Abstract
Bacterial lipases play important roles in bacterial metabolism and environmental response. Our laboratory recently discovered that a novel lipoprotein lysophospholipase, VolA, localizes on the surface of the Gram-negative aquatic pathogen Vibrio cholerae. VolA functions to cleave exogenous lysophosphatidylcholine, freeing the fatty acid moiety for use by V. cholerae. This fatty acid is transported into the cell and can be used as a nutrient and, more importantly, as a way to alter the membrane architecture via incorporation into the phospholipid biosynthesis pathway. There are few examples of Gram-negative, surface-exposed lipoproteins, and VolA is unique, as it has a previously undercharacterized function in V. cholerae membrane remodeling. Herein, we report the biochemical characterization of VolA. We show that VolA is a canonical lipoprotein via mass spectrometry analysis and demonstrate the in vitro activity of VolA under a variety of conditions. Additionally, we show that VolA contains a conserved Gly-Xaa-Ser-Xaa-Gly motif typical of lipases. Interestingly, we report the observation of VolA homologs in other aquatic pathogens. An Aeromonas hydrophila VolA homolog complements a V. cholerae VolA mutant in growth on lysophosphatidylcholine as the sole carbon source and in enzymatic assays. These results support the idea that the lipase activity of surface-exposed VolA likely contributes to the success of V. cholerae, improving the overall adaptation and survival of the organism in different environments.
INTRODUCTION
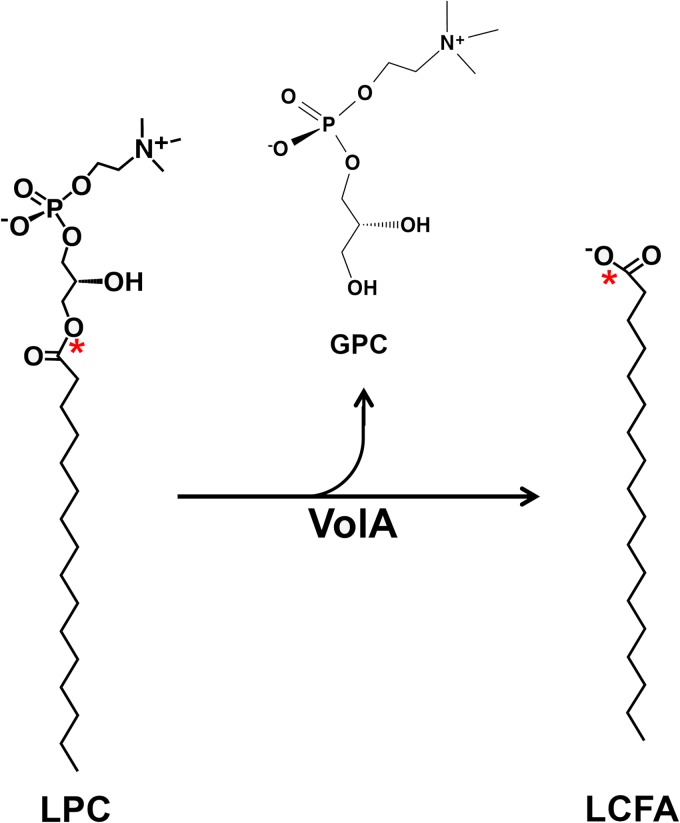
Recent work from our laboratory revealed the presence of a unique membrane-anchored lipase, VolA, localized on the surface of Vibrio cholerae cells (1). We demonstrated that VolA is required for growth when lysophosphatidylcholine (LPC) (Fig. 1) serves as the sole carbon source. We hypothesized that VolA could cleave exogenous lysophosphatidylcholine into long-chain fatty acid (LCFA) derivatives (Fig. 1), allowing them to be brought into the cell via a coexpressed fatty acid transporter, FadL. These data, taken together with the presence of a conserved lipase domain identified in V. cholerae (2) and the inability of wild-type V. cholerae to grow on phosphatidylcholine as a sole carbon source, suggest that VolA acts as a lysophospholipase in vivo.
FIG 1.
Proposed reaction scheme for VolA. Structures of lipids and proposed reaction for the lysophospholipase activity of VolA. VolA is predicted to act as a lysophospholipase A, hydrolyzing the ester bond between the fatty acid moiety and the glycerophosphocholine (GPC) head group. The asterisks indicate the position of the 14C label on the LPC substrate used during in vitro assays.
Phospholipases are members of the acylhydrolase family of enzymes, acting on ester bonds in phospholipid targets (3). The structure and function of such acylhydrolases are conserved and have been well studied. All contain a characteristic fold (4, 5), sharing a conserved lipase motif (2). In our previous work (1), we noted that VolA contains this conserved motif, which strongly implicates it as a lipase. Phospholipases vary in terms of their site of action on the phospholipid. Some target phosphate bonds, while others hydrolyze bonds between the glycerol moiety and the acyl chains. Most microbial lysophospholipases hydrolyze the ester bond between the acyl chain and the polar head group, releasing a free fatty acid (6). It is likely that VolA functions similarly to existing bacterial lysophospholipases, given that V. cholerae can survive on lysophosphatidylcholine, but not phosphatidylcholine, as the sole carbon source. It is unknown whether the enzymatic activity of VolA requires a cofactor, as both metal-dependent and metal-independent phospholipases have been identified in a variety of bacterial species (7, 8).
VolA contains a predicted N-terminal lipoprotein signal peptide. Previously, we demonstrated that VolA localizes to the surfaces of V. cholerae cells (1), suggesting that VolA could be anchored to the outer leaflet of the outer membrane by N-terminal lipidation. However, physical characterization of VolA is required to confirm its status as a lipoprotein.
Herein, we describe the biochemical characterization of VolA. Elucidation of the enzymatic activity of VolA will aid our understanding of how it contributes to bacterial fitness in both aquatic and host environments. This work confirms that VolA both is a lipoprotein and hydrolyzes lysophosphatidylcholine substrates in a metal-independent manner. We show that a conserved serine within a conserved Gly-Xaa-Ser-Xaa-Gly lipase motif is required for enzymatic activity. Additionally, we demonstrate that a homolog of VolA found in the aquatic pathogen Aeromonas hydrophila is able to complement a VolA mutant of V. cholerae. This suggests that lipase activity may be conserved across the VolA homologs found in various Gram-negative aquatic organisms. VolA is a novel lysophospholipase, the first surface-anchored lipase identified in Gram-negative organisms. Characterization of such an enzyme will reveal its larger role in microbial metabolism and survival as well as describing the features of a potentially new lipase class.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains were generally grown at 37°C on LB broth with the exception of LPC growth experiments. Growth experiments using LPC as the sole carbon source were conducted in modified M9 minimal media (1.7 mM MgSO4, 0.117 mM CaCl2, 0.03 mM FeSO4) using 2 mM C16:0 LPC as the carbon source and 6 mM Brij-56 for lipid stability in solution (9). To establish bacterial growth, readings of the optical density at 600 nm (OD600) were taken either once at 24 h for single-read experiments or every hour for multiple reads.
Site-directed mutagenesis of volA and generation of expression plasmids.
Site-directed mutagenesis of volA was performed using the QuikChange primer design kit (Agilent). Briefly, primers (see Table S2 in the supplemental material) were used to amplify a copy of the pVolA plasmid (1) containing a single amino acid change. This amplification was digested with DpnI to remove any plasmid carrying a wild-type copy of volA, and the resulting mutant plasmid was used to transform E. coli BL21Star (DE3). The A. hydrophila pla gene was cloned into pWSK29 as previously described (1).
Preparation of cell extracts and washed membrane.
A 200-ml portion of culture was induced with 0.5 mM IPTG upon inoculation and grown at 37°C to an OD600 of ∼1.0 and harvested via centrifugation at 10,000 × g for 10 min. Samples were prepared at 4°C as previously described (10). Protein concentration was established by bicinchoninic acid (BCA) assay using bovine serum albumin as a standard. Purified cell extracts and membranes were stored at −20°C.
Expression and purification of V. cholerae VolA.
Escherichia coli strain BL21(DE3) containing pVolA-8H (see Table S1 in the supplemental material) was grown overnight in 50 ml of LB broth at 37°C. This overnight was used to inoculate a 1-liter culture of LB broth with 100 μg ml−1 ampicillin and induced with 500 μM IPTG (isopropyl-β-d-thiogalactopyranoside). The culture was harvested at an OD600 of ∼1.0 via centrifugation at 10 000 × g for 10 min. The resulting cell pellet was washed in 40 ml 50 mM HEPES, pH 7.5. The pellet was then resuspended in ∼20 ml of buffer A (250 mM NaCl, 20 mM imidazole, 10 mM HEPES [pH 7.5]), containing either 2% Triton X-100 (for enzymatic assays) or 7 M urea (for mass spectrometry) and lysed via sonication in a Branson Sonifier 250. Cell debris was removed from the sample via centrifugation at 10,000 × g for 20 min. A GE Life Sciences AKTA fast protein liquid chromatograph (FPLC) was used to load the sample on a 1-ml His-Trap FF column charged with Ni-Sepharose 6 Fast Flow, and the column was washed with 20 column volumes of buffer A. Protein was purified via a 30-ml continuous gradient from 100% buffer A to 50% buffer B (250 mM NaCl, 500 mM imidazole, 10 mM HEPES [pH 7.5], containing either 2% Triton X-100 or 7 M urea), applied at 0.5 ml min−1. The protein was eluted at ∼80 mM imidazole. SDS-PAGE was used to confirm which fractions contained VolA, which were pooled and concentrated in a 15-ml Amicon Ultra concentrator (Millipore). Concentrated VolA was further purified using the AKTA FPLC and a 24 ml Superdex 200 gel filtration column equilibrated with buffer C (150 mM NaCl, 10 mM HEPES [pH 7.5], 2% Triton X-100 or 7 M urea) at 1 ml min−1. Purified protein was collected in 500-μl fractions, and VolA purity was established via SDS-PAGE. The molecular weight determination of VolA was performed via mass spectrometry by the ICMB Protein and Metabolite Analysis Facility using a 4000 QTRAP instrument (AB Sciex).
Assay of VolA lysophospholipase activity.
Lysophospholipase activity was assayed under in a 22.5-μl reaction mixture containing 50 mM HEPES (pH 7.5), 0.4% Triton X-100, and 20 μM C16:0 l-1-[palmitoyl-1-14C] (111,000 cpm nmol−1) (PerkinElmer). For membrane assays, protein was added at 0.1 mg ml−1 as the enzyme source; for pure-protein assays, enzyme was added at a concentration of 0.002 mg ml−1 (24 nM VolA). In assay mixtures supplemented with metal ions or metal ion chelators, CaCl2, MgCl2, EDTA, or EGTA was added to a final concentration of 10 mM. Reaction mixtures were incubated at 30°C for 15 min and terminated by spotting 10-μl portions on silica gel 60 thin-layer chromatography (TLC) plates. The products of the reaction were separated using a chloroform–methanol–28% ammonia hydroxide (65:25:8, vol/vol) solvent system. Temperature dependence was assayed by using standard enzymatic conditions and incubating the reaction mixture at 4 to 42°C. Dependence on pH was established via a stepwise gradient from pH 4 to 9 using a modified tribuffer system (11), with 100 mM sodium acetate, 50 mM 2-(bis-2(hydroxyethyl)imino-2-(hydroxymethyl))-1,3-propanediol, and 50 mM Tris. TLC plates were analyzed and quantitative densitometry was performed via Quantity One software and a personal molecular imager phosphorimager (Bio-Rad).
RESULTS
Membranes containing VolA are capable of hydrolyzing lysophosphatidylcholine.
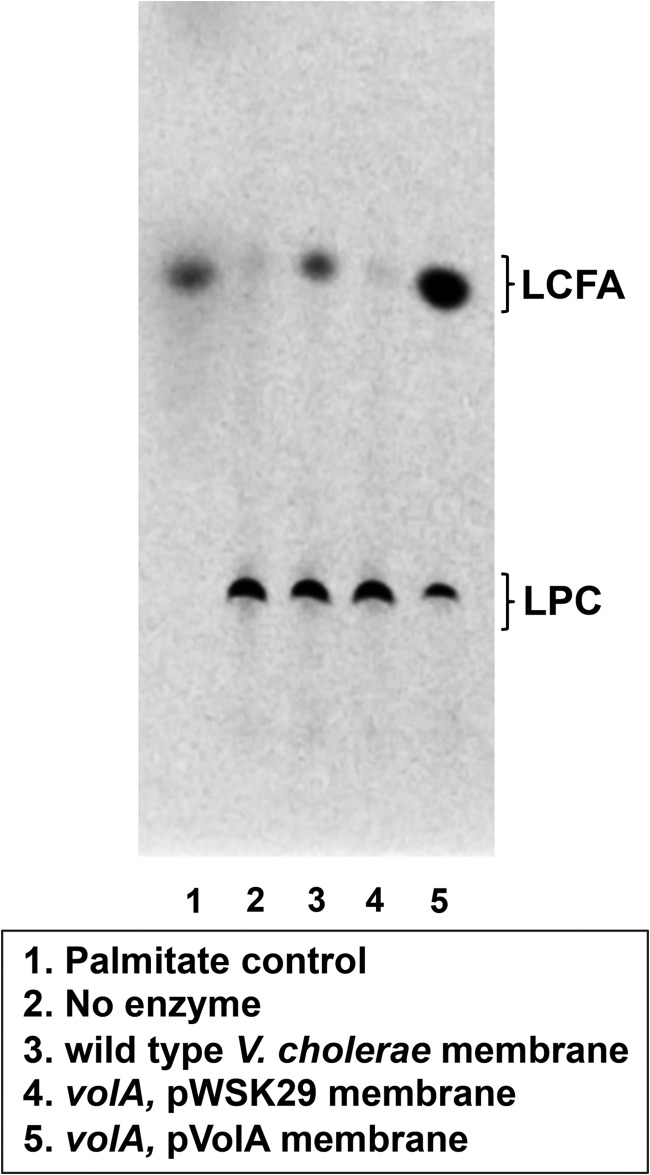
To determine if VolA displays lysophospholipase activity, bacteria (strains listed in Table S1 in the supplemental material) were grown in LB, and the washed membrane fraction was isolated as previously described (10). Membranes were assayed for possible phospholipase activity using C16:0 L-1-[palmitoyl-1-14C] (111,000 cpm nmol−1) (PerkinElmer) as the substrate, and the reaction products were analyzed by thin-layer chromatography (Fig. 2). When wild-type or VolA-complemented V. cholerae membranes were assayed, the TLC results showed two spots, one with an Rf of 0.09, matching that of intact (C16:0)-LPC, and one with an Rf of 0.49, running equidistant with a 3H-palmitate control. We consider this spot to be freed C16:0 LCFA. Membranes derived from bacteria expressing VolA display the released fatty acid, whereas membranes isolated from a volA mutant fail to hydrolyze LPC. Also, membranes isolated from the complemented mutant showed increased production of the free fatty acid reaction product with wild-type membranes yielding a specific activity of 3.1 nmol min−1 mg−1 and membranes overexpressing VolA showing a 5-fold increase in lipase activity (15.1 nmol min−1 mg−1). Together, these data confirm the expected the reaction scheme for VolA lysophospholipase activity displayed in Fig. 1.
FIG 2.
Lysophospholipase activity of VolA-containing membranes. V. cholerae membranes cleaved 14C-labeled C16:0 lysophosphatidylcholine in a VolA expression-dependent manner. The lysophospholipid substrate and the LCFA reaction product were separated by TLC. 3H-palmitate was used as a TLC standard.
Purification of VolA lipoprotein.
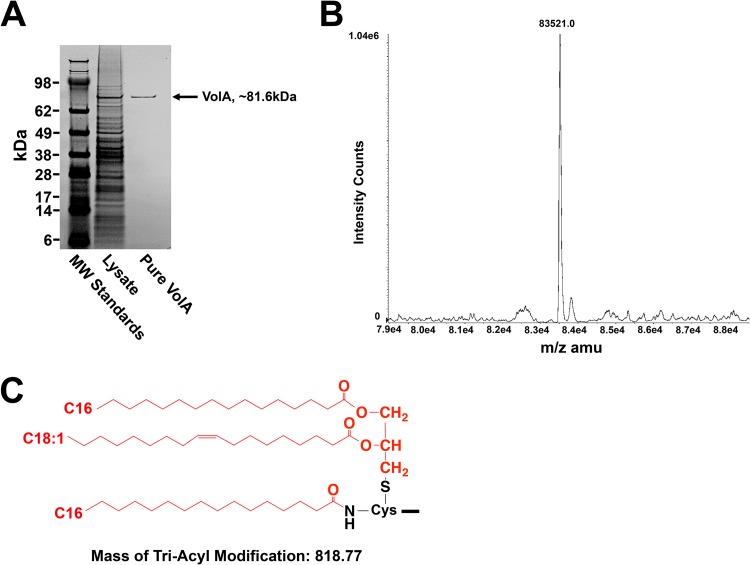
Bioinformatic analysis revealed a putative lipoprotein signal sequence within VolA. To investigate the physical and biochemical properties of purified VolA, the vca0863 gene, encoding VolA, was cloned into pET21a with a C-terminal 8× histidine fusion and expressed in E. coli BL21(DE3). The protein was solubilized and initially purified using affinity chromatography, followed by a second round of purification using a gel filtration column. A purified protein of ∼81 kDa was observed on SDS-PAGE, corresponding to VolA (Fig. 3A). In order to establish VolA as a lipoprotein, purified protein was analyzed via liquid chromatography/tandem mass spectrometry. The expected mass of 8×-His-tagged VolA as predicted by the Scripps Protein Calculator is 82,702.5 m/z (http://protcalc.sourceforge.net/). The deconvoluted spectra showed a predominant peak with a mass of 83,521.0 m/z (Fig. 3B), a difference of ∼818.53 m/z. This mass difference is likely due to the triacyl modification (predicted mass of 818.77 m/z) that is covalently added to the N terminus of lipoproteins in Gram-negative organisms. Prediction of the VolA N-terminal cysteine modification (Fig. 3C) shows that addition of a C16:0/C18:1 diacylglycerol via a thioether bond and a C16:0 fatty acid via an amide linkage would result in the observed mass difference.
FIG 3.
Purification and mass spectrometry of the VolA lipoprotein. (A) SDS-PAGE of purified VolA (∼82-kDa protein). (B) Mass spectrometry analysis showed a mature protein with a predominant mass of 83,521.0, a difference of 818.53 from the expected mass. (C) Predicted N-terminal modification of the VolA lipoprotein. Triacylation with two C16:0 and one C18:1 fatty acids (predicted mass, 818.77) would result in the observed mass of mature VolA. The lipid moiety is in red.
VolA acts as a lysophospholipase in vitro in a metal-independent manner.
In order to establish if VolA was responsible for the lysophospholipase activity observed in the membranes of wild-type and complemented strains, purified VolA was assayed for lysophospholipase activity using 14C-labeled C16:0 LPC. Similar to the TLC data obtained by assaying isolated membranes, there were two reaction products (Fig. 4). One, with an Rf of 0.09, ran equidistant with the no-enzyme LPC control. The second reaction product, with an Rf of 0.49, was liberated C16:0 LCFA, confirming that purified VolA acts as a lysophospholipase in vitro. The purified enzyme had a specific activity of 404 nmol min−1 mg−1, an ∼128-fold increase over the in vitro activity of purified wild-type membranes.
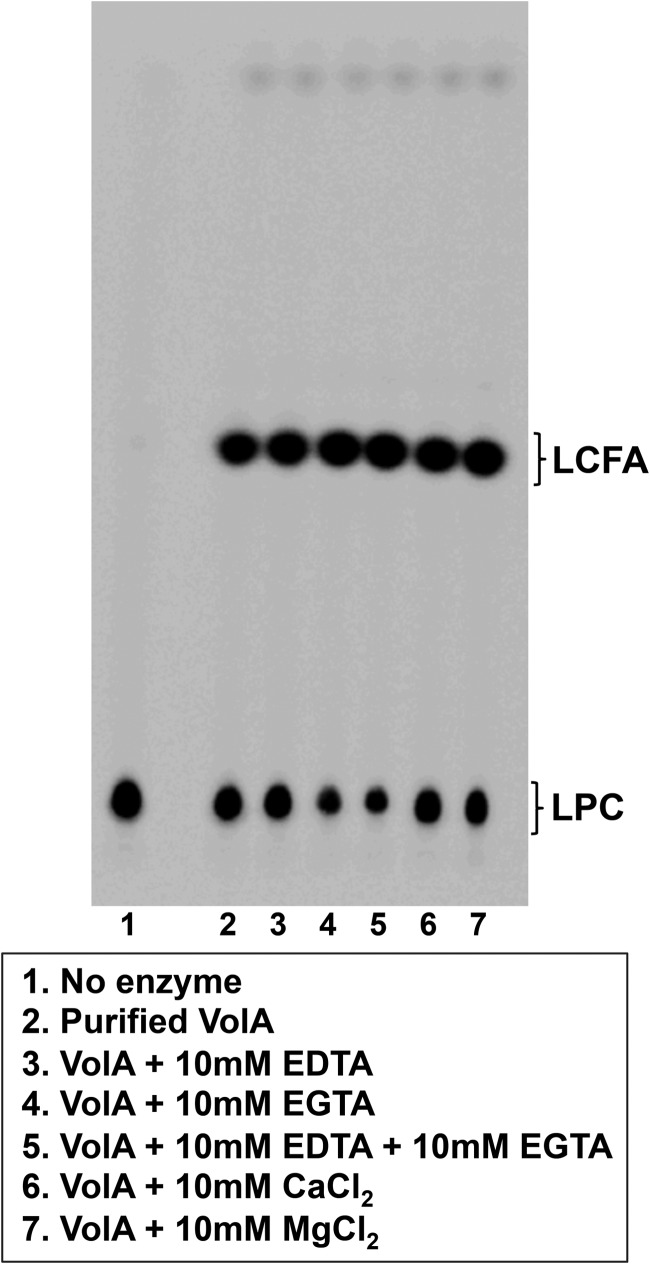
FIG 4.
VolA is a metal-independent lysosphospholipase. Purified VolA retained wild-type activity in the presence of the metal ion chelators EDTA and EGTA. Addition of supplemental metal ions (Ca2+ and Mg2+) had no effect on the activity of VolA.
Because many bacterial phospholipases are metal dependent, with Ca2+ or Mg2+ being a common requirement, we sought to determine whether VolA lipase activity displayed similar requirements. To further characterize the lysophospholipase activity of purified VolA, the enzymatic assay conditions were altered to include either metal ions or metal ion chelators. Both EDTA and EGTA were added to the reaction mixture, either separately or together, to test whether loss of metal ions in the active site of VolA might have an adverse effect on the activity of the enzyme (Fig. 4). While the addition of EDTA alone had no effect on the lipase activity of VolA, addition of EGTA by itself or in combination with EDTA caused a slight increase in the conversion of LPC to the LCFA product. When metal ions were used to supplement the reaction mixture, there was no effect on the conversion of LPC to LCFA compared to enzyme alone (Fig. 4). Taken together, these results suggest that VolA lysophospholipase activity does not require the presence of metal ions.
Enzymatic parameters of purified VolA.
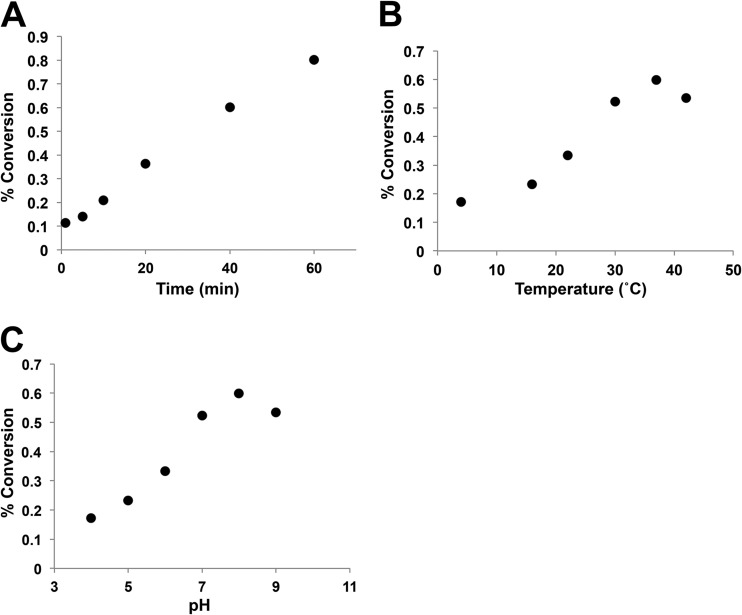
In vitro assays were performed to establish the parameters of VolA activity (Fig. 5). Enzymatic activity was measured via quantitative TLC. Percent conversion of LPC to LCFA was calculated using densitometry to quantitate the LPC and LCFA spots. We show that VolA activity is linear with time (Fig. 5A), which is characteristic of an enzyme. We also assessed the temperature and pH dependence of VolA. Dependence on temperature was established via a step gradient from 4°C to 42°C (Fig. 5B), with VolA showing the largest amount of LPC conversion at 37°C. Finally, the pH dependence of VolA was determined by varying the pH (Fig. 5C). The pH was varied between 4 and 9; VolA showed optimal enzymatic activity at a pH of 8.0.
FIG 5.
Enzymatic parameters of VolA in vitro. (A) Conversion of 14C-LPC into C16:0 LCFA at different times under standard conditions. (B) Assay of VolA activity at various temperatures. (C) VolA activity as a function of pH.
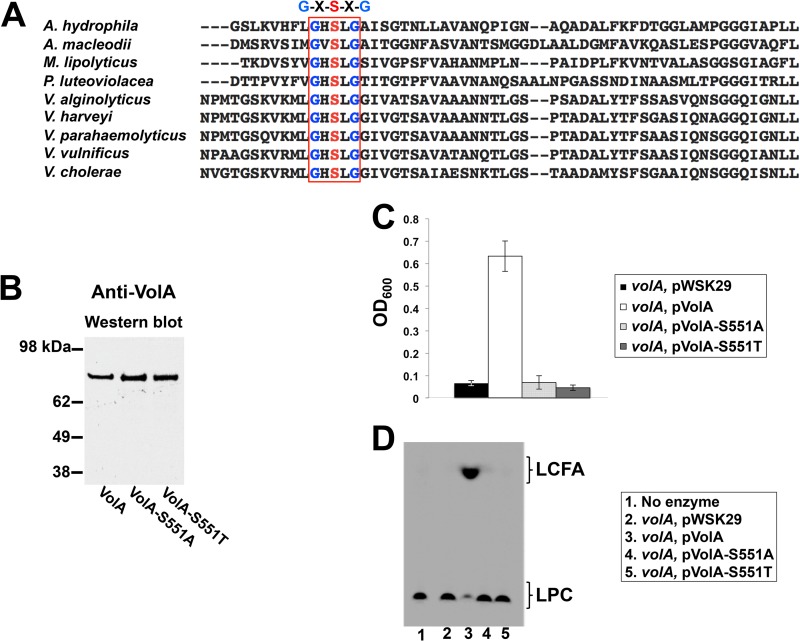
A catalytic serine within a conserved lipase domain is required for VolA activity.
Previously, a well-conserved Gly-Xaa-Ser-Xaa-Gly lipase domain was identified within the sequence of VolA (1). When the sequences of VolA homologs were analyzed, this domain was invariably conserved (Fig. 6A); the central serine is presumed to function in a catalytic Asp-His-Ser triad (12). We sought to determine if this serine was essential to the enzymatic activity of VolA. Wild-type VolA was mutagenized as described in Materials and Methods, and the putative catalytic serine was altered to either an alanine (VolA-S551A) or a threonine (VolA-S551T). Alanine was selected because it can maintain the protein carbon backbone while eliminating any potential side chain interactions, thus eliminating any active-site participation; threonine was selected because it most closely resembles the biochemical character of serine. Mutant protein expression was confirmed by Western blotting using an anti-VolA polyclonal antibody (Fig. 6B). Initially, volA mutant strains of V. cholerae carrying a vector control, pVolA, pVolA-S551A, or pVolA-S551T were grown in the presence of 2 mM C16:0 LPC. When the strains were grown with LPC as the sole carbon source (Fig. 6C), only the mutant complemented with the wild-type pVolA plasmid was able to grow. This indicates that the presence of a serine within the Gly-Xaa-Ser-Xaa-Gly motif is required for the catabolism of LPC.
FIG 6.
VolA requires a catalytic serine within a conserved lipase motif for activity. (A) Alignment of the protein region containing the conserved lipase motif Gly-Xaa-Ser-Xaa-Gly (residues 549 to 553 in V. cholerae VolA). Homologs of VolA were identified in Vibrio species and other marine organisms. (B) Anti-VolA Western blot of cell lysate expressing wild-type VolA or one of the VolA mutants (S551A and S551T). Mutant proteins were strongly expressed despite mutation of the active-site serine. (C) Growth of the V. cholerae volA mutant expressing either wild-type VolA or VolA catalytic serine mutants on LPC as the sole carbon source. Both S551A and S551T mutants failed to grow using LPC. (D) Lysophospholipase assay of membranes expressing either wild-type VolA or VolA S551 or S551T mutants.
To confirm that the conserved serine is required for proper active-site function, membranes isolated from the V. cholerae volA mutant expressing mutagenized copies of volA were assayed for lipase activity (Fig. 6D). Both the S551A and S551T mutants fail to cleave the labeled LPC molecule. These data, combined with the failure of the serine mutants to complement the volA mutant growth phenotype, strongly suggest that serine 551 acts as a nucleophile within the VolA active site.
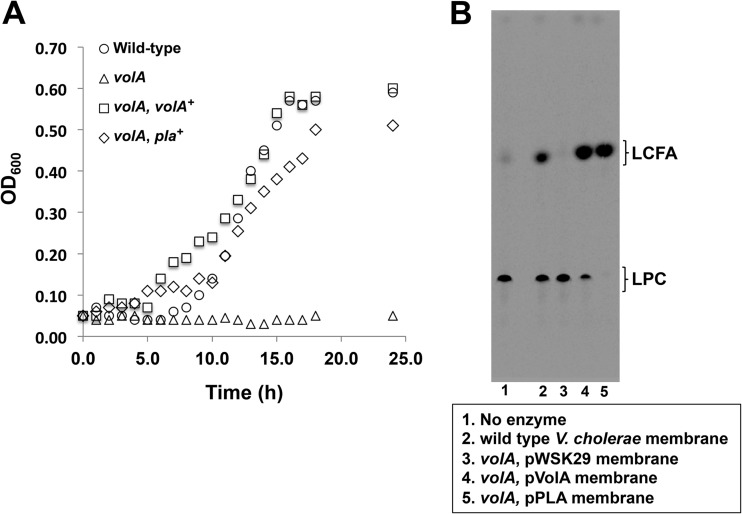
A VolA homolog from the aquatic pathogen Aeromonas hydrophila can complement a V. cholerae volA mutant.
In our previous study (1), homologs of VolA were identified in various Vibrio species; further exploration revealed homologs in several other marine bacteria (Fig. 6A; Table 1). Each of these homologs contains the conserved lipase motif found within VolA and a predicted lipoprotein signal sequence (13). The lipoprotein signal sequences all include a nonaspartate amino acid in the +2 position after the cleavage site, and based upon the rules of lipoprotein sorting, this predicts an outer-membrane association (14) similar to that of VolA. The Pla-1 protein found in the aquatic pathogen Aeromonas hydrophila displays the closest identity to VolA (BLAST E value = 5e-100; 33% identity) and was predicted to be a secreted lipase based on bioinformatic analysis (15). We wanted to determine if Aeromonas Pla-1 could functionally complement the Vibrio volA mutant (Fig. 7). Interestingly, expression of Pla-1 restored growth of V. cholerae on LPC to wild-type levels (Fig. 7A). Also, membranes isolated from the pla-complemented volA mutant showed robust levels of lysophospholipase activity (Fig. 7B), indicating that the Aeromonas enzyme remains associated with the bacterial surface, similar to what occurs in Vibrio.
TABLE 1.
Homologs of VolA
| Species | Protein | VolA |
Lipase motifa | |
|---|---|---|---|---|
| BLAST score | % identity | |||
| Vibrio cholerae | VolA | 0.0 | 100 | G-H-S-L-G |
| Aeromonas hydrophila | Pla-1 | 5e-100 | 33 | G-H-S-L-G |
| Alteromonas macleodii | MASE_09195 | 2e-94 | 29 | G-V-S-L-G |
| Marinobacter lipolyticus | MARLIPOL_11920 | 2e-12 | 31 | G-H-S-L-G |
| Pseudoalteromonas haloplanktis | PH_1344 | 3e-56 | 29 | G-H-S-L-G |
| Shewanella baltica | Pla-1 | 4e-61 | 29 | G-L-S-L-G |
A lipase signal was detected in all proteins.
FIG 7.
Expression of Aeromonas hydrophila Pla is capable of complementing the V. cholerae volA mutant. (A) Expression of A. hydrophila pla successfully complemented the growth phenotype of the V. cholerae volA mutant when LPC was used as the sole carbon source. (B) Lysophospholipase activity of membranes of the V. cholerae volA mutant expressing A. hydrophila Pla. TLC results show that expression of Pla in the volA mutant successfully restores lysophospholipase activity.
DISCUSSION
Very few surface-anchored lipoproteins have been identified in Gram-negative bacteria (16–19). Pullulanase, expressed by Klebsiella pneumoniae, is the first example of such a protein acting as an enzyme (20). VolA is a novel, as it is the first example of a surface-anchored lipoprotein in a Gram-negative bacterium functioning as a lipase. VolA plays a role not only in the metabolism of extracellular lipid substrates (i.e., lysophospholipids) but also in the incorporation of environmental lipids into the bacterial membrane. Previously, our laboratory demonstrated that V. cholerae was capable of incorporating a diverse array of LCFAs from the extracellular milieu into phospholipids, thereby influencing the bacterial membrane architecture (21). V. cholerae was able to incorporate very long (≥22-carbon) host-derived acyl chains containing multiple unsaturated positions, while other Gram-negatives, including E. coli and Salmonella enterica could only process shorter (≤20-carbon) acyl chains. This expanded uptake profile could be attributed to the fact that V. cholerae contains three homologs of FadL, an outer membrane fatty acid transporter (22). Indeed, our recent work showed that VolA is coexpressed with a FadL homolog. It is likely that the lysophospholipase activity of VolA is coupled to the uptake of LPC-derived LCFAs, which can then be incorporated into the bacterial membrane. Bacteria have been shown to use membrane remodeling as a survival adaptation, making it likely that VolA plays a similar role during infection of the host (23, 24). We observed that VolA is well conserved across many Vibrio species, prompting us to further characterize this intriguing protein. The enzyme is conserved not only in other Vibrio species but also in other Gram-negative bacteria (Table 1). Here, we confirm that the homolog produced by Aeromonas hydrophila, which has the closest identity to VolA among the different marine homologs, can successfully complement a volA V. cholerae mutant.
In this work, we confirm that VolA is indeed a lipoprotein. Mass spectrometry results show VolA is modified with both a diacylglycerol and a fatty acid, with saturated acyl chains of C16 and C18:1 (Fig. 3); this follows the canonical modification of bacterial lipoproteins (25, 26). The predicted modification is likely one of a mixture of species, following the distribution of acyl chains found in the phospholipids of E. coli (27). V. cholerae shows a similar fatty acid profile, with the predominant phospholipids bearing C16:0 and C18:1 fatty acyl chains (28). Bacterial lipoproteins are sorted based on the residue in the +2 position that occurs after the N-terminal peptide has been cleaved (14). VolA has a nonaspartate residue in the +2 position (1) and therefore is sorted to the outer membrane. However, how surface-anchored lipoproteins like VolA are localized to the outer leaflet of the outer membrane is currently unknown. It remains a possibility that transport machinery, such as the PulD-PulS secretin channel found in Klebsiella that functions to export pullulanase (29), the Bam complex involved with the transport of outer membrane β-barrels (30), or an unknown dedicated chaperone, exists in the outer membrane to fully process surface-exposed lipoproteins.
We also show that pure VolA possesses enzymatic activity acting as a lysophospholipase. Previous studies have shown that a conserved serine commonly involved in a Asp-His-Ser triad (31) acts as the catalytic residue (32). We show a conserved serine at position 551 within VolA is absolutely essential to its activity (Fig. 6), implying that this residue functions as the active-site nucleophile. VolA displays other potential lipase-related features. Many lipases have a motif that indicates the structure of the oxyanion hole. A conserved Gly-Gly-Gly-Xaa or Gly-Xaa sequence exists between secondary structural elements of the lipase, where the first Gly residue or the Xaa residue, respectively, functions as the oxyanion hole, contributing a hydrogen atom to help stabilize the reaction intermediate (33). In VolA, the Gly-Gly-Gly-Xaa motif is absent. Previously it was observed that lipases with the Gly-Gly-Gly-Xaa motif act upon short-chain substrates, while Gly-Xaa motif-containing lipases act on medium- and long-chain substrates (32). VolA follows this convention, being capable of hydrolyzing C16:0 and C18:0 lysophospholipids.
In vitro assays were used to establish the parameters for VolA enzymatic activity. Initially, we wanted to establish if VolA had metal ion dependence. While both metal-dependent and metal-independent bacterial phospholipases have been characterized (7, 8), there is a strong preponderance of metal-dependent enzymes. Enzymatic assays showed that VolA is capable of cleaving LPC into free C16:0 fatty acid and glycerophosphocholine in the presence of the metal chelators EDTA and EGTA (Fig. 4). This, along with data showing no appreciable increase when metal ions are added, implies that VolA is a metal-independent lysophospholipase. This is intriguing because while several mammalian metal-independent phospholipases with lysophospholipase activity have been identified (34), no such metal-independent lysophospholipases have been identified in bacteria.
In addition to determining metal independence, we also assayed other parameters of VolA enzymatic activity (Fig. 5). VolA shows both temperature and pH dependence. Interestingly, VolA showed a preference for increased temperatures, performing optimal conversion of LPC to LCFA at 37°C. This optimal activity implies that VolA may play a role during infection of the host. VolA demonstrated a partiality for alkaline pH. It is possible that at an alkaline pH, the active-site residues of VolA exist in a negatively charged state, allowing efficient release of the LCFA reaction product through electrostatic repulsion (35).
Our previous work identified several homologs of VolA in other Vibrio species (1), including V. alginolyticus, V. vulnificus, V. parahaemolyticus, and V. harveyi. All were capable of growth using LPC as the sole carbon source, while other Vibrio species without a VolA homolog, such as V. fischeri, lacked this ability. In the present work, we identifies VolA homologs in several other Gram-negative organisms—specifically, bacteria found in marine environments (Table 1). BLAST analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi) showed that Aeromonas, Alteromonas, and Marinobacter species all encode a homolog of VolA. In lipases, the catalytic serine is encoded by a specific sequence: either AGY, where Y is any pyrimidine, or TCN, where N is any nucleotide. Lipases that share the same serine codon may come from a common ancestor, as two distinct changes are required to change AGY to TCN and vice versa (2, 36). VolA and its homologs from marine organisms all use a TCN codon to encode their catalytic serine, suggesting that they have a shared origin. Notably, homologs of VolA are absent in terrestrial Gram-negative organisms, indicating that VolA may increase the fitness of bacteria living in aquatic environments. A homolog identified in Aeromonas hydrophila, encoded by the pla gene (15), showed the highest sequence identity to V. cholerae VolA. Expression of the Aeromonas homolog was able to complement the Vibrio volA mutant, restoring lysophospholipase activity and growth on LPC as the sole carbon source (Fig. 7), suggesting that VolA likely plays a similar role in other aquatic organisms.
Although the complete role that VolA plays in the life cycle of V. cholerae remains to be determined, there are many ways such an enzyme could confer advantages in both the marine and host environments. A surface-anchored lysophospholipase could have potential benefits over a secreted copy. A secreted enzyme could quickly diffuse away from the cell of origin, particularly in aquatic environments, generating valuable lipid substrates too distant from the cell to be used. By anchoring the enzyme to the outer surface of the cell, V. cholerae could produce a local increase in the concentration of free fatty acids, preventing any loss of potential substrate. Additional characterization of VolA and its effects on the life cycle of V. cholerae remain under study.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants AI064184 and AI76322 from the National Institutes of Health (NIH) and by grant W911NF-12-1-0390 from the Army Research Office to M.S.T.
Footnotes
Published ahead of print 14 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01281-13.
REFERENCES
- 1.Pride AC, Herrera CM, Guan Z, Giles DK, Trent MS. 2013. The outer surface lipoprotein VolA mediates utilization of exogenous lipids by Vibrio cholerae. mBio 4:e00305–13. 10.1128/mBio.00305-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner S. 1988. The molecular evolution of genes and proteins: a tale of two serines. Nature 334:528–530. 10.1038/334528a0 [DOI] [PubMed] [Google Scholar]
- 3.Verheij H, Dijkstra BW. 1994. Phospholipase A2: mechanism and structure, p 119–138 In Woolley P, Peterson SV. (ed), Lipases, their structure, biochemistry and application. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 4.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al. 1992. The alpha/beta hydrolase fold. Protein Eng. 5:197–211. 10.1093/protein/5.3.197 [DOI] [PubMed] [Google Scholar]
- 5.Nardini M, Dijkstra BW. 1999. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 9:732–737. 10.1016/S0959-440X(99)00037-8 [DOI] [PubMed] [Google Scholar]
- 6.Wilton DC, Waite M. 2002. Phospholipases, p 291–314 In Vance DE, Vance JE. (ed), Biochemistry of lipids lipoproteins and membranes, 4th ed. Elsevier Science B.V., Amsterdam, The Netherlands [Google Scholar]
- 7.Griffith OH, Ryan M. 1999. Bacterial phosphatidylinositol-specific phospholipase C: structure, function, and interaction with lipids. Biochim. Biophys. Acta 1441:237–254. 10.1016/S1388-1981(99)00153-5 [DOI] [PubMed] [Google Scholar]
- 8.Grant KA, Belandia IU, Dekker N, Richardson PT, Park SF. 1997. Molecular characterization of pldA, the structural gene for a phospholipase A from Campylobacter coli, and its contribution to cell-associated hemolysis. Infect. Immun. 65:1172–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 10.Tran AX, Karbarz MJ, Wang X, Raetz CR, McGrath SC, Cotter RJ, Trent MS. 2004. Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A. J. Biol. Chem. 279:55780–55791. 10.1074/jbc.M406480200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metzger LE, IV, Raetz CR. 2010. An alternative route for UDP-diacylglucosamine hydrolysis in bacterial lipid A biosynthesis. Biochemistry 49:6715–6726. 10.1021/bi1008744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaeger KE, Ransac S, Dijkstra BW, Colson C, van Heuvel M, Misset O. 1994. Bacterial lipases. FEMS Microbiol. Rev. 15:29–63. 10.1111/j.1574-6976.1994.tb00121.x [DOI] [PubMed] [Google Scholar]
- 13.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652–1662. 10.1110/ps.0303703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okuda S, Tokuda H. 2011. Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65:239–259. 10.1146/annurev-micro-090110-102859 [DOI] [PubMed] [Google Scholar]
- 15.Merino S, Aguilar A, Nogueras MM, Regue M, Swift S, Tomas JM. 1999. Cloning, sequencing, and role in virulence of two phospholipases (A1 and C) from mesophilic Aeromonas sp. serogroup O:34. Infect. Immun. 67:4008–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummelsmith J, Whitfield C. 2000. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 19:57–66. 10.1093/emboj/19.1.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manning PA, Beutin L, Achtman M. 1980. Outer membrane of Escherichia coli: properties of the F sex factor traT protein which is involved in surface exclusion. J. Bacteriol. 142:285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowles CE, Li YF, Semmelhack MF, Cristea IM, Silhavy TJ. 2011. The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol. Microbiol. 79:1168–1181. 10.1111/j.1365-2958.2011.07539.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. 2006. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59:870–881. 10.1111/j.1365-2958.2005.04997.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pugsley AP, Chapon C, Schwartz M. 1986. Extracellular pullulanase of Klebsiella pneumoniae is a lipoprotein. J. Bacteriol. 166:1083–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giles DK, Hankins JV, Guan Z, Trent MS. 2011. Remodelling of the Vibrio cholerae membrane by incorporation of exogenous fatty acids from host and aquatic environments. Mol. Microbiol. 79:716–728. 10.1111/j.1365-2958.2010.07476.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black PN, Said B, Ghosn CR, Beach JV, Nunn WD. 1987. Purification and characterization of an outer membrane-bound protein involved in long-chain fatty acid transport in Escherichia coli. J. Biol. Chem. 262:1412–1419 [PubMed] [Google Scholar]
- 23.Li Y, Powell DA, Shaffer SA, Rasko DA, Pelletier MR, Leszyk JD, Scott AJ, Masoudi A, Goodlett DR, Wang X, Raetz CR, Ernst RK. 2012. LPS remodeling is an evolved survival strategy for bacteria. Proc. Natl. Acad. Sci. U. S. A. 109:8716–8721. 10.1073/pnas.1202908109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meniche X, Labarre C, de Sousa-d'Auria C, Huc E, Laval F, Tropis M, Bayan N, Portevin D, Guilhot C, Daffe M, Houssin C. 2009. Identification of a stress-induced factor of Corynebacterineae that is involved in the regulation of the outer membrane lipid composition. J. Bacteriol. 191:7323–7332. 10.1128/JB.01042-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sankaran K, Wu HC. 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269:19701–19706 [PubMed] [Google Scholar]
- 26.Nakayama H, Kurokawa K, Lee BL. 2012. Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J. 279:4247–4268. 10.1111/febs.12041 [DOI] [PubMed] [Google Scholar]
- 27.Oursel D, Loutelier-Bourhis C, Orange N, Chevalier S, Norris V, Lange CM. 2007. Lipid composition of membranes of Escherichia coli by liquid chromatography/tandem mass spectrometry using negative electrospray ionization. Rapid Commun. Mass Spectrom. 21:1721–1728. 10.1002/rcm.3013 [DOI] [PubMed] [Google Scholar]
- 28.Guckert JB, Hood MA, White DC. 1986. Phospholipid ester-linked fatty acid profile changes during nutrient deprivation of Vibrio cholerae: increases in the trans/cis ratio and proportions of cyclopropyl fatty acids. Appl. Environ. Microbiol. 52:794–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nouwen N, Ranson N, Saibil H, Wolpensinger B, Engel A, Ghazi A, Pugsley AP. 1999. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl. Acad. Sci. U. S. A. 96:8173–8177. 10.1073/pnas.96.14.8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagan CL, Silhavy TJ, Kahne D. 2011. Beta-barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80:189–210. 10.1146/annurev-biochem-061408-144611 [DOI] [PubMed] [Google Scholar]
- 31.Brady L, Brzozowski AM, Derewenda ZS, Dodson E, Dodson G, Tolley S, Turkenburg JP, Christiansen L, Huge-Jensen B, Norskov L, et al. 1990. A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature 343:767–770. 10.1038/343767a0 [DOI] [PubMed] [Google Scholar]
- 32.Casas-Godoy L, Duquesne S, Bordes F, Sandoval G, Marty A. 2012. Lipases: an overview. Methods Mol. Biol. 861:3–30. 10.1007/978-1-61779-600-5_1 [DOI] [PubMed] [Google Scholar]
- 33.Pleiss J, Fischer M, Peiker M, Thiele C, Schmid RD. 2000. Lipase engineering database: understanding and exploiting sequence-structure-function relationships. J. Mol. Catal. B Enzym. 10:491–508. 10.1016/S1381-1177(00)00092-8 [DOI] [Google Scholar]
- 34.Winstead MV, Balsinde J, Dennis EA. 2000. Calcium-independent phospholipase A(2): structure and function. Biochim. Biophys. Acta 1488:28–39. 10.1016/S1388-1981(00)00107-4 [DOI] [PubMed] [Google Scholar]
- 35.Neves Petersen MT, Fojan P, Petersen SB. 2001. How do lipases and esterases work: the electrostatic contribution. J. Biotechnol. 85:115–147. 10.1016/S0168-1656(00)00360-6 [DOI] [PubMed] [Google Scholar]
- 36.Peterson SB, Drablos F. 1994. A sequence analysis of lipases, esterases, and related proteins, p 23–48 In Woolley P, Peterson SB. (ed), Lipases: their structure, biochemistry, and application. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.