FMN/heme | TIM_barrel (FMN_dh) | Mito. intermembr. sp. | cyb2 | YML054C |
| 2 | 1.1.2.4 | d-Lactate dehydrogenase | FAD/heme | – | I. mito. membr. | dld1 | YDL174C |
| Mito. matrix | dld2 | YDL178W | |||||
| Cytoplasm | dld3 | YEL071W | |||||
| 3 | 1.1.3.37 | d-Arabino-1,4-lactone oxidase | 8α-(N3-His) -FAD |
FAD_PCMH | O. mito. membr. | alo1 | YML086C |
| 4 | 1.1.5.3 | Glycerol-3-phosphate dehydrogenase | FAD | NADP_Rossmann (DAO) | I. mito. membr. | gut2 | YIL155C |
| 5 | 1.3.1.90 | tRNA dihydrouridine synthase | FMN | TIM_barrel (Dus) | Nucleus | dus1 | YML080W |
| Nucleus/cytoplasm | dus2 | YNR015W | |||||
| Nucleus/cytoplasm | dus3 | YLR401C | |||||
| – | dus4 | YLR405W | |||||
| 6 | 1.3.3.1 | Dihydroorotate dehydrogenase | FMN | TIM_barrel (DHO_dh) | Cytoplasm | ura1 | YKL216W |
| 7 | 1.3.3.4 | Protoporphyrinogen IX oxidase | FAD | NADP_Rossmann (Amino_oxidase) | I. mito. membr. | hem14 | YER014W |
| 8 | 1.3.3.6 | Acyl-CoA oxidase | FAD | Acyl-CoA_dh (ACOX, acyl-CoA_dh_1) | Peroxisome | pox1 | YGL205W |
| 9 | 1.3.5.1 | Succinate dehydrogenase | 8α-(N3-His) | NAPH_Rossmann | I. mito. membr. | sdh1 | YKL148C |
| Flavoprotein subunit A | -FAD/2Fe-2S/ | (FAD_binding_2) | sdh1b | YJL045W | |||
| – | Protein required for flavinylation of sdh | – | – | I. mito. membr. | emi5 | YOL071W | |
| 10 | 1.4.1.14 | NAD-dependent glutamate synthase | FMN/3Fe-4S | Glu_synthase/Glu_syn_central | Mito. matrix | glt1 | YDL171C |
| 11 | 1.4.3.5 | Pyridoxal 5′-phosphate oxidase | FMN | FMN-binding | Mito. intermembr. sp. | pdx3 | YBR035C |
| Pyridoxine 5′-phosphate oxidase | (Pyridox_oxidase) | ||||||
| 12 | 1.4.3.17 | Polyamine oxidase | FAD | NADP_Rossmann | cytoplasm | fms1 | YMR020W |
| (FAD_binding_2) | |||||||
| 13 | 1.5.1.20 | Methylenetetrahydrofolate reductase | FAD | FAD_oxidored (MTHFR) | – | met12 | YPL023C |
| Mito. | met13 | YGL125W | |||||
| 14 | 1.5.5.1 | Electron-transferring flavoprotein-ubiquinone oxidoreductase | FAD/4Fe–4S | 4Fe–4S (ETF_QO) | I. mito. membr. | cir2 | YOR356W |
| 15 | – | Electron transferring flavoprotein | FAD | FAD_DHS (ETF_alpha) | Mito. matrix | aim45 | YPR004C |
| 16 | 1.5.99.8 | Proline dehydrogenase | FAD | FAD_oxidored (Pro_dh) | Mito. matrix | put1 | YLR142W |
| 17 | 1.6.2.2 | Cytochrome-b5 reductase | FAD | FAD_Lum_binding | ER & o. mito. membr. | cbr1 | YIL043C |
| (FAD_binding_6) | ER & plasma membr. | pga3 | YML125C | ||||
| 18 | 1.6.2.4 | NADPH-hemoprotein reductase | FMN/heme | Flavoprotein (Flavodoxin_1) | O. mito. membr., | ncp1 | YHR042W |
| (cytochrome P450 reductase) | FAD | FAD_Lum_binding | ER & plasma membr. | ||||
| (FAD_binding_1) | |||||||
| 19 | 1.6.5.2 | NAD(P)H quinone oxidoreductase | FMN | Flavoprotein (Flavodoxin_2) | cytoplasm | lot6 | YLR011W |
| 20 | 1.6.5.9 | NADH-ubiquinone oxidoreductase (rotenone-insensitive) | FAD/Fe-S | NADP_Rossmann (Pyr_redox_2) | I. mito. membr. | ndi1 | YML120C |
| 21 | 1.6.99.1 | NADPH dehydrogenase | FMN | TIM_barrel (Oxidored_FMN) | Cytoplasm | oye2 | YHR179W |
| oye3 | YPL171C | ||||||
| 22 | 1.–.–.– | External NADH dehydrogenase | FAD | – | I. mito. membr. | nde1 | YMR145C |
| I. mito. membr. | nde2 | YDL085W | |||||
| 23 | 1.6.–.– | NADPH-dep. diflavin oxidoreductase | FMN | Flavoprotein (Flavodoxin_1) | Mito. matrix | tah18 | YPR048W |
| FAD | FAD_Lum_binding | ||||||
| (FAD_binding_1) | |||||||
| 24 | – | 5-Carboxymethylaminomethylation of uridine (heterodimer with Mss1p) | FAD | GIDA | Mito. | mto1 | YGL236C |
| 25 | – | Wybutosine biosynthesis, a tRNA-modification | FMN/4Fe–4S | Flavoprotein | ER | tyw1 | YPL207W |
| 26 | 1.8.1.2 | Sulphite reductase (beta subunit) | FMN/heme | Flavoprotein (Flavodoxin_1) | Cytoplasm | met5 | YJR137C |
| FAD | FAD_Lum_binding | ||||||
| (FAD_binding_1) | |||||||
| 27 | 1.8.1.4 | Dihydrolipoyl dehydrogenase | FAD | NADP_Rossmann | Mito. matrix | lpd1 | YFL018C |
| (Pyr_redox_2) | |||||||
| 28 | 1.8.1.7 | Glutathione-disulfide reductase | FAD | NADP_Rossmann (Pyr_redox_2) | Cytoplasm & mito. | glr1 | YPL091W |
| 29 | 1.8.1.9 | Thioredoxin-disulfide reductase | FAD | NADP_Rossmann (Pyr_redox_2) | Cytoplasm & mito. intermembr. sp. | trr1 | YDR353W |
| Cytoplasm & mito. | trr2 | YHR106W | |||||
| 30 | – | Microtubule associated protein | FAD | NADP_Rossmann? | cytoplasm | irc15 | YPL017C |
| (Pyr_redox_2) | microtubule | ||||||
| 31 | 1.8.3.2 | Sulfhydryl oxidase | FAD/Fe–S cluster/ | Erv1_Alr | Mito. intermembr. sp. | erv1 | YGR029W |
| Heme | ER membr. | erv2 | YPR037C | ||||
| 32 | 1.8.4.- | Endoplasmic oxidoreductin 1 | FAD | Ero1 | ER & ER membr.e | ero1 | YML130C |
| 33 | 1.14.12.17 | Nitric oxide oxidoreductase | FAD/heme | FAD_Lum_Binding | Cytoplasm | yhb1 | YGR234W |
| (flavohemoglobin) | (FAD_binding_6) | Mito. matrix | |||||
| 34 | 1.14.13.– | Oxidase of thiols in the ER | FAD | – | ER membr. | fmo1 | YHR176W |
| 35 | 1.14.13.9 | Kynurenine 3-monooxygenase | FAD | NADP_Rossmann (FAD_Binding_3) | O. mito. membr. | bna4 | YBL098W |
| 36 | 1.14.99.7 | Squalene monooxygenase | FAD | – | ER membr. | erg1 | YGR175C |
| 37 | 1.14.99.- | Monooxygenase in coenzyme Q biosyn. | FAD | – | I. mito. membr. | coq6 | YGR255C |
| 38 | 1.–.–.– | Ferric reductase | FAD/heme | – | Plasma membr. | fre1 | YLR214W |
| Plasma membr. | fre2 | YKL220C | |||||
| Plasma membr. | fre3 | YOR381W | |||||
| Plasma membr. | fre4 | YNR060W | |||||
| Plasma membr. | fre5 | YOR384W | |||||
| Vacuole membr. | fre6 | YLL051C | |||||
| Plasma membr. | fre7 | YOL152W | |||||
| – | fre8 | YLR047C | |||||
| 39 | 1.–.–.– | NADPH oxidase | FAD/heme | – | Perinucl. ER membr. | aim14 | YGL160W |
| 40 | 1.–.–.– | NAD(P)H-dep. heme reductase | FAD | – | Inner mito. membr. | cyc2 | YOR037W |
| 41 | 2.2.1.6 | Acetolactate synthase | FAD/TPP | FAD_DHS (TPP_enzyme_M) | Mito. | ilv2 | YMR108W |
| 42 | 2.3.1.86 | Fatty acid synthase, subunit β, chain I | FMN | Not reported | Cytoplasm & mito. | fas1 | YKL182W |
| 43 | 4.1.1.36 | 4′-Phosphopantothenoylcysteine decarboxylase (forms a heterotrimeric complex with Sis2p and Vhs3p) | FMN | Flavoprotein | Cytoplasm | cab3 | YKL088W |
| 44 | 4.1.99.3 | Deoxyribodipyrimidine photo-lyase | FAD | HUP (DNA_photolyase) | Cytoplasm, mito. & nucleus | phr1 | YOR386W |
| 45 | 4.2.3.5 | Chorismate synthase | FMN | Chorismate_synt | Cytoplasm | aro2 | YGL148W |
| 46 | – | Flavodoxin-like protein | FMN | Flavoprotein (Flavodoxin_1) | Cytoplasm, mito. | pst2 | YDR032C |
| – | Flavodoxin-like protein | FMN | Flavoprotein (Flavodoxin_1) | Cytoplasm | rfs1 | YBR052C | |
| – | flavodoxin-like protein | FMN | Flavoprotein (Flavodoxin_1) | Cytoplasm, mito. | ycp4 | YCR004C | |
| 47 | – | Apoptosis-inducing factor | FAD | – | O. mito. membr., plasma membr. & nucleus | aif1 | YNR074C |
Abbreviations used in Table 1: biosyn., biosynthesis; dh, dehydrogenase; degr., degradation; dep., dependent; i., inner; ER, endoplasmic reticulum; mito., mitochondrion; o., outer; perinucl., perinuclear; red., reductase; sp., space.
Pfam classification given in plain text is for yeast proteins and those in italics are for homologs from other species.
Structural information through X-ray crystallographic analysis is available for about one third of yeast flavoproteins listed in Table 1 (plain font in column “Structure clan/family”). In addition, the three-dimensional structure can be inferred from the known structure of homologs from other species (italics in column “Structure clan/family”). In some cases the structure of yeast flavoproteins served as paradigms for a family of enzymes, for example yeast OYEs [6], [29] and more recently kynurenine monooxygenase [30]. The general difficulties to elucidate the structure of integral or membrane associated proteins is also seen for flavoproteins (see Table 1).
Table 1 also provides information on the localisation of flavoproteins in the yeast cell. More than half of yeast flavoproteins (36 entries in Table 1) operate in the mitochondrion. Many of these are directly participating in redox reactions connected to the electron transport chain (ETC) (see also next section). Seventeen flavoproteins are located in the cytoplasm and only a few in the nucleus (Dus1-4p, Phr1p, Aif1p), endoplasmic reticulum (Aim14p, Cbr1p, Erg1p, Ero1p, Erv2p, Fmo1p, Ncp1p, Pga3p, Tyw1p) or the peroxisome (Pox1p). Requirement for the same flavoenzyme activity in different cellular compartments is either satisfied by expression of isozymes (e.g. Dld1-3p are found in either the cytoplasm, the inner mitochondrial membrane or the matrix) or the same flavoenzyme is present in multiple compartments (e.g. Cbr1p, Pga3p, Ncp1p, Trr1/2p, Yhb1p, Fas1p, Phr1p, Aif1p).
3. Flavoproteins, the stewards of iron
A remarkable result of our analysis concerns the multi-layered relationship between flavins and iron. As discussed in more detail in the next section, flavin-dependent ferric reductases (Fre1p-8p) are essential for reduction of ferric iron (and copper), which is prerequisite for iron (and copper) uptake by yeast transporters (permeases). Moreover, in several flavoproteins the flavin is responsible for the reduction of either heme iron or an iron–sulfur cluster (Table 1 and Scheme 1). Prominent examples include succinate dehydrogenase (Sdh1p), (Aim1p), nitric oxide oxidoreductase and l-lactate:cytochrome c oxido reductase (flavocytochrome b2). In addition to this intramolecular electron transfer, two yeast flavoproteins, Cyc2p and Tah18p, are involved in intermolecular electron transfer. In the case of Cyc2p, which is located in the inner mitochondrial membrane with its FAD-containing active site exposed to the intermembrane space, the enzyme reduces Fe(III) and participates in the incorporation of the heme prosthetic group into apocytochrome c and c1 [31], [32]. Similarly, the diflavin reductase Tah18p forms a complex with Dre2p and provides electrons derived from NADPH to support the biosynthesis of two iron–sulfur clusters [33]. Recently, Tah18p was reported to be involved in NO generation in yeast and hence it is conceivable that it has more clients than Dre2p [34]. Thus flavoproteins fulfill various crucial tasks ranging from iron uptake, delivery of electrons to the mitochondrial electron transport chain, reduction of cytochrome-dependent reductases, biogenesis of iron–sulfur clusters and insertion of the heme cofactor into apocytochromes.
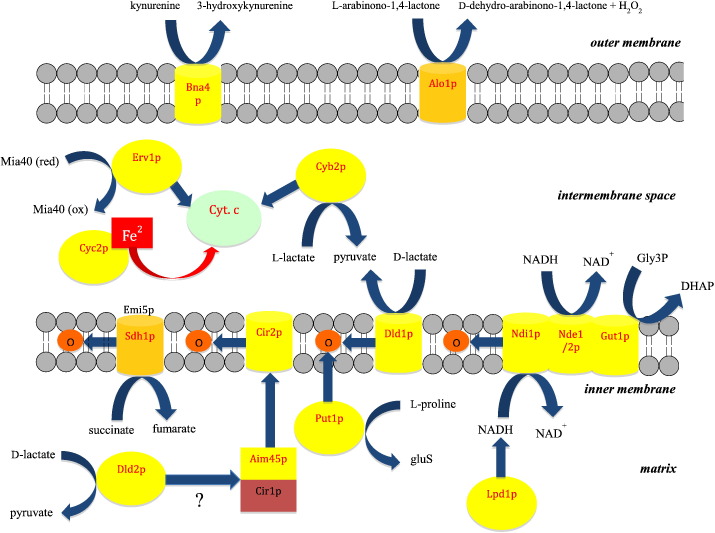
Scheme 1.
Flavoproteins in mitochondrial redox processes.
Flavoproteins are represented by yellow spheres (soluble flavoproteins in the matrix or intermembrane space) or barrels (inner or outer mitochondrial membrane). Flavoproteins with a covalently bound FAD (Sdh1p in the inner and Alo1p in the outer membrane) are shown in light orange. Cytochrome c is shown as a light green sphere. Curved blue arrows indicate redox reactions and straight arrows electron transfer. For further explanations and comments see main text. Several flavoproteins appear to participate in a multi-protein complex in the inner mitochondrial membrane [57]. For clarity, we have shown only a complex consisting of Ndi1p, Nde1/2p and Gut1p. Abbreviations used are: DHAP, dihydroxy acetone phosphate; Gly3p, glycerol 3-phosphate; GluSA, γ-glutamic acid semialdehyde; Mia(40), mitochondrial intermembrane space import and assay/oxidoreductase 40 (ox, oxidized; red, reduced); Q, ubiquionone.
4. Flavoprotein families in yeast
As mentioned above, the yeast flavoproteome contains several families of flavoproteins, which catalyze identical or similar reactions. The largest group are the ferric/cupric reductases encoded by fre1-8 (see Table 1, entry 38). Fre1 and fre2 are metalloregulated by either iron or copper availability and the encoded metalloreductases reduce Fe(III) and Cu(II) at the expense of NADPH [35]. In addition to fre1 and fre2, the yeast genome contains six homologous genes, termed fre3-8. Fre3-6 are regulated by iron whereas fre7 is copper-regulated [36], [37]. Fre8 and the homologous aim14 are not regulated by iron or copper suggesting a different role for these proteins [36]. Recently, it was demonstrated that aim14 encodes an NADPH-oxidase, which produces superoxide in the endoplasmatic reticulum [38] and it is thus conceivable that fre8 also encodes an enzyme with similar properties. This notion is also supported by pair-wise sequence alignments showing the highest identity (30.6%) and similarity (56.5%) on the amino acid level between fre8 and aim14 within this family of flavoproteins [36]. Again, these functions highlight the importance of flavoenzymes for iron uptake and reduction as introduced in the previous section.
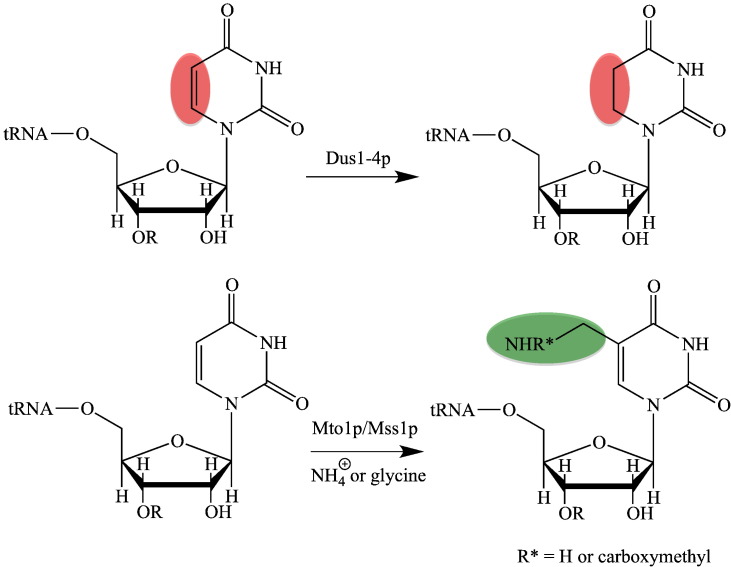
The second largest family comprise four tRNA-dihydrouridine synthases encoded by dus1-4 (Table 1, entry 5). The reduction of uracils to dihydrouridines in tRNA is one of the most common modifications of nucleosides in tRNA in all kingdoms of life [39]. A recent mechanistic study employing Dus2p revealed that reduction of uracil to dihydrouracil (see Scheme 2, top) is promoted by other tRNA modifications suggesting that tRNA maturation may occur in an ordered fashion [40]. Cytoplasmic tRNA in S. cerevisiae contains dihydrouridine in the D loop at positions 16, 17, 20, 20A, 20B and at the base of the variable arm at position 47. The four yeast enzymes exclusively reduce uracils in specific positions in tRNA: Dus1p reduces uracils in positions 16 and 17, Dus2p in position 20, Dus3p in position 47 and Dus4p in positions 20a and 20b. Thus these four enzymes are sufficient to generate all dihydrouridine modifications known in yeast [41]. In humans, only one dihydrouridine synthase homologous to yeast Dus2p (42% identity) was identified so far, which reduces uracil in tRNA for phenylalanine [28], [42]. The human enzyme appears to be upregulated in malignant tissues resulting in higher levels of dihydrouridine [42]. Despite its putative role in malignancy the specificity and exact role of the human enzyme remains unclear.
Scheme 2.
Flavoproteins in tRNA-modification.
Top, reaction catalyzed by tRNA-dihydrouridine synthase (Dus1-4p); Bottom, reaction catalyzed by Mto1p/Mss1p. Depending on the co-substrate, ammonia or glycine, the side chain in position 5′ of the uracil base is aminomethyl or carboxymethylaminomethyl. “R” represents the next 3′-nucleotide in the tRNA molecule.
In addition to reduction of uracil, the flavoenzyme Mto1p (also termed GidA) is involved in the biosynthesis of modifications at the C5-position of the uracil base in tRNAs [43], [44]. Depending on the nitrogen source, ammonia or glycine, this reaction leads to the formation of either 5-aminomethyl- or 5-carboxymethylaminomethyluridine (Scheme 2, bottom). This modification occurs at the wobble position in mitochondrial tRNAs for lysine, glutamate and glutamine [44]. Detailed characterization of the bacterial protein complex of MnmE and MnmG (homolog of Mto1p) led to a mechanistic proposal in which the FAD-dependent MnmG serves a dual function during the reaction [43], [45]. In this model, methylene tetrahydrofolate bound to MnmE reacts with either ammonia or glycine to form a methylene amino group at N-5 of the tetrahydrofolate cofactor. Then, FAD oxidizes the carbon–nitrogen bond to yield an imine, which is then nucleophilically attacked by the uracil base of the tRNA substrate. In the next step the reduced FAD transfers a hydride to the imine to reduce the carbon–nitrogen double bond thus completing the biosynthesis of the C-5 side chain (see Scheme 2). Thus MnmG combines two canonical flavin-dependent reactions – oxidation of amines and reduction of double bonds – to catalyze the biosynthesis of the amino methyl or carboxymethylaminomethyl side chain.
Recently, yet another flavin-dependent enzyme encoded by tyw1 (Table 1, entry 25) was discovered that catalyzes the second step in the biosynthesis of wybutosine-modified tRNA [46]. This enzyme belongs to the radical SAM superfamily characterized by the presence of a [4Fe–4S] cluster and a S-adenosylmethionine (SAM) domain [47]. Catalytic activity requires reduction of the [4Fe–4S] cluster in order to initiate one-electron transfer for reductive cleavage of SAM to generate the 5′-deoxyadenosyl radical [48]. In vivo, flavodoxins or ferredoxins might act as potential electron donors to convert [4Fe–4S]2 + to [4Fe–4S]+ and therefore it is conceivable that the N-terminal flavodoxin domain of Tyw1p relays electrons from an external electron donor such as NAD(P)H or an electron transfer protein to the [4Fe–4S] cluster. Such a functional role is supported by the finding that deletion of the flavodoxin domain abolishes TYW1p activity [49].
Interestingly, the yeast genome contains three homologous genes, pst1, rfs1 and ycp4 encoding three highly similar flavodoxin-like proteins (Table 1, entry 46) [50]. Although none of the proteins was functionally characterized with respect to their electron transfer properties and physiological redox partners they were found to act as transcriptional regulators of spi1, a gene responding to various environmental stimuli [51]. The lack of information on yeast flavodoxins is very surprising in view of the abundance of structural (148 structures of wild-type and variants in the pdb) and biochemical studies available for bacterial flavodoxins. Therefore, the current state of affairs for yeast flavodoxin-like proteins is very unsatisfactory and clearly warrants further investigations to define their biochemical and structural properties.
The family of d-lactate dehydrogenases comprising three enzymes, Dld1-3p, will be discussed in the context of redox processes in the next section.
5. Yeast flavoproteins in redox balancing
More than a quarter of yeast flavoproteins listed in Table 1 participate in redox reactions in the mitochondrion. As shown in Scheme 1, transfer of electrons into the ETC can either occur through electron donation to cytochrome c (cyt. c) in the intermembrane space or directly by reduction of ubiquinone to ubiquinol in the inner mitochondrial membrane. The latter route is clearly the dominating process in yeast mitochondria. Electrons transferred to NAD+ in the isocitrate, α-ketoglutarate and malate dehydrogenase reactions of the tricarboxylic acid cycle enter the ETC. via the NADH:ubiquinone oxidoreductase Ndi1p (“internal NADH dehydrogenase”). In contrast to complex I of higher eukaryotes this membrane-bound enzyme does not engage in proton translocation resulting in lower phosphorylation efficiency. The oxidation of succinate to fumarate is catalyzed by a canonical membrane-bound succinate dehydrogenase in which the covalently linked FAD becomes reduced by the substrate and the electrons are passed on to ubiquinone via an iron–sulfur cluster and heme relay system. The Sdh1p subunit of yeast succinate dehydrogenase (complex II) is one of only two flavin-dependent proteins exhibiting a covalent linkage (see Table 1, entry 9). Typically, covalent flavinylation is a spontaneous co- or posttranslational process. However, in the case of Sdh1p the assistance of Emi1p (Sdh5p) is required, which is conserved in higher eukaryotes and hence appears to be essential for complex II assembly [52], [53]. Yeast also possesses a heterodimeric electron transfer flavoprotein (Aim45p/Cir1p) located in the mitochondrial matrix, which communicates with a membrane-bound electron transferring flavoprotein ubiquinone oxidoreductase (Cir2p). The latter flavoprotein feeds electrons received from Aim45p/Cir1p into the ETC. The clients for Aim45p/Cir1p, however, remain elusive as most electron donor proteins, such as the acyl-Co dehydrogenases involved in β-oxidation or amino acid degradation, are not present in S. cerevisiae. A potential candidate, l-proline dehydrogenase (Put1p), evidently feeds electrons from l-proline oxidation directly into the ETC. by reduction of ubiquinone to ubiquinol (see Scheme 1) [54].
Cytosolic NADH generated for example by the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase is oxidized by either Nde1p or Nde2p (“external NADH:ubiquinone oxidoreductases”) and the electrons serve to reduce ubiquinone (Scheme 1). The mitochondrial ETC. can also be fuelled by the glycerol 3-phosphate shuttle: glycerol is first phosphorylated by glycerol kinase (Gut1p) in the cytosol and then transported to the intermembrane space to become oxidized by the membrane-bound glycerol 3-phosphate dehydrogenase (Gut2p) [55], [56]. Several of the membrane-bound flavoproteins involved in substrate oxidation and electron transfer form a large supramolecular complex containing Nde1p, Nde2p and Gut2p and therefore inter-protein electron transfer may also occur prior to ubiquinone reduction [57].
Yeast possesses three d-lactate dehydrogenases (Dldp1-3, see Table 1), which operate in different compartments of the cell: Dld1p is located in the inner mitochondrial membrane, Dld2p in the matrix and Ddl3 in the cytosol [58], [59]. d-lactate is produced by the glyoxalase pathway that detoxifies methylglyoxal adventitiously generated by non-enzymatic elimination of hydrogen and phosphate from the enediol intermediate of triose phosphates [60]. Since this detoxification pathway is active in the cytosol and the mitochondrial matrix d-lactate dehydrogenase activity is required in these compartments to oxidize d-lactate to pyruvate (Scheme 1). Oxidation of d-lactate by the d-lactate dehydrogenase (Dld2p) localized in the mitochondrial matrix is coupled to ATP synthesis and therefore Dld2p apparently donates substrate-derived electrons to the ETC [61]. However, it is currently unknown whether electrons are directly used to reduce ubiquinone or transferred to the heterodimeric Aim45p/Cir2p electron transfer complex (Scheme 1).
The involvement of flavoproteins in central mitochondrial redox processes is also reflected by the fact that the flavin oxidation state oscillates in synchronized aerobically grown yeast cultures. During the oxidative phase of the culture the increase of flavin fluorescence indicates that more flavoproteins become oxidized whereas in the reductive phase of the internal rhythm a decrease of flavin fluorescence indicates a shift to the reduced state [62], [63].
6. Flavin biosynthesis and transport
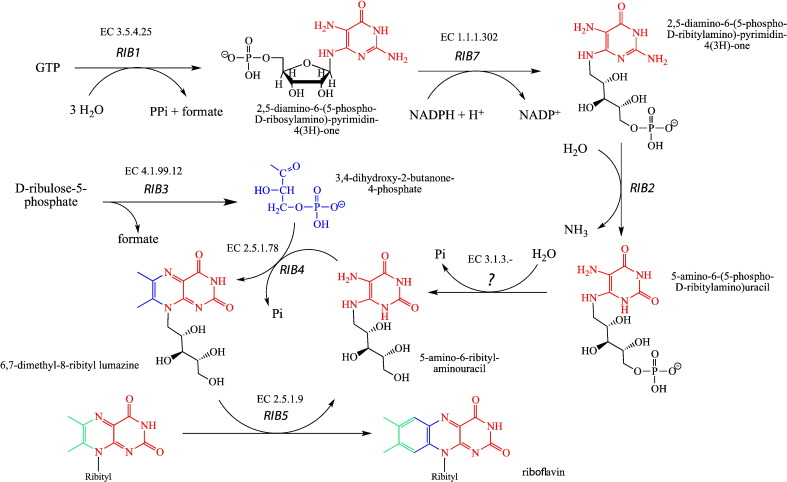
The biosynthesis of riboflavin in S. cerevisiae utilizes the canonical precursors, GTP and ribulose 5-phosphate. However, riboflavin biosynthesis deviates from the bacterial pathway in that deamination and reduction of the initial metabolite, 2,5-diamino-6-(ribosylamino)-4-(3H)-pyrimidinone 5′-phosphate (DRAP), take place in reverse order (www.kegg.jp). Briefly, in the first step GTP is converted by GTP cyclohydrolase II, encoded by rib1, to DRAP, which is reduced by Rib7p to 2,5-diamino-6-(ribitylamino)-4-(3H)-pyrimidinone 5′-phosphate (Table 2 and Scheme 3). This reaction is followed by deamination to 5-amino-6-ribitylamino-2,4-(1H,3H)-pyrimidinedione 5′-phosphate catalyzed by Rib2p [64]. After dephosphorylation by an unidentified phosphatase condensation with 3,4-dihydroxy-2-butanone-4-phosphate (DHAB) occurs. The latter metabolite is synthesized from ribulose 5-phosphate by DHBP synthase (encoded by rib3). The condensation reaction is catalyzed by lumazine synthase (encoded by rib4) and yields 6,7-dimethyl-8-(1-d-ribityl)lumazine [65], [66]. In the final reaction, riboflavin synthase (encoded by rib5) uses two molecules of 6,7-dimethyl-8-(1-d-ribityl)-lumazine where one acts as donor and the other as acceptor of four carbon atoms leading to the generation of the isoalloxazine ring of one molecule of riboflavin [67]. The coenzyme forms of riboflavin, FMN and FAD, are synthesized from riboflavin by riboflavin kinase (Fmn1p) and FAD synthetase (Fad1p), respectively [68], [69].
Table 2.
Yeast flavin transporters and biosynthesis.
| No. | E.C. | Protein/enzyme | Substrate/ligand | Structure clan (family)a | Abbrev. | Syst. name |
|---|---|---|---|---|---|---|
| Transporters | ||||||
| 1 | – | FAD transmembrane transporter | FAD | – | flx1 | YIL134W |
| 2 | – | FAD transporter (into ER) | FAD | – | flc1 | YPL221W |
| 3 | – | FAD transporter (into ER) | FAD | – | flc2 | YAL053W |
| 4 | – | FAD transporter (into ER) | FAD | – | flc3 | YGL139W |
| 5 | – | Plasma-membrane riboflavin transporter | Riboflavin | – | mch5 | YOR306C |
| Biosynthesis of riboflavin, FMN and FAD | ||||||
| 1 | 3.5.4.25 | GTP cyclohydrolase II (1st step) | – | GTP cyclohydrolase II | rib1 | YBL033C |
| 2 | 1.1.1.302 | DRAP reductase (2nd step) | – | DHFred (RibD_C) | rib7 | YBR153W |
| 3 | – | Deaminase (3rd step) | – | DHFred (RibD_C) | rib2 | YOL066C |
| 4 | 4.1.99.12 | DHBP synthase (4th step) | – | DHBP_synthase | rib3 | YDR487C |
| 5 | 2.5.1.78 | Lumazine synthase (5th step) | – | DMRL_synthase | rib4 | YOL143C |
| 6 | 2.5.1.9 | Riboflavin synthase (6th step) | – | FAD_Lum_binding (Lum_binding) | rib5 | YBR256C |
| 7 | 2.7.1.26 | Riboflavin kinase | Riboflavin | Flavokinase | fmn1 | YDR236C |
| 8 | 2.7.7.2 | FAD-adenylyl transferase (synthetase) | FMN | HUP (PAPS_reduct) | fad1 | YDL045C |
Abbreviations used in Table 2 are: DHBP, 3,4-dihydroxy-2-butanone-4-phosphate; DRAP, 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate.
Pfam classification given in plain text is for yeast proteins and those in italics are for homologs from other species.
Scheme 3.
Biosynthesis of riboflavin in S. cerevisiae.
GTP and d-ribulose-5-phosphate serve as building blocks for the biosynthesis of 5-amino-6-ribityl-aminouracil and 3,4-dihydroxy-2-butanone-4-phosphate, respectively. These two compounds are then used by Rib4p to synthesize 6,7-dimethyl-8-ribityl lumazine. Two molecules 6,7-dimethyl-8-ribityl lumazine areconverted by Rib5p to riboflavin and 5-amino-6-ribityl-aminouracil which serves again as substrate for Rib4p. This way all atoms of the dimethylbenzene moiety are derived from 3,4-dihydroxy-2-butanone-4-phosphate (colored in blue and green) while remainder of riboflavin is derived from GTP (colored in red).
In addition to de novo biosynthesis, yeast is also capable of riboflavin uptake from the medium and it was shown that a plasma membrane flavin transporter, encoded by mch5, is regulated by the proline-dependent transcription factor Put3p [70], [71]. Since proline utilization depends on the FAD-dependent proline dehydrogenase Put1p (Table 1) upregulation of Mch5p suggests that riboflavin uptake is necessary under these conditions to meet the cellular demand for flavin coenzymes.
Despite the wealth of genetic and biochemical information available on riboflavin biosynthesis in the cytosol, transport to other compartments, in particular the mitochondrion as the dominant organelle for flavoenzyme catalyzed reactions, remains controversial [53]. Based on the finding that yeast mitochondria possess riboflavin kinase but no FAD synthetase activity, Tzagaloff et al. [72] proposed a model according to which the carrier protein Flx1p acts as a “flavin antiporter” by exchanging FMN from the mitochondrial matrix with FAD from the cytosol. In contrast to this model, Barile and coworkers claim that riboflavin is transported into mitochondria where both FMN and FAD can be synthesized and are even exported back to the “extramitochondrial phase” [73], [74]. More recent data from Pallotta indicated that mitochondria can also hydrolyze FAD and FMN to riboflavin and are thus capable of balancing the pools of riboflavin, FMN and FAD [75]. Yeast FAD synthetase (Fad1p) is essential and deletion of fad1 makes yeast unviable. The localization of yeast FAD1p is still unclear, although recent studies on human FAD synthetase isoform 1 (hFADS1) suggest a mitochondrial localization in eukaryotes [76].
In addition to flx1 and mch5, yeast possesses three flc genes encoding putative transporters of flavins (Table 2). These transporters are responsible for FAD transport into the endoplasmic reticulum (ER) where several flavoenzymes (e. g. Ero1p, Erv2p and Fmo1p) are involved in the redox balance of thiols and disulfide linkages [77]. However, the exact role and localisation of Flc1-3p in yeast are currently not fully understood.
An alternative mechanism for assembling the holo-flavoenzyme is realized for the sole peroxisomal flavoenzyme, acyl-CoA oxidase (Pox1p). In this case, the holo-enzyme is formed in the cytosol, then binds to the import receptor Pex5p and, following an unknown import pathway, is transported into the peroxisome [78], [79].
7. Yeast flavoproteins as models for human diseases
The yeast S. cerevisiae has been used as a model organism for studying fundamental biological processes for some time [80]. In 1997, Botstein and colleagues showed that nearly 31% of yeast open reading frames (ORF) have a homologue in mammalian genomes [81]. Since the number of annotated ORFs has almost doubled since 1997 this percentage is likely to have risen significantly. Moreover, an estimated 30% of human genes implicated in human diseases have a yeast homologue [82]. In a recent review, we have documented that fifty human flavoproteins are implicated in human diseases [28]. As shown in Table 3, nearly half of the disease-related human flavoproteins possess a yeast homologue. Interestingly, the majority of disease-related human flavoproteins operate in the mitochondrion [28]. Owing to the similarity of mitochondrial processes in eukaryotes it is conceivable that the yeast homologs located in mitochondria (see Table 3) may be particularly suitable as models for an improved understanding of human mitochondrial diseases.
Table 3.
Yeast flavoproteins as human disease models.
| No. | E.C. | Human enzyme | Disease | OMIM | Yeast homolog | E valuea |
|---|---|---|---|---|---|---|
| 1 | 1.1.5.3 | Glycerol 3-phosphate dh | Diabetes mellitus, type 2 | 138430 | Gut2p | 7.3 e − 124 |
| 2 | 1.1.99- | d-2-Hydroxyglutarate dh | d-2-Hydroxyglutaric aciduria | 605176 | Dld1p | 1.9 e − 39 |
| Dld2p | 8.7 e − 128 | |||||
| Dld3p | 3.3 e − 112 | |||||
| 3 | 1.3.1.2 | Dihydropyrimidine dh | Deficiency | 612779 | Glt1p | 2.7 e − 14 |
| 4 | 1.3.5.2 | Dihydroorotate dh | Miller syn. | 126064 | Ura1p | 2.0 e − 6 |
| 5 | 1.3.3.4 | Protoporphyrinongen IX ox. | Variegate porphyria | 600923 | Hem1p | 7.2 e − 20 |
| 6 | 1.3.3.6 | Acyl-CoA ox. | Deficiency | 609751 | Pox1p | 2.0 e − 45 |
| 7 | 1.3.5.1 | Succinate dh | Complex II deficiency, | 600857 | Sdh1p | 3.9 e − 219 |
| Flavoprotein subunit A | Leigh syn., paraganglioma 5 | Sdh1bp | 3.0 e − 214 | |||
| 8 | 1.4.3.4 | Monoamine ox | Brunner syn., antisocial behavior, autism | 309850 | Fms1p | 7.8 e − 11 |
| 9 | 1.4.3.5 | Pyridoxine 5′-phosphate ox. | Encephalopathy | 603287 | Pdx3p | 5.1 e − 36 |
| 10 | 1.5.1.20 | Methylenetetrahydofolate red. | Homocystinuria, neural tube | 607093 | Met12p | 2.9 e − 98 |
| Defects, schizophrenia | Met13p | 7.5 e − 122 | ||||
| 11 | 1.5.5.1 | Electron-transferring flavo-protein ubiquinone oxidored. | Glutaric academia IIC | 231675 | Cir2p | 8.0 e − 157 |
| 12 | – | Electron-transferring flavoprot. | Glutaric acidemia IIA | 608053 | Aim45p | 8.5 e − 66 |
| Glutaric acidemia IIB | 130410 | |||||
| 13 | 1.5.99.8 | Proline dh | Hyperprolinemia type I, schizophrenia | 606810 | Put1p | 8.7 e − 12 |
| 14 | 1.6.2.2 | Cytochrome-b5 red. | Methemoglobinemia types I & II | 613213 | Cbr1p | 1.9 e − 30 |
| 15 | 1.6.2.4 | NADPH-hemoprotein red. (cytochrome P450 red.) | Antley–Bixler syn., | 124015 | Ncp1p | 2.4 e − 86 |
| 16 | 1.6.5.2 | NAD(P)H:quinone oxidored. | Benzene toxicity, breast cancer | 125860 | Lot6p | 1 |
| 17 | 1.8.1.4 | Dihydrolipoyl dh | Leigh syn., maple syrup urine disease | 238331 | Lpd1p | 2.8 e − 147 |
| 18 | 1.8.1.7 | Glutathione-disulfide red. | Hemolytic anemia | 138300 | Glr1p | 1.4 e − 104 |
| 19 | 1.14.13.8 | Flavin-containing monooxy. | Trimethylaminuria | 136132 | Fmo1p | 4.8 e − 27 |
| 20 | 1.14.13.39 | Nitric-oxide synthase | Hypertension | 163729 | Tah18p | 9.6 e − 27 |
| 163730 | ||||||
| 21 | 1.14.99.- | Monooxy. in coenzyme Q | Deficiency, nephrotic syn. | 614647 | Coq6p | 7.0 e − 55 |
| 22 | 1.16.1.8 | Methionine synthase red. | Homocystinuria, neural tube | 602568 | Met5p | 0.24 |
| 23 | 2.5.1.26 | Alkyldihydroxyacetone | Rhizomelic chondrodysplasia | 603051 | Dld1p | 2.1 e − 25 |
| Phosphate synthase | Punctata type 3 | Dld2p | 4.4 e − 16 | |||
| Dld3p | 1.1 e − 15 | |||||
| 24 | – | Apoptosis inducing protein | Combined oxidative phosphorylation deficiency | 300169 | Aif1p | 8.8 e − 4 |
Abbreviations used in Table 3: dh, dehydrogenase; flavoprot., flavoprotein; monooxy., monooxygenase; ox, oxidase; oxidored., oxidoreductase; red., reductase; syn., syndrome.
E value; expect value, was generated by searching of Saccharomyces Genome Database (SGD; http://www.yeastgenome.org) open reading frames (DNA or protein) against human protein sequences using the SGD WU_Blast2 program.
Functional assignments based on sequence similarity generated ambiguities for several flavoproteins. For example, the yeast d-lactate dehydrogenases (Dld1-3p) show remarkable similarity to human d-2-hydroxyglutarate dehydrogenase and to a much lesser degree to alkyldihydroxyacetone phosphate synthase. Similarly, the yeast glutamate synthase, Glt1p, exhibits similarity to human dihydropyrimidine dehydrogenase (see Table 3). On the other hand the yeast NAD(P)H:quinone oxidoreductase Lot6p shows only a very low similarity to the human ortholog (P = 1) although it possesses a similar structure and function [83], [84], [85], [86]. These examples illustrate the need for biochemical characterization to provide a solid basis for comparative functional studies.
Yeast deletion strains were also used as convenient models to investigate the impact of mutations discovered in human genes. Examples are deletions of the genes ura1, sdh1, ldp1 and coq6, leading to auxotrophic yeast strains, which were complemented with the orthologous human gene to investigate the functional impairment of mutations [87], [88], [89], [90]. Yeast was also utilized as a host for heterologous expression of the human gene encoding 3β-hydroxysterol Δ24-reductase (DHCR24). Desmosterolosis, a rare autosomal recessive disorder is caused by mutations in the gene encoding DHCR24. Heterologous expression of the human DHCR24 gene bearing different missense mutations confirmed their role in desmosterolosis [91].
A genetic screen in yeast suggested that kynurenine 3-monooxygenase may be a useful therapeutic target for Huntington disease [92]. This has prompted structural studies with the yeast enzyme leading to the elucidation of its X-ray crystal structure, which may serve as a model to investigate the structural basis of inhibitor binding [30]. Similarly, the crystal structure of the yeast flavin-containing monooxygenase Fmo1p proved useful as a model to understand the effect of mutations in human FMO3 that cause trimethylaminuria (“fish-odor” syndrome) [93].
8. Concluding remarks
Our analysis of the yeast flavoproteome has highlighted the importance of flavin-dependent enzymes in mitochondrial redox processes. Many of these mitochondrial enzymes have human homologs involved in diseases and thus genetically manipulated yeast strains (e.g. gene deletions) have potential as convenient model systems. On the other hand, many yeast flavoenzymes are barely characterized with regard to their biochemical properties, such as substrate specificity, kinetic parameters and reaction partners. This deficit is clearly illustrated by the yeast flavodoxin-like proteins and the electron-transferring flavoprotein, none of which were characterized in any biochemical or structural detail. Similarly, our understanding of riboflavin uptake and trafficking between cellular compartments as well as flavin homeostasis are at an early stage necessitating further studies. Since these processes are also poorly understood in humans, yeast lends itself as a valuable model organism to gain insight into uptake, transport and trafficking of this vital vitamin.
9. Methods
The names and gene abbreviations of yeast flavoproteins were recently compiled for a review article [27]. This list of flavoproteins was updated using the information available in the Saccharomyces genome database (http://www.yeastgenome.org/). This database was also used to extract information on viability of gene knock-outs and localisation of flavoproteins in the yeast cell. Structural information was obtained from the protein database (http://www.pdb.org).
Acknowledgments
We thank the Austrian Research Fund (FWF) for financial support through project P22361 and the PhD program “Molecular Enzymology” (W901).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Warburg O., Christian W. Über das gelbe Ferment und seine Wirkungen. Biochem. Z. 1933;266:377–392. [Google Scholar]
- 2.Theorell H., Warburg O. Reindarstellung (Kristallisation) des gelben Atmungsfermentes und die reversible Spaltung desselben. Biochem. Z. 1934;272:155–156. [Google Scholar]
- 3.Haas E., Warburg O. Isolierung eines neuen gelben Ferments. Biochem. Z. 1938;298:378–390. [Google Scholar]
- 4.Massey V., Schopfer L.M. Reactivity of Old Yellow Enzyme with alpha-NADPH and other pyridine nucleotide derivatives. J. Biol. Chem. 1986;261:1215–1222. [PubMed] [Google Scholar]
- 5.Haarer B.K., Amberg D.C. Old Yellow Enzyme protects the actin cytoskeleton from oxidative stress. Mol. Biol. Cell. 2004;15:4522–4531. doi: 10.1091/mbc.E04-06-0445. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Fox K.M., Karplus P.A. Old Yellow Enzyme at 2 A resolution: overall structure, ligand binding, and comparison with related flavoproteins. Structure. 1994;2:1089–1105. [PubMed] [Google Scholar]
- 7.Niino Y.S., Chakraborty S., Brown B.J., Massey V. A new Old Yellow Enzyme of Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:1983–1991. doi: 10.1074/jbc.270.5.1983. [DOI] [PubMed] [Google Scholar]
- 8.Stott K., Saito K., Thiele D.J., Massey V. Old Yellow Enzyme. The discovery of multiple isozymes and a family of related proteins. J. Biol. Chem. 1993;268:6097–6106. [PubMed] [Google Scholar]
- 9.Meah Y., Massey V. Old Yellow Enzyme: stepwise reduction of nitro-olefins and catalysis of aci-nitro tautomerization. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10733–10738. doi: 10.1073/pnas.190345597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaz A.D., Chakraborty S., Massey V. Old Yellow Enzyme: aromatization of cyclic enones and the mechanism of a novel dismutation reaction. Biochemistry. 1995;34:4246–4256. doi: 10.1021/bi00013a014. [DOI] [PubMed] [Google Scholar]
- 11.Hall M., Stueckler C., Kroutil W., Macheroux P., Faber K. Asymmetric bioreduction of activated alkenes using cloned 12-oxophytodienoate reductase isoenzymes OPR-1 and OPR-3 from Lycopersicon esculentum (tomato): a striking change of stereoselectivity. Angew. Chem. Int. Ed. 2007;46:3934–3937. doi: 10.1002/anie.200605168. [DOI] [PubMed] [Google Scholar]
- 12.Muller A., Sturmer R., Hauer B., Rosche B. Stereospecific alkyne reduction: novel activity of Old Yellow Enzymes. Angew. Chem. Int. Ed. 2007;46:3316–3318. doi: 10.1002/anie.200605179. [DOI] [PubMed] [Google Scholar]
- 13.Stuermer R., Hauer B., Hall M., Faber K. Asymmetric bioreduction of activated C C bonds using enoate reductases from the Old Yellow Enzyme family. Curr. Opin. Chem. Biol. 2007;11:203–213. [Google Scholar]
- 14.Williams R.E., Bruce N.C. ‘New uses for an old enzyme’ — the Old Yellow Enzyme family of flavoenzymes. Microbiology. 2002;148:1607–1614. doi: 10.1099/00221287-148-6-1607. [DOI] [PubMed] [Google Scholar]
- 15.Williams R.E., Rathbone D.A., Scrutton N.S., Bruce N.C. Biotransformation of explosives by the Old Yellow Enzyme family of flavoproteins. Appl. Environ. Microbiol. 2004;70:3566–3574. doi: 10.1128/AEM.70.6.3566-3574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaller F., Weiler E.W. Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana. Structural and functional relationship to yeast Old Yellow Enzyme. J. Biol. Chem. 1997;272:28066–28072. doi: 10.1074/jbc.272.44.28066. [DOI] [PubMed] [Google Scholar]
- 17.Strassner J., Fürholz A., Macheroux P., Amrhein N., Schaller A. A homolog of Old Yellow Enzyme in tomato: Spectral properties and substrate specificity of the recombinant protein. J. Biol. Chem. 1999;274:35067–35073. doi: 10.1074/jbc.274.49.35067. [DOI] [PubMed] [Google Scholar]
- 18.French C.E., Bruce N.C. Bacterial morphinone reductase is related to Old Yellow Enzyme. Biochem. J. 1995;312:671–678. doi: 10.1042/bj3120671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitzpatrick T.B., Amrhein N., Macheroux P. Characterization of YqjM, an Old Yellow Enzyme homolog from Bacillus subtilis. J. Biol. Chem. 2003;278:19891–19897. doi: 10.1074/jbc.M211778200. [DOI] [PubMed] [Google Scholar]
- 20.Adachi O., Matsushita K., Shinagawa E., Ameyama M. Occurrence of old yellow enzyme in Gluconobacter suboxydans, and the cyclic regeneration of NADP. J. Biochem. 1979;86:699–709. doi: 10.1093/oxfordjournals.jbchem.a132574. [DOI] [PubMed] [Google Scholar]
- 21.French C.E., Nicklin S., Bruce N.C. Sequence and properties of pentaerythritol tetranitrate reductase from Enterobacter cloacae PB2. J. Bacteriol. 1996;178:6623–6627. doi: 10.1128/jb.178.22.6623-6627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snape J.R., Walkley N.A., Morby A.P., Nicklin S., White G.F. Purification, properties, and sequence of glycerol trinitrate reductase from Agrobacterium radiobacter. J. Bacteriol. 1997;179:7796–7802. doi: 10.1128/jb.179.24.7796-7802.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blehert D.S., Fox B.G., Chambliss G.H. Cloning and sequence analysis of two Pseudomonas flavoprotein xenobiotic reductases. J. Bacteriol. 1999;181:6254–6263. doi: 10.1128/jb.181.20.6254-6263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strassner J., Schaller F., Frick U., Howe G.A., Weiler E.E., Amrhein N., Macheroux P., Schaller A. Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J. 2002;32:585–601. doi: 10.1046/j.1365-313x.2002.01449.x. [DOI] [PubMed] [Google Scholar]
- 25.Stintzi A., Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10625–10630. doi: 10.1073/pnas.190264497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Louis E.J., Mewes H.W., Murakami Y., Philippsen P., Tettelin H., Oliver S.G. Life with 6000 genes. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. (563–547) [DOI] [PubMed] [Google Scholar]
- 27.Macheroux P., Kappes B., Ealick S.E. Flavogenomics—a genomic and structural view of flavin-dependent proteins. FEBS J. 2011;278:2625–2634. doi: 10.1111/j.1742-4658.2011.08202.x. [DOI] [PubMed] [Google Scholar]
- 28.Lienhart W.D., Gudipati V., Macheroux P. The human flavoproteome. Arch. Biochem. Biophys. 2013;535:150–162. doi: 10.1016/j.abb.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox K.M., Karplus P.A. Crystallization of Old Yellow Enzyme illustrates an effective strategy for increasing protein crystal size. J. Mol. Biol. 1993;234:502–507. doi: 10.1006/jmbi.1993.1604. [DOI] [PubMed] [Google Scholar]
- 30.Amaral M., Levy C., Heyes D.J., Lafite P., Outeiro T.F., Giorgini F., Leys D., Scrutton N.S. Structural basis of kynurenine 3-monooxygenase inhibition. Nature. 2013;496:382–385. doi: 10.1038/nature12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernard D.G., Quevillon-Cheruel S., Merchant S., Guiard B., Hamel P.P. Cyc2p, a membrane-bound flavoprotein involved in the maturation of mitochondrial c-type cytochromes. J. Biol. Chem. 2005;280:39852–39859. doi: 10.1074/jbc.M508574200. [DOI] [PubMed] [Google Scholar]
- 32.Corvest V., Murrey D.A., Hirasawa M., Knaff D.B., Guiard B., Hamel P.P. The flavoprotein Cyc2p, a mitochondrial cytochrome c assembly factor, is a NAD(P)H-dependent haem reductase. Mol. Microbiol. 2012;83:968–980. doi: 10.1111/j.1365-2958.2012.07981.x. [DOI] [PubMed] [Google Scholar]
- 33.Soler N., Delagoutte E., Miron S., Facca C., Baille D., d'Autreaux B., Craescu G., Frapart Y.M., Mansuy D., Baldacci G., Huang M.E., Vernis L. Interaction between the reductase Tah18 and highly conserved Fe–S containing Dre2 C-terminus is essential for yeast viability. Mol. Microbiol. 2011;82:54–67. doi: 10.1111/j.1365-2958.2011.07788.x. [DOI] [PubMed] [Google Scholar]
- 34.Nishimura A., Kawahara N., Takagi H. The flavoprotein Tah18-dependent NO synthesis confers high-temperature stress tolerance on yeast cells. Biochem. Biophys. Res. Commun. 2013;430:137–143. doi: 10.1016/j.bbrc.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 35.Dancis A., Roman D.G., Anderson G.J., Hinnebusch A.G., Klausner R.D. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc. Natl. Acad. Sci. U. S. A. 1992;89:3869–3873. doi: 10.1073/pnas.89.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georgatsou E., Alexandraki D. Regulated expression of the Saccharomyces cerevisiae Fre1p/Fre2p Fe/Cu reductase related genes. Yeast. 1999;15:573–584. doi: 10.1002/(SICI)1097-0061(199905)15:7<573::AID-YEA404>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 37.Martins L.J., Jensen L.T., Simon J.R., Keller G.L., Winge D.R. Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:23716–23721. doi: 10.1074/jbc.273.37.23716. [DOI] [PubMed] [Google Scholar]
- 38.Rinnerthaler M., Buttner S., Laun P., Heeren G., Felder T.K., Klinger H., Weinberger M., Stolze K., Grousl T., Hasek J., Benada O., Frydlova I., Klocker A., Simon-Nobbe B., Jansko B., Breitenbach-Koller H., Eisenberg T., Gourlay C.W., Madeo F., Burhans W.C., Breitenbach M. Yno1p/Aim14p, a NADPH-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8658–8663. doi: 10.1073/pnas.1201629109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sprinzl M., Steegborn C., Hubel F., Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1996;24:68–72. doi: 10.1093/nar/24.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rider L.W., Ottosen M.B., Gattis S.G., Palfey B.A. Mechanism of dihydrouridine synthase 2 from yeast and the importance of modifications for efficient tRNA reduction. J. Biol. Chem. 2009;284:10324–10333. doi: 10.1074/jbc.M806137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xing F., Hiley S.L., Hughes T.R., Phizicky E.M. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J. Biol. Chem. 2004;279:17850–17860. doi: 10.1074/jbc.M401221200. [DOI] [PubMed] [Google Scholar]
- 42.Kato T., Daigo Y., Hayama S., Ishikawa N., Yamabuki T., Ito T., Miyamoto M., Kondo S., Nakamura Y. A novel human tRNA-dihydrouridine synthase involved in pulmonary carcinogenesis. Cancer Res. 2005;65:5638–5646. doi: 10.1158/0008-5472.CAN-05-0600. [DOI] [PubMed] [Google Scholar]
- 43.Armengod M.E., Moukadiri I., Prado S., Ruiz-Partida R., Benitez-Paez A., Villarroya M., Lomas R., Garzon M.J., Martinez-Zamora A., Meseguer S., Navarro-Gonzalez C. Enzymology of tRNA modification in the bacterial MnmEG pathway. Biochimie. 2012;94:1510–1520. doi: 10.1016/j.biochi.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 44.Wang X., Yan Q., Guan M.X. Mutation in MTO1 involved in tRNA modification impairs mitochondrial RNA metabolism in the yeast Saccharomyces cerevisiae. Mitochondrion. 2009;9:180–185. doi: 10.1016/j.mito.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moukadiri I., Prado S., Piera J., Velazquez-Campoy A., Bjork G.R., Armengod M.E. Evolutionarily conserved proteins MnmE and GidA catalyze the formation of two methyluridine derivatives at tRNA wobble positions. Nucleic Acids Res. 2009;37:7177–7193. doi: 10.1093/nar/gkp762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L., Jia X., Ward D.M., Kaplan J. Yap5 protein-regulated transcription of the TYW1 gene protects yeast from high iron toxicity. J. Biol. Chem. 2011;286:38488–38497. doi: 10.1074/jbc.M111.286666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sofia H.J., Chen G., Hetzler B.G., Reyes-Spindola J.F., Miller N.E. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S.C., Frey P.A. S-adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem. Sci. 2007;32:101–110. doi: 10.1016/j.tibs.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki Y., Noma A., Suzuki T., Senda M., Senda T., Ishitani R., Nureki O. Crystal structure of the radical SAM enzyme catalyzing tricyclic modified base formation in tRNA. J. Mol. Biol. 2007;372:1204–1214. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 50.Grandori R., Carey J. Six new candidate members of the alpha/beta twisted open-sheet family detected by sequence similarity to flavodoxin. Protein Sci. 1994;3:2185–2193. doi: 10.1002/pro.5560031204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cardona F., Orozco H., Friant S., Aranda A., del Olmo M. The Saccharomyces cerevisiae flavodoxin-like proteins Ycp4 and Rfs1 play a role in stress response and in the regulation of genes related to metabolism. Arch. Microbiol. 2011;193:515–525. doi: 10.1007/s00203-011-0696-7. [DOI] [PubMed] [Google Scholar]
- 52.Hao H.X., Khalimonchuk O., Schraders M., Dephoure N., Bayley J.P., Kunst H., Devilee P., Cremers C.W., Schiffman J.D., Bentz B.G., Gygi S.P., Winge D.R., Kremer H., Rutter J. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim H.J., Winge D.R. Emerging concepts in the flavinylation of succinate dehydrogenase. Biochim. Biophys. Acta. 2013;1827:627–636. doi: 10.1016/j.bbabio.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopes J., Pinto M.J., Rodrigues A., Vasconcelos F., Oliveira R. The Saccharomyces cerevisiae genes, AIM45, YGR207c/CIR1 and YOR356w/CIR2, Are Involved in Cellular Redox State Under Stress Conditions. Open Microbiol. J. 2010;4:75–82. doi: 10.2174/1874285801004010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ansell R., Granath K., Hohmann S., Thevelein J.M., Adler L. The two isoenzymes for yeast NAD +-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J. 1997;16:2179–2187. doi: 10.1093/emboj/16.9.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grauslund M., Lopes J.M., Ronnow B. Expression of GUT1, which encodes glycerol kinase in Saccharomyces cerevisiae, is controlled by the positive regulators Adr1p, Ino2p and Ino4p and the negative regulator Opi1p in a carbon source-dependent fashion. Nucleic Acids Res. 1999;27:4391–4398. doi: 10.1093/nar/27.22.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grandier-Vazeille X., Bathany K., Chaignepain S., Camougrand N., Manon S., Schmitter J.M. Yeast mitochondrial dehydrogenases are associated in a supramolecular complex. Biochemistry. 2001;40:9758–9769. doi: 10.1021/bi010277r. [DOI] [PubMed] [Google Scholar]
- 58.Chelstowska A., Liu Z., Jia Y., Amberg D., Butow R.A. Signalling between mitochondria and the nucleus regulates the expression of a new d-lactate dehydrogenase activity in yeast. Yeast. 1999;15:1377–1391. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1377::AID-YEA473>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 59.Rojo E.E., Guiard B., Neupert W., Stuart R.A. Sorting of d-lactate dehydrogenase to the inner membrane of mitochondria. Analysis of topogenic signal and energetic requirements. J. Biol. Chem. 1998;273:8040–8047. doi: 10.1074/jbc.273.14.8040. [DOI] [PubMed] [Google Scholar]
- 60.Penninckx M.J., Jaspers C.J., Legrain M.J. The glutathione-dependent glyoxalase pathway in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1983;258:6030–6036. [PubMed] [Google Scholar]
- 61.Pallotta M.L., Valenti D., Iacovino M., Passarella S. Two separate pathways for d-lactate oxidation by Saccharomyces cerevisiae mitochondria which differ in energy production and carrier involvement. Biochim. Biophys. Acta. 2004;1608:104–113. doi: 10.1016/j.bbabio.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 62.Murray D.B., Haynes K., Tomita M. Redox regulation in respiring Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2011;1810:945–958. doi: 10.1016/j.bbagen.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 63.Murray D.B., Roller S., Kuriyama H., Lloyd D. Clock control of ultradian respiratory oscillation found during yeast continuous culture. J. Bacteriol. 2001;183:7253–7259. doi: 10.1128/JB.183.24.7253-7259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Urban A., Ansmant I., Motorin Y. Optimisation of expression and purification of the recombinant Yol066 (Rib2) protein from Saccharomyces cerevisiae. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003;786:187–195. doi: 10.1016/s1570-0232(02)00742-0. [DOI] [PubMed] [Google Scholar]
- 65.Jin C., Barrientos A., Tzagoloff A. Yeast dihydroxybutanone phosphate synthase, an enzyme of the riboflavin biosynthetic pathway, has a second unrelated function in expression of mitochondrial respiration. J. Biol. Chem. 2003;278:14698–14703. doi: 10.1074/jbc.M300593200. [DOI] [PubMed] [Google Scholar]
- 66.Garcia-Ramirez J.J., Santos M.A., Revuelta J.L. The Saccharomyces cerevisiae RIB4 gene codes for 6,7-dimethyl-8-ribityllumazine synthase involved in riboflavin biosynthesis. Molecular characterization of the gene and purification of the encoded protein. J. Biol. Chem. 1995;270:23801–23807. doi: 10.1074/jbc.270.40.23801. [DOI] [PubMed] [Google Scholar]
- 67.Santos M.A., Garcia-Ramirez J.J., Revuelta J.L. Riboflavin biosynthesis in Saccharomyces cerevisiae. Cloning, characterization, and expression of the RIB5 gene encoding riboflavin synthase. J. Biol. Chem. 1995;270:437–444. doi: 10.1074/jbc.270.1.437. [DOI] [PubMed] [Google Scholar]
- 68.Santos M.A., Jimenez A., Revuelta J.L. Molecular characterization of FMN1, the structural gene for the monofunctional flavokinase of Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:28618–28624. doi: 10.1074/jbc.M004621200. [DOI] [PubMed] [Google Scholar]
- 69.Wu M., Repetto B., Glerum D.M., Tzagoloff A. Cloning and characterization of FAD1, the structural gene for flavin adenine dinucleotide synthetase of Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:264–271. doi: 10.1128/mcb.15.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reihl P., Stolz J. The monocarboxylate transporter homolog Mch5p catalyzes riboflavin (vitamin B2) uptake in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:39809–39817. doi: 10.1074/jbc.M505002200. [DOI] [PubMed] [Google Scholar]
- 71.Spitzner A., Perzlmaier A.F., Geillinger K.E., Reihl P., Stolz J. The proline-dependent transcription factor Put3 regulates the expression of the riboflavin transporter MCH5 in Saccharomyces cerevisiae. Genetics. 2008;180:2007–2017. doi: 10.1534/genetics.108.094458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tzagoloff A., Jang J., Glerum D.M., Wu M. FLX1 codes for a carrier protein involved in maintaining a proper balance of flavin nucleotides in yeast mitochondria. J. Biol. Chem. 1996;271:7392–7397. doi: 10.1074/jbc.271.13.7392. [DOI] [PubMed] [Google Scholar]
- 73.Pallotta M.L., Brizio C., Fratianni A., De Virgilio C., Barile M., Passarella S. Saccharomyces cerevisiae mitochondria can synthesise FMN and FAD from externally added riboflavin and export them to the extramitochondrial phase. FEBS Lett. 1998;428:245–249. doi: 10.1016/s0014-5793(98)00544-4. [DOI] [PubMed] [Google Scholar]
- 74.Bafunno V., Giancaspero T.A., Brizio C., Bufano D., Passarella S., Boles E., Barile M. Riboflavin uptake and FAD synthesis in Saccharomyces cerevisiae mitochondria: involvement of the Flx1p carrier in FAD export. J. Biol. Chem. 2004;279:95–102. doi: 10.1074/jbc.M308230200. [DOI] [PubMed] [Google Scholar]
- 75.Pallotta M.L. Evidence for the presence of a FAD pyrophosphatase and a FMN phosphohydrolase in yeast mitochondria: a possible role in flavin homeostasis. Yeast. 2011;28:693–705. doi: 10.1002/yea.1897. [DOI] [PubMed] [Google Scholar]
- 76.Torchetti E.M., Brizio C., Colella M., Galluccio M., Giancaspero T.A., Indiveri C., Roberti M., Barile M. Mitochondrial localization of human FAD synthetase isoform 1. Mitochondrion. 2010;10:263–273. doi: 10.1016/j.mito.2009.12.149. [DOI] [PubMed] [Google Scholar]
- 77.Protchenko O., Rodriguez-Suarez R., Androphy R., Bussey H., Philpott C.C. A screen for genes of heme uptake identifies the FLC family required for import of FAD into the endoplasmic reticulum. J. Biol. Chem. 2006;281:21445–21457. doi: 10.1074/jbc.M512812200. [DOI] [PubMed] [Google Scholar]
- 78.Klein A.T., van den Berg M., Bottger G., Tabak H.F., Distel B. Saccharomyces cerevisiae acyl-CoA oxidase follows a novel, non-PTS1, import pathway into peroxisomes that is dependent on Pex5p. J. Biol. Chem. 2002;277:25011–25019. doi: 10.1074/jbc.M203254200. [DOI] [PubMed] [Google Scholar]
- 79.Subramani S. Hitchhiking fads en route to peroxisomes. J. Cell Biol. 2002;156:415–417. doi: 10.1083/jcb.200112122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Drubin D. The yeast Saccharomyces cerevisiae as a model organism for the cytoskeleton and cell biology. Cell Motil. Cytoskeleton. 1989;14:42–49. doi: 10.1002/cm.970140110. [DOI] [PubMed] [Google Scholar]
- 81.Botstein D., Chervitz S.A., Cherry J.M. Yeast as a model organism. Science. 1997;277:1259–1260. doi: 10.1126/science.277.5330.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Foury F. Human genetic diseases: a cross-talk between man and yeast. Gene. 1997;195:1–10. doi: 10.1016/s0378-1119(97)00140-6. [DOI] [PubMed] [Google Scholar]
- 83.Li R., Bianchet M.A., Talalay P., Amzel L.M. The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: Mechanism of the two-electron reduction. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8846–8850. doi: 10.1073/pnas.92.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liger D., Graille M., Zhou C.-Z., Leulliot N., Quevillon-Cheruel S., Blondeau K., Janin J., van Tilbeurgh H. Crystal structure and functional characterization of yeast YLR011wp, an enzyme with NAD(P)H-FMN and ferric iron reductase activities. J. Biol. Chem. 2004;279:34890–34897. doi: 10.1074/jbc.M405404200. [DOI] [PubMed] [Google Scholar]
- 85.Sollner S., Nebauer R., Ehammer H., Prem A., Deller S., Palfey B.A., Daum G., Macheroux P. Lot6p from Saccharomyces cerevisiae is a FMN-dependent reductase with a potential role in quinone detoxification. FEBS J. 2007;274:1328–1339. doi: 10.1111/j.1742-4658.2007.05682.x. [DOI] [PubMed] [Google Scholar]
- 86.Sollner S., Macheroux P. New roles of flavoproteins in molecular cell biology: an unexpected role for quinone reductases as regulators of proteasomal degradation. FEBS J. 2009;276:4313–4324. doi: 10.1111/j.1742-4658.2009.07143.x. [DOI] [PubMed] [Google Scholar]
- 87.Burnichon N., Briere J.J., Libe R., Vescovo L., Riviere J., Tissier F., Jouanno E., Jeunemaitre X., Benit P., Tzagoloff A., Rustin P., Bertherat J., Favier J., Gimenez-Roqueplo A.P. SDHA is a tumor suppressor gene causing paraganglioma. Hum. Mol. Genet. 2010;19:3011–3020. doi: 10.1093/hmg/ddq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heeringa S.F., Chernin G., Chaki M., Zhou W., Sloan A.J., Ji Z., Xie L.X., Salviati L., Hurd T.W., Vega-Warner V., Killen P.D., Raphael Y., Ashraf S., Ovunc B., Schoeb D.S., McLaughlin H.M., Airik R., Vlangos C.N., Gbadegesin R., Hinkes B., Saisawat P., Trevisson E., Doimo M., Casarin A., Pertegato V., Giorgi G., Prokisch H., Rotig A., Nurnberg G., Becker C., Wang S., Ozaltin F., Topaloglu R., Bakkaloglu A., Bakkaloglu S.A., Muller D., Beissert A., Mir S., Berdeli A., Varpizen S., Zenker M., Matejas V., Santos-Ocana C., Navas P., Kusakabe T., Kispert A., Akman S., Soliman N.A., Krick S., Mundel P., Reiser J., Nurnberg P., Clarke C.F., Wiggins R.C., Faul C., Hildebrandt F. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J. Clin. Invest. 2011;121:2013–2024. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rainger J., Bengani H., Campbell L., Anderson E., Sokhi K., Lam W., Riess A., Ansari M., Smithson S., Lees M., Mercer C., McKenzie K., Lengfeld T., Gener Querol B., Branney P., McKay S., Morrison H., Medina B., Robertson M., Kohlhase J., Gordon C., Kirk J., Wieczorek D., Fitzpatrick D.R. Miller (Genee-Wiedemann) syndrome represents a clinically and biochemically distinct subgroup of postaxial acrofacial dysostosis associated with partial deficiency of DHODH. Hum. Mol. Genet. 2012;21:3969–3983. doi: 10.1093/hmg/dds218. [DOI] [PubMed] [Google Scholar]
- 90.Vaubel R.A., Rustin P., Isaya G. Mutations in the dimer interface of dihydrolipoamide dehydrogenase promote site-specific oxidative damages in yeast and human cells. J. Biol. Chem. 2011;286:40232–40245. doi: 10.1074/jbc.M111.274415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Waterham H.R., Koster J., Romeijn G.J., Hennekam R.C., Vreken P., Andersson H.C., FitzPatrick D.R., Kelley R.I., Wanders R.J. Mutations in the 3beta-hydroxysterol Delta24-reductase gene cause desmosterolosis, an autosomal recessive disorder of cholesterol biosynthesis. Am. J. Hum. Genet. 2001;69:685–694. doi: 10.1086/323473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giorgini F., Guidetti P., Nguyen Q., Bennett S.C., Muchowski P.J. A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nat. Genet. 2005;37:526–531. doi: 10.1038/ng1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yeung C.K., Adman E.T., Rettie A.E. Functional characterization of genetic variants of human FMO3 associated with trimethylaminuria. Arch. Biochem. Biophys. 2007;464:251–259. doi: 10.1016/j.abb.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]