Significance
Identifying proteins localized on the surface and envelope of Gram-negative bacterial cells is an important problem in vaccine development and antibiotic target discovery. We show that the characterization of proteins associated with outer membrane vesicles (OMVs) released by Gram-negative cells provides a solution in that contamination with abundant cytoplasmic proteins (caused by cell lysis) can be avoided. Integrated at a systems level with other transcription and proteomic data sets, our research provides a view of the surface architecture of a pathogen undergoing host-programmed changes in gene expression. Also provided is the first evidence to our knowledge that secreted protein-folding quality control (a property of the DegP protease) influences the composition of OMVs and bacterial virulence, validating DegP as a target for virulence-blocking drugs.
Keywords: HtrA family, in-solution digestion, biofilm formation, CTXϕ phage
Abstract
Outer membrane vesicles (OMVs) produced by Gram-negative bacteria provide an interesting research material for defining cell-envelope proteins without experimental cell disruption. OMVs are also promising immunogenic platforms and may play important roles in bacterial survival and pathogenesis. We used in-solution trypsin digestion coupled to mass spectrometry to identify 90 proteins present in OMVs of Vibrio cholerae when grown under conditions that activate the TCP pilus virulence regulatory protein (ToxT) virulence regulon. The ToxT expression profile and potential contribution to virulence of these proteins were assessed using ToxT and in vivo RNA-seq, Tn-seq, and cholera stool proteomic and other genome-wide data sets. Thirteen OMV-associated proteins appear to be essential for cell growth, and therefore may represent antibacterial drug targets. Another 12 nonessential OMV proteins, including DegP protease, were required for intestinal colonization in rabbits. Comparative proteomics of a degP mutant revealed the importance of DegP in the incorporation of nine proteins into OMVs, including ones involved in biofilm matrix formation and various substrates of the type II secretion system. Taken together, these results suggest that DegP plays an important role in determining the content of OMVs and also affects phenotypes such as intestinal colonization, proper function of the type II secretion system, and formation of biofilm matrix.
The Gram-negative bacterium Vibrio cholerae is the etiologic agent of cholera, an acute and often fatal diarrheal disease (1, 2). The 2010–2012 epidemic in Haiti provides proof that this devastating disease remains an ongoing public health threat (3). Strains belonging to the O1 serogroup of V. cholerae are the major cause of epidemic and pandemic cholera, and these isolates can be further classified as belonging to either the classical or the El Tor biotypes (4, 5). Although classical biotype V. cholerae strains are thought to have caused the first six pandemics, strains of the seventh pandemic El Tor biotype have now become dominant since their emergence ∼50 y ago (6). Virulence gene expression by classical and El Tor biotype strains requires different in vitro growth conditions, and these parameters may reflect signals that exist within the human small intestine (7, 8).
V. cholerae virulence gene expression is controlled by a transcriptional regulatory cascade that includes TCP pilus virulence regulatory protein (ToxT), a positive regulatory protein (9) as well as a small regulatory RNA and cyclic dinucleotides (10, 11). Recent studies have shown that sodium bicarbonate can induce virulence gene expression by enhancing ToxT-activated gene expression particularly in El Tor strains (12, 13). ToxT controls the expression of genes for cholera toxin (CTX) and toxin-coregulated pili (TCP), which are encoded by the bacteriophage CTXϕ (14) and the TCP chromosomal island, respectively (15). Interestingly, small molecules that target ToxT are promising candidates for V. cholerae specific antivirulence drugs (16, 17). Recently emerged strains of V. cholerae isolated from Bangladesh and Haiti express much more cholera toxin and TcpA (major pilin subunit that plays a significant role in microcolony formation by enabling pilus–pilus interactions) under laboratory conditions (18, 19). However, in vitro conditions that allow clinical El Tor isolates to express morphologically polymerized TCP pili that can also function as receptors for CTXϕ phage have not been reported.
Gram-negative bacteria, including V. cholerae, use different types of secretion systems to transport important virulence factors to the cell envelope and the extracellular milieu. Outer membrane vesicles (OMVs) may also serve a function analogous to secretion systems in that they provide a means to transport envelope proteins beyond the cell surface (20–22). Unlike other secretion systems, OMVs can carry insoluble membrane proteins, proteolytically unstable enzymes, and other nonprotein molecules (e.g., innate immune agonists such as lipopolysaccharide), all within particles that may stabilize and concentrate them until they can interact with host cell receptors. The surface of OMVs is thought to reflect the outer membrane composition of the bacterial cells whereas the lumen is predicted to contain mainly periplasmic components. Because some of these proteins have critical roles in host colonization, immune evasion, nutrient uptake, and tissue damage, it is thought that OMV production may be a property that is important to pathogenesis (23, 24). Recently, it was reported that toxins produced by Escherichia coli and V. cholerae were carried by native OMVs (25, 26). Additionally, OMVs may provide a survival advantage between competing species by virture of their bacteriocidal activity (27–29). OMVs also show promise as vaccine antigen platforms, given their composition and physico-chemical properties (30–34). For example, OMVs of V. cholerae have been shown to induce protective immunity in experimental animals (35–41). Although these immunization studies demonstrate the potential of OMVs as novel vaccine immunogens, such investigations have not been fully integrated into a comprehensive proteomic analysis of the OMV protein content.
Various approaches have been used to characterize the protein content of OMVs from different human pathogens, including 1- and 2-dimensional electrophoresis (1-DE and 2-DE) (30, 32, 33) coupled with mass spectrometry (MS) (25, 31, 34). Liquid chromatography coupled with MS (LC-MS/MS) has also been applied to define proteins associated with OMVs (42) but only in a limited fashion and never fully coupled with systematic genetic analysis for the role of OMV proteins in virulence or bacterial growth and survival.
In this study, we analyzed the protein content of OMVs derived from V. cholerae El Tor strain C6706 grown under conditions that active the ToxT regulon. By this approach, we identified 90 proteins, most of which are predicted to be outer membrane and periplasmic proteins. We have determined the extent of overlap between this group of OMV proteins with those encoded by genes that are (i) required for in vitro growth, (ii) required for efficient intestinal colonization of experimental animals, and (iii) known to be expressed in vivo by V. cholerae cells recovered from cholera patients and infected animals or activated by overexpression of the virulence regulator ToxT. This systematic approach identified a subset of interesting genes including VC0566, which encodes a DegP ortholog (43, 44). We show that DegP is required for the secretion of biofilm matrix components and certain substrates of the type II secretion system. Furthermore, the activity of this enzyme strongly influences biofilm formation of V. cholerae, thereby associating the action of this enzyme with a process that is known to influence infectivity and the intestinal colonization process.
Results
Sodium Bicarbonate Treatment of El Tor C6706 Strain Causes in Vitro TCP Formation.
To induce virulence gene expression and enrich the encoded proteins in the OMVs of V. cholerae El Tor C6706 strain, we used a recently described sodium bicarbonate induction method (45). To determine whether this method, a derivative of the earlier “AKI” method of Iwanaga et al. (46), activated ToxT gene and protein expression, we assessed assembly of TCP pili. These organelles are presumably expressed by El Tor strains in vivo, given that CTXϕ can infect such strains during intestinal infections (14) and that gene expression studies show strong up-regulation of TCP genes in both rabbit and mouse infections using El Tor strain C6706 (40, 47).
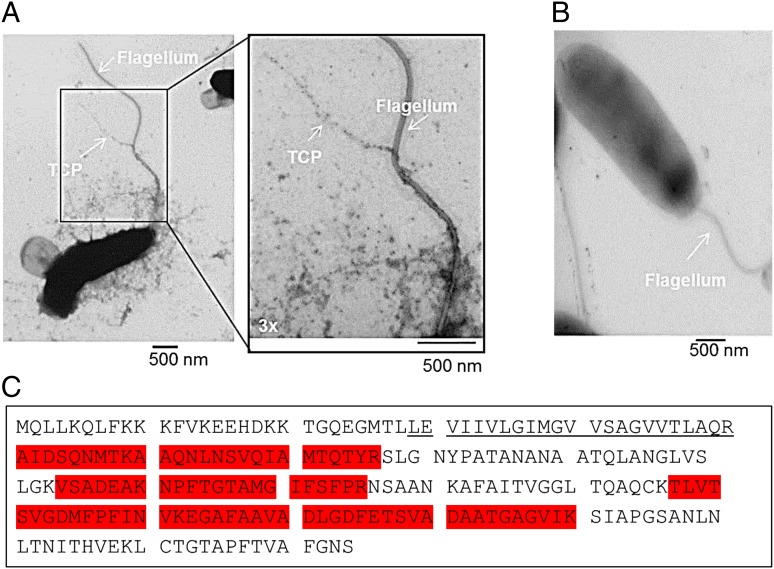
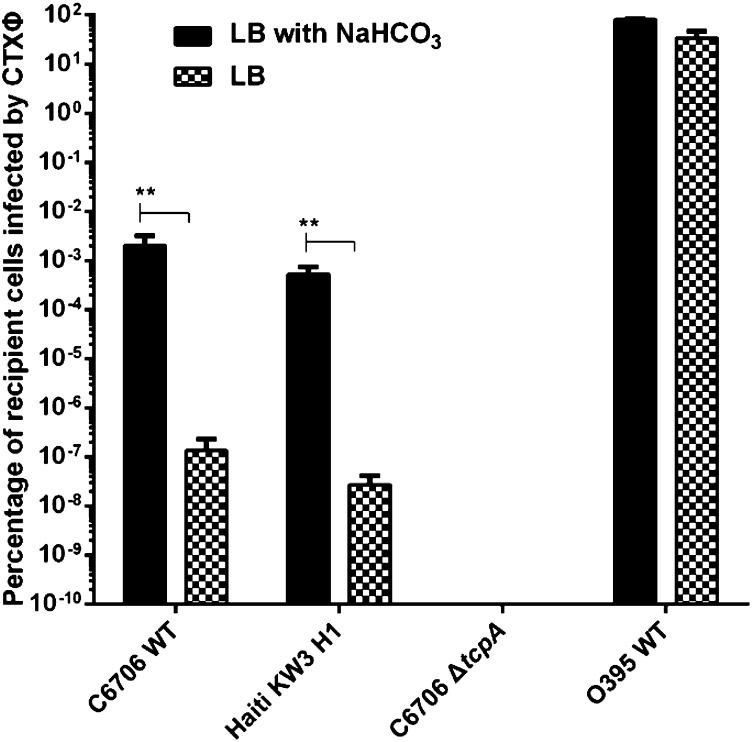
We observed formation of TCP on C6706 cells grown in the presence of bicarbonate by viewing them using an immunogold-labeled α-TcpA monoclonal antibody and transmission electron microscopy (TEM) (Fig. 1 A and B). Additional evidence of TCP formation was obtained when we identified eight peptides of TcpA in our OMV preparation (Fig. 1C and Table S1). To confirm that functional TCP pili were being expressed by the wild-type El Tor strain C6706 grown under these in vitro conditions, we performed transduction assays using CTX-Kmϕ, a derivative of CTXϕ that uses TCP pili as receptors (14). The efficiency of phage transduction of a recipient strain can be measured by determining the frequency of acquisition of the kanamycin (Km) resistance gene encoded by CTX-Kmϕ (14). We found that both C6706 and a 2010 isolate from Haiti (strain H1) were infected by CTX-Kmϕ when sodium bicarbonate was added to growth medium whereas infection was greatly reduced in noninduced LB conditions (Fig. 2). As expected, the classical strain 0395 was infected by the phage in the presence and absence of bicarbonate, and a ΔtcpA derivative of C6706 was completely resistant to phage infection under both conditions. These results confirmed that TCP expression and assembly occurs under these bicarbonate-treated culture conditions and prompted us to examine OMVs purified from cultures of strain C6706 grown in this manner.
Fig. 1.
TcpA was identified in OMVs produced by V. cholerae El Tor C6706. El Tor C6706 cells were grown in the presence (A) or absence (B) of sodium bicarbonate induction and imaged by transmission electron microscopy with an α-TcpA monoclonal antibody. The white arrow indicates TCP and flagella. (“3x” indicates 3× zoom into the selected region). (C) Amino acid sequence of TcpA highlighting the transmembrane domain (underlined) and peptides identified by tryptic digestion of OMVs (in red).
Fig. 2.
CTXΦ-Km phage-infected assays. Susceptibility of V. cholerae O1 El Tor and classical strains to CTX-KmΦ under ToxT-induced in vitro condition (LB with sodium bicarbonate) and normal laboratory condition (LB). El Tor strains C6706 wild type, Haiti H1 wild type, C6706 Δ tcpA, and classical strain O395 were tested. Values represent the averages of three independent observations. Significance was determined by t test: **P < 0.01. Mean with stand error of mean (SEM) is shown.
Proteomics Analysis Reveals the Presence of 90 OMV-Associated Proteins.
Nielsen et al. (45) reported that TcpA gene expression was significantly induced in cells 30 min after the addition of bicarbonate to an early exponential phase culture. Accordingly for OMV preparations, we added bicarbonate at OD600 ∼0.4, incubated cells statically for 30 min, and then grew cultures in rapidly shaking flasks for 30–60 min. During this period we removed samples at 30, 40, 50, and 60 min and prepared OMVs from culture filtrates using an ultracentrifugation method (Materials and Methods). Because avoiding cell lysis is essential for producing high-quality OMV samples, we carefully examined the protein content of OMVs made at these four time points from the same culture. Fig. S1 illustrates the experimental approach used. In brief, purified OMVs were directly digested with trypsin, and the released peptides were identified by LC-MS/MS. Although the apparent protein content of the 40-, 50-, and 60-min samples was similar to the 30-min sample in terms of the most abundant proteins identified based on peptide count (Table S2), we did detect some evidence of cell lysis at time points beyond 30 min based on the increasing prevalence of peptides in our tryptic digests that mapped to known abundant cytoplasmic proteins (Table S2). Accordingly, we used only the 30-min time point to determine the protein content of our highest quality V. cholerae OMVs.
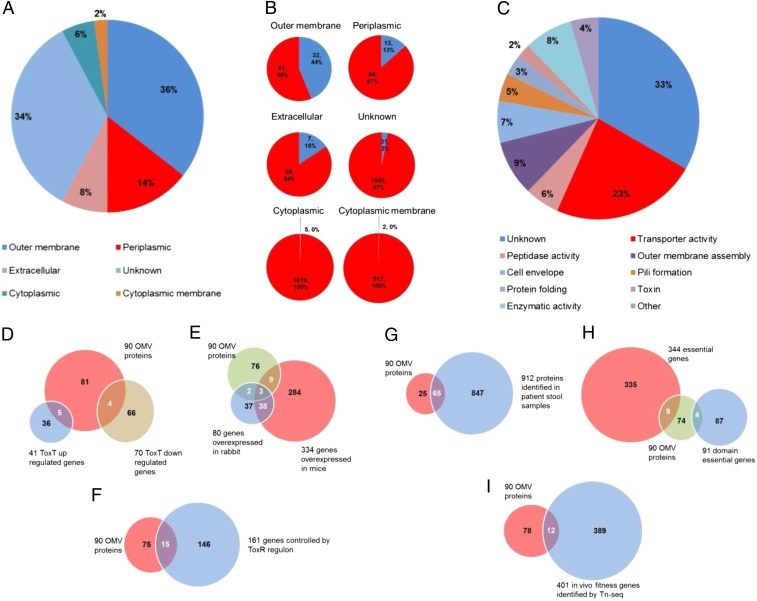
LC-MS/MS analysis of the OMVs collected at the 30-min time point identified 3,310 peptides encoded by the V. cholerae genome. By setting a cutoff of 2 or more predicted gene-encoded, protein-mapped peptides per individual protein identified, 629 peptides were mapped uniquely to 116 individual proteins (Table S3). Because 15 of these proteins were flagellar proteins and 11 were identified with only one peptide, these 15 candidates were eliminated from our final list of probable OMV proteins (Table S3). In total, 90 proteins were designated OMV proteins through this process, and this protein content was determined to be consistent with the theoretical mechanism of OMV formation in that Psort predicted that many (50%) of these 90 proteins were outer membrane (32 proteins) or periplasmic proteins (13 proteins) (Fig. 3A). Of the 90 OMV proteins identified, 7 were predicted to be secreted, which is consistent with the periplasm location of many substrates of the type II secretion system (T2SS) (53, 54) (Fig. 3A and Table S3). Compared with the theoretical proteome of V. cholerae, 45 outer membrane and periplasmic proteins were significantly enriched in the OMVs whereas only 7 of 2,536 cytoplasmic proteins were identified (Fig. 3B). We classified the identified OMV proteins based on their predicted functions using The Institute for Genomic Research or UniProt databases (Fig. 3C). Transporter proteins (n = 21) were the most abundant class represented within 10 functional groups, followed by outer membrane assembly proteins (n = 8), proteins with peptidase activity (n = 7), and proteins with different enzymatic activities (n = 5). Thirty other proteins identified on OMVs did not have predicted or characterized function.
Fig. 3.
(A–C) Venn diagrams showing the cellular location and putative function for identified OMV proteins. The 90 proteins identified on the outer membrane vesicles of V. cholerae grouped into families according to (A) their predicted subcellular localization, (B) proportion of identified proteins to the whole V. cholerae theoretical proteome and (C) function. (D–I) Venn diagrams comparing OMV proteins with previous genomics and proteomics research of V. cholerae. (D) OMVs and ToxT-induced transcriptome (10). (E) OMVs and in vivo-overexpressed genes (48). (F) OMVs and ToxR regulon (49). (G) OMVs and proteins identified in patient stool samples (50). (H) OMVs and V. cholerae essential genes (51). (I) OMVs and important in vivo fitness genes (52).
The identified OMV proteins included many proteins that had known or putative roles in pathogenicity, including TcpA and TcpC (55, 56), OmpU (57), accessory colonization factor AcfA (58), hemolysin (59), neuraminidase (60), cytolysin (61), serine protease (62), MSHA biogenesis protein MshL (63), TolB (64), TolC (65), and biofilm matrix proteins RbmA, RbmC, and Bap1 (47, 66, 67). The B-subunit of CTX was also identified in our OMVs, which is consistent with its being a periplasmic substrate for the T2SS (53, 54), its presence in previously characterized OMVs of V. cholerae (26, 37), and other studies that have detected surface-bound toxin subunits (68).
Comparative Analysis of OMV Protein Content with Other Genome-Wide Gene and Protein Expression Data Sets.
The presence of TCP and CTX in the 30-min postbicarbonate OMV preparations that we characterized suggested that ToxT activation was indeed achieved under these culture conditions. Accordingly, we compared the protein content of these OMVs to the proteins that were predicted to be expressed based on RNA-seq data obtained from cultures where ToxT was ectopically overexpressed in strain C6706 (10). Of five proteins identified in both groups, two were TCP-associated proteins, one was the B-subunit of CTX, and the last two were involved in maltose transport (Fig. 3D and Table S4, column A). These same five proteins were identified as the only overlapping OMV proteins compared with in vivo up-regulated genes in infected rabbits (Fig. 3E and Table S4, column B), and three of these were also transcriptionally up-regulated in V. cholerae during infection of infant mice (48). Three of of these five proteins were shown to be also transcriptionally up-regulated in a different El Tor V. cholerae strain (N16961) when measured using microarray analysis and somewhat different, but AKI-related, conditions for ToxR activation (49) (Fig. 3F and Table S4, column F). These results suggest that the OMV preparation characterized here was produced under conditions that correspond to in vitro ToxT activation using several different analytical tools and strains of V. cholerae.
We further asked whether the 90 OMV proteins identified were also among the proteins detected in samples prepared from the watery stools of patients infected with El Tor O1 V. cholerae in Dhaka, Bangladesh (50). Indeed, 65 of the 90 OMV proteins that we identified here were also detected in these clinical samples collected from cholera patients (Fig. 3G and Table S4, column C). Thus, the protein content of our OMV preparation appears to largely reflect the pattern of protein expression that can be measured in V. cholerae cells that are being shed by human cholera victims.
Comparative Analysis of OMV Protein Content with Other Genome-Wide Gene Functional Analysis Data Sets.
The preparation of an ordered transposon insertion library of strain C6706 as well as Tn-seq analysis has recently facilitated functional studies on the genes required for growth in vitro (51) and in vivo in experimental animals (52). Accordingly, we asked whether OMV proteins were among the gene products that had been identified as being essential under these growth conditions. Surprisingly, of 344 gene products that had been designated as essential for in vitro growth, only 9 were present in OMVs (Fig. 3H and Table S4, column E). Nonetheless, these proteins might represent interesting candidates for drug targets, given their association with the accessible outer membrane or cell periplasm. Consistent with this idea, orthologs of these proteins have been validated as drug targets, given their essential roles in either lipopolysaccharide or outer membrane protein transport (69–74).
Similarly, of 401 gene products that were found to be important for efficient intestinal colonization in rabbits (52), only 12 were detected as OMV proteins. (Fig. 3I and Table S4, column D). Although some of these proteins are known components of essential virulence factors (e.g., TcpA and TcpC), the OMV location of others may reflect their participation in outer membrane modifications that are specifically important to interacting with the host environment.
DegP Is Essential for Colonization of Mouse and Infant Rabbit Intestine.
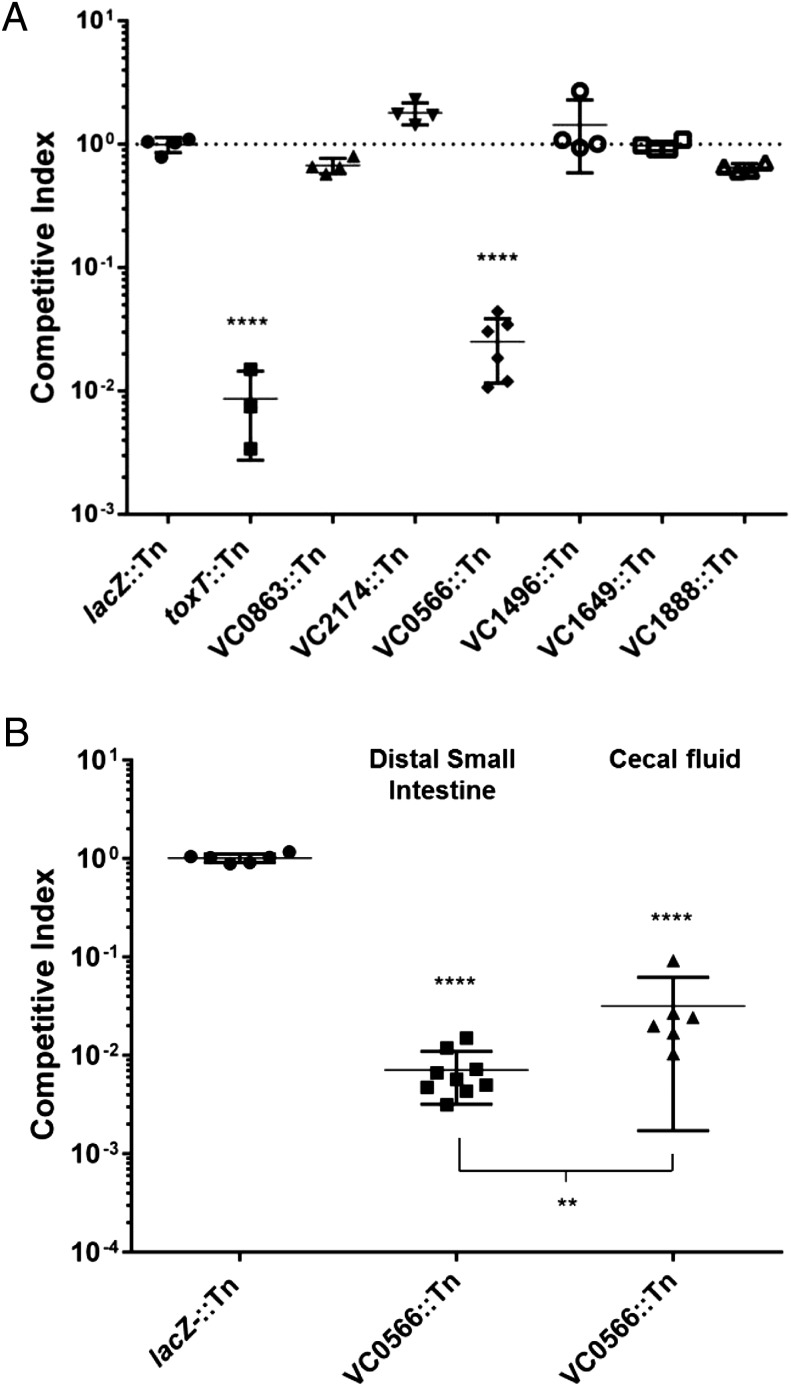
To further identify potential anti-infective targets, we focused on OMV proteins that were likely to encode enzymes because these proteins might be more susceptible to inhibition by small molecules. Disruption of five of the six genes encoding OMV proteins predicted to be enzymes did not alter the colonization fitness of C6706 in infant mice (Fig. 4A). These five genes included genes encoding a lipase (VC0863), 5′-nucleotidase (VC2174), tail-specific protease (VC1496), serine protease (VC1649), and biofilm-associated extracellular matrix protein (VC1888) (62, 75). However, inactivation of VC0566 (encoding a DegP ortholog) caused an ∼50-fold decrease in colonization relative to its wild-type (WT) parental strain. The magnitude of this defect was similar to that observed for the negative control (a toxT null mutant), suggesting that DegP is likely an essential determinant of intestinal colonization of this rodent species by V. cholerae.
Fig. 4.
Competition of V. cholerae strains using infant mouse and rabbit colonization models. (A) Ability of six different selected mutant strains to colonize the infant mouse intestine was compared with the parental strain. The dotted line indicates a competitive index of 1. Error bars represent SDs from at least four mice. The major virulence regulatory protein ToxT mutant was used as a negative control. Significance was determined by t test relative to colonization ratio of parental strains WT C6706 versus C6706 lacZ:Tn. ****P < 0.0001. Mean with SEM is shown. (B) Ability of degP mutant (EC956) strain to colonize the infant rabbit intestine was compared with the parental strain. The competitive index was determined for a phenotypically LacZ+ TnFGL3 insertion in lacZ (lacZ::Tn) when competed against its parental strain ΔlacZ WT strain (WT). The competitive index was determined for a phenotypically LacZ+ TnFGL3 insertion in degP mutant (EC956) when competed against its ΔlacZ parental strain (WT) either from the cecal fluid (Center) or distal small intestine (Right). ****P value < 0.0001 and **P < 0.01. Mean with SEM is shown.
Infection of infant rabbits provides arguably the best model for human cholera, given that oral-gastrically challenged animals exhibit the hallmark cholera clinical symptoms of watery diarrhea, dehydration, and death (76). To further investigate the role of DegP during in vivo colonization of V. cholerae, we challenged infant rabbits with a degP insertion mutant (EC956) in competition with its WT parental strain. The EC956 mutant was recovered from our defined transposon insertion library (77). Consistent with infant mouse competition assays, the degP mutant EC956 showed a significant colonization defect in infant rabbits (Fig. 4B). After 18 h of infection, the degP mutant was ∼100-fold less abundant in the distal small intestine and ∼30-fold reduced in colonization in cecal fluids. It is also worth noting that the degP mutant EC956 was one of the 401 mutants that was identified as colonization defective through an in vivo Tn-seq analysis that involved infant rabbit infections (52). More recently, another study showed that both V. cholerae C6706 insertion and deletion mutants in degP were colonization-defective in infant rabbits (78). Because degP appears to be a monocistronic gene in V. cholerae (as it apparently is in many other bacterial species), we elected to forgo attempts to complement EC956 in vivo, a procedure that frequently fails because of in vivo plasmid instability. EC956 also displayed normal growth kinetics compared with wild-type strains, no detectable auxotrophic phenotype, and a similar motility phenotype on agar swarm plates as its WT C6706 parental strain (Fig. S2 A and B). Together, these results indicate that DegP is likely an important factor contributing to intestinal colonization but is not required for all cell-envelope–dependent processes (such as those required for optimal growth or motility and chemotaxis by V. cholerae).
DegP Is Important for Biogenesis of Biofilm-Formation Proteins.
DegP is predicted to be a protein of 456 amino acids in length that is composed of a trypsin serine protease domain (PF00089) and two PDZ domains (PF13180) (44). In other organisms, DegP family proteases function as periplasmic chaperones involved in the quality control, processing, and maturation of extracellular and outer membrane proteins (43). To understand the colonization defect caused by disruption of DegP, we scored the WT and degP mutant strain EC956 for TCP pili production. TEM results showed that loss of DegP did not have an obvious effect on the extracellular appearance of TCP pili (Fig. S2C). Next we explored whether DegP was important to the production of other proteins detectable in the OMV fraction of V. cholerae. In total, 139 proteins were identified, including 18 flagella subunits from OMVs prepared from the degP mutant strain EC956 and its WT parental strain in two different experiments for each strain (Table S5). Using this comparative screen, we identified nine proteins (with ≥5 unique peptides each) present in OMVs produced by WT cells that were absent in OMVs produced by the degP mutant EC956 (Table 1). These proteins were classified into three groups according to their known or predicted functional categories with some known overlap: (i) biofilm formation (VC0928-RmbA, VC0930-RmbC, VC1888-Bap1, VCA0865-HAP), (ii) chitin utilization (VCA0140, VC1280 and VC0769), and (iii) proteins with unknown functions (VC0157, VC1280). The function of VC0157 is not characterized although its annotation suggests that it is a secreted protease. Additionally, five of these nine proteins are secreted by a type II secretion system (Table S6). These results suggest that DegP has an important function as a chaperone for secreted proteins found in OMVs and for proteins that contribute to chitin utilization and biofilm formation.
Table 1.
Nine proteins whose abundance in vesicles is influenced by DegP
| No. of unique peptides |
|||||||
| No. | Gene ID | Annotation | Subcellular localization | WT experiment 1 | WT experiment 2 | degP experiment 1 | degP experiment 2 |
| 1 | VC0930 | Matrix biofilm protein, RmbC | Extracellular | 20 | 24 | 0 | 1 |
| 2 | VC1888 | Biofilm-associated protein 1, Bap1 | Extracellular | 14 | 13 | 0 | 0 |
| 3 | VC0928 | Matrix biofilm protein, RmbA | Extracellular | 11 | 11 | 1 | 0 |
| 4 | VC1950 | Biotin sulfoxidereductase | Periplasmic | 9 | 7 | 0 | 0 |
| 5 | VCA0140 | Spindolin-related | Unknown | 7 | 7 | 0 | 0 |
| 6 | VC0157 | Alkalineserine protease | Extracellular | 6 | 5 | 0 | 0 |
| 7 | VC1280 | Hypothetical protein | Unknown | 5 | 4 | 0 | 0 |
| 8 | VCA0865 | Hemagglutinin/protease | Extracellular | 5 | 6 | 0 | 0 |
| 9 | VC0769 | Chitinase | Unknown | 5 | 3 | 0 | 0 |
The results of comparative proteomic experiments on OMVs of the degP mutant (EC956) and its parental strain revealed nine proteins the abundance of which in vesicles is influenced by DegP. These proteins are classified according to the number of identified unique peptides. The following information is reported for each protein: National Center for Biotechnology Information gene ID, annotation, predicted subcellular localization (PsortB), and unique numbers of identified peptides.
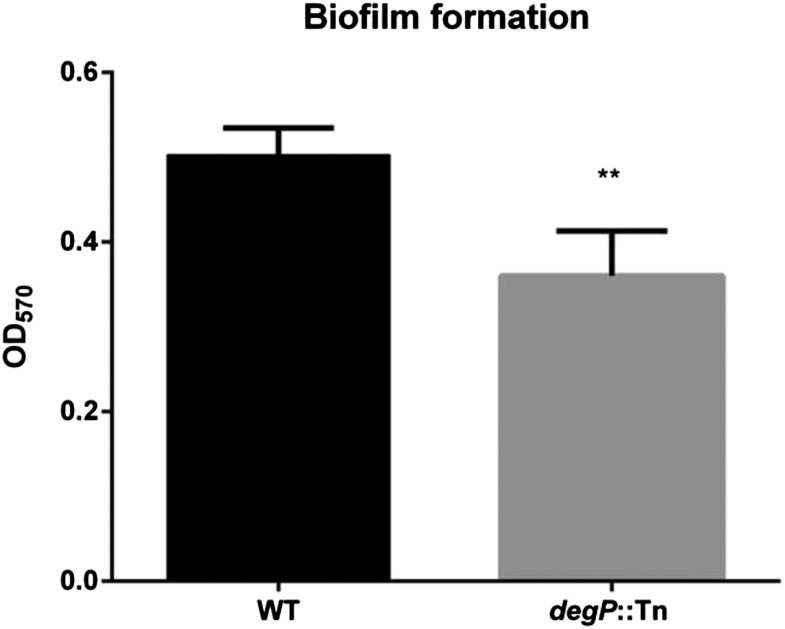
To test whether DegP is required for biofilm formation by V. cholerae, we measured the biofilm mass generated by wild-type and mutant strains under static culture conditions (Fig. 5). These results indicate that DegP does enhance biofilm formation perhaps by facilitating the secretion of RmbA, RmbB, Bap1, and hemagglutinin protease (HAP) into the maturing biofilm matrix. It has previously been observed that these proteins contribute to the architecture of V. cholerae biofilms (53). However, this insight does not explain the rabbit colonization defect of our degP mutant because individual mutants inactivated in genes encoding any one of these four DegP-dependent OMV proteins display no detectable colonization defect in infant rabbits (52). Because some of these proteins are likely or known substrates of the T2SS (e.g., HAP) (53, 54), we checked for a defect in T2SS function and for sensitivity of the degP mutant (EC956) to bile salts (Fig. S3); the latter is a known phenotype of mutants that have lost T2SS function (54, 52). When we checked the 19 proteins reported to be secreted by T2SS (54, 79), 11 of them were identified in OMVs of V. cholerae (Table S6). Although inactivation of the degP gene caused the loss of six of these proteins in OMV preparations (Table S6), inactivation of degP did not affect secretion of other five T2SS substrates present in OMVs. Thus, DegP depletion does not affect the function of the T2SS apparatus but simply its recognition of a subset of T2SS substrate proteins that may need DegP for their folding, stability, or as a transport chaperone. Mutants carrying insertion genes encoding any one of these six T2SS-dependent and DegP-dependent proteins did not produce an intestinal colonization defect in infant rabbits (52). Furthermore, the degP mutant EC956 displayed normal resistance to bile salts (Fig. S3), suggesting that its colonization defect could not be explained by loss of T2SS function and resultant sensitivity to bile (52).
Fig. 5.
Effect of degP mutation on biofilm formation in V. cholerae. The wild-type and degP mutant (EC956) strains were inoculated into culture tubes and allowed to grow without shaking at room temperature for 24 h. Data presented are averages of three replicates and error bars represent calculated SDs (OD570 of the crystal violet-stained biofilm).
Discussion
The bacterial outer membrane is the envelope layer of Gram-negative cells that is exposed to the extracellular environment and thus serves as the essential scaffold for extracellular organelles and polymers (e.g., lipopolysaccharide and capsules) as well as a barrier to the diffusion of toxic molecules. The extracellular processes that depend on the integrity of the outer membrane include those involved in pathogenesis such as assembly of adhesive organelles and protein secretion systems. Thus, a comprehensive understanding of the protein composition of bacterial outer membranes could influence efforts to develop anti-infective drugs as well as vaccines that target pathogenic Gram-negative bacteria. Here we show that proteomic analysis of OMVs of V. cholerae grown under virulence-inducing conditions can efficiently provide insights into the protein composition of this organism’s outer membrane and, to a lesser degree, its periplasmic space. In-solution protease digestion coupled to mass spectrometry allowed us to identify 90 proteins present in OMV preparations of V. cholerae (Table S3). The presence of certain hallmark proteins such as TcpA, TcpC, and CtxB in this group of 90 proteins further suggests that the composition of the OMVs analyzed here reflects the state of the expression of the V. cholerae genome under ToxT-activation conditions and thus, to some degree, the physiological state of V. cholerae cells in the gastrointestinal tract of humans and experimental animals.
By performing systematic comparisons with various functional genomic data bases, we also showed that a moderate number of the OMV-associated proteins were apparently also involved in processes that were essential for cell growth in vitro or in gastrointestinal colonization in experimental animals in vivo. OMV proteins encoded by genes that are essential for cell growth represent intriguing drug targets simply because small molecules that interfere with their function may be less susceptible to cytosolic membrane processes such as drug efflux or impermeability. OMV proteins that are essential for host colonization included components of known virulence factors such as TCP but also novel proteins whose functions are not fully understood. For example, disruption of the gene coding a DegP ortholog caused a severe defect in V. cholerae intestinal colonization of both infant mice and rabbits (Fig. 4). DegP also stands out as an interesting OMV protein because its protease function may make it a reasonable drug target, given that peptidomimic chemistry has led to drugs against various viral proteases (80, 81). DegP is also a conserved protein in both Gram-negative and Gram-positive organisms, and this enhances its appeal as potentially a broad-spectrum antibacterial target.
The high-temperature requirement A (HtrA) family proteins (serine proteases) are involved in protein quality control in stress conditions. Among these, periplasmic DegP combines both protease and chaperone activities (43). It behaves like a protein-packaging device that is adaptable to size and concentration of the substrate (44). The fundamental role of DegP in guiding OMPs through the periplasm was recently shown (44). The folded protomers of OMPs are encapsulated by DegP, and this entrapment is thought to provide protection during their trafficking through periplasm. The function of DegP proteins in bacterial pathogenesis has been reported for different pathogens. For example, a degP mutant of Streptococcus pyogenes was unable to efficiently process cysteine protease SpeB and the hemolysin streptolysin, two important virulence factors of this organism (82). In a different study, extended filamentous hemagglutinin polypeptide of Bordetella pertussis was shown to be protected by DegP chaperone activity (83). The secreted DegP of Helicobacter pylori cleaves an important host factor (E-cadherin) to disrupt intercellular adhesion (84). DegP of Chlamydia trachomatis was one of many immunogenic proteins in a microarray study based on human sera. Moreover, a vaccine study showed that the immunization with homolog of DegP confers protection against Vibrio harveyi in a fish model (85). However, the function of DegP was not previously reported in V. cholerae.
In an effort to understand DegP function in V. cholerae and why loss of this protein causes a colonization defect, we performed comparative proteomic experiments on OMVs purified from the degP mutant EC956 and its parental strain. This analysis revealed nine proteins whose abundance in vesicles is influenced by DegP (Table 1). Among these nine proteins, RmbA, RmbC, and Bap1 are major components of biofilm matrix and are known to enhance biofilm formation to different levels (86). In a recent study, investigators concluded that Bap1 helps biofilms to adhere to surfaces whereas RmbC and and Bap1 encapsulate cell clusters attached to those surfaces (87). Moreover, we found that HAP was also dependent on DegP for its OMV association (Table 1). This protein has also been shown to be associated with biofilms of V. cholerae (53). Thus, we performed biofilm studies, and, as expected, disruption of degP caused a significant biofilm defect for V. cholerae (Fig. 5). A role for other degP orthologs in biofilm formation has been recently reported for Streptococcus mutans and Porphyromonas gingivalis (88, 89). Similarly, Pseudomonas aeruginosa MucD (a DegP homolog) has been shown to respond to signals that lead to the degradation of MucA, with subsequent alginate overproduction driving biofilm formation (90, 91).
It is unclear why the V. cholerae degP mutant characterized here displays an infant rabbit intestinal colonization defect, given that the loss of individual proteins that depend on DegP for their OMV localization did not show similar colonization defects. We propose that it is likely the cumulative effect of depletion of DegP on several important outer membrane functions that causes this virulence defect in infant rabbits and mice. One such function may be DegP’s effect on in vivo biofilm formation. Recent results suggest that in vivo-formed biofilms of V. cholerae have enhanced infectivity at least in infant mice (92, 93). However, mutations that affect in vitro biofilm formation have been shown to have variable effects on host intestinal colonization. For example, although a mutation in the rbmA gene caused a defect in intestinal colonization in infant mice, the double-mutant rbmC bap1 did not show a significant defect (94). Zhu et al. also showed that a mutation at major biofilm component Vibrio polysaccharide synthesis (vps) gene (VC0920) did not affect the long-term persistence and colonization in infant mouse model (95). It is also possible that the colonization defect of degP mutant EC956 may be related in part to its effect on Bap1 expression. Duperthuy et al. suggested a role for Bap1 in resistance to antimicrobial peptides; however, the role of these host innate immune factors during mucosal colonization of infant animals has not been fully evaluated (75). Thus, understanding how the function of DegP influences intestinal colonization and biofilm formation will require additional studies that address both bacterial and host factors. Nonetheless, the work presented here suggests that small molecules that inhibit DegP function might be promising compounds for evaluation as anti-infectives for V. cholerae and other Gram-negative bacterial species. Although four DegP orthologs are encoded in the human genome (so-called HtrA proteins), these proteins are sufficiently diverged in primary sequence that they may not be susceptible to drugs that target bacterial DegP proteases, particularly if such hypothetical drugs target the PDZ2 domain, which is involved in recognition of unfolded bacterial proteins and is a domain that is not highly conserved in eukaryotic HtrA proteins (43, 44). Further work will be needed to test the hypothesis that DegP is a valid target for the development of antibacterials that do not also target its eukaryotic orthologs.
Materials and Methods
Ethics Statement.
We performed all animal experiments according to protocols approved by Harvard Medical School Office for Research Protection Standing Committee on Animals. The Harvard Medical School animal management program meets National Institutes of Health standards as set forth in the Guide for the Care and Use of Laboratory Animals. The institution is also accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care, International. Public Health Service Policy on Humane Care and Use of Laboratory Animals by AWARDEE Institutions and National Institutes of Health Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training are accepted as mandatory. There is on file with the Office of Laboratory Animal Welfare an approved Assurance of Compliance (A3431-01).
Bacterial Strains.
V. cholerae strains used in this work are listed in Table S7. V. cholerae El Tor biotype strain C6706 and a spontaneous lacZ derivative of C6706 were used as parental (WT) strains. Streptomycin (Sm; 100 µg/mL), kanamycin (50 µg/mL), and chloramphenicol (2.5 µg/mL) were included as needed. LB was used for normal growth conditions [10 g/L of tryptone (Bacto), 5 g/L of yeast extract (Bacto), and 5 g/L of NaCl] and was supplemented with 16 g/L of agar (Bacto) for growth on plates. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used at 40 mg/mL.
Transduction Assays.
Preparations of phage and transduction assays were conducted according to protocols described by Faruque et al. (96, 97). CTX-Km used in this study was prepared from the culture strain O395, which carries the replicative form of the phage genome (6). Recipient cells were grown in ToxT-activated conditions (adding sodium bicarbonate to LB growth culture as described above) or in normal LB growth culture. El Tor strains C6706 and H1 were tested for their ability to express TCP in sodium bicarbonate-added conditions whereas the 0395 classical strain and ΔtcpA (C6706) mutant were used as positive and negative controls, respectively. Approximately 105 cells were mixed with 10 µL of the phage preparation inoculated into 5 mL LB and incubated 24 h at 30 °C. The aliquots of the cultures were plated on LB plates containing kanamycin (50 µg/mL) or on plates with streptomycin (100 µg/mL). The ratio of Km-transduced colonies to the total number of colonies derived from the recipient strain was calculated and expressed as the percentage of recipient cells infected (6).
Bicarbonate Induction of Virulence Genes and Preparation of OMVs.
We used sodium bicarbonate to induce virulence gene expression by activating major virulence regulator ToxT (12, 45). We optimized the method suggested by Nielsen et al. (45) to avoid cell lysis. Briefly, 5 mL of stationary-phase culture of a V. cholerae El Tor biotype strain C6706 LB was used to inoculate 445 mL of Hepes-buffered LB (50 mM Hepes, 1 µg/µL streptomycin). The cells were grown at 37 °C with aeration until they reached an OD600 of 0.4. The culture was induced by addition of 50 mL of sodium bicarbonate-buffered LB (1 M sodium bicarbonate), and the cells were incubated at 37 °C for 30 min without aeration and another 30, 40, 50, or 60 min with aeration. OMVs for comparative proteomics experiments were prepared from LB cultures without bicarbonate induction (OD600 of ∼0.9). The bacterial cells were removed by centrifugation (15 min, 4,500 × g, 4 °C), and supernatant was filtered through 0.22-µm pore-size filters (Corning). Protease inhibitors (complete EDTA free protease inhibitor mixture, Roche) were added to filtrates to inhibit protein degradation. The filtrate was subjected to high-speed centrifugation (12 h, 100,000 × g, 4 °C). Pellets containing OMVs were suspended with 300 µL of 5 mM of ammonium bicarbonate. Purified OMVs were stored at −80 °C.
In-Solution Digestion and Protein Sequence Analysis by LC-MS/MS.
In-solution digestions of OMVs were carried out overnight at 37 °C by adding 5 ng/µL modified sequencing-grade trypsin (Promega) in 5 mM of ammonium bicarbonate. The standard protocol of the Taplin Mass Spectrometry Facility was used for mass spectrometry. Briefly, peptides were dried in a speed-vac (∼1 h), and samples were reconstituted in 5–10 µL of HPLC solvent A [2.5% (vol/vol) acetonitrile, 0.1% formic acid]. A nano-scale reverse-phase HPLC capillary column was created by packing 5 µm C18 spherical silica beads into a fused silica capillary (125-µm inner diameter × ∼20-cm length) with a flame-drawn tip. After equilibrating the column, each sample was loaded via a Famos auto sampler (LC Packings). A gradient was formed and peptides were eluted with increasing concentrations of solvent B [97.5% (vol/vol) acetonitrile, 0.1% formic acid]. As peptides eluted, they were subjected to electrospray ionization and then entered an LTQ Velos ion-trap mass spectrometer (ThermoFisher). Peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide.
Bioinformatics.
Peptide sequences (protein identity) were determined by matching protein databases with the acquired fragmentation pattern with the software program Sequest (ThermoFisher). Sequest was run on a database containing protein sequences of the V. cholerae 01 (El Tor Inaba N16961) genome downloaded from NCBInr. A computational analysis of each identified protein was performed by PsortB to predict the subcellular localization (www.psort.org/psortb/). A computational analysis of each identified protein sequence was performed with the PSORTb v.3.0 package.
Infant Mouse Colonization Assays.
Suckling mouse colonization competition assays were performed according to the protocol described by Davies et al. (10) for infection and recovery of C6706 strains. All strains were grown on LB–agar plates with streptomycin overnight at 37 °C. Equal amounts of WT C6706 strain (carrying a neutral lacZ allele for identification) and transposon mutant strains were mixed together in LB. This competition mixture (50 µL or ∼50,000 bacteria) was inoculated into a 5-d-old CD1 mouse pup (Charles River Company). Serial dilutions of the competition mixture were plated in LB + Sm100 + X-Gal and quantified to determine the input ratio of wild-type and mutant strains. After incubation at 30 °C for 18 h, the mouse pups were killed and small intestines were removed and homogenized in 10 mL of LB. Serial dilutions were plated in LB + Sm100 + X-Gal and quantified to determine the output ratio of wild-type and mutant strains. The competitive index for each mutant is defined as the input ratio of mutant/wild-type strain divided by the output ratio of mutant/wild-type strain. Statistical significance was determined by comparing the resulting ratio to the ratio of WT versus WT lacZ−. A minimum of four mice were assayed for each mutant strain.
Infant Rabbit Colonization Competition Assays.
V. cholerae C6706 derivatives were initially grown overnight on LB–agar plates at 37 °C. A fresh colony of each strain was inoculated into LB and incubated at 250 × g at 37 °C for several hours until the OD600 was ∼1.0. Wild-type (lacZ−) and test strains (lacZ+) were mixed together (1:1) in 2.5% (wt/vol) sodium bicarbonate buffer and inoculated into 2-d-old New Zealand White rabbit pups (total cfu ∼109 bacteria/animal). After 18 h or when symptoms were apparent, infant rabbits were killed, dissected, and a 1-cm sample of the distal small intestine was removed and homogenized in 1 mL of PBS, and cecal fluid was directly collected. Serial dilutions were plated in LB with Sm and X-Gal to enumerate the output ratio of the wild-type and mutant strain. The competitive index for each mutant is defined as the input ratio of mutant/WT strain divided by the output ratio of mutant/WT strain. A minimum of six rabbits were assayed for each mutant strain (52).
Motility Assay.
The bacteria were grown on a LB agar plate and inoculated into LB. Strains were grown at 37 °C until exponential phase and diluted to an OD600 of 0.1. Two microliters of this culture was daubed onto motility plates containing 1% tryptone, 0.5% NaCl, and 0.3% bactoagar. After incubation at 30 °C for 48 h, motility zones were recorded with a digital camera.
Negative-Staining Electron Microscopy.
A drop of OMV suspension was placed on Formvar/carbon-coated grids and adsorbed for 5 min. Grids were washed with distilled water and blotted with filter paper. For negative staining, grids were treated with 2% (wt/vol) uranyl acetate for 1 min, air-dried, and viewed with a Jeol JEM 1200 EXII electron microscope operating at 80 kV.
Biofilm Assay.
The wild-type and transposon mutant V. cholerae strains were grown overnight on LB agar plates. Few colonies from each strain were resuspended in LB broth and incubated until the absorbance (OD600) reached 0.6. A 1:100 dilution of this suspension was inoculated in LB broth into 10- × 75-mm borosilicate glass test tubes and incubated for 24 h at room temperature (66). Subsequently, tubes were rinsed with distilled water and then filled with 1% crystal violet stain. After 15 min of incubation, the tubes were rinsed, and biofilm-associated crystal violet was suspended with dimethyl sulphoxide. The OD570 of the resulting suspension was measured to evaluate biofilm formation. All experiments were performed at least three independent times and samples were also performed in triplicate. Error bars represent SDs.
Bile Resistance Assay.
Overnight-cultured strains were subcultured to fresh LB medium and grown to OD600 1.0 in LB with Sm. Strains were serial-diluted in LB. Five microliters of each dilution was daubed onto LB–ager containing 0.5% crude bile (bovine) and incubated overnight at 37 °C. Each colony was counted.
Supplementary Material
Acknowledgments
We thank Dr. Ronald Taylor for providing α-TcpA monoclonal antibodies; Dr. Ewen Cameron for the V. cholera-defined TnFGL3 transposon library; the J.J.M. group for helpful discussion; Dr. William Robins and Dr. Andrew McCluskey for their comments and critical reading of this manuscript; and Ross Tomaino (Taplin Mass Spectrometry Facility) and Maria Ericsson (Cell Biology Conventional Electron Microscopy Facility) for their technical contributions. This work was supported by National Institute of Allergy and Infectious Diseases Grant AI-01845 (to J.J.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403683111/-/DCSupplemental.
References
- 1.Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB. Cholera. Lancet. 2012;379(9835):2466–2476. doi: 10.1016/S0140-6736(12)60436-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson EJ, Harris JB, Morris JG, Jr, Calderwood SB, Camilli A. Cholera transmission: The host, pathogen and bacteriophage dynamic. Nat Rev Microbiol. 2009;7(10):693–702. doi: 10.1038/nrmicro2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farmer P, et al. Meeting cholera’s challenge to Haiti and the world: A joint statement on cholera prevention and care. PLoS Negl Trop Dis. 2011;5(5):e1145. doi: 10.1371/journal.pntd.0001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beyhan S, Tischler AD, Camilli A, Yildiz FH. Differences in gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infect Immun. 2006;74(6):3633–3642. doi: 10.1128/IAI.01750-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dziejman M, et al. Comparative genomic analysis of Vibrio cholerae: Genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci USA. 2002;99(3):1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62(4):1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin CS, et al. The origin of the Haitian cholera outbreak strain. N Engl J Med. 2011;364(1):33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lombardo MJ, et al. An in vivo expression technology screen for Vibrio cholerae genes expressed in human volunteers. Proc Natl Acad Sci USA. 2007;104(46):18229–18234. doi: 10.1073/pnas.0705636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matson JS, Withey JH, DiRita VJ. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect Immun. 2007;75(12):5542–5549. doi: 10.1128/IAI.01094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149(2):358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richard AL, Withey JH, Beyhan S, Yildiz F, DiRita VJ. The Vibrio cholerae virulence regulatory cascade controls glucose uptake through activation of TarA, a small regulatory RNA. Mol Microbiol. 2010;78(5):1171–1181. doi: 10.1111/j.1365-2958.2010.07397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abuaita BH, Withey JH. Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect Immun. 2009;77(9):4111–4120. doi: 10.1128/IAI.00409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cobaxin M, et al. Cholera toxin expression by El Tor Vibrio cholerae in shallow culture growth conditions. Microb Pathog. 2014;66:5–13. doi: 10.1016/j.micpath.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272(5270):1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 15.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84(9):2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anthouard R, DiRita VJ. Small-molecule inhibitors of toxT expression in Vibrio cholerae. MBio. 2013;4(4) doi: 10.1128/mBio.00403-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science. 2005;310(5748):670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 18.Son MS, Megli CJ, Kovacikova G, Qadri F, Taylor RK. Characterization of Vibrio cholerae O1 El Tor biotype variant clinical isolates from Bangladesh and Haiti, including a molecular genetic analysis of virulence genes. J Clin Microbiol. 2011;49(11):3739–3749. doi: 10.1128/JCM.01286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jude BA, Taylor RK. The physical basis of type 4 pilus-mediated microcolony formation by Vibrio cholerae O1. J Struct Biol. 2011;175(1):1–9. doi: 10.1016/j.jsb.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005;19(22):2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- 21.Schertzer JW, Whiteley M. Bacterial outer membrane vesicles in trafficking, communication and the host-pathogen interaction. J Mol Microbiol Biotechnol. 2013;23(1-2):118–130. doi: 10.1159/000346770. [DOI] [PubMed] [Google Scholar]
- 22.Scanlan D. Ecology. Bacterial vesicles in the ocean. Science. 2014;343(6167):143–144. doi: 10.1126/science.1248566. [DOI] [PubMed] [Google Scholar]
- 23.Amano A, Takeuchi H, Furuta N. Outer membrane vesicles function as offensive weapons in host-parasite interactions. Microbes Infect. 2010;12(11):791–798. doi: 10.1016/j.micinf.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74(1):81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berlanda Scorza F, et al. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli DeltatolR IHE3034 mutant. Mol Cell Proteomics. 2008;7(3):473–485. doi: 10.1074/mcp.M700295-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Chatterjee D, Chaudhuri K. Association of cholera toxin with Vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett. 2011;585(9):1357–1362. doi: 10.1016/j.febslet.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 27.MacDonald KL, Beveridge TJ. Bactericidal effect of gentamicin-induced membrane vesicles derived from Pseudomonas aeruginosa PAO1 on gram-positive bacteria. Can J Microbiol. 2002;48(9):810–820. doi: 10.1139/w02-077. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Clarke AJ, Beveridge TJ. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J Bacteriol. 1998;180(20):5478–5483. doi: 10.1128/jb.180.20.5478-5483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadurugamuwa JL, Beveridge TJ. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: Conceptually new antibiotics. J Bacteriol. 1996;178(10):2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avila-Calderón ED, et al. Characterization of outer membrane vesicles from Brucella melitensis and protection induced in mice. Clin Dev Immunol. 2012;2012:352493. doi: 10.1155/2012/352493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrari G, et al. Outer membrane vesicles from group B Neisseria meningitidis delta gna33 mutant: Proteomic and immunological comparison with detergent-derived outer membrane vesicles. Proteomics. 2006;6(6):1856–1866. doi: 10.1002/pmic.200500164. [DOI] [PubMed] [Google Scholar]
- 32.McCaig WD, Koller A, Thanassi DG. Production of outer membrane vesicles and outer membrane tubes by Francisella novicida. J Bacteriol. 2013;195(6):1120–1132. doi: 10.1128/JB.02007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park SB, et al. Outer membrane vesicles as a candidate vaccine against edwardsiellosis. PLoS ONE. 2011;6(3):e17629. doi: 10.1371/journal.pone.0017629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uli L, et al. Outer membrane vesicles of the VA-MENGOC-BC vaccine against serogroup B of Neisseria meningitidis: Analysis of protein components by two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2006;6(11):3389–3399. doi: 10.1002/pmic.200500502. [DOI] [PubMed] [Google Scholar]
- 35.Bishop AL, Schild S, Patimalla B, Klein B, Camilli A. Mucosal immunization with Vibrio cholerae outer membrane vesicles provides maternal protection mediated by antilipopolysaccharide antibodies that inhibit bacterial motility. Infect Immun. 2010;78(10):4402–4420. doi: 10.1128/IAI.00398-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bishop AL, et al. Immunization of mice with vibrio cholerae outer-membrane vesicles protects against hyperinfectious challenge and blocks transmission. J Infect Dis. 2012;205(3):412–421. doi: 10.1093/infdis/jir756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatterjee D, Chaudhuri K. Vibrio cholerae O395 outer membrane vesicles modulate intestinal epithelial cells in a NOD1 protein-dependent manner and induce dendritic cell-mediated Th2/Th17 cell responses. J Biol Chem. 2013;288(6):4299–4309. doi: 10.1074/jbc.M112.408302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leitner DR, et al. Lipopolysaccharide modifications of a cholera vaccine candidate based on outer membrane vesicles reduce endotoxicity and reveal the major protective antigen. Infect Immun. 2013;81(7):2379–2393. doi: 10.1128/IAI.01382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy N, et al. Immunogenicity and protective efficacy of Vibrio cholerae outer membrane vesicles in rabbit model. FEMS Immunol Med Microbiol. 2010;60(1):18–27. doi: 10.1111/j.1574-695X.2010.00692.x. [DOI] [PubMed] [Google Scholar]
- 40.Schild S, Nelson EJ, Bishop AL, Camilli A. Characterization of Vibrio cholerae outer membrane vesicles as a candidate vaccine for cholera. Infect Immun. 2009;77(1):472–484. doi: 10.1128/IAI.01139-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schild S, Nelson EJ, Camilli A. Immunization with Vibrio cholerae outer membrane vesicles induces protective immunity in mice. Infect Immun. 2008;76(10):4554–4563. doi: 10.1128/IAI.00532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mullaney E, et al. Proteomic and functional characterization of the outer membrane vesicles from the gastric pathogen Helicobacter pylori. Proteomics Clin Appl. 2009;3(7):785–796. doi: 10.1002/prca.200800192. [DOI] [PubMed] [Google Scholar]
- 43.Clausen T, Kaiser M, Huber R, Ehrmann M. HTRA proteases: Regulated proteolysis in protein quality control. Nat Rev Mol Cell Biol. 2011;12(3):152–162. doi: 10.1038/nrm3065. [DOI] [PubMed] [Google Scholar]
- 44.Krojer T, et al. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008;453(7197):885–890. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- 45.Nielsen AT, et al. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog. 2010;6(9):e1001102. doi: 10.1371/journal.ppat.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwanaga M, et al. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol. 1986;30(11):1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 47.Fong JC, Yildiz FH. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol. 2007;189(6):2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandlik A, et al. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe. 2011;10(2):165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bina J, et al. ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc Natl Acad Sci USA. 2003;100(5):2801–2806. doi: 10.1073/pnas.2628026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LaRocque RC, et al. Proteomic analysis of Vibrio cholerae in human stool. Infect Immun. 2008;76(9):4145–4151. doi: 10.1128/IAI.00585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chao MC, et al. High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res. 2013;41(19):9033–9048. doi: 10.1093/nar/gkt654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu Y, Waldor MK, Mekalanos JJ. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe. 2013;14(6):652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Absalon C, Van Dellen K, Watnick PI. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. 2011;7(8):e1002210. doi: 10.1371/journal.ppat.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sikora AE, Zielke RA, Lawrence DA, Andrews PC, Sandkvist M. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J Biol Chem. 2011;286(19):16555–16566. doi: 10.1074/jbc.M110.211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krebs SJ, Taylor RK. Protection and attachment of Vibrio cholerae mediated by the toxin-coregulated pilus in the infant mouse model. J Bacteriol. 2011;193(19):5260–5270. doi: 10.1128/JB.00378-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogierman MA, Manning PA. TCP pilus biosynthesis in Vibrio cholerae O1: Gene sequence of tcpC encoding an outer membrane lipoprotein. FEMS Microbiol Lett. 1992;76(1-2):179–184. doi: 10.1016/0378-1097(92)90383-y. [DOI] [PubMed] [Google Scholar]
- 57.Nakasone N, Iwanaga M. Characterization of outer membrane protein OmpU of Vibrio cholerae O1. Infect Immun. 1998;66(10):4726–4728. doi: 10.1128/iai.66.10.4726-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hughes KJ, Everiss KD, Kovach ME, Peterson KM. Isolation and characterization of the Vibrio cholerae acfA gene, required for efficient intestinal colonization. Gene. 1995;156(1):59–61. doi: 10.1016/0378-1119(95)00054-a. [DOI] [PubMed] [Google Scholar]
- 59.Cinar HN, et al. Vibrio cholerae hemolysin is required for lethality, developmental delay, and intestinal vacuolation in Caenorhabditis elegans. PLoS ONE. 2010;5(7):e11558. doi: 10.1371/journal.pone.0011558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galen JE, et al. Role of Vibrio cholerae neuraminidase in the function of cholera toxin. Infect Immun. 1992;60(2):406–415. doi: 10.1128/iai.60.2.406-415.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Debellis L, et al. The Vibrio cholerae cytolysin promotes chloride secretion from intact human intestinal mucosa. PLoS ONE. 2009;4(3):e5074. doi: 10.1371/journal.pone.0005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Syngkon A, et al. Studies on a novel serine protease of a ΔhapAΔprtV Vibrio cholerae O1 strain and its role in hemorrhagic response in the rabbit ileal loop model. PLoS ONE. 2010;5(9):e13122. doi: 10.1371/journal.pone.0013122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsiao A, Liu Z, Joelsson A, Zhu J. Vibrio cholerae virulence regulator-coordinated evasion of host immunity. Proc Natl Acad Sci USA. 2006;103(39):14542–14547. doi: 10.1073/pnas.0604650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heilpern AJ, Waldor MK. CTXphi infection of Vibrio cholerae requires the tolQRA gene products. J Bacteriol. 2000;182(6):1739–1747. doi: 10.1128/jb.182.6.1739-1747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bina JE, Mekalanos JJ. Vibrio cholerae tolC is required for bile resistance and colonization. Infect Immun. 2001;69(7):4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hung DT, Zhu J, Sturtevant D, Mekalanos JJ. Bile acids stimulate biofilm formation in Vibrio cholerae. Mol Microbiol. 2006;59(1):193–201. doi: 10.1111/j.1365-2958.2005.04846.x. [DOI] [PubMed] [Google Scholar]
- 67.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol. 2004;53(2):497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 68.Blanco LP, DiRita VJ. Bacterial-associated cholera toxin and GM1 binding are required for transcytosis of classical biotype Vibrio cholerae through an in vitro M cell model system. Cell Microbiol. 2006;8(6):982–998. doi: 10.1111/j.1462-5822.2005.00681.x. [DOI] [PubMed] [Google Scholar]
- 69.Chimalakonda G, et al. Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2011;108(6):2492–2497. doi: 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henderson DP, Payne SM. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: Sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J Bacteriol. 1994;176(11):3269–3277. doi: 10.1128/jb.176.11.3269-3277.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jain S, Goldberg MB. Requirement for YaeT in the outer membrane assembly of autotransporter proteins. J Bacteriol. 2007;189(14):5393–5398. doi: 10.1128/JB.00228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kenyon WJ, Humphreys S, Roberts M, Spector MP. Periplasmic peptidyl-prolyl isomerases SurA and FkpA play an important role in the starvation-stress response (SSR) of Salmonella enterica serovar Typhimurium. Antonie van Leeuwenhoek. 2010;98(1):51–63. doi: 10.1007/s10482-010-9428-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rossiter AE, et al. The essential β-barrel assembly machinery complex components BamD and BamA are required for autotransporter biogenesis. J Bacteriol. 2011;193(16):4250–4253. doi: 10.1128/JB.00192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sherman DJ, Okuda S, Denny WA, Kahne D. Validation of inhibitors of an ABC transporter required to transport lipopolysaccharide to the cell surface in Escherichia coli. Bioorg Med Chem. 2013;21(16):4846–4851. doi: 10.1016/j.bmc.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Duperthuy M, et al. Role of the Vibrio cholerae matrix protein Bap1 in cross-resistance to antimicrobial peptides. PLoS Pathog. 2013;9(10):e1003620. doi: 10.1371/journal.ppat.1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ritchie JM, Rui H, Bronson RT, Waldor MK. Back to the future: Studying cholera pathogenesis using infant rabbits. MBio. 2010;1(1) doi: 10.1128/mBio.00047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cameron DE, Urbach JM, Mekalanos JJ. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci USA. 2008;105(25):8736–8741. doi: 10.1073/pnas.0803281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 2013;9(12):e1003800. doi: 10.1371/journal.ppat.1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sikora AE. Proteins secreted via the type II secretion system: Smart strategies of Vibrio cholerae to maintain fitness in different ecological niches. PLoS Pathog. 2013;9(2):e1003126. doi: 10.1371/journal.ppat.1003126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hammamy MZ, Haase C, Hammami M, Hilgenfeld R, Steinmetzer T. Development and characterization of new peptidomimetic inhibitors of the West Nile virus NS2B-NS3 protease. ChemMedChem. 2013;8(2):231–241. doi: 10.1002/cmdc.201200497. [DOI] [PubMed] [Google Scholar]
- 81.Qiu X, Liu ZP. Recent developments of peptidomimetic HIV-1 protease inhibitors. Curr Med Chem. 2011;18(29):4513–4537. doi: 10.2174/092986711797287566. [DOI] [PubMed] [Google Scholar]
- 82.Lyon WR, Caparon MG. Role for serine protease HtrA (DegP) of Streptococcus pyogenes in the biogenesis of virulence factors SpeB and the hemolysin streptolysin S. Infect Immun. 2004;72(3):1618–1625. doi: 10.1128/IAI.72.3.1618-1625.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baud C, et al. Role of DegP for two-partner secretion in Bordetella. Mol Microbiol. 2009;74(2):315–329. doi: 10.1111/j.1365-2958.2009.06860.x. [DOI] [PubMed] [Google Scholar]
- 84.Hoy B, et al. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 2010;11(10):798–804. doi: 10.1038/embor.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang WW, Sun K, Cheng S, Sun L. Characterization of DegQVh, a serine protease and a protective immunogen from a pathogenic Vibrio harveyi strain. Appl Environ Microbiol. 2008;74(20):6254–6262. doi: 10.1128/AEM.00109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Giglio KM, Fong JC, Yildiz FH, Sondermann H. Structural basis for biofilm formation via the Vibrio cholerae matrix protein RbmA. J Bacteriol. 2013;195(14):3277–3286. doi: 10.1128/JB.00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Berk V, et al. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337(6091):236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Biswas S, Biswas I. Role of HtrA in surface protein expression and biofilm formation by Streptococcus mutans. Infect Immun. 2005;73(10):6923–6934. doi: 10.1128/IAI.73.10.6923-6934.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan L, Rodrigues PH, Bélanger M, Dunn WA, Jr, Progulske-Fox A. Porphyromonas gingivalis htrA is involved in cellular invasion and in vivo survival. Microbiology. 2008;154(Pt 4):1161–1169. doi: 10.1099/mic.0.2007/015131-0. [DOI] [PubMed] [Google Scholar]
- 90.Damron FH, Goldberg JB. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol Microbiol. 2012;84(4):595–607. doi: 10.1111/j.1365-2958.2012.08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Damron FH, Yu HD. Pseudomonas aeruginosa MucD regulates the alginate pathway through activation of MucA degradation via MucP proteolytic activity. J Bacteriol. 2011;193(1):286–291. doi: 10.1128/JB.01132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Faruque SM, et al. Transmissibility of cholera: In vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc Natl Acad Sci USA. 2006;103(16):6350–6355. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tamayo R, Patimalla B, Camilli A. Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect Immun. 2010;78(8):3560–3569. doi: 10.1128/IAI.00048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fong JC, Syed KA, Klose KE, Yildiz FH. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology. 2010;156(Pt 9):2757–2769. doi: 10.1099/mic.0.040196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5(4):647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 96.Faruque SM, et al. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXPhi: Molecular basis for origination of new strains with epidemic potential. Infect Immun. 1998;66(12):5819–5825. doi: 10.1128/iai.66.12.5819-5825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Faruque SM, et al. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect Immun. 1998;66(8):3752–3757. doi: 10.1128/iai.66.8.3752-3757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.