Abstract
Prolyl 3-hydroxylation is a rare but conserved post-translational modification in many collagen types and, when defective, may be linked to a number of human diseases with musculoskeletal and potentially ocular and renal pathologies. Prolyl 3-hydroxylase-1 (P3H1), the enzyme responsible for converting proline to 3-hydroxyproline (3Hyp) in type I collagen, requires the coenzyme CRTAP for activity. Mass spectrometric analysis showed that the Crtap−/− mouse was missing 3-hydroxyproline in type I collagen α-chains. This finding led to the discovery mutations in genes encoding the P3H1 complex as a cause of recessively inherited osteogenesis imperfecta (brittle bone disease). Since then, many additional 3Hyp sites have been identified in various collagen types and classified based on observed substrate and tissue specificity. P3H1 is part of a family of gene products that also includes isoenzymes P3H2 and P3H3 as well as CRTAP and Sc65. It is believed these isoenzymes and coenzymes have evolved different collagen substrate site and tissue specificities in their activities. The post-translational fingerprinting of collagens will be essential in understanding the basic role and extent of regulated variations of prolyl 3-hydroxylation in collagen. We believe that prolyl 3-hydroxylation is a functionally significant collagen post-translational modification and can be a cause of disease when absent.
Keywords: 3-hydroxyproline, fibrillar collagen, post-translational modification, prolyl 3-hydroxylase
Introduction
Collagen post-translational modifications have evolved to support animal life on land. The fibril-forming collagens (types I, II, III, V and XI) are characterized by three polypeptide α-chains (over 1000 residues each) coiled into triple helices (1). The triple-helical domain is defined by a repeating Gly-X-Y triplet, where X and Y are commonly proline (Pro) and 4R,2S-hydroxyproline (4Hyp), respectively. Prolyl 4-hydroxylation is the single most prevalent post-translational modification in humans. Indeed, a quarter of the amino acids in mammalian fibrillar collagen α-chains are prolyl residues, approximately 50% of which are 4-hydroxylated. Additionally, lysine residues can be hydroxylated to 5-hydroxylysine, which in turn can be glycosylated to galactosylhydroxylysine and glucosylgalactosylhydroxylysine. Furthermore, all fibril-forming collagens in higher vertebrates are covalently cross-linked through the lysine aldehyde or the hydroxylysine aldehyde pathway (2). It is well established that these post-translational modifications enhance collagen structural stability (1). For example, it is thought that 4Hyp residues stabilize the triple helix through either water-bridged hydrogen bonding or stereoelectronic effects (3,4). Five decades ago a new amino acid, 3-hydroxyproline (3S, 2S-hydroxyproline; 3Hyp), was identified in collagen (5). 3Hyp is a rare amino acid, with an occurrence of two residues per α-chain in collagen types I and II, between three to six residues per α-chain of collagen types V and XI, and over 10 residues per α-chain of collagen type IV (6-8). It is established that the modified Pro residues occur within the sequence Gly-Pro-4Hyp (5,9) (Figure 1A). This residue adds to the already large number of collagen post-translational modifications. Yet a half century after its discovery, a function for 3Hyp is still unclear (6).
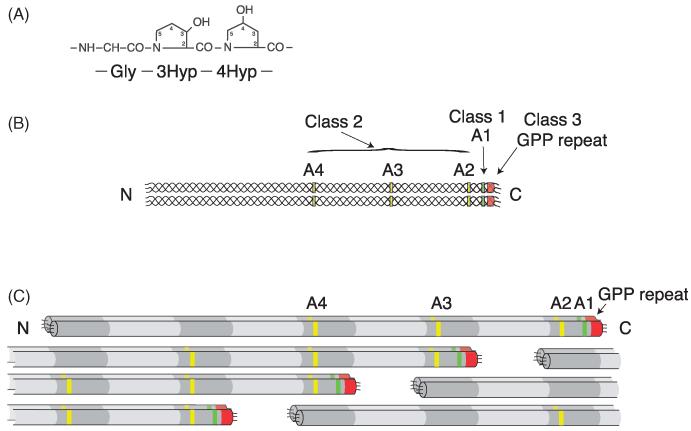
Figure 1.
Model of the three classes of 3Hyp substrate sites identified in A-clade collagen. 3Hyp always occurs in the sequence Gly-3Hyp-4Hyp, A. We propose the Class 1 3Hyp (Pro986) functions in collagen fibril molecular packing, in which the subunits are in-register dimers staggered axially by D-periods, B. D-periodic spacing is evident between the Class 2 3Hyp residues (Pro470, Pro707 and Pro944) in the triple helix, C. Occurrence of the Class 3 3Hyp residues in the (GPP)n motif exhibited clear tissue specificity, with the modification occurring almost exclusively in tendon.
Prolyl 3-hydroxylase family
Prolyl 3-hydroxylase-1 (P3H1), the enzyme responsible for catalyzing the conversion of proline to 3Hyp in type I collagen, was shown to form a complex with cartilage-associated protein (CRTAP) and cyclophlin B (PPIB) in a 1:1:1 ratio (10). It has been suggested that the P3H1 complex may function as both an enzyme and a chaperone in collagen molecular assembly, possibly by preventing premature aggregation of collagen chains during protein synthesis (11). P3H1 and 3Hyp have experienced a surge of interest lately due largely to a connection with autosomal recessive forms of osteogenesis imperfecta (OI), in several genetic variants of which the prolyl 3-hydroxylation modification is missing. Gene mutations that disrupt expression of any protein in the P3H1 complex (P3H1, CRTAP and PPIB) have been shown to cause recessive OI (12-17). Analysis of collagen from tissue and cultured fibroblasts from these OI cases revealed a lack of or decrease in 3-hydroxylation at proline residues, α1(I) Pro986 and α2(I) Pro707 (12,14,18). OI, a rare genetic disorder affecting normal type I collagen synthesis, is phenotypically characterized by brittle bones. In contrast to autosomal dominant variants of OI, which typically result from mutations in the type I collagen genes COL1A1 and COL1A2, a genotypic spectrum of recessively inherited OI has recently emerged (19). Mutations in genes such as SERPINH1 (encodes heat shock protein HSP47) (20), SERPINF1 (pigment epithelium-derived factor, PEDF) (21), FKBP10 (FKBP65) (22), SP7 (osterix) (23), PLOD2 (lysyl hydroxylase-2) (24), BMP1 (collagen C-propeptidase) (25), TMEM38B (trimeric intracellular cation channel) (26), IFITM5 (interferon-induced transmembrane protein 5) (27) and most recently WNT1 (Wnt1) (28) have also been identified as causes of OI. The recessively inherited variants of OI account for less than 10% of all known individuals with the disease, most cases being caused by dominant mutations in type I collagen genes (29). The pathophysiology of recessive OI can be attributed to at least four basic mechanisms, which affect either collagen post-translational modifications (loss of 3Hyp, loss of Hyl, over-hydroxylation of Hyl, or over-glycosylation of Hyl), collagen processing (deficient C-propeptide cleavage) (25,30), collagen trafficking (defects in ER chaperone proteins) and/or osteoblast differentiation. Whether the brittle bone phenotype of those recessive OI variants with reduced prolyl 3-hydroxylation is a direct result of the missing modification itself or another consequence of the disrupted enzyme complex has yet to be determined.
The role of CRTAP and P3H1 in collagen homeostasis and OI pathology was defined in the last decade. For example, the Crtap−/− mouse was extensively characterized and observed to have a generalized connective tissue phenotype that was consistent with the systemic failure of collagen homeostasis, affecting bone, cartilage, skin, lung and kidney (31). The Crtap−/− mouse also exhibited complete loss of 3-hydroxylation at Pro986 of α1(I) and α1(II) and α2(V) (12,31). Similarly, this 3Hyp residue was absent in the P3H1-null mouse, which presented with hearing loss (32) and an abnormal skin, tendon and bone phenotype due to collagen fibril disturbances (33). CRTAP and P3H1 are part of a family of genes that includes three isoenzymes in the P3H family, P3H1 (LEPRE1), P3H2 (LEPREL1) and P3H3 (LEPREL2), as well as two homologous proteins and potential coenzymes, CRTAP (CRTAP) and Sc65 (LEPREL4) (34). These different proteins have likely evolved unique but complimentary functions affecting a wide-range of organ systems. For example, a single point mutation in P3H2 has been implicated in high myopia in humans (35), and a recent study has cited Sc65 as an autoimmune target in membranous nephropathy (36).
Methods for mapping 3Hyp
3Hyp was originally discovered through the amino acid analysis of collagen hydrolysates (5). Automated Edman degradation later became the standard for amino acid sequence analysis of this modification, which gives a unique PTH-derivative signature on HPLC analysis (37,38). Today, mass spectrometric approaches have largely replaced these earlier methods for the initial identification of 3Hyp within collagen α-chains (6,39-42). This approach typically involves analyzing collagen peptides from SDS-PAGE gels or from reverse phase column HPLC fractions after protease digestion (usually trypsin). Mass spectrometry is carried out on the tryptic peptides, and search software is used for peptide identification. Larger collagenous peptides are identified manually by calculating the theoretical MS/MS ions and matching these to the actual MS/MS spectrum. Hydroxyl differences (±16Da) are probed manually by averaging the full scan over several minutes of the LCMS elution profile so that all the post-translational variations of a given peptide are combined in the full scan. A limitation of the mass spectrometric approach is the inability to distinguish between 3Hyp and 4Hyp residues; however, 4Hyp only occurs in the Y position of the (Gly-X-Y)n repeat, with the rare exception of 4Hyp in the X position of certain GPA triplets (43). This finding supports the labelling of hydroxylated prolines in the X position of a Gly-Pro-4Hyp sequence as 3Hyp.
Novel sites of 3Hyp in fibrillar collagen
We recently identified several novel molecular sites of 3Hyp in fibrillar collagen using peptide mass spectrometry (6). A-clade chains (α1(I), α2(I), α1(II), α1(III) and α2(V)) and B-clade chains (α1(V), α1(XI) and α2(XI)) were examined from human and bovine tissues. In fibril-forming A-clade collagens, these sites have been divided into three classes: the original 3Hyp site at residue Pro986 termed A1 (Class 1), three new sites Pro944, Pro707 and Pro434 termed A2, A3 and A4 (Class 2) and the C-terminal (GPP)n motif (Class 3). In the B-clade collagens, three high occupancy sites (B1, B2 and B3) were originally identified in bone type V collagen and cartilage type XI collagen (6); however, multiple additional low occupancy sites have since been detected in bovine placental α1(V) by high-sensitivity mass spectrometric methods (40).
Variability in 3Hyp site occupancy suggests local sequence substrate-specificity. Notably, almost complete 3-hydroxylation of Pro986 is seen in α1(I) and α1(II) but not in type III collagen, where the site is unoccupied in mammals. Curiously, this site is 100% occupied in chicken and xenopus type III collagen (41). Based on sequence data from the genomic database (Ensembl), a local sequence motif containing a histidine six residues immediately upstream from the A1 site proline was determined to preclude 3-hydroxylation of mammalian type III collagen. Available avian, amphibian and reptilian sequences all have a tyrosine instead of histidine predicting a distinction between type III collagen of mammals compared with other vertebrates. Also, although the A-clade 3Hyp sites are specific to A-clade collagens, only α2(V) hosts some occupancy at all four A-clade sites (see Table 1 for details).
Table 1.
Substrate specificity of 3Hyp sites in A-clade collagen α-chains.
|
Mammal (human)
| ||||
| Type | A4 Site | A3 Site | A2 Site | A1 Site |
| α1(I) | 0% | 0% | 0% | 99% |
| α2(I) | 0% | 80% | 0% | n/a |
| α1(II) | 0% | 0% | 10-87% | 99% |
| α1(III) | n/a | 0% | 0% | 0% |
| α2(V) | 13% | 80% | 60% | 99% |
|
| ||||
|
Avian (chicken)
| ||||
| Type | A4 Site | A3 Site | A2 Site | A1 Site |
| α1(I) | n/a | 5-10% | 0% | 99% |
| α2(I) | n/a | 95% | n/a | n/a |
| α1(II) | 0% | 18% | 0% | 99% |
| α1(III) | n/a | 0% | 0% | 100% |
| α2(V) | 18% | 40% | 28% | 99% |
|
| ||||
|
Amphibian (xenopus)
| ||||
| Type | A4 Site | A3 Site | A2 Site | A1 Site |
| α1(I) | 0% | 12% | 0% | 99% |
| α2(I) | n/a | n/a | n/a | n/a |
| α1(II) | 0% | n/a | 0% | 99% |
| α1(III) | n/a | 0% | 0% | 100% |
Molecular locations and abundance of 3Hyp sites within several vertebrates were identified using mass spectrometry. 3Hyp content was determined by scrolling the full scan to include all post-translational modification variations (n/a represents no substrate GPP in the known sequence). Note that only the A1 site is fully 3-hydroxylated across species.
Pronounced tissue specificity was also observed for several sites (Table 2). For example, in type II collagen, the A2 site ranged in occupancy from 80% in vitreous humour to 20% in cartilage (6). 3Hyp was originally found at the A3 site only in α2(I) and α2(V) from multiple tissue sources (6), but was also later detected in α1(I) from skin and tendon (39,41). Tissue specificity is particularly striking for the Class 3 site, with the C-terminal (GPP)n motif of type I collagen containing 3Hyp exclusively in tendon (42). Such collagen substrate and tissue specificity in the degree of 3Hyp site occupancy is likely a function of variable expression of the distinct P3H isoenzymes and coenzymes (44). Indeed, each P3H has been shown to exhibit distinct tissue specificity in its expression, which can vary between species and with development (45,46). P3H1 mRNA expression was found in all fetal human tissues tested, but no expression was observed in adult human skeletal muscle and brain, or mouse renal tissue (46). P3H2, which is highly expressed in kidney, placenta and lung, has been proposed to be the active 3-hydroxylase for basement membrane collagens (46). Amino acid analysis of bovine type I collagen revealed higher levels of prolyl 3-hydroxylation in kidney (seven 3Hyp residues per α-chain) than other tissues (one 3Hyp residue per α-chain in skin and bone) (47). In support of this trend, mass spectrometry of the (GPP)n motif from bovine kidney type I collagen showed higher 3Hyp levels than from any other bovine tissue examined (Hudson et al., unpublished).
Table 2.
Tissue specificity of 3Hyp occupancy in type I and type II collagen α-chains in mammals.
| Bone | Tendon | Cartilage | Vitreous | |||
|---|---|---|---|---|---|---|
| Sites | α1(I) | α2(I) | α1(I) | α2(I) | α1(II) | α1(II) |
| A1 (P986) | +++ | 0 | +++ | 0 | +++ | +++ |
| A2 (P944) | 0 | 0 | 0 | 0 | + | ++ |
| A3 (P707) | 0 | ++ | + | ++ | 0 | 0 |
| A4 (P470) | 0 | 0 | 0 | 0 | 0 | 0 |
| (GPP)n | 0 | 0 | +++ | +++ | + | ++ |
Relative abundance of 3Hyp sites in α1(I), α2(I) and α1(II) (A1, A2, A3, A4 and (GPP)n) in different tissues. Symbols (+++, ++, +, 0) represent relative abundance of prolyl 3-hydroxylation, ranging from fully occupied to unoccupied (approximate 3Hyp occupancy estimates: +++ = 99%–70%; ++ = 69%–30%; + = 29%–5%). This data was obtained from human, bovine and mouse samples.
Evolutionary origins of 3Hyp
The 3-hydroxylation of proline is a highly conserved modification in many types of collagens throughout the animal kingdom (6,41). Evolutionarily, this ancient and ubiquitous collagen modification is found as far back as porifera, the most primitive extant multicellular animal (48). It seems unlikely that a post-translational modification as rare but conserved as 3Hyp would not contribute basically to collagen structure and function. At which point in evolution 3-hydroxylation first appeared in fibrillar collagens, and specifically at P986, is unknown. According to the Ensembl database, the earliest recognizable A-clade fibrillar collagen (CIFCOL1; Ensembl gene ID: ENSCING00000006961) is found in the pre-vertebrate chordate, Ciona intestinalis. Many of the known A-clade 3Hyp sites lack the GPP motif at the homologous site in the CIFCOL1 gene sequence. For example, the A1 site (Pro986), highly conserved across chordate species in α1(I) as GPIGPPGPR, is replaced with GPIGATGPR. The genome of C. intestinalis does contain a gene for a prolyl 3-hydroxylase (49,50), but the collagen substrates of this enzyme are unknown. We have located several novel 3Hyp sites in the CIFCOL1 gene product of C. intestinalis using mass spectrometry, however, these did not follow the known A-clade or B-clade site patterns (Hudson et al., unpublished). The expression of a fibril-forming collagen gene perhaps resulted in a gain of function for the lone P3H enzyme from C. intestinalis, which likely evolved initially to act on basement membrane collagens. As such, the site sequences of 3Hyp in the CIFCOL1 gene product may have diverged from those of early type IV collagens. The occurrence of 3Hyp potentially impacted collagen fibril assembly at the threshold of vertebrate evolution in a way that enhanced the properties of skeletal tissues and their development in particular. Genomic duplications in early chordates gave rise to the three P3H isoenzymes now found in higher vertebrates (50). Understanding the mode of action of this ancient P3H enzyme from C. intestinalis may provide important clues to differential functions of P3H1, P3H2 and P3H3. Although the main substrate of P3H2 is thought to be basement membrane collagens (46), the best characterized activity is that of the P3H1 enzyme complex, responsible for converting Pro986 to 3Hyp in collagen types I, II and V (15,31).
Potential function
Subtle variations in local proline hydroxylation chemistry can have considerable effects on collagen peptide stability. For example, 4Hyp can both stabilize and destabilize the triple helix depending on its location in the Gly-X-Y motif (51), whereas the non-physiological diastereoisomer 4S-hydroxy-2S-proline has a destabilizing effect regardless of position (52). Initial studies employing 3Hyp-containing synthetic collagen-like polypeptides have suggested a possible destabilization of the triple helix by 3Hyp (53), but further work revealed a marginal added stability (54). It seems unlikely that one or two 3Hyp residues/α-chain (in comparison to more than 100 4Hyp residues/α-chain) have evolved solely to contribute additional thermal stability to the collagen triple helix. No function is known for prolyl 3-hydroxylation; however, we speculate that the different classes of 3Hyp sites may each serve a unique purpose.
Class 1: Based on the stabilizing role of 4Hyp residues through water-bridged hydrogen bonds, we proposed that Pro986 (Class 1) functions as a local site of interaction that aids in collagen fibril assembly (6). The best arrangement of molecules for efficient mature trivalent cross-links would be for lateral pairs of collagen molecules in register to pack into a quarter-staggered collagen lattice (Figure 1B). This alignment could be aided through short-range hydrogen-bonding interactions or stereoelectronic effects between 3Hyp residues. We propose that the 3-hydroxyl of Pro986 and a peptide backbone carbonyl of an adjacent triple helix could perhaps interact via one or more water molecules analogous to the mechanism of triple helix stabilization by 4Hyp (55,56). This hypothesis is supported, theoretically, by the crystal structure of a 3Hyp-containing collagen-like synthetic polypeptide (57), in which the 3-hydroxyl extended outward from the triple helix axis, thereby allowing potential intermolecular interactions to occur. Moreover, in this crystal structure, direct inter-helical hydrogen bonds were identified between the hydroxyls of two 3Hyp side chains (57). Many short-range stabilizing effects brought about by prolyl hydroxyl moieties are thought to result from either favorable electrostatic dipole–dipole interactions or steric clashes within the collagen triple helix (58).
Solid-phase binding studies revealed that 3Hyp could potentially function in interchain and so potentially intermolecular recognition and binding (59). The physical binding properties of 3Hyp in collagen chains were investigated using synthetic collagen-like peptides. Evidence of self-association was observed between a 3Hyp-containing synthetic peptide and the CB6 domain of the α1(I) chain from tissue collagen, which contains the fully single 3-hydroxylated proline. It was clear that the interaction was highly dependent upon the 3-hydroxylation of Pro986. Indeed, using collagen from a case of severe recessive OI with a CRTAP mutation, in which Pro986 was minimally 3-hydroxylated, such binding was not observed (59). These findings suggested that the brittle bone phenotype observed in OI could be in part caused by a missing fundamental role of prolyl 3-hydroxylation. For example, a fundamental short-range order in the molecular and/or supramolecular assembly of collagen may be disordered in severe OI.
Early collagen X-ray diffraction data support a packing arrangement of in-register dimers. A structural model consisting of collagen dimers packed into a tetragonal lattice was proposed by Woodhead–Galloway almost four decades ago (60). Such a placement of collagen molecules would result in a regular array of discrete holes within the gap region of the fibril, which could theoretically provide the spacing required for bone mineralization. As such it is possible that disrupting the alignment of collagen molecules could also disrupt the fibril framework necessary for ordered hydroxyapatite nanocrystal growth, potentially resulting in brittle bones.
Class 2: The function of the Class 2 sites may also be involved with collagen fibril assembly. The D-periodic spacing (234 ± 3 residues) between the A2, A3 and A4 3Hyp sites in A-clade collagen chains and between the B2 and B3 3Hyp sites in B-clade collagen chains suggests a role for 3Hyp in orienting fibril formation (6) (Figure 1C). Notably, in the supramolecular structure, all the Class 2 3Hyp sites are aligned within the molecular overlap region of the fibril.
Class 3: The occurrence of the Class 3 tendon-specific 3Hyp sites appears to have arisen with early vertebrates (Hudson et al., unpublished). Type I collagen in tendon has several distinctive properties, including its manner of cellular assembly, cross-linking and material properties (61). We suspect that this third class of 3Hyp motif is functionally regulated during synthesis in the ER and is a potential determinant of the intramolecular collagen α-chain registry or other interactions in the assembly of a fibrillar matrix.
3Hyp in basement membrane collagen
Type IV collagen has the highest concentration of 3Hyp among collagens (7), and is both ancient and structurally quite distinct from the fibrillar collagens. Type IV collagen α-chains contain an amino-terminal triple-helical 7S domain, a central triple-helical region and a carboxy-terminal non-collagenous domain (NC1 domain) (62). The 7S domain is reported to contain as many as half the total 3Hyp residues in type IV collagen (8). Their molecular location and function remain undefined. The 7S domain is composed of four antiparallel triple helices that are largely brought together by hydrophobic and ionic interactions and held by disulfide bonds (62). When self-assembly proceeds into an extended supramolecular structure, the resulting open mesh network is responsible for the tensile framework and characteristic architecture of all basement membranes. The 7S domain of type IV collagen therefore provides a unique window to study the evolutionary origins and relationship between the different isoenzymes (P3H1, P3H2 and P3H3) and potential coenzymes (CRTAP and Sc65), as it likely contains several distinct substrate motifs.
Future directions
In addition to a role in collagen assembly, 3Hyp domains may provide sites of interaction for fibril-binding non-collagenous proteins, for example, the small leucine-rich proteoglycans (SLRPs) and other proteins perhaps involved in mineralization. The function of 3Hyp is clearly important to understand given its significance in recessive OI and heritable high myopia (19,35). Point mutations causing single amino acid substitutions in collagen type I (osteogenesis imperfecta), type II (chondrodysplasias), type III/V (Ehlers-Danlos syndrome), and type IV (Alport’s syndrome) have all been linked to diseases affecting multiple tissues and organ systems. It seems likely that, with further investigation, more disorders based on altered collagen post-translational modifications will be revealed.
Acknowledgements
We thank Rachel Werther and MaryAnn Weis for their helpful comments, suggestions and critical review of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Work described in this publication from the authors’ laboratory was supported by the National Institutes of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Numbers AR37318 and AR36794.
Footnotes
Declaration of interest
There is no conflict of interest whatsoever or financial relationships that could affect the presentation of the results or bias their interpretation.
References
- 1.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends in genetics: TIG. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Eyre DR, Paz MA, Gallop PM. Cross-linking in collagen and elastin. Ann Rev Biochem. 1984;53:717–48. doi: 10.1146/annurev.bi.53.070184.003441. [DOI] [PubMed] [Google Scholar]
- 3.Berg RA, Prockop DJ. The thermal transition of a non-hydroxylated form of collagen. Evidence for a role for hydroxyproline in stabilizing the triple-helix of collagen. Biochem Biophys Res Commun. 1973;52:115–20. doi: 10.1016/0006-291x(73)90961-3. [DOI] [PubMed] [Google Scholar]
- 4.Kotch FW, Guzei IA, Raines RT. Stabilization of the collagen triple helix by O-methylation of hydroxyproline residues. J Am Chem Soc. 2008;130:2952–3. doi: 10.1021/ja800225k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogle JD, Arlinghaus RB, Logan MA. 3-Hydroxyproline, a new amino acid of collagen. J Biol Chem. 1962;237:3667–73. [PubMed] [Google Scholar]
- 6.Weis MA, Hudson DM, Kim L, et al. Location of 3-hydroxyproline residues in collagen types I, II, III, and V/XI implies a role in fibril supramolecular assembly. J Biol Chem. 2010;285:2580–90. doi: 10.1074/jbc.M109.068726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean DC, Barr JF, Freytag JW, Hudson BG. Isolation of type IV procollagen-like polypeptides from glomerular basement membrane. Characterization of pro-alpha 1(IV) J Biol Chem. 1983;258:590–6. [PubMed] [Google Scholar]
- 8.Risteli J, Bachinger HP, Engel J, et al. 7-S collagen: characterization of an unusual basement membrane structure. Eur J Biochem/FEBS. 1980;108:239–50. doi: 10.1111/j.1432-1033.1980.tb04717.x. [DOI] [PubMed] [Google Scholar]
- 9.Gryder RM, Lamon M, Adams E. Sequence position of 3-hydroxyproline in basement membrane collagen. Isolation of glycyl-3-hydroxyprolyl-4-hydroxyproline from swine kidney. J Biol Chem. 1975;250:2470–4. [PubMed] [Google Scholar]
- 10.Vranka JA, Sakai LY, Bachinger HP. Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J Biol Chem. 2004;279:23615–21. doi: 10.1074/jbc.M312807200. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa Y, Wirz J, Vranka JA, et al. Biochemical characterization of the prolyl 3-hydroxylase 1.cartilage-associated protein.cyclophilin B complex. J Biol Chem. 2009;284:17641–7. doi: 10.1074/jbc.M109.007070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morello R, Bertin TK, Chen Y, et al. CRTAP is required for prolyl 3-hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006;127:291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 13.Barnes AM, Chang W, Morello R, et al. Deficiency of cartilage-associated protein in recessive lethal osteogenesis imperfecta. New England J Med. 2006;355:2757–64. doi: 10.1056/NEJMoa063804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabral WA, Chang W, Barnes AM, et al. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat Genet. 2007;39:359–65. doi: 10.1038/ng1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baldridge D, Schwarze U, Morello R, et al. CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Human Mutat. 2008;29:1435–42. doi: 10.1002/humu.20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Dijk FS, Nesbitt IM, Zwikstra EH, et al. PPIB mutations cause severe osteogenesis imperfecta. Am J Human Genet. 2009;85:521–7. doi: 10.1016/j.ajhg.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes AM, Carter EM, Cabral WA, et al. Lack of cyclophilin B in osteogenesis imperfecta with normal collagen folding. New England J Med. 2010;362:521–8. doi: 10.1056/NEJMoa0907705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pyott SM, Schwarze U, Christiansen HE, et al. Mutations in PPIB (cyclophilin B) delay type I procollagen chain association and result in perinatal lethal to moderate osteogenesis imperfecta phenotypes. Human Molec Genet. 2011;20:1595–609. doi: 10.1093/hmg/ddr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cundy T. Recent advances in osteogenesis imperfecta. Calcified Tissue Int. 2012;90:439–49. doi: 10.1007/s00223-012-9588-3. [DOI] [PubMed] [Google Scholar]
- 20.Christiansen HE, Schwarze U, Pyott SM, et al. Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am J Human Genet. 2010;86:389–98. doi: 10.1016/j.ajhg.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker J, Semler O, Gilissen C, et al. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am J Human Genet. 2011;88:362–71. doi: 10.1016/j.ajhg.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alanay Y, Avaygan H, Camacho N, et al. Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am J Human Genet. 2010;86:551–9. doi: 10.1016/j.ajhg.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapunzina P, Aglan M, Temtamy S, et al. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. Am J Human Genet. 2010;87:110–14. doi: 10.1016/j.ajhg.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puig-Hervas MT, Temtamy S, Aglan M, et al. Mutations in PLOD2 cause autosomal-recessive connective tissue disorders within the Bruck syndrome – osteogenesis imperfecta phenotypic spectrum. Human Mutat. 2012;33:1444–9. doi: 10.1002/humu.22133. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Glez V, Valencia M, Caparros-Martin JA, et al. Identification of a mutation causing deficient BMP1/mTLD proteolytic activity in autosomal recessive osteogenesis imperfecta. Human Mutat. 2012;33:343–50. doi: 10.1002/humu.21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaheen R, Alazami AM, Alshammari MJ, et al. Study of autosomal recessive osteogenesis imperfecta in Arabia reveals a novel locus defined by TMEM38B mutation. J Med Genet. 2012;49:630–5. doi: 10.1136/jmedgenet-2012-101142. [DOI] [PubMed] [Google Scholar]
- 27.Cho TJ, Lee KE, Lee SK, et al. A single recurrent mutation in the 5′-UTR of IFITM5 causes osteogenesis imperfecta type V. Am J Human Genet. 2012;91:343–8. doi: 10.1016/j.ajhg.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fahiminiya S, Majewski J, Mort J, et al. Mutations in WNT1 are a cause of osteogenesis imperfecta. J Med Genet. 2013;2:2. doi: 10.1136/jmedgenet-2013-101567. Epub 2013/02/26. [DOI] [PubMed] [Google Scholar]
- 29.Schwarze U, Cundy T, Pyott SM, et al. Mutations in FKBP10, which result in Bruck syndrome and recessive forms of osteogenesis imperfecta, inhibit the hydroxylation of telopeptide lysines in bone collagen. Human Molec Genet. 2013;22:1–17. doi: 10.1093/hmg/dds371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindahl K, Barnes AM, Fratzl-Zelman N, et al. COL1 C-propeptide cleavage site mutations cause high bone mass osteogenesis imperfecta. Human Mutat. 2011;32:598–609. doi: 10.1002/humu.21475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldridge D, Lennington J, Weis M, et al. Generalized connective tissue disease in Crtap−/− mouse. PloS one. 2010;5:e10560. doi: 10.1371/journal.pone.0010560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pokidysheva E, Tufa S, Bresee C, et al. Prolyl 3-hydroxylase-1 null mice exhibit hearing impairment and abnormal morphology of the middle ear bone joints. Matrix Biol. 2013;32:39–44. doi: 10.1016/j.matbio.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vranka JA, Pokidysheva E, Hayashi L, et al. Prolyl 3-hydroxylase 1 null mice display abnormalities in fibrillar collagen-rich tissues such as tendons, skin, and bones. J Biol Chem. 2010;285:17253–62. doi: 10.1074/jbc.M110.102228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarnum S, Kjellman C, Darabi A, et al. LEPREL1, a novel ER and Golgi resident member of the Leprecan family. Biochem Biophys Res Commun. 2004;317:342–51. doi: 10.1016/j.bbrc.2004.03.060. [DOI] [PubMed] [Google Scholar]
- 35.Mordechai S, Gradstein L, Pasanen A, et al. High myopia caused by a mutation in LEPREL1, encoding prolyl 3-hydroxylase 2. Am J Human Genet. 2011;89:438–45. doi: 10.1016/j.ajhg.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavazzini F, Magistroni R, Furci L, et al. Identification and characterization of a new autoimmune protein in membranous nephropathy by immunoscreening of a renal cDNA library. PloS one. 2012;7:e48845. doi: 10.1371/journal.pone.0048845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuppan D, Glanville RW, Timpl R. Covalent structure of mouse type-IV collagen. Isolation, order and partial amino-acid sequence of cyanogen-bromide and tryptic peptides of pepsin fragment P1 from the alpha 1(IV) chain. Eur J Biochem/FEBS. 1982;123:505–12. [PubMed] [Google Scholar]
- 38.Fietzek PP, Rexrodt FW, Wendt P, et al. The covalent structure of collagen. Amino-acid sequence of peptide 1-CB6-C2. Eur J Biochem/FEBS. 1972;30:163–8. doi: 10.1111/j.1432-1033.1972.tb02083.x. [DOI] [PubMed] [Google Scholar]
- 39.Ishikawa Y, Vranka JA, Boudko SP, et al. Mutation in cyclophilin B that causes hyperelastosis cutis in American Quarter Horse does not affect peptidylprolyl cis-trans isomerase activity but shows altered cyclophilin B-protein interactions and affects collagen folding. J Biol Chem. 2012;287:22253–65. doi: 10.1074/jbc.M111.333336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang C, Park AC, Davis NA, et al. Comprehensive mass spectrometric mapping of the hydroxylated amino acid residues of the alpha1(V) collagen chain. J Biol Chem. 2012;287:40598–610. doi: 10.1074/jbc.M112.406850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudson DM, Weis M, Eyre DR. Insights on the evolution of prolyl 3-hydroxylation sites from comparative analysis of chicken and Xenopus fibrillar collagens. PloS one. 2011;6:e19336. doi: 10.1371/journal.pone.0019336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eyre DR, Weis M, Hudson DM, et al. A novel 3-hydroxyproline (3Hyp)-rich motif marks the triple-helical C terminus of tendon type I collagen. J Biol Chem. 2011;286:7732–6. doi: 10.1074/jbc.C110.195768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura T, Cheah KS, Chan SD, et al. The human alpha 2(XI) collagen (COL11A2) chain. Molecular cloning of cDNA and genomic DNA reveals characteristics of a fibrillar collagen with differences in genomic organization. J Biol Chem. 1989;264:13910–16. [PubMed] [Google Scholar]
- 44.Fernandes RJ, Farnand AW, Traeger GR, et al. A role for prolyl 3-hydroxylase 2 in post-translational modification of fibril-forming collagens. J Biol Chem. 2011;286:30662–9. doi: 10.1074/jbc.M111.267906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vranka J, Stadler HS, Bachinger HP. Expression of prolyl 3-hydroxylase genes in embryonic and adult mouse tissues. Cell Struct Funct. 2009;34:97–104. doi: 10.1247/csf.09002. [DOI] [PubMed] [Google Scholar]
- 46.Tiainen P, Pasanen A, Sormunen R, Myllyharju J. Characterization of recombinant human prolyl 3-hydroxylase isoenzyme 2, an enzyme modifying the basement membrane collagen IV. J Biol Chem. 2008;283:19432–9. doi: 10.1074/jbc.M802973200. [DOI] [PubMed] [Google Scholar]
- 47.Fujiwara S, Nagai Y. Bovine renal cortex type I collagen: high contents of 3- and 4-hydroxyprolines. J Biochem. 1981;89:1397–401. doi: 10.1093/oxfordjournals.jbchem.a133331. [DOI] [PubMed] [Google Scholar]
- 48.Ehrlich H, Deutzmann R, Brunner E, et al. Mineralization of the metre-long biosilica structures of glass sponges is templated on hydroxylated collagen. Nat Chem. 2010;2:1084–8. doi: 10.1038/nchem.899. [DOI] [PubMed] [Google Scholar]
- 49.Dunn MP, Di Gregorio A. The evolutionarily conserved leprecan gene: its regulation by Brachyury and its role in the developing Ciona notochord. Develop Biol. 2009;328:561–74. doi: 10.1016/j.ydbio.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Capellini TD, Dunn MP, Passamaneck YJ, et al. Conservation of notochord gene expression across chordates: insights from the Leprecan gene family. Genesis (New York, NY: 2000) 2008;46:683–96. doi: 10.1002/dvg.20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inouye K, Kobayashi Y, Kyogoku Y, et al. Synthesis and physical properties of (hydroxyproline-proline-glycine)10: hydroxyproline in the X-position decreases the melting temperature of the collagen triple helix. Arch Biochem Biophys. 1982;219:198–203. doi: 10.1016/0003-9861(82)90149-7. [DOI] [PubMed] [Google Scholar]
- 52.Inouye K, Sakakibara S, Prockop DJ. Effects of the stereo-configuration of the hydroxyl group in 4-hydroxyproline on the triple-helical structures formed by homogenous peptides resembling collagen. Biochim Biophys Acta. 1976;420:133–41. doi: 10.1016/0005-2795(76)90352-4. [DOI] [PubMed] [Google Scholar]
- 53.Jenkins CL, Bretscher LE, Guzei IA, Raines RT. Effect of 3-hydroxyproline residues on collagen stability. J Am Chem Soc. 2003;125:6422–7. doi: 10.1021/ja034015j. [DOI] [PubMed] [Google Scholar]
- 54.Mizuno K, Peyton DH, Hayashi T, et al. Effect of the -Gly-3(S)-hydroxyprolyl-4(R)-hydroxyprolyl-tripeptide unit on the stability of collagen model peptides. FEBS J. 2008;275:5830–40. doi: 10.1111/j.1742-4658.2008.06704.x. [DOI] [PubMed] [Google Scholar]
- 55.Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 56.Kramer RZ, Venugopal MG, Bella J, et al. Staggered molecular packing in crystals of a collagen-like peptide with a single charged pair. J Molec Biol. 2000;301:1191–205. doi: 10.1006/jmbi.2000.4017. [DOI] [PubMed] [Google Scholar]
- 57.Schumacher MA, Mizuno K, Bachinger HP. The crystal structure of a collagen-like polypeptide with 3(S)-hydroxyproline residues in the Xaa position forms a standard 7/2 collagen triple helix. J Biol Chem. 2006;281:27566–74. doi: 10.1074/jbc.M602797200. [DOI] [PubMed] [Google Scholar]
- 58.Vitagliano L, Berisio R, Mazzarella L, Zagari A. Structural bases of collagen stabilization induced by proline hydroxylation. Biopolymers. 2001;58:459–64. doi: 10.1002/1097-0282(20010415)58:5<459::AID-BIP1021>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 59.Hudson DM, Kim LS, Weis M, et al. Peptidyl 3-hydroxyproline binding properties of type I collagen suggest a function in fibril supramolecular assembly. Biochemistry. 2012;51:2417–24. doi: 10.1021/bi2019139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woodhead-Galloway J. Structure of the collagen fibril: some variations on a theme of tetragonally packed dimers. Proc Royal Soc B. 1980;209:275–97. [Google Scholar]
- 61.Canty EG, Kadler KE. Collagen fibril biosynthesis in tendon: a review and recent insights. Comparative biochemistry and physiology Part A. Molec Integrat Physiol. 2002;133:979–85. doi: 10.1016/s1095-6433(02)00212-x. [DOI] [PubMed] [Google Scholar]
- 62.Hudson BG, Tryggvason K, Sundaramoorthy M, Neilson EG. Alport’s syndrome, Goodpasture’s syndrome, and type IV collagen. New England J Med. 2003;348:2543–56. doi: 10.1056/NEJMra022296. [DOI] [PubMed] [Google Scholar]