Abstract
The global spread of Klebsiella pneumoniae carbapenemase (KPC) is predominately associated with K. pneumoniae strains genotyped as sequence type 258 (ST258). The first ST258-associated plasmid, pKpQIL, was described in Israel in 2006, but its history in the northeastern United States remains unknown. Six pKpQIL-like plasmids from four K. pneumoniae isolates (three ST258 and one ST234), one Escherichia coli isolate, and one Enterobacter aerogenes isolate, collected from 2003 to 2010 in New York (NY) and New Jersey (NJ) hospitals, were completely sequenced. The sequences and overall sizes of the six plasmids are highly similar to those of pKpQIL; the major difference is that five of six NJ/NY strains harbor blaKPC-2, while pKpQIL contains blaKPC-3. Moreover, a 26.7-kb fragment was inverted in pKpQIL-234 (from ST234 K. pneumoniae), while a 14.5-kb region was deleted in pKpQIL-Ec (from ST131 E. coli). PCR screening of 284 other clinical K. pneumoniae isolates identified 101 (35.6%) harboring pKpQIL-like plasmids from 9 of 10 surveyed hospitals, demonstrating the wide dissemination of pKpQIL in this region of endemicity. Among the positive isolates, 87.1% were typed as ST258 and 88.1% carried blaKPC-2. The finding of pKpQIL-like plasmid in this study from strains that predate the initial report of KPC in Israel provides evidence that pKpQIL may have originated in the United States. Our findings demonstrate that pKpQIL plasmids are both spreading clonally in ST258 strains and spreading horizontally to different sequence types and species, further highlighting the clinical and public health concerns associated with carbapenem resistance.
INTRODUCTION
Infection with carbapenem-resistant Enterobacteriaceae, in particular, Klebsiella pneumoniae, has emerged as a significant global public health dilemma (1). K. pneumoniae carbapenemase (KPC), first reported in 2001 (2), is currently the most common carbapenemase in the United States. During the last 10 years, hospitals in the New York (NY) metropolitan area, which define an “epicenter of KPC,” have experienced aggressive dissemination and clinical failures associated with KPC-producing K. pneumoniae in high-risk patients (3).
The widespread dissemination of KPC-producing K. pneumoniae is largely associated with a single clone, defined as multilocus sequence type 258 (ST258), although a number other STs have also been found harboring KPC (3–6). The gene coding for KPC, blaKPC, located on a Tn3-like transposon (Tn4401), has been identified in various transferable plasmids which show the ability to transfer between different strains and various Gram-negative species (3, 7). Recently, blaKPC has been identified in plasmids of different incompatibility (Inc) groups, including IncFII, IncI2, IncA/C, IncN, IncX, IncR, and ColE (8–12). The first ST258-associated plasmid, pKpQIL, a blaKPC-3-harboring IncFIIK2 plasmid, was initially identified in 2006 in K. pneumoniae ST258 strains from Israel and subsequently spread to Poland, Italy, and other countries (10, 13–16). Intriguingly, genotyping showed that the blaKPC-3-harboring pKpQIL plasmids found in Israel were different from the blaKPC-3-harboring plasmids carried by genetically related ST258 strains from United States (17). In contrast, Kitchel et al. found that a blaKPC-3-harboring pKpQIL plasmid from Israel had restriction and Tn4401 profiles similar to those of the blaKPC-2-harboring plasmids from ST258 isolates from the United States (18).
Although the spread of pKpQIL in Israel and Poland appears to be endemic, the distribution of pKpQIL-like plasmids in the New York metropolitan area is unknown (16, 19, 20). In this study, we determined the DNA sequence of six pKpQIL-like plasmids isolated from six strains of three different Enterobacteriaceae species (K. pneumoniae, Escherichia coli, and Enterobacter aerogenes) from New Jersey (NJ) and New York city (NYC) hospitals and compared their plasmid structures with those of pKpQIL plasmids from Israel and Italy. The sequence data provided the information to design a PCR scheme to explore the distribution of pKpQIL-like plasmids in 284 clinical K. pneumoniae isolates from 10 hospitals in NJ and NYC collected between 2002 and 2012.
(This work was presented in part at the 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy.)
MATERIALS AND METHODS
Bacterial strains.
Four KPC-producing K. pneumoniae isolates (13001, 15720, 26633, and 30799), one Escherichia coli isolate (28960), and one Enterobacter aerogenes isolate (28944) were selected from a retrospective collection of carbapenem-resistant clinical isolates. K. pneumoniae 13001 and 15720 were identified at the same hospital in NYC in 2003 and 2004, respectively, while 30799 was recovered from a second NYC hospital in 2010. K. pneumoniae 26633 was collected from a central New Jersey hospital in 2009, and E. aerogenes 28944 and E. coli 28960 were identified at a northern New Jersey hospital in 2009 and 2010, respectively. An additional 284 clinical KPC-producing K. pneumoniae isolates, collected from 10 hospitals in the New York and New Jersey areas between 2002 and 2012, were screened for the presence of pKpQIL-like plasmid by PCR (described below).
Genotyping and plasmid characterization.
Plasmid DNA was extracted using a Qiagen Plasmid Midi kit (Qiagen, Valencia, CA), followed by electroporation into E. coli DH10B (Invitrogen) using a Gene Pulser II instrument (Bio-Rad Laboratories). E. coli DH10B transformants were selected on lysogeny broth (LB) agar plates containing 100 μg/ml ampicillin or 0.5 μg/ml imipenem and then screened by multiplex real-time PCR for the presence of blaKPC genes (21). Plasmid size and number were estimated by S1 nuclease digestion of plasmid DNA, followed by pulsed-field gel electrophoresis (S1-PFGE) (22). Transformants with single plasmids were then selected for plasmid sequencing and subjected to susceptibility testing according to Clinical and Laboratory Standards Institute methods and interpretations (23, 24). Conjugation experiments were carried out in LB broth with E. coli J53AzR as the recipient using a method described previously (25).
Multilocus sequence typing (MLST) of the four K. pneumoniae strains (13001, 15720, 26633, and 30799) and one E. coli strain (28960) was performed using previously described methods for K. pneumoniae and E. coli, respectively (26, 27).
Plasmid sequencing and bioinformatics.
Plasmid DNA from E. coli DH10B transformants of six isolates (13001, 15720, 26633, 30799, 28944, and 28960) was extracted as described above and sequenced using a Roche 454 GS-FLX or Illumina MiSeq system. Sequencing reads were de novo assembled using the Roche Genome Sequencer FLX software GSA assembler or Velvet algorithm (28). Gaps between contigs were closed by PCR followed by standard Sanger sequencing. Open reading frames (ORFs) were predicted and annotated using the RAST (rast.nmpdr.org) server (29), followed by manual comparative curation. Mauve 2.3.1 was used to perform comparative genome alignment for different plasmids (30).
PCR screening for pKpQIL-like plasmids.
Complete-genome sequencing of the pKpQIL-like plasmids analyzed in this study as well as the pKpQIL and pKpQIL-IT plasmids sequenced previously (GenBank accession numbers GU595196 and JN233705) (10, 15) provided the information to develop a PCR strategy to detect this family of plasmids. The scheme includes six individual PCRs. PCR-I and PCR-VI were designed to target the IncFIIK-specific repA gene (shared by all IncFIIK plasmids) and the IncFIIK2 plasmid-specific repFIB gene, respectively. PCR-II and PCR-IV were designed to target the upstream and downstream junctions between neighboring Tn4401 and pKpQIL regions. PCR-III was used to detect the pKpQIL-associated Tn4401a isoform (with a 99-bp-deletion upstream blaKPC gene in comparison to Tn4401b isoform), and PCR-V was designed to target the pKpQIL-like plasmid backbone gene hsdR. The combination of PCR-I to -VI is able to identify pKpQIL-like plasmids with the exception that pKpQIL-Ec (from E. coli strain 28960) is positive for PCR-I to -IV and -VI but negative for -V, due to a 14.5-kb deletion (described below). The primer targeting regions for the identification of pKpQIL-like plasmids are shown in Fig. 1, and primer sequences are listed in Table 1. The PCR cycling conditions were as follows: an initial denaturation step of 95°C for 4 min, followed by 35 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 1 min (PCR-I, -III, -V, and -VI) or 2 min (PCR-II and -IV) and a final extension step of 72°C for 7 min.
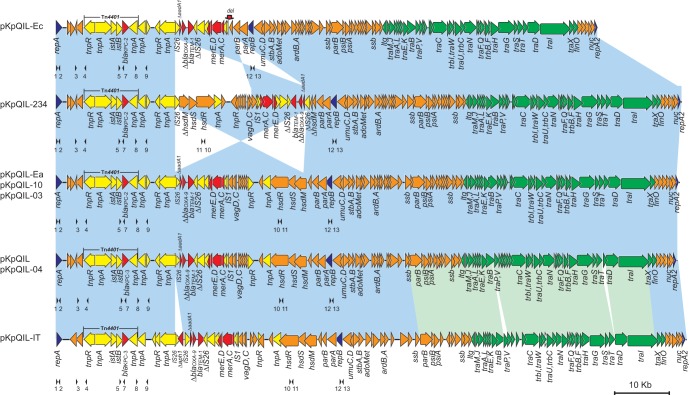
FIG 1.
Comparative analysis of pKpQIL-like plasmids. Light-blue shading denotes shared regions of homology with >99% identities, while light-green shading indicates homologies with ∼97% identities. ORFs are portrayed by arrows and colored based on predicted gene function. Orange arrows indicate plasmid scaffold regions. The genes associated with the tra locus are indicated by green arrows, and replication-associated genes are denoted as dark-blue arrows. Antimicrobial and mercury resistance genes are indicated by red arrows, while the accessory genes are indicated by yellow arrows. We note that a 24-kb region is inversely displayed in pKpQIL-234, in contrast to other pKpQIL-like plasmids. A red arrow above plasmid pKpQIL-Ec illustrates the deletion site of 14-kb sequence. Small black arrowheads beneath the plasmids indicate the locations of primers used for PCR screening; primer sequences are shown in Table 1.
TABLE 1.
Primers used for screening of pKpQIL-like plasmids in this study
| PCR | Primer no.a | Name | Sequence | Size (bp) | Target | Reference |
|---|---|---|---|---|---|---|
| PCR-I | 1 | FIIK-repA-F1 | CTTCACGTCCCGTTTTGATT | 657 | IncFII repA gene | 34 |
| 2 | FIIK-repA-R1 | CGCTTCAGCGCTTCTTTATC | 34 | |||
| PCR-II | 3 | QIL-F1 | ACAGGGAGTGCCAGGAAAG | 2,001 | Junction between Tn4401 tnpR and upstream IncFIIK2 backbone gene | This study |
| 4 | QIL-R1 | TGTATTTGCATGGCGATGAG | This study | |||
| PCR-III | 5 | Tn4401v-F(3098U) | TGACCCTGAGCGGCGAAAGC | 604b | pKpQIL-associated Tn4401a isoform | 6 |
| 6 | Tn4401v-R1 | GCAAGCCGCTCCCTCTCCAG | 35 | |||
| 7 | Tn4401v-R(3781L) | CACAGCGGCAGCAAGAAAGC | 6 | |||
| PCR-IV | 8 | QIL-F2 | GCCTCAGATAGATGCGGTAGC | 1,831 | Junction between Tn4401 ISKpn6 and downstream gene | This study |
| 9 | QIL-R2 | AAGCTGGAGACATGGAATGG | This study | |||
| PCR-V | 10 | QIL-hsdR-F1 | GGGTCGTTCACAAAGTCGAT | 498 | IncFIIK2-associated type I restriction modification system hsdR gene | This study |
| 11 | QIL-hsdR-R1 | CGTTGAGCACTTCACCAAAA | This study | |||
| PCR-VI | 12 | K2-repB-F1 | CCATTCCGATCCTTTTCTGA | 395 | IncFIIK2 repFIB gene | This study |
| 13 | K2-repB-R1 | AACGCTACTGTCCAGCCTGT | This study |
The locations of the primers are illustrated in Fig. 1.
The sizes are variable for different Tn4401 isoforms. Isoform b, 703bp, 382bp; a, 604bp; c, 487bp; d, 635bp, 314bp; e, 448bp.
In this study, a total of 284 other clinical K. pneumoniae isolates were analyzed using the PCR screening scheme described above. Identification of the epidemic K. pneumoniae ST258 clone from these clinical isolates was performed using a multiplex real-time PCR method described previously (5). The presence and allele types of blaKPC were determined by a multiplex real-time PCR method (21).
A subset (n = 10) of pKpQIL-like plasmids identified by the PCR screening approach were subjected to conjugation using E. coli J53 AzR as the recipient and following the method described above. Single blaKPC-harboring plasmids isolated from E. coli J53 transformants were digested with restriction endonuclease EcoRV (New England BioLabs, Boston, MA), and their restriction patterns were compared to those of sequenced pKpQIL plasmids in this study (9).
Statistical analysis.
The statistical analysis regarding the differences in the distributions of IncFIIK and pKpQIL-like plasmids was performed using the chi-square test (Prism, GraphPad, San Diego, CA). The differences were considered statistically significant at P ≤ 0.05.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of pKpQIL-234 (pBK26633), pKpQIL-Ec (pBK28960), and pKpQIL-10 (pBK30799) were deposited in GenBank under accession numbers KJ146689, KJ146688, and KJ146687, respectively.
RESULTS
Characterization of pKpQIL-like plasmid-harboring isolates.
Antibiotic susceptibility testing revealed that the six parental clinical isolates were resistant to all β-lactams and β-lactamase inhibitors tested in this study, including four carbapenem antimicrobial agents (imipenem, meropenem, ertapenem, and doripenem), except that 26633 was determined to be intermediate with respect to ceftazidime and cefepime (Table 2). All isolates were susceptible to tigecycline, colistin, and polymyxin B but showed differing patterns of resistance to other antimicrobial agents, including doxycycline, amikacin, gentamicin, tobramycin, ciprofloxacin, levofloxacin, and cotrimoxazole, suggesting that they carry different antimicrobial resistance determinants. The E. coli DH10B transformants displayed β-lactam resistance profiles similar to those of the parental isolates (Table 2). The overall susceptibility profiles of the parental isolates and transformants are similar to those of the pKpQIL-harboring strains and their transformants originally identified in Israel (19, 20).
TABLE 2.
Characteristics of pKpQIL-like plasmid-harboring isolates and their E. coli DH10 transformants
| Isolatea | Species | STb | MIC (μg/ml)c |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPM | ERT | MEM | DOR | CTX | CAZ | CFP | ATM | TIC/CLA | SXT | GEN | AMK | TOB | CIP | LVX | DOX | TGC | CST | PMB | |||
| 13001 | K. pneumoniae | 258 | >8 | >4 | >8 | ≥4 | >32 | >16 | ≥32 | >16 | >128/2 | >4/76 | 2 | 32 | ≥16 | ≥4 | >8 | 4 | 0.5 | 0.5 | 1 |
| 15720 | K. pneumoniae | 258 | >8 | >4 | >8 | ≥4 | >32 | >16 | ≥32 | >16 | >128/2 | >4/76 | >16 | 32 | ≥16 | ≥4 | >8 | ≤2 | ≤0.25 | 0.5 | 1 |
| 26633 | K. pneumoniae | 234 | 8 | >4 | >8 | ≥4 | 8 | 8 | 4 | >16 | >128/2 | ≤0.5/9.5 | ≤1 | ≤4 | ≤1 | ≤0.25 | ≤1 | 4 | ≤0.25 | 0.5 | 1 |
| 28611 | K. pneumoniae | 258 | >8 | >4 | >8 | ≥4 | >32 | >16 | ≥32 | >16 | >128/2 | >4/76 | 2 | ≥64 | ≥16 | ≥4 | >8 | 4 | 0.5 | 0.5 | 1 |
| 30799 | K. pneumoniae | 258 | >8 | >4 | >8 | ≥4 | >32 | >16 | ≥32 | >16 | >128/2 | >4/76 | 4 | ≤4 | ≥16 | ≥4 | >8 | 8 | 0.5 | 0.5 | 1 |
| 28944 | E. aerogenes | — | >8 | >4 | >8 | ≥4 | >32 | >16 | ≥32 | >16 | >128/2 | ≤0.5/9.5 | ≤1 | ≤4 | ≤1 | ≤0.25 | ≤1 | 8 | 1 | 0.5 | 1 |
| 28960 | E. coli | 131 | 8 | >4 | >8 | ≥4 | >32 | 16 | ≥32 | >16 | >128/2 | >4/76 | ≤1 | ≤4 | 2 | ≥4 | >8 | >16 | 0.5 | 1 | 1 |
| 13001T | E. coli | — | 4 | 4 | 8 | ≥4 | >32 | >16 | ≥32 | >16 | >128/2 | ≤0.5/9.5 | ≤1 | ≤4 | ≤1 | ≤0.25 | ≤1 | ≤2 | ≤0.25 | 0.5 | 1 |
| 15720T | E. coli | — | 8 | >4 | 8 | ≥4 | >32 | >16 | ≥32 | >16 | >128/2 | ≤0.5/9.5 | 2 | ≤4 | ≤1 | ≤0.25 | ≤1 | ≤2 | ≤0.25 | 0.5 | 1 |
| 26633T | E. coli | — | 4 | 2 | 2 | 2 | 8 | 8 | 4 | >16 | >128/2 | ≤0.5/9.5 | ≤1 | ≤4 | ≤1 | ≤0.25 | ≤1 | ≤2 | ≤0.25 | ≤0.25 | ≤0.25 |
| 28611T | E. coli | — | 8 | >4 | 8 | ≥4 | >32 | >16 | ≥32 | >16 | >128/2 | ≤0.5/9.5 | ≤1 | ≤4 | ≤1 | ≤0.25 | ≤1 | ≤2 | ≤0.25 | 0.5 | 1 |
| 30799T | E. coli | — | 4 | 2 | 4 | 2 | 16 | 16 | 8 | >16 | >128/2 | ≤0.5/9.5 | ≤1 | ≤4 | ≤1 | ≤0.25 | ≤1 | ≤2 | ≤0.25 | 0.5 | ≤0.25 |
| 28944T | E. coli | — | 8 | >4 | 8 | ≥4 | >32 | >16 | ≥32 | >16 | >128/2 | ≤0.5/9.5 | ≤1 | ≤4 | ≤1 | ≤0.25 | ≤1 | ≤2 | ≤0.25 | 0.5 | 1 |
| 28960T | E. coli | — | 4 | 1 | 2 | 2 | >32 | >16 | 16 | >16 | >128/2 | ≤0.5/9.5 | ≤1 | ≤4 | ≤1 | ≤0.25 | ≤1 | ≤2 | ≤0.25 | 0.5 | 1 |
T, E. coli DH10B transformant.
—, not tested.
MIC values were determined using broth microdilution; resistance values are indicated in boldface. IPM, imipenem; MEM, meropenem; ERT, ertapenem; DOR, doripenem; CAZ, ceftazidime; CTX, cefotaxime; CFP, cefepime; ATM, aztreonam; TIC/CLA, ticarcillin/clavulanate; AMK, amikacin; GEN, gentamicin; TOB, tobramycin; DOX, doxycycline; CIP, ciprofloxacin; LVX, levofloxacin; SXT, trimethoprim-sulfamethoxazole; TGC, tigecycline; CST, colistin; PMB, polymyxin B.
MLST showed that isolates 13001, 15720, and 30799 belonged to the epidemic K. pneumoniae ST258 clone, while 26633 was ST234 (27), and isolate 28960 is an example of the epidemic E. coli ST131 clone (26). A conjugation test showed that all the blaKPC-bearing plasmids in the six isolates were successfully transferred to E. coli J53 strains, lending credence to the hypothesis that these plasmids are conjugative in nature. S1-PFGE experiments indicated the six blaKPC-bearing plasmids were of similar sizes (∼100 kb).
Comparison of pKpQIL-like plasmids.
Six pKpQIL-like plasmids were sequenced in this study. These plasmids were named based on the isolation time, genetic background, or parental species. Accordingly, the plasmids from strains 13001 (isolated in 2003), 15720 (isolated in 2004), and 30799 (isolated in 2010) were named pKpQIL-03, -04, and -10, respectively. The plasmids from strains 26633 (from ST234 K. pneumoniae), 28944 (from E. aerogenes), and 28960 (from E. coli) were named pKpQIL-234, -Ea, and -Ec, respectively (Fig. 1).
pKpQIL-03, -04, -10, and -Ea are identical in size (113,639 bp), have G+C content of 53.9%, and harbor 125 predicted open reading frames (ORFs). This matches the published pKpQIL sequence, which is 113,637 bp in length (10). pKpQIL-234 and -Ec are, respectively, 114,464 and 99,142 bp in length, with G+C content of 53.9% and 54.0%, and harbor 125 and 110 predicted ORFs, respectively. A BLAST search of all six plasmid genomes against the GenBank database (http://blast.ncbi.nlm.nih.gov) showed they have a high degree of similarity to that of the previously published pKpQIL plasmid from Israel (10), with 100% query coverage and overall >99.9% nucleotide identity.
Mauve analysis revealed the high conservation of these plasmids, as fewer than 10 single nucleotide polymorphisms (SNPs) were identified, covering over 100 kb of plasmid genomes in comparisons of the six study plasmids to pKpQIL. The results showed that the SNP variations between pKpQIL and pKpQIL-03 (4 SNPs), -04 (4 SNPs), -10 (7 SNPs), -234 (6 SNPs), -Ec (9 SNPs), and -Ea (7 SNPs) are minimal. The majority of these SNPs are located in the intergenic regions, and the only nonsynonymous mutation found among the seven pKpQIL-like plasmids is that pKpQIL and pKpQIL-04 carry blaKPC-3 whereas the other five plasmids (pKpQIL-03, -10, -234, -Ec, and -Ea) carry blaKPC-2. To our knowledge, this is the first report showing that nearly identical plasmids carry different blaKPC variants. In contrast, the pKpQIL-IT plasmid isolated in Italy is divergent from the pKpQIL plasmids from Israel and NJ/NY, with 1,661 to 1,666 differing SNPs. Further inspection of the SNP locations revealed that >99% are located in an ∼47-kb fragment from gene ssb to gene traI, encompassing nearly the entire tra operon. The remaining sequence in pKpQIL-IT is nearly identical to the sequences of the other seven pKpQIL plasmids (>99.9% similarity) (Fig. 1).
pKpQIL, -03, -04, -10, -234, -Ec, and -Ea carry the same number of antimicrobial and mercury resistance genes, including blaKPC-2/-3, blaTEM-1, ΔblaOXA-9, ΔaadA1, merA, merC, merD, and merE (Fig. 1). pKpQIL-IT has an additional resistance gene, aphA1, which is carried on a composite transposon-like element, IS26-aphA1-IS26, located downstream of Tn4401 (15).
Interestingly, analysis of the plasmid genome of pKpQIL-234 revealed a 26.7-kb inversion flanked by two inverted IS26 elements. This inversion ranges from the IS26 downstream of Tn4401 to the type I restriction-modification DNA-methyltransferase hsdM gene. The inversion presumably originated from an IS26 insertion in hsdM at nucleotide (nt) 215, resulting in the splitting of hsdM, followed by an inversion between two oppositely oriented IS26 elements (Fig. 1). Moreover, examination of the pKpQIL-Ec plasmid genome identified a 14.5-kb deletion, spanning the region between the downstream portion of the mer operon and the upstream portion of the parAB operon and encompassing the entire type I restriction-modification system (hsdRSM) and putative virulence genes vagC-vagD (31).
Distribution of pKpQIL-like plasmids in NJ/NY hospitals.
As part of an ongoing surveillance project, hospitals in NY and NJ (The CRE Hospital Network) routinely submit carbapenem-resistant and -susceptible K. pneumoniae isolates to the Public Health Research Institute TB Center for genotyping. A total of 284 clinical KPC-producing K. pneumoniae isolates collected between 2002 and 2012 from 10 hospitals were evaluated by PCR for the presence of pKpQIL-like plasmid markers. Among them, 148 (52.1%) were blaKPC-2 positive, while 136 (47.9%) were blaKPC-3 positive and 245 (86.3%) belonged to the ST258 clone (Table 3).
TABLE 3.
Distribution of pKpQIL-like plasmids in clinical K. pneumoniae isolates
| Comparison | No. (%) ofa: |
Total distribution (n) | |
|---|---|---|---|
| IncFIIK plasmids | pKpQIL-like plasmids | ||
| KPC-2 vs KPC-3 | |||
| KPC-2 | 128 (86.5) | 89 (60.1) | 148 |
| KPC-3 | 48 (35.3)* | 12 (8.8)* | 136 |
| ST258 vs non-ST258 | |||
| Non-ST258 | 26 (66.7) | 13 (33.3) | 39 |
| ST258 | 150 (61.2) | 88 (35.9) | 245 |
*, P < 0.01 (compared with KPC-2-bearing isolates).
Among these 284 K. pneumoniae isolates, 62.0% (n = 176) were positive for IncFIIK repA PCR (PCR-1), showing the wide spread of this group of plasmids in KPC-bearing K. pneumoniae. Interestingly, the prevalence of IncFIIK plasmids in KPC-2-bearing isolates (86.5%) was significantly higher than that in KPC-3-bearing isolates (35.3%) (P < 0.01). Among the 284 blaKPC-positive K. pneumoniae isolates, pKpQIL-like plasmids were identified in 101 (35.6%) found in 9 of the 10 submitting hospitals. Among the 101 pKpQIL-like plasmids, 89 (88.1%) carry blaKPC-2, while only 12 (11.9%) harbor blaKPC-3 (P < 0.01). pKpQIL-like plasmids were identified in 35.9% of ST258 and in 33.3% of non-ST258 K. pneumoniae isolates (Table 3).
Ten (9.9%) of the 101 pKpQIL-like plasmids were selected and successfully transferred to E. coli J53 by conjugation. Comparative restriction enzyme digests of the 10 plasmids using EcoRV displayed profiles that were indistinguishable from or highly similar to the pKpQIL-10 profile (data not shown). This finding showed that the PCR screening method, described in this study and used on the parental K. pneumoniae isolates, was highly specific in correctly identifying those strains that harbor pKpQIL-like plasmids.
DISCUSSION
In this study, we sequenced six blaKPC-harboring plasmids from clinical isolates collected from 2003 to 2010 in NYC and NJ hospitals. The sequenced plasmids displayed high identities to the prototype pKpQIL plasmid, originally isolated from an ST258 strain in Israel (10). Comparative plasmid genome analysis suggested that the six NY/NJ plasmids and pKpQIL are virtually the same plasmid that evolved from a common ancestor. pKpQIL initially emerged in Israel in 2006, carrying blaKPC-3 and blaTEM-1 and associated with K. pneumoniae clone Q based on its PFGE pattern (and was later assigned the designation ST258 by MLST) (32).
PFGE typing showed that Israeli pKpQIL-harboring K. pneumoniae clone Q isolates were genetically related to several ST258 isolates collected in NYC in 2000 and in NJ and Arizona in 2006 (17). Interestingly, plasmid characterization showed that the blaKPC-3-harboring plasmids from the U.S. ST258 isolates (from New York, New Jersey, and Arizona) were distinct from blaKPC-3-harboring pKpQIL collected in Israel (17). The results suggested that the Israeli ST258 strain may have originated in the United States and spread to Israel as a result of international travel; however, the origin of pKpQIL remains intriguing, as the same K. pneumoniae strains carry different blaKPC-3-harboring plasmids.
Part of the mystery appears to have been unraveled in this current study. Among the six completely sequenced pKpQIL-like plasmids, two (pKpQIL-03 and -04) predate (2003 and 2004) the emergence of the pKpQIL in Israel (2006). pKpQIL-03 harbors blaKPC-2, while pKpQIL-04 contains blaKPC-3, which is the same blaKPC variant identified in the Israeli pKpQIL prototype plasmid (Fig. 1). More importantly, the PCR screening of pKpQIL-like plasmid in 10 hospitals in NY/NJ of isolates collected from 2002 to 2012 supported the idea that this plasmid is more significantly associated with KPC-2 than with KPC-3 in this area (Table 3). This finding is in agreement with results of previous plasmid restriction digestion analysis indicating that a blaKPC-3-harboring pKpQIL-like plasmid collected in Israel had a restriction profile similar to those of blaKPC-2-harboring plasmids collected in the United States (18). The most plausible explanation is that a single KPC-3-positive pKpQIL-harboring ST258 strain (e.g., pKpQIL-04-harboring ST258) was transferred from the United States into Israel, where it became the predominant clone Q. The paradoxical plasmid profiles differences between the U.S. and Israeli blaKPC-3-harboring ST258 strains were probably due to a strain selection bias, since ∼90% of pKpQIL-like plasmids in NY/NJ carry blaKPC-2 instead of blaKPC-3. However, the reason for the disproportionate distribution of blaKPC-2 and blaKPC-3-harboring pKpQIL plasmids in our area still needs further study.
In this study, the six sequenced pKpQIL-like plasmids are highly similar to the prototype pKpQIL plasmid, with less than 10 SNP differences in more than100 kb of plasmid sequence. The stability is more striking given that the pKpQIL plasmids analyzed in this study were collected more than 7 years ago and from different species and STs. The one noticeable outlier is pKpQIL-IT, isolated from Italy in 2009 and harboring a variable 47-kb fragment that contains more than 1,600 SNPs in comparison to the other pKpQIL plasmids (Fig. 1). The reason for the sequence variability in pKpQIL-IT is still not clear. It is likely the 47-kb region was acquired from other IncFII plasmids through homologous recombination, as large-fragment recombination among different plasmids commonly occurs. Another possible explanation might be that the variability is the result of a high mutation rate due to a different selection pressure of Italian isolates, as the pKpQIL-IT-harboring ST258 strain also carries porin mutants with OmpK35 and OmpK36 dysfunction (15), which could confer resistance to carbapenems and other antimicrobial agents.
The plasticity of the genome of pKpQIL-Ec is observed in the 14.5-kb deletion compared to other pKpQIL-like plasmids, although the mechanism for this deletion remains unknown as no significant repeated sequences are found on either side of the junctions (data not shown). A similar deletion has previously been reported in pKpQIL-like plasmids by Villa et al. (33), who described a carbapenem-resistant ST258 strain that became susceptible due to the large deletion of a 36-kb fragment, including the full length of a blaKPC-3-harboring Tn4401a transposon. In that instance, those authors suggested that the deletion was the result of an IS26-mediated loop out (33). Similarly, the 24-kb inversion in pKpQIL-234, flanked by two IS26 elements, is also likely the result of an intramolecular transposition mediated by IS26. Our results and those of studies by others indicate that pKpQIL-like plasmids are in constant genetic flux (15, 33), and we have seen examples of insertions (IS26-aphA1-IS26 in pKpQIL-IT), deletions (pKpQIL-Ec), and reversions (pKpQIL-234). It is evident that insertion elements, such as IS26, play an important role in creating the plasmid architecture of new pKpQIL plasmid variants.
In this study, pKpQIL-like plasmids were identified in different K. pneumoniae clones (STs) and in different species (K. pneumoniae, E. aerogenes, and E. coli). Our surveillance of 284 KPC-producing K. pneumoniae isolates identified this plasmid in more than one-third of the collection and in isolates from 9 of the 10 submitting hospitals, highlighting its success in the epicenter of the KPC epidemic. More importantly, 33.3% of non-ST258 KPC-producing K. pneumoniae strains carry this plasmid, indicating that pKpQIL-like plasmids have spread beyond the K. pneumoniae ST258 clone into other STs. Collectively, the distribution of plasmids in different species and STs suggests that interstrain and interspecies transfer significantly contributed to the spread of this plasmid in our area.
In conclusion, we sequenced six pKpQIL-like plasmids from different STs and species collected in NY/NJ hospitals from 2003 to 2010. The epidemic pKpQIL-like plasmids are very prevalent among the resistant strains in this region. The finding of pKpQIL-like plasmid in this study from isolates that predate the initial report of KPC in Israel provides evidence that this resistance plasmid may have originated in the United States. pKpQIL plasmids are spreading clonally in ST258 strains as well as horizontally to different sequence types and species. The disproportionate distribution of blaKPC-2 and blaKPC-3-harboring pKpQIL plasmids in the epicenter northeastern region of the United States warrants further study.
ACKNOWLEDGMENTS
This study was supported by a grant (to B.N.K.) from the National Institutes of Health (1R01AI090155). This work was also supported by Public Health Service grant R01AI072219 and R01AI063517 (to R.A.B.) from the National Institutes of Health and funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program, and the Geriatric Research Education and Clinical Center (VISN 10) to R.A.B.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
B.N.K. discloses that he holds two patents that focus on using DNA sequencing to identify bacterial pathogens.
Footnotes
Published ahead of print 10 March 2014
REFERENCES
- 1.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13:785–796. 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151–1161. 10.1128/AAC.45.4.1151-1161.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682–707. 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giakkoupi P, Papagiannitsis CC, Miriagou V, Pappa O, Polemis M, Tryfinopoulou K, Tzouvelekis LS, Vatopoulos AC. 2011. An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009–10). J. Antimicrob. Chemother. 66:1510–1513. 10.1093/jac/dkr166 [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Chavda KD, Mediavilla JR, Zhao Y, Fraimow HS, Jenkins SG, Levi MH, Hong T, Rojtman AD, Ginocchio CC, Bonomo RA, Kreiswirth BN. 2012. Multiplex real-time PCR for detection of an epidemic KPC-producing Klebsiella pneumoniae ST258 clone. Antimicrob. Agents Chemother. 56:3444–3447. 10.1128/AAC.00316-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, Carmeli Y, Gales AC, Venezia SN, Quinn JP, Nordmann P. 2010. Worldwide diversity of Klebsiella pneumoniae that produce b-lactamase blaKPC-2 gene. Emerg. Infect. Dis. 16:1349–1356. 10.3201/eid1609.091389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18:263–272. 10.1016/j.molmed.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 8.Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Huard RC. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 53:1998–2004. 10.1128/AAC.01355-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob. Agents Chemother. 57:269–276. 10.1128/AAC.01648-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob. Agents Chemother. 54:4493–4496. 10.1128/AAC.00175-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Chavda KD, Al Laham N, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob. Agents Chemother. 57:5019–5025. 10.1128/AAC.01397-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant KA, Van Schooneveld TC, Thapa I, Bastola D, Williams LO, Safranek TJ, Hinrichs SH, Rupp ME, Fey PD. 2013. KPC-4 is encoded within a truncated Tn4401 in an IncL/M plasmid, pNE1280, isolated from Enterobacter cloacae and Serratia marcescens. Antimicrob. Agents Chemother. 57:37–41. 10.1128/AAC.01062-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hidalgo-Grass C, Warburg G, Temper V, Benenson S, Moses AE, Block C, Strahilevitz J. 2012. KPC-9, a novel carbapenemase from clinical specimens in Israel. Antimicrob. Agents Chemother. 56:6057–6059. 10.1128/AAC.01156-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warburg G, Hidalgo-Grass C, Partridge SR, Tolmasky ME, Temper V, Moses AE, Block C, Strahilevitz J. 2012. A carbapenem-resistant Klebsiella pneumoniae epidemic clone in Jerusalem: sequence type 512 carrying a plasmid encoding aac(6′)-Ib. J. Antimicrob. Chemother. 67:898–901. 10.1093/jac/dkr552 [DOI] [PubMed] [Google Scholar]
- 15.García-Fernández A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 56:2143–2145. 10.1128/AAC.05308-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baraniak A, Grabowska A, Izdebski R, Fiett J, Herda M, Bojarska K, Zabicka D, Kania-Pudlo M, Mlynarczyk G, Zak-Pulawska Z, Hryniewicz W, Gniadkowski M. 2011. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008–2009. Antimicrob. Agents Chemother. 55:5493–5499. 10.1128/AAC.05118-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navon-Venezia S, Leavitt A, Schwaber MJ, Rasheed JK, Srinivasan A, Patel JB, Carmeli Y. 2009. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob. Agents Chemother. 53:818–820. 10.1128/AAC.00987-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365–3370. 10.1128/AAC.00126-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leavitt A, Carmeli Y, Chmelnitsky I, Goren MG, Ofek I, Navon-Venezia S. 2010. Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob. Agents Chemother. 54:3002–3006. 10.1128/AAC.01818-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S. 2010. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J. Antimicrob. Chemother. 65:243–248. 10.1093/jac/dkp417 [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, Kreiswirth BN. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (blaKPC) variants. J. Clin. Microbiol. 49:579–585. 10.1128/JCM.01588-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240. 10.1006/abio.1995.1220 [DOI] [PubMed] [Google Scholar]
- 23.CLSI. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—ninth edition (M07-A9). CLSI, Wayne, PA [Google Scholar]
- 24.CLSI. 2014. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement, M100-S24. CLSI, Wayne, PA [Google Scholar]
- 25.Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, Hooper DC. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242–2248. 10.1128/AAC.47.7.2242-2248.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182. 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pullinger GD, Lax AJ. 1992. A Salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol. Microbiol. 6:1631–1643. 10.1111/j.1365-2958.1992.tb00888.x [DOI] [PubMed] [Google Scholar]
- 32.Leavitt A, Navon-Venezia S, Chmelnitsky I, Schwaber MJ, Carmeli Y. 2007. Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob. Agents Chemother. 51:3026–3029. 10.1128/AAC.00299-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villa L, Capone A, Fortini D, Dolejska M, Rodriguez I, Taglietti F, De Paolis P, Petrosillo N, Carattoli A. 2013. Reversion to susceptibility of a carbapenem-resistant clinical isolate of Klebsiella pneumoniae producing KPC-3. J. Antimicrob. Chemother. 68:2482–2486. 10.1093/jac/dkt235 [DOI] [PubMed] [Google Scholar]
- 34.Chen L, Chavda KD, Melano RG, Jacobs MR, Levi MH, Bonomo RA, Kreiswirth BN. 2013. Complete sequence of a blaKPC-2-harboring IncFIIK1 plasmid from a Klebsiella pneumoniae sequence type 258 strain. Antimicrob. Agents Chemother. 57:1542–1545. 10.1128/AAC.02332-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Chavda KD, Melano RG, Hong T, Rojtman AD, Jacobs MR, Bonomo RA, Kreiswirth BN. 3 February 2014. A molecular survey of the dissemination of two blaKPC-harboring IncFIA plasmids in New Jersey and New York hospitals. Antimicrob. Agents Chemother. 10.1128/AAC.02749-13 [DOI] [PMC free article] [PubMed] [Google Scholar]