Noncoding sequences conserved over long evolutionary distances can be used to identify DNA regions functioning in gene regulation. This report identifies a set of 211 conserved noncoding sequences found throughout eudicots, a subset of which are found in all flowering plants. These deeply conserved sequences are much shorter than their vertebrate counterparts, yet serve similar functions.
Abstract
In vertebrates, conserved noncoding elements (CNEs) are functionally constrained sequences that can show striking conservation over >400 million years of evolutionary distance and frequently are located megabases away from target developmental genes. Conserved noncoding sequences (CNSs) in plants are much shorter, and it has been difficult to detect conservation among distantly related genomes. In this article, we show not only that CNS sequences can be detected throughout the eudicot clade of flowering plants, but also that a subset of 37 CNSs can be found in all flowering plants (diverging ∼170 million years ago). These CNSs are functionally similar to vertebrate CNEs, being highly associated with transcription factor and development genes and enriched in transcription factor binding sites. Some of the most highly conserved sequences occur in genes encoding RNA binding proteins, particularly the RNA splicing–associated SR genes. Differences in sequence conservation between plants and animals are likely to reflect differences in the biology of the organisms, with plants being much more able to tolerate genomic deletions and whole-genome duplication events due, in part, to their far greater fecundity compared with vertebrates.
INTRODUCTION
DNA sequences conserved in cross-species alignments, known as phylogenetic footprints, are under negative (purifying) selection because functional sequences change at a lower rate over evolutionary time compared with functionless sequences. Previous studies comparing vertebrate genomes find that whereas transcription factor binding motifs themselves are too short and variable to identify functional regulatory sequences, evolutionarily conserved noncoding sequences (CNSs) are enriched in transcription factor binding sites (Blanchette et al., 2006; Pennacchio et al., 2007) and can be used to identify novel regulatory sequences. In several cases, conserved noncoding elements (CNEs) have been shown to be modules composed of multiple transcription factor binding sites (Panne et al., 2007; Strähle and Rastegar, 2008).
Mammalian CNEs make up roughly 3% of the human (Homo sapiens) genome. Mammalian CNEs can be both very long and very highly conserved. Among placental mammals, there are ∼14,000 CNEs that are at least 100 bp long and share 100% sequence identity (Stephen et al., 2008). Mammalian CNEs are frequently found in gene-poor regions of the genome that are associated with developmental genes that may be megabases away (Bejerano et al., 2004). For instance, over 1000 CNEs (>100 bp, >70% identity) have been found associated with the human nuclear regulatory gene DACH1 within a 2630-kb window (Nobrega et al., 2003). Why such surprisingly large and highly conserved sequences exist is a mystery. They are not simply mutational cold spots (Drake et al., 2006; Sakuraba et al., 2008) but are under even greater negative selection than nonsynonymous sites from coding sequences (Katzman et al., 2007). Their extreme conservation may be instead due to overlapping functions for the same noncoding sequence and/or the presence of multiple, closely spaced, or overlapped transcription factor binding sites (Siepel et al., 2005; Feng et al., 2006; Panne et al., 2007).
While enhancer activity has been shown for a number of mammalian CNEs (Loots et al., 2000), a study examining a random set of human-mouse (Mus musculus) CNEs for enhancer or promoter activity showed activity for only 15 to 20% of CNEs across a wide range of human cell types and revealed no enrichment for DNase I hypersensitive sites (Attanasio et al., 2008). Two approaches have been taken to improve the likelihood of finding functional enhancers among the many thousands of mammalian CNEs. In the first approach, ultraconserved regions are chosen for enhancer analysis. For instance, by choosing to assay ultraconserved CNEs (>200 bp with 100% identity) or highly constrained CNEs, Visel et al. (2008) found that 50% showed enhancer activity in a transgenic mouse reporter assay. The second approach focuses on deeply conserved CNEs, particularly those conserved between mammals and fish (Woolfe and Elgar, 2008). By testing only CNEs conserved between humans and fugu, a type of puffer fish (Fugu rubripes), Nobrega et al. (2003) found that seven out of nine DACH1 CNE sequences acted as enhancers. In the largest study conducted to date, more than 40% of 150 human-fugu CNEs tested had enhancer activity versus only 5% of human-rodent CNEs (Visel et al., 2007). Similarly, 45% of 22 CNEs conserved between the invertebrate amphioxus (Branchiostoma floridae) and vertebrates (mouse, fugu, and zebra fish [Danio rerio]) had enhancer activity in a transgenic zebra fish assay (Hufton et al., 2009).
Plant genomes differ markedly from animal genomes in not having long CNSs with high sequence identity. When plant and vertebrate genomes were searched genome-wide for long identical multispecies elements (LIMEs), sequences identical over at least 100 bp between genomes having diverged more than 50 million years ago (mya), the results differed so greatly between clades to suggest that plant CNSs and vertebrate CNEs have very little in common (Reneker et al., 2012). Vertebrates had 1.8 million LIMEs with 99% of them conserved syntenically (retained with their flanking genes). Plants, on the other hand, had many fewer LIMEs (∼26,000) and, with the exception of telomeric repeats, all of them were nonsyntenic, with 95% of these being repetitive sequences. Freeling and Subramaniam (2009) concluded that, if the definition of plant-conserved noncoding elements were the same as that used for vertebrates, plants would have no such sequences. Therefore, there must be something very different about the biology and evolution of plants versus animals that accounts for these dramatic differences.
In fact, plants do have CNSs, but they differ markedly from animal CNEs. They are shorter, less well-conserved, and found in much greater proximity to their associated genes (Kaplinsky et al., 2002; Guo and Moose, 2003). Intragenomic CNSs between Arabidopsis thaliana homoeologous gene pairs, for instance, have a median CNS size of only 25 bp (Thomas et al., 2007). These are CNSs conserved since the most recent Arabidopsis whole-genome duplication (WGD) event ∼43 mya (α-CNSs) (Fawcett et al., 2009). Despite their small size, CNSs likely represent functional sequences because they are often found clustered adjacent to transcription factor and signal transduction genes and are enriched in motifs such as the G-box motif (Freeling et al., 2007). Although plant CNSs are small in size, they are extremely abundant in the plant genome, with almost 15,000 CNSs detected among homoeologous Arabidopsis genes (Thomas et al., 2007), over 90,000 among crucifers (Haudry et al., 2013), and 1865 between Arabidopsis and the promoter regions of noncruciferous eudicots (Baxter et al., 2012). To identify CNSs to test for functional activity, it would be useful to narrow this list to the CNSs most likely to be functional.
In this article, we identify the most deeply conserved eudicot CNSs and compare them to the previously reported set of the most deeply conserved CNSs in commelinids (D’Hont et al., 2012), the superorder clade within the monocots that includes the grasses. Almost all flowering plants (angiosperms) can be separated into monocots or eudicots, lineages that diverged ∼140 mya (Moore et al., 2007). In the process of identifying eudicot and commelinid deep CNSs, we found 59 deep CNSs that are conserved between monocots and eudicots, 35 of which are conserved throughout angiosperms. We also show that deeply conserved plant CNSs share a number of fundamental properties with deeply conserved animal CNEs, in particular, their strong association with core developmental genes and, when present intragenically, with genes whose protein products interact with RNA.
RESULTS
Identification of 211 Deep Eudicot CNSs
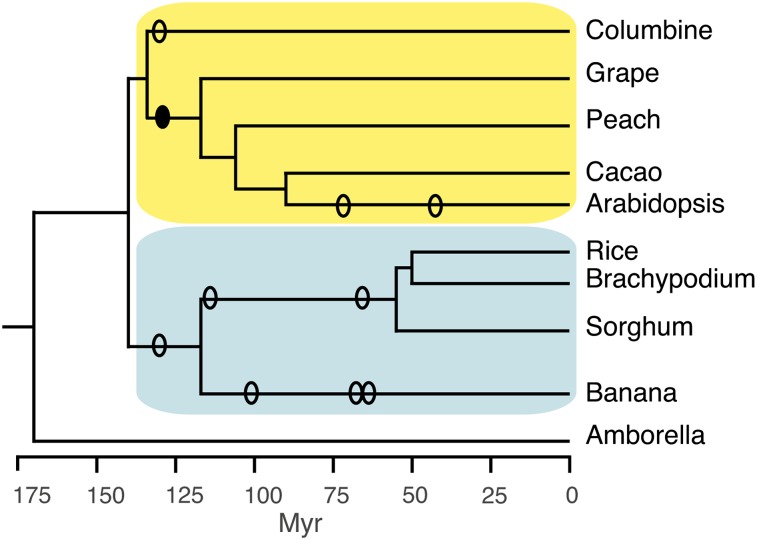
To identify deeply conserved eudicot CNSs, we looked for noncoding sequences conserved between columbine (Aquilegia coerulea), a member of an early diverging eudicot clade (Kramer, 2009), and Arabidopsis, a rosid (Figure 1). Arabidopsis and columbine diverged from each other ∼135 mya (Wikström et al., 2001) (modal Ks = 2.26, Ks measuring the number of synonymous substitutions per synonymous site), a time frame even longer than the ∼117 million year divergence between rice (Oryza sativa) and banana (Musa acuminata) (Janssen and Bremer, 2004), the subject of a previous study in which we identified 116 deeply conserved pan-commelinid CNSs (D’Hont et al., 2012).
Figure 1.
Phylogenetic Relationships among Genomes Used in Identifying Pan-Eudicot (Yellow) and Pan-Commelinid (Blue) CNSs.
Approximate points of divergence and WGD events (ellipses) are shown in million years (Myr), with a filled-in ellipse indicating the paleohexaploidy event that occurred in the ancestor of most eudicots.
Since the small size of plant CNSs makes the identification of deeply conserved CNSs particularly challenging, we utilized the visual power of GEvo panels, an application within the CoGe toolbox (Lyons and Freeling, 2008) that can be used to study genome evolution. GEvo allowed us to detect, using a BLASTN search (http://blast.ncbi.nlm.nih.gov/Blast.cgi), high-scoring segment pairs (HSPs) between pairs of syntenous genomic regions from Arabidopsis, columbine, grape (Vitis vinifera), peach (Prunus persica), and cacao (Theobroma cacao) (Supplemental Figure 1). To be as thorough as possible, we used two different approaches to identify deeply conserved CNSs. In the first approach, we started with a set of 5778 previously identified α-CNSs between Arabidopsis homoeologous gene pairs. Based on BLASTN results of α-CNSs queried to grape, peach, and columbine genomes, 380 homoeologous gene pairs (approximately one-third of the α-CNSs) were manually analyzed on GEvo panels, resulting in the identification of 116 deeply conserved eudicot CNSs (Supplemental Data Set 1). It is likely that additional deeply conserved CNSs would have been identified if the remaining homoeologous gene pairs had been analyzed.
In the second approach to detecting deeply conserved CNSs, peach-cacao orthologous CNSs identified using the PL3.0 automated CNS Discovery pipeline (Turco et al., 2013) were used to search for BLASTN hits present in the 10,000-bp region surrounding columbine and Arabidopsis orthologous genes. This approach allowed CNSs to be identified from Arabidopsis genes in which one of the original duplicate genes had not been retained. Using this stepping-stone approach, 112 deep CNSs were identified (Supplemental Data Set 1), of which only 17 had been independently identified starting with Arabidopsis α-CNSs.
Plant Deep CNSs Are Small Compared with Vertebrate Deep CNSs
The standard definition of a vertebrate CNE is 70% identity over at least 100 bp. Only eight of the pan-commelinid CNSs and one of the eudicot CNSs fit this definition, and the majority of these are intronic CNSs. The median deep plant CNS length is only 38 bp in commelinids and 27 bp in eudicots. While few genes retain more than two deeply conserved CNSs, duplicate pairs of Arabidopsis BEL1-like homeodomain transcription factor genes (AT2G23760/AT4G36870) and bHLH transcription factor genes (AT3G25710/AT1G68810) each have four deep CNSs, and the rice MYB transcription factor gene LOC_Os01g08160 has six CNSs (D’Hont et al., 2012).
Deeply Conserved Plant CNSs Are Located Closer to Their Cognate Genes Than Are Deeply Conserved Vertebrate CNEs
Deeply conserved vertebrate CNEs are commonly located in gene-poor regions that are megabases away from their cognate genes. In contrast, the most distantly located deeply conserved plant CNS is just under 11 kb away from its cognate gene. The average distance is 1.4 kb in Arabidopsis and 2.5 kb in rice, with the distribution difference reflecting the more compact genome of Arabidopsis (Wilcoxon test, P = 1.4E-05). Because each CNS gene pair was manually compared in GEvo, more distant CNSs would have been expected to be observed had they existed.
Both vertebrate and plant CNSs may be located either in intergenic regions 5′ or 3′ to their cognate gene or in the 5′ untranslated region (UTR), 3′ UTR, or intron. A similar percentage of deep CNSs are located in the 5′ intergenic region in rice and Arabidopsis (59 and 67%, respectively). Rice has a greater percentage of deep CNSs present in intronic regions compared with Arabidopsis (16 versus 8%, respectively; P = 6.5E-05, binomial test), but percentages present in the 5′ UTR (17 and 14%, respectively) and 3′ UTR (10 and 11%, respectively) are similar in both species. A slightly greater proportion of deep CNSs are found in the 3′ intergenic region in rice compared with Arabidopsis (12 and 8%, respectively; P = 0.044, binomial test), and the majority of 3′ intergenic CNSs are 3′ distal in rice but 3′ proximal in Arabidopsis.
59 Ultra-Deep CNSs Are Conserved in Both Rice and Arabidopsis
Animal CNEs can be detected between mammals and the early diverging chordate amphioxus, lineages that diverged ∼520 mya (Hufton et al., 2009). To test whether any of the deep CNSs discovered in eudicots are also conserved in monocots (diverged ∼140 mya), we added orthologous rice genes to GEvo panels containing deep eudicot CNSs (Supplemental Figure 1). Altogether, 23% of deep eudicot CNSs were found to be conserved in rice (Supplemental Data Set 2), similar to the 15% pan-commelinid CNSs found to be conserved in Arabidopsis (D’Hont et al., 2012). Combining both studies, we found 59 ultra-deep CNSs conserved between commelinid monocots and eudicots.
To determine whether any of the deep CNSs span all angiosperms, orthologous genes from the basal angiosperm Amborella trichopoda (Amborella Genome Project, 2013) were added to GEvo panels. Of the 51 CNSs for which an A. trichopoda ortholog could be identified, 35 (68%) were also present in the expected syntenic position in A. trichopoda, a lineage diverging from higher plants ∼170 mya (Moore et al., 2007) (Table 1).
Table 1. CNSs Found in All Angiosperms.
| Arabidopsis Genes | Gene Function | CNS Location | Comments | |
|---|---|---|---|---|
| AT1G01030/AT4G01500 | TF | ABI3VP1 | 5′ Proximala | |
| AT2G20100/AT4G29100 | TF | bHLH | 3′ UTR | Secondary structure |
| AT3G25710/AT1G68810 | TF | bHLH | 5′ Distal | |
| AT2G46270/AT4G01120 | TF | bZIP | 5′ Proximal | G-box motif |
| AT1G25250/AT1G68130 | TF | C2H2 | 5′ Proximal | |
| AT4G04890/AT4G21750 | TF | HB | 5′ Proximal | L1 motif |
| 3′ UTRb | Secondary structure | |||
| 3′ UTRb | Secondary structure | |||
| AT5G06710 | TF | HB | 5′ Distal | |
| AT1G73410/AT1G17950 | TF | MYB | 5′ Proximal | |
| AT2G16720/AT4G34990 | TF | MYB | 5′ Proximalc | |
| AT4G21440/AT4G05100 | TF | MYB | 5′ Proximal | |
| AT1G12260/AT1G62700 | TF | NAC | 5′ Distal | |
| AT1G14440/AT2G02540 | TF | zf-HD | 5′ Proximal | |
| AT3G53340 | TF | CCAAT | 5′ UTR/5′ proximal | |
| AT1G46264 | TF | HSF | 5′ Distal | |
| AT2G12646 | TF | PLATZ | 5′ Proximal | |
| AT4G26480/AT5G56140 | RNA binding protein | Intron | ||
| Intron | ||||
| AT4G27000/AT5G54900 | RNA binding protein | Intron | Secondary structure | |
| AT2G46610/AT3G61860 | SR splicing factor | 5′ UTR/intron | AS | |
| AT2G37340/AT3G53500 | SR splicing factor | 5′ Proximal/intron | AS | |
| AT2G33440 | RRM splicing factor | Intron | ||
| AT5G55100 | RNA processing | Intron | ||
| AT5G14610/AT3G01540 | RNA helicase | 5′ UTR/intron | Secondary structure | |
| AT4G14465 | DNA binding protein | 5′ Proximal | ||
| AT1G57680 | G-protein receptor | 5′ UTR/5′ proximal | Secondary structure | |
| AT5G17420 | Cellulose synthase | 5′ Proximal | ||
| AT1G13940/AT1G26620 | Unknown | 5′ Proximal | Secondary structure | |
| AT2G29630 | Thiamine biosynthesis | 3′ UTR | Riboswitch | |
| AT4G37580/AT2G23060 | N-acetyltransferase HLS1 | 5′ Proximal | GCC-box | |
| AT5G64040 | PSI reaction center | 5′ UTR/5′ proximal | ||
| AT4G13250 | Chlorophyll b reductase | 5′ Proximal | ||
| AT3G09050/AT5G01175 | Unknown | 3′ UTR/intron | Secondary structure | |
| AT2G39950 | Unknown | 5′ UTR/intron | AS | |
TF, transcription factor; AS, alternative splicing.
Also AT2G46870 and AT3G61970.
Also AT1G05230, AT2G32370, AT1G17920, AT1G73360, AT4G17710, AT5G46880, AT3G61150, and AT4G00730.
Also AT4G38620.
Deep Eudicot CNSs Are Strongly Associated with Genes Encoding Transcription Factors, Especially Those Regulating Development
Deeply conserved animal CNEs are associated with genes involved in transcriptional regulation and development (trans-dev genes) (Woolfe et al., 2005). Arabidopsis genes associated with deeply conserved eudicot CNSs (deep eudicot genes) (Supplemental Data Set 3) are also highly associated with trans-dev genes, with half of Arabidopsis deep eudicot genes being annotated with the gene ontology (GO) term “transcription factor activity” (P value adjusted for multiple testing using the false discovery rate method, 1.5E-99), as are almost one-quarter of deep commelinid rice genes (3.8E-21 P value) (Figure 2). The GO term “multicellular organismal development” appears in the annotation of one-quarter of Arabidopsis deep eudicot genes (2.4E-30 P value) and “response to hormone stimulus” appears in 14% of these genes (4.6E-18 P value).
Figure 2.
Deep Eudicot CNS-Associated Genes Are Primarily Involved in Transcriptional Regulation, Development, and Response to Hormones and Salt.
Shown in ranked order of their false discovery rate–corrected Fisher's P values are 289 deep CNS-associated genes versus all 31,819 Arabidopsis genes (left) and 289 deep CNS-associated genes versus 3681 α-CNS–associated genes (right). Transcription-related GO term rows are filled in blue, development GO term rows in green, and response GO term rows in orange.
Because α-CNS genes are themselves enriched in transcription and “response to” GO terms (Thomas et al., 2007), we compared GO term enrichment between these two groups and found that deep eudicot genes are significantly more enriched in transcription-related GO terms (5.4E-40 P value for “transcription factor activity”) and development-related GO terms (8.4E-14 for “multicellular organismal development”) compared with α-CNS genes. Conversely, many GO terms highly enriched in α-CNS genes such as “response to stress” (P 3.8E-08), “protein kinase activity” (P 1.7E-19), and “catalytic activity” (P 1.9E-13) are not enriched in deep eudicot CNS genes (Supplemental Data Set 4). GO term enrichment, for the most part, is not dependent on the position of the deep CNS with respect to its cognate gene. Even when deep CNSs are found within UTRs or introns, the cognate genes are still highly enriched in “transcription factor activity” (P 1.0E-17) and “multicellular organismal development” (P 3.1E-07) GO terms, although they are also enriched in the term “RNA binding” (P 2.1E-05), which is not found for genes associated with intergenic CNSs (Supplemental Data Set 5).
α-CNSs from CNS-Rich Genes Are More Likely to Be Deeply Conserved
Deep CNSs comprise a special subset of α-CNSs as evidenced by their strong association with genes enriched in transcription factor and developmental GO terms. α-CNS genes present in chromosomal regions devoid of other genes (Bigfoot genes) are CNS-rich and highly enriched in transcription factor and “regulation of…” GO terms relative to other α-CNS genes (Freeling et al., 2007). To determine whether deep CNSs are more likely to be associated with CNS-rich genes, we compared the number of deep CNSs in four different bins (<6, 6 to 10, 11 to 15, and >15 α-CNSs) to the number expected based on the distribution of α-CNS number per gene. The overall distribution is highly biased (P = 3.15E-12, χ2), with almost twice as many deep CNSs as expected present in the most CNS-rich category. Deep CNSs are also similar to Bigfoot genes, plant genes that cover more than 4 kb of noncoding chromosome, in being present in intergenic regions that are larger than the average intergenic region in Arabidopsis (3260 bp versus 1672 bp; P < 1.4E=10, Wilcoxon test). A comparison of GO terms associated with deep CNS genes and GO terms associated with Bigfoot genes found no statistically significant differences.
Deep CNSs Are Enriched for Predicted Transcription Factor Binding Sites
Transcription factor binding sites are generally short (4 to 12 bp), with specificity arising from the binding of multiple factors in complexes to cis-regulatory modules. Animal CNEs are known to be enriched for transcription factor binding sites, and the combination of transcription factor binding sites has been successfully used to assign tissue-specific predictions (Pennacchio et al., 2007). To determine whether plant deep CNSs are enriched in transcription factor binding sites, deep CNSs were scanned for the presence of known motifs. We found 12 motifs that were statistically enriched compared with control sequences (intergenic and intronic nonconserved sequences) (Tables 2 and 3). Most of the overrepresented motifs have been implicated in stress responses. Four of the overrepresented motifs are versions of the WRKY-box (abiotic and biotic stress), two are related to abscisic acid response elements (abiotic stress), and two are related to MYC2 binding sites (jasmonic acid response). Also overrepresented are the GCC-box (ethylene response), the G-box (light response), and WLE1-box (salicylic acid inducibility). Only the cell cycle–related MSA box is not involved in stress responses. WRKY and MYC2 motifs are also overrepresented among deep rice CNSs.
Table 2. Transcription Factor Binding Sites Enriched in Deeply Conserved CNSs: Arabidopsis.
| TFBS | Motif | Adjusted P Value | Expected | Observed | Conserved in Eudicots |
|---|---|---|---|---|---|
| WRKY | TGAC | 0.0026 | 39.4 | 66 | 37 |
| GCC-box | GCCGCC | 0.0050 | 0.6 | 5 | 3 |
| WLE | TGTCA | 0.0250 | 14.1 | 27 | 11 |
| G-box | CACGTG | 0.0280 | 0.9 | 5 | 4 |
| MYC2 | CACATG | 0.0280 | 3.4 | 10 | 8 |
| MSA | AACGG | 0.0320 | 5.4 | 13 | 7 |
| ABRE | MACGYGB | 0.0360 | 2.6 | 8 | 6 |
| WRKY | TGACY | 0.0380 | 19.7 | 33 | 21 |
| ABRE | ACGTG | 0.0380 | 5.2 | 12 | 7 |
| MYC2 | TCACATG | 0.0380 | 1.2 | 6 | 4 |
| WRKY | TTGAC | 0.0390 | 14.8 | 26 | 17 |
| WRKY | CTGACY | 0.0440 | 2.9 | 8 | 4 |
P values were calculated using Fisher's exact test and corrected for multiple testing using the Benjamini-Hochberg procedure. Motifs conserved in eudicots were required to be present in at least one CNS sequence each from Arabidopsis, grape, peach, chocolate, and columbine. TFBS, transcription factor binding site.
Table 3. Transcription Factor Binding Sites Enriched in Deeply Conserved CNSs: Rice.
| TFBS | Motif | Adjusted P Value | Expected | Observed | Conserved in Commelinids |
|---|---|---|---|---|---|
| WRKY | TGACY | 0.0097 | 16.3 | 32 | 26 |
| WRKY | TTGACY | 0.0097 | 5.4 | 15 | 11 |
| MYB | CCWACC | 0.0097 | 4.8 | 14 | 4 |
| MYC2 | TCACATG | 0.0200 | 1.2 | 6 | 5 |
| WRKY | TGAC | 0.0370 | 36.1 | 55 | 41 |
| WRKY | TTGACTT | 0.0390 | 1.0 | 5 | 3 |
P values were calculated using Fisher's exact test and corrected for multiple testing using the Benjamini-Hochberg procedure. Motifs conserved in commelinids had to be present in at least one CNS sequence each from rice, sorghum, and banana. TFBS, transcription factor binding site.
To substantiate the significance of these overrepresented motifs, Arabidopsis and rice motifs were tested for their phylogenetic conservation in other eudicot or commelinid CNS sequences, and overrepresented motifs were found to coincide with ultraconserved islands within the aligned sequences (Tables 2 and 3). For instance, the motif 5′-TCACATG-3′, an extended version of a MYC2 binding site (Godoy et al., 2011), appears six times in Arabidopsis deep CNSs, and five of these occurrences are conserved in Arabidopsis, grape, peach, cacao, and columbine (Figure 3). In the case of the CNS associated with NGATHA, conservation of this motif extends to A. trichopoda. Four G-box motifs are conserved in eudicot genomes, and all four CNSs with phylogenetically conserved G-box motifs (CACGTG) are associated with genes shown to bind the bZIP transcription factor HY5 in an in vitro assay (Lee et al., 2007b). GCC-boxes are phylogenetically conserved in three of their five occurrences (Supplemental Figure 2B), and the GCC-box present in the promoter of SHI/STY RING-like zinc finger genes has been experimentally verified to be functional (Eklund et al., 2011). Whereas the overrepresented MSA box is only a 5-bp sequence (5′AACGG), over half of its occurrences are phylogenetically conserved throughout eudicots (Supplemental Figure 2A). The MSA box is bound by c-MYB transcription factors that regulate expression of genes at the G2/M transition in the cell cycle (Ito et al., 2001), and three of the phylogenetically conserved CNSs are associated with genes explicitly involved in cell cycle regulation, two CNSs associated with cyclin-dependent protein kinases, and the CNS associated with c-MYB transcription factors responsible for repressing G2/M phase–specific genes. Interestingly, the CNS associated with the cyclin-dependent protein kinase genes, which has two MSA motifs, was found incorporated into a columbine MULE element, an example of this transposable element's capacity to acquire gene fragments (Jiang et al., 2004), in this case one with the potential of tethering transcription to the cell cycle.
Figure 3.
TCACATG Motifs in Deep CNSs Are Phylogenetically Conserved.
Five of the six TCACATG motifs appearing in Arabidopsis deep CNSs are conserved in grape, peach, cacao, and columbine sequences. TF, transcription factor. See Supplemental Table 1 for genes associated with these CNS sequences.
Due to the short size and degenerate nature of transcription factor binding sites, many are false positives. Detection of multiple transcription factor binding sites has been shown to increase the probability of predicting functional sites (Lifanov et al., 2003; Gómez-Porras et al., 2007). To see if any specific motif was overrepresented in duplicate in deep CNSs, z-test statistics were used to look at the four motifs present in duplicate in 10 or more deep CNSs, and all were found to be significantly overrepresented at the 5% significance level. Three have a core sequence recognized by DOF (DNA binding with one finger) transcription factors (AAAG, AAAGH, and WAAAG), and the fourth has a WRKY binding site core (TGAC). Both DOF and WRKY binding sites have been shown to cluster and act either additively or synergistically (Eulgem et al., 2000; Cominelli et al., 2011). The total number of transcription factor binding sites present in duplicate was also significantly higher in deep CNSs compared with matched sets of control sequences (126 versus 84.6 ± 11.5; z = 1.5 × 10−4).
A Subset of Intragenic CNSs Have Conserved RNA Secondary Structure
Some of the vertebrate CNEs with the most extreme conservation are found in 3′ UTRs, and 3′ UTRs are statistically enriched in secondary structure (Siepel et al., 2005). To determine whether the function of plant intragenic deep CNSs may in some cases be mediated through RNA secondary structure formation, multiple alignments of eudicot and commelinid CNS sequences were used to search for conserved RNA secondary structure on the RNAz Web server (Washietl et al., 2005). Structures with an RNAz-defined RNA class probability of 0.5 or greater were manually inspected, and 14 CNSs were identified as having a high likelihood of forming RNA structures, with the great majority of them being conserved in both eudicot and commelinid alignments (Supplemental Table 3). This group includes a CNS in the 3′ UTR of THIC that acts as a riboswitch that changes conformation upon binding the metabolite thiamine pyrophosphate, thereby altering splicing and transcript stability (Wachter et al., 2007). A CNS in the 3′ UTR of the bHLH family members AT2G20100 and AT4G29100 has a predicted RNA structure that is conserved in eudicots, commelinids, and A. trichopoda (Figure 4B). The significance of this structure is supported by multiple double-stranded RNA (dsRNA) sequencing reads coinciding with the predicted base-paired region (Zheng et al., 2010). dsRNA sequencing reads were also found in the folded structure predicted to be present in the 3′ UTR of EBF2 in both eudicots and commelinids (Figure 4A). Interestingly, it is likely that the 3′ UTR of EBF2 is required for maintaining proper expression levels of EBF2, a gene encoding a F-box protein that regulates ethylene signaling via targeting EIN3 for degradation (Konishi and Yanagisawa, 2008). Potential 3′ UTR RNA folds are present among numerous members of the HD-ZIPIV family, including 10 Arabidopsis and eight rice HD-ZIPIV genes (Javelle et al., 2011) (Supplemental Figure 3). The predicted RNA structures are anchored by almost invariant 15- and 16-bp sequences enclosing RNA structures of varying length. Corroborating the existence of these RNA structures are numerous dsRNA reads coinciding with predicted regions of secondary structure in all but two of the nine genes that are expressed in unopened flower buds, the organ that was used to make the dsRNA library. Potential conserved RNA structures are also present in 5′ UTRs and introns, including a CNS present in the 5′ UTR/intron of alternative isoforms of a RNA helicase gene (Figure 4C). It is intriguing to speculate that regulation of stability or translation of the RNA helicase transcript itself might involve unwinding by the protein product of the transcript.
Figure 4.
Intragenic CNSs with Conserved RNA Secondary Structure.
(A) RNAz-predicted consensus RNA structure of the 3′ UTR of the EBF2 family based on the multiple alignment of sequences from rice (LOC_Os06g40360 and LOC_Os02g10700) and sorghum (Sb04g006870).
(B) RNAz-predicted consensus RNA structure of the 3′ UTR from a multiple alignment of sequences from a bHLH gene family (Supplemental Table 2). The region corresponding to dsRNA reads in AT4G29100 is indicated by arrows.
(C) RNAz-predicted consensus RNA structure from a multiple alignment of sequences from a DEAD-box RNA helicase family (Supplemental Table 2). Multiple isoforms for AT3G01540 and LOC_Os01g36860 are shown with the position of the CNS indicated by the green bar. dsRNA reads are depicted as screenshots from the IGB browser (http://bioviz.org/igb/). RNAz-predicted base-paired regions conserved throughout the multiple alignment are colored blue, with lighter shades of blue depicting the presence of incompatible pairs within the alignment; positions in which consistent or compensatory mutations have taken place are colored green, with lighter shades depicting the presence of incompatible pairs within the alignment.
An unusual CNS with conserved secondary structure was found in the 3′ UTR of the chloroplast thylakoid membrane gene, AT3G09050. While all other CNSs could be assigned with high confidence to a cognate gene based on retention of that gene in all genomes examined, the two flanking genes present in grass, eudicot, and A. trichopoda genomes have been lost in one of the Arabidopsis homoeologous regions, and the CNS was present in the intron of the noncoding gene AT5G01175. In both rice and peach, the CNS has been duplicated, and in maize (Zea mays) there are 15 tandem copies.
A Subset of Deep CNSs Is Associated with Alternative Splicing
Alternative splicing of RNA splicing–associated SR genes in vertebrates results in the inclusion of stop codon–containing exons that target the resulting transcripts for degradation by the nonsense-mediated decay pathway. These stop codon–containing exons are enriched in ultraconserved elements, and eight of the nine ultraconserved elements that overlap stop codon–containing exons are associated with RNA splicing genes (Bejerano et al., 2004; Lareau et al., 2007; Ni et al., 2007). Among the 59 ultra-deep plant CNSs (Supplemental Data Set 2), 12 are intragenic CNSs in genes whose products interact with RNA, including six of the 19 Arabidopsis SR genes. For instance, the CNS in AT3G53500 and AT2G37340 (encoding SR proteins RSZ32 and RSZ33) is just upstream of an alternative exon that carries a premature stop codon whose retention subjects the transcript to nonsense-mediated decay (Reddy and Ali, 2011; Kalyna et al., 2012). The RNA binding KH domain–containing genes LOC_Os08g01930, AT1G33680, and AT4G10070 have an ultra-deep CNS present in their largest intron. This CNS has been co-opted in the chimeric gene LOC_Os12g16350, which is a scrambled fusion between a KH domain gene and an enoyl-CoA hydratase gene flanked by a retrotransposon.
Deep CNSs with Experimentally Validated Functions
A number of CNSs were found to overlap with experimentally validated cis-regulatory regions (Supplemental Table 4). These include a GCC-box present in the SHI/STY family of transcriptional activators of auxin biosynthetic genes (Eklund et al., 2011) (Supplemental Figure 2A), a CArG3-box required for the negative autoregulation of AGL15, a MADS-domain transcription factor active during embryogenesis (Zhu and Perry, 2005), and a GATA-box found in the light-regulated chloroplast glyceraldehyde-3-phosphate dehydrogenase gene (Jeong and Shih, 2003). These transcription factor binding sites coincide with exceptionally conserved regions within the CNS, as can be seen in Supplemental Figure 3 for the L1-box conferring epidermal expression in HD-ZIP IV genes ATML1 and PDF2 (Abe et al., 2001). In addition to CNSs functioning in transcriptional regulation, two CNSs function in posttranscriptional regulation: the CNS in THIC that regulates transcript stability (Wachter et al., 2007) and a microRNA-responsive element that affects translation of a family of squamosa transcription factors known to activate floral meristem identity genes (Gandikota et al., 2007).
DISCUSSION
While deeply conserved CNEs between mammals and teleost fish are easily detectable due to their large size and degree of conservation, identification of deeply conserved CNSs in plants has been more problematic. Reineke et al. (2011) estimated that identification of plant CNSs would be limited to those diverging <100 mya but recently Baxter et al. (2012) showed that CNSs can be detected between the eudicots grape and Arabidopsis, which diverged ∼108 to 117 mya (Wikström et al., 2001). Using an algorithm that scores 60-bp windows of aligned sequence between orthologous promoters, they identified 1851 CNSs conserved in eudicots, with 910 of them conserved in grape. However, only 20% of the deep eudicot CNSs we identified (74/364) overlap with their Arabidopsis-grape CNSs. This is largely due to their algorithm being limited to the 2-kb 5′ upstream region of Arabidopsis genes. Using whole-genome BLAST searches between Arabidopsis and grape, Kritsas et al. (2012) identified 29 orthologous hits with at least 85% identity over a span of 56 bp or more. Only three of the ultraconserved-like elements (ULEs) overlap deep CNSs. We examined the nonoverlapping ULEs and found that 18 of them were conserved in columbine. Thus, the 211 deep CNSs we identified comprise only a subset of deep CNSs. While ULEs are almost exclusively associated with Arabidopsis genes lacking a homoeolog (singletons), only 13% of our deep CNSs are associated with singletons. This is not surprising since we used α-CNSs as the starting material in one of our two approaches. The strong bias toward singleton association among ULEs, which by definition have vertebrate-like conservation, may be as a result of CNSs that are associated with duplicate genes being under reduced selective constraint (Subramaniam et al., 2013).
By using Arabidopsis-columbine and rice-banana CNSs as intermediaries, we were able to identify 59 ultra-deep CNSs conserved in both commelinid and eudicot lineages, 39 of which were also conserved in A. trichopoda. Besides being associated with transcription factor genes, the most deeply conserved CNSs were associated with genes whose products interact with RNA (genes encoding RNA binding proteins, splicing factors, a Dicer, a helicase, and a MIR). These CNSs generally reside at intragenic positions, frequently show conserved secondary structure, and are often associated with alternative splicing.
We assembled a gold standard set of deeply conserved CNSs to use in comparing regulation of gene expression in plant versus vertebrate genomes. One likely function for CNSs is to serve as scaffolds organizing the binding of multiple transcription factors (Kaplinsky et al., 2002; Guo and Moose, 2003), and we show here that not only are deep CNSs enriched in several characterized transcription factor binding sites, but these motifs often coincide with phylogenetically conserved sequences within CNSs. This enrichment is corroborated by studies showing that regions bound by transcription factors are overrepresented in both plant and vertebrate CNSs (Lee et al., 2007a; Haudry et al., 2013). In plants, the G-box 5′-CACGTG-3′ is reported in several studies as being enriched in CNS sequences (Freeling et al., 2007; Baxter et al., 2012; Haudry et al., 2013). This motif is bound by bHLH and bZIP transcription factors and in vertebrates by bHLH transcription factors, including c-MYC, whose binding sites have been shown to be overrepresented in human-fugu CNEs associated with transcription factor genes (Lee et al., 2007a). However, motifs that are overrepresented in CNSs do not generally overlap between plants and vertebrates, both due to differential expansion of transcription factor families and to plant-specific families (DOF, WRKY, and AP2/ERF).
Both animal and plant deep CNSs are strongly associated with trans-dev genes. Human-fugu deep CNEs are almost all present in clusters of five or more, with 93% of these clusters being associated with trans-dev genes (Woolfe et al., 2005). While CNEs present since the beginning of vertebrate evolution are highly enriched in trans-dev genes, CNEs emerging more recently are instead enriched in receptor binding and posttranslational protein modification GO terms (Lowe et al., 2011). Plant deep CNSs are similarly associated with trans-dev genes (e.g., abaxial/adaxial polarity genes), with 52% of CNS-associated genes annotated with the GO term “transcription regulator activity” (as opposed to 9% of all Arabidopsis genes and 14% of Arabidopsis genes with α-CNSs). Similarly, the most highly conserved human-fugu CNEs are associated with master regulator genes that function during embryo development (Sandelin et al., 2004). In addition to genes involved in transcriptional regulation and development, plant deep CNSs are also enriched in “response to” GO terms, this fitting a plant's sessile nature.
It is not only CNSs present in the 5′ flanking region that are associated with trans-dev genes. Half of 3′ flanking CNSs and one-third of intragenic CNSs are associated with genes annotated with the GO term “transcription factor activity” (Supplemental Data Set 5). CNSs in plant introns are found in a duplicate pair of BEL1-like homeobox genes, an HD-ZIPIV homeobox gene, and KAN2, which encodes a homeodomain-like transcription factor. These intronic CNSs are present in the middle of large introns among clusters of less deeply conserved CNSs, suggesting that they function as transcription factor binding sites rather than in alternative splicing. Transcription factor binding sites are known to be present in both plant and vertebrate introns (Haerry and Gehring, 1996; Morello and Breviario, 2008). Transcription factor binding sites can also be present in UTRs. Clusters of human transcription factor binding sites (cis-regulatory modules) were found to peak on both sides of transcription start sites and 3′ UTRs to be approximately as enriched in transcription factor binding site clusters as 5′ UTRs (Blanchette et al., 2006). Plant CNSs in the 5′ UTR of the chloroplast glyceraldehyde-phosphate dehydrogenase gene and the gene encoding transcription factor SHI both contain experimentally verified transcription factor binding sites (Jeong and Shih, 2003; Eklund et al., 2011).
In addition to trans-dev functions, intragenic deep CNSs are associated with posttranscriptional regulation, such as mRNA stability and localization, alternative splicing, and translational control. A deeply conserved CNS in the 3′ UTR of SPL4 and SPL5 coincides with a miRNA-responsive element that confers translational repression on a transcription factor family that mediates flowering time (Gandikota et al., 2007). The ultra-deep CNS in the 3′ UTR of THIC is capable of folding into an RNA structure that can bind thiamine pyrophosphate and thereby exert feedback control on the stability of its transcript via alternative splicing (Wachter et al., 2007). Several other deep plant CNSs also have the potential to form secondary structure (Supplemental Table 3) as do intragenic vertebrate CNEs. The most conserved 100 deep vertebrate CNEs are relatively enriched in 3′ UTR sequences (14.3% of bases versus 5.6% in all CNEs), and these tend to have potential secondary structure (Siepel et al., 2005).
In vertebrates, ultraconserved CNEs overlapping stop codon exons are frequently found in genes encoding activators of alternative splicing, especially members of the SR family of splicing regulators. Long regions (118 to 618 nucleotides in length) within these CNEs are 100% identical between human and mouse orthologs (Lareau et al., 2007). Intragenic CNSs in plants are also particularly prevalent in genes encoding RNA binding and processing proteins. Eighteen eudicot or commelinid CNSs are found in this class of genes, with 12 of them being conserved in both eudicot and commelinid lineages. They include some of the longest CNSs we found, including the 309-bp CNS in the intron of PRP39, which has been implicated in the processing of flowering time genes (Wang et al., 2007), and the 112-bp CNS in the SR splicing factor genes RSZ32 and RSZ33. The CNS in RSZ32/RSZ33 is adjacent to a poison cassette exon whose presence subjects the transcript to nonsense-mediated degradation (Reddy and Ali, 2011). A deep CNS in the SR34 gene is associated with a temperature-dependent alternative splicing event in which some isoforms lack the PSK kinase domain (Lazar and Goodman, 2000).
Functionally, plant and vertebrate deep CNSs are very similar. They occur at both 5′ and 3′ flanking positions, and intragenically. They are enriched in conserved transcription factor binding sites and are highly associated with trans-dev genes. Some of the most conserved CNSs in both plants and animals are present flanking alternatively spliced stop codon exons in genes that function in alternative splicing. While functionally similar, plant and vertebrate CNSs differ both in their degree of conservation and in the distance separating them from their cognate genes. While 11% of human-fugu CNEs are 1 to 2 Mb from their human target gene (Woolfe and Elgar, 2008), the deep plant CNSs described in this article are all within 11 kb. Even when non-synteny-based approaches are employed, plant CNSs are not found far from their target genes. Reneker et al. (2012) used a whole-genome search to look in parallel for ultraconserved elements in plant and animal genomes (LIMEs). While they found 489 syntenic LIMEs in animal genomes with 100% identity over 200 bp, they did not find a single syntenic LIME in plants (100% identity over 100 bp). Using less stringent conditions (85% identity over 56 bp), Kritsas et al. (2012) found CNSs between Arabidopsis and grape; however, every CNS found was within 2 kb of its gene.
We now know that ultra-deep CNSs exist in plants and function similarly to their vertebrate cognates. However, they are shorter and less conserved over time. Why is this? Vertebrate CNEs may be more conserved than plant CNSs because they are required to fulfill multiple, physiologically diverse roles. That is, they might encode overlapping functions, such as the binding of multiple transcription factors or the capacity to function at both transcriptional and posttranscriptional levels. The evolutionarily conserved interferon-β-enhanceosome is a 55-bp sequence bound by eight proteins, with the presence of all eight factors required to activate transcription (Panne et al., 2007). Synergistic binding can occur when transcription factors bind to overlapping bases on opposite strands or grooves (Guturu et al., 2013). However, binding a greater number of transcription factors does not necessarily lead to greater conservation since cooperative binding tends to result in shorter and less distinct binding sites (Bilu and Barkai, 2005). Vertebrate CNEs that are deeply conserved or ultraconserved are often associated with genes that function in development of the nervous system (Pennacchio et al., 2006), structures where regulation would be expected to be the most complex. The ability of vertebrate CNEs to function megabases away from their target gene likely places additional constraints on them, requiring them to be present not just within the appropriate chromatin structures but also within the appropriate chromatin domains at the appropriate time during development (Harmston and Lenhard, 2013). For instance, the spatial localization of a gene within the nucleus is likely regulated by enhancer sequences. Recently, a subset of vertebrate enhancers has been found to be transcribed (eRNA), particularly those enhancers which physically interact with their target promoter via looping. A large fraction of ultraconserved CNEs were found to be transcribed at some stage in mouse development, including a majority of CNEs with enhancer activity (Licastro et al., 2010).
Perhaps the most likely explanation of the mutation frequency difference (base changes/million years) between vertebrate and plant CNSs involves differences in the organisms themselves and how they have evolved to remove functionless DNA. Plants differ from animals in being able to efficiently remove redundant genes and CNSs using a deletion mechanism as opposed to the slow pseudogene pathway used by animals (Woodhouse et al., 2010; Subramaniam et al., 2013). The mechanism likely involves some type of recombination because the deleted DNA lies between short tandem repeats, with only one repeat remaining at the site of deletion. Another major difference between plants and vertebrates is that plant genomes have undergone repeated WGD events (Murat et al., 2012). For instance, four WGD events separate Columbine from Arabidopsis. By contrast, there has been only one WGD event between vertebrates and teleost fish, and this single WGD event coincides with a marked increase in the rate of CNE, but not coding sequence, substitution rate, resulting in human-fugu CNEs being less conserved than the more anciently diverged human-elephant shark (Callorhinchus milii) CNEs (Wang et al., 2009; Lee et al., 2011). This seems to be the case also for deep plant CNSs. The best alignments between intragenic grape deep CNSs and peach, cacao, or columbine sequences are significantly longer and more conserved than the corresponding alignments between Arabidopsis and grape, peach, or cacao sequences (paired t test: P = 0.002 [length] and P = 0.0003 [conservation]), despite the latter having diverged at a similar or more recent time. In fact, when no intervening WGD events have taken place, plant CNSs can be both fairly long and highly conserved. For instance, the CUC2 deep CNS includes a 21-bp sequence required for cotyledon boundary determination (Larsson et al., 2012); the grape-peach alignment of this CNS is 99% identical over a 99-bp region in contrast with the Arabidopsis-grape alignment of 87% identity over 70 bp. We think it probable that polyploidy leads to a relaxation of the constraints on CNS sequence conservation.
Perhaps the most important difference between plant and animal CNS divergence rates lies in the biology of the organisms themselves and not in the nature of CNSs themselves (Freeling et al., 2012). A typical single higher plant develops hundreds or thousands of diploid seeds and also gametophytes (haploid stages of the plant life cycle), developing into millions of pollen grains. Compare this to fecundity in vertebrates: a few offspring and no gametophytes. This abundance of mutational targets/selectable units in plants permits a strength of purifying selection that could, in theory, underlie a process of massive trial and error followed by massive death. Since deletion mechanisms are more damaging than point mutational mechanisms, and plants seem to be built to tolerate damage (since it only take one surviving pollen grain to effect one fertilization), the quantitative differences in sequence conservation between plant and animal CNSs could reduce to basic biology.
Taken together, plant and vertebrate CNSs differ greatly in rates of divergence and length, but deep CNSs in plants are qualitatively similar to deep CNSs. Quantitatively, however, plants and animals have evolved quite different mechanisms for DNA removal, mechanisms that correspond to differences in the strength of purifying selection, representing the very different life styles of plants and vertebrates. Thus, the conserved stretches of plant CNSs are shorter and evolve more quickly than do CNEs of vertebrates simply because purifying selection is killing many more progeny of an individual plant than of an individual vertebrate each generation, and mutational mechanisms have coevolved to fit this stronger purifying selection. CNS decay in detectability is thus mainly a spandrel of the organism’s optimal DNA loss mechanisms.
METHODS
Genomes Used
Sources of genomic data used in this study are described in the Accession Numbers section at the end of Methods: Arabidopsis thaliana, columbine (Aquilegia coerulea Goldsmith), peach (Prunus persica), grape (Vitis vinifera), cacao (Theobroma cacao), and Amborella trichopoda.
The phylogenetic relationships among the higher plants used in this study are from the Model Organism Tree (http://www.mobot.org/mobot/research/apweb/trees/modeltreemap.html) (Figure 1). Divergence times shown are only approximate, reflecting a lack of fossil calibration points and heterogeneous evolutionary rates. The more basal branch estimates (A. trichopoda and the monocot-eudicot split) are from Moore et al. (2007). Approximate divergence times for eudicot species are from Wikström et al. (2001), the divergence time for banana (Musa acuminata; Commelinales) is from Janssen and Bremer (2004), and the grass divergence dates are from the International Brachypodium Initiative (2010). WGD dates are also approximate. In eudicots, separate ancient WGD events occurred in the Ranunculales and core eudicot clades (Cui et al., 2006; Jaillon et al., 2007). Subsequently, Arabidopsis underwent two additional WGD events (Fawcett et al., 2009). An ancient WGD event also occurred early in the monocot lineage (Tang et al., 2010), with subsequent events occurring in the lineages leading to banana and to the grasses (Fawcett et al., 2009; D’Hont et al., 2012).
Detecting Deeply Conserved Noncoding Sequences between Arabidopsis and Columbine
Deeply conserved eudicot CNSs were identified starting with CNSs conserved between intragenomic Arabidopsis genes and, in parallel, starting with CNSs conserved between orthologous peach and cacao genes. In the first approach, the starting material was 5578 CNSs obtained via manual inspection of Arabidopsis homoeologous gene pairs (Thomas et al., 2007; Subramaniam et al., 2013) derived from the most recent WGD event. Homoeologous Arabidopsis gene pairs have a modal Ks value of 0.76. CNS sequences associated with each homoeologous gene were used to query columbine, grape, and peach genomes using automated BLASTN searches (CoGeBlast, http://genomevolution.org/CoGe/CoGeBlast.pl set at wordsize 7, default settings, e-value cutoff 1). CNSs with best-hit e-values to columbine of ≤0.001 or best hits to grape or peach of ≤0.0001 were chosen for closer inspection on GEvo (http://genomevolution.org/CoGe/GEvo.pl) panels (Lyons and Freeling, 2008) populated with the corresponding homoeologous Arabidopsis gene pairs and orthologous genes from grape, peach, columbine, and cocoa (Supplemental Figure 1) and using the BLASTN algorithm under default parameters. Orthologous genes were identified, whenever possible, using Synfind (http://genomevolution.org/CoGe/SynFind.pl; Woodhouse et al., 2011). When the gap between syntenic retained genes was too large, BLASTN and TBLASTN were used within the CoGe platform to find the most homologous sequences that were then checked for synteny using BLASTZ on GEvo panels set to a window size of at least 400,000 bp. Using these criteria, 210 of the original 1879 homoeologous gene pairs were analyzed, yielding 39 deeply retained CNSs. An additional 11 deeply retained CNSs were identified by analyzing CNSs associated with homoeologous gene pairs in which CNS sequences derived from each homoeologous gene differed yet had identical BLASTN best hits to columbine. Finally, four deeply retained CNSs were identified by analyzing CNSs associated with homoeologous gene pairs in which one of the CNSs hit both grape and peach with e-values ≤0.001. For each visually inspected GEvo panel, all CNSs associated with a homoeologous gene pair were checked for retention in grape, peach, and columbine, resulting in an additional 49 deeply conserved CNSs being identified. The final 13 CNSs identified were found through inspection of homoeologous gene pairs that did not meet the above requirements, with four of the CNSs corresponding to best hits. Altogether 34% of Arabidopsis α-CNSs were analyzed (1894 CNSs associated with 380 homoeologous gene pairs).
In a second approach to finding deeply conserved CNSs, we started with a set of orthologous CNSs (cocoa-peach) and analyzed only BLASTN hits to other genomes in the region surrounding orthologous genes. Cocoa-peach CNS sequences identified using CNS Discovery PL3.0 automated pipeline (Turco et al., 2013) were padded with 10 bp of sequence, and the automated pipeline was used to search for BLASTN hits in the 10,000-bp region upstream and downstream from columbine orthologous genes, identified using SynMap set to 1:1 QuotaAlign and standard GEvo settings (http://genomevolution.org/CoGe/SynMap.pl; Lyons and Freeling, 2008; Tang et al., 2011). CNS sequences corresponding to the 1083 positive hits were then used to search for BLASTN hits in the 10,000-bp region upstream and downstream from Arabidopsis-orthologous genes, resulting in 321 potential hits. The orthologous genes from cocoa, peach, and Arabidopsis, as well as the orthologous grape gene and, when present, the homoeologous Arabidopsis gene, were analyzed on GEvo panels; 112 of them were confirmed as deeply conserved CNSs, of which 17 had been independently identified starting with CNSs from Arabidopsis homoeologous gene pairs.
The genomic region corresponding to each Arabidopsis CNS was analyzed on TAIR10 Vista tracks (http://tairvm17.tacc.utexas.edu/cgi-bin/gb2/gbrowse/Arabidopsis/) for evidence of transcription (Expression and Sequence Similarity tracks), presence of open reading frames (Six Frame Translation and Community/Alternative Annotation tracks), or presence of transposable elements or noncoding RNAs (Gene tracks). Each putative CNS was examined for matches to noncoding RNAs in the Rfam database (Gardner et al., 2011) (http://rfam.sanger.ac.uk), as well as to matches in the snoRNA (Ellis et al., 2010) and microRNA (Zhang et al., 2010) databases. CNSs were also checked for coding potential, particularly in the region flanking intron/exon junctions and in the 5′ UTR. Twelve CNSs were flagged as potential upstream open reading frames and were not included in the final CNS list. With the exception of CP45-1, the remaining CNSs were ruled unlikely to be coding due to the presence of out-of-frame indels and/or the absence of RNA sequencing reads. Although CP45-1 lacks out-of-frame indels and coincides with mRNA sequencing reads, the mRNA sequencing reads are in the antisense orientation relative to AT5G18930 and do not lead to a conserved open reading frame.
Retrieval and Sequence Alignment of Deeply Conserved CNS Sequences
Sequences from HSPs between Arabidopsis, grape, peach, cacao, and columbine CNSs were retrieved from GEvo panels. Overlapping HSP sequences from each genome were assembled together and imported into the Muscle multiple sequence alignment tool (Edgar, 2004). Sequences from each genome were retrieved from the aligned region conserved between Arabidopsis and columbine. For determination of CNS length and percentage of identity, Arabidopsis and columbine sequences were aligned using Muscle, and for each Arabidopsis-columbine pair, the percent matches, mismatches, and gaps were calculated from the first to the last exact matches. A quality score for each pair was determined by multiplying the percentage of identity (minus 25) by the CNS length. A similar scoring procedure was used for rice (Oryza sativa)–banana CNSs.
Detecting Deeply Conserved Noncoding Sequences between Arabidopsis and Rice
In order to identify any CNS sequences conserved since the divergence of the monocot and dicot lineages, rice genes belonging to the same putative orthologous group (http://pogs.uoregon.edu; Walker et al., 2007) and clustering more closely to the Arabidopsis genes carrying the CNS than to Arabidopsis genes lacking the CNS were added to GEvo panels for manual inspection.
Enrichment for GO Terms
Enrichment for GO terms was performed using agriGO Singular Enrichment Analysis (http://bioinfo.cau.edu.cn/agriGO/) (Du et al., 2010) using TAIR9 and MSU6.1 annotations, a minimum of five mapping entries, and significance set at 0.05 using the Fisher’s exact test corrected for multiple comparisons using the Benjamini-Yekutieli method., which controls for false discovery rate, rather than family-wise error rate, when multiple, and dependent, hypotheses are tested, as occurs in GO enrichment analysis (Benjamini and Hochberg, 1995). For CNSs assigned to duplicate Arabidopsis genes, both homoeologs were included in the analysis. agriGO outputs significant terms in a hierarchical tree format. To reduce redundant terms, GO significant terms of a parent were not reported when the number of query hits to a child term (a more specialized term) was at least 80% the number of query hits to the parent term.
To ensure that GO enrichment does not simply reflect an increased statistical likelihood for genes with more CNSs to include a deeply conserved CNS, a control experiment was performed in which the genes associated with a random set of α-CNSs were compared with an identical number of genes associated with deeply conserved α-CNSs for annotation with the GO term 0003700, sequence-specific DNA binding transcription factor activity. On a per CNS basis, 23% of α-CNS genes are annotated with GO:0003700 compared with 50% of deep α-CNS genes. In 10,000 repetitions, the maximum reached in the control set, 37%, was far below 50% (z = 2.3E-14).
Enrichment of Transcription Factor Binding Sites
CNS sequences were analyzed for transcription factor binding sites using a custom perl script to search both strands for nonoverlapping matches to 321 motifs compiled from Agris (http://Arabidopsis.med.ohio-state.edu/AtcisDB/bindingsites.html), PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE/), and PlantPan (http://PlantPAN.mbc.nctu.edu.tw) databases and from 7-bp sequences overrepresented in CNSs from Bigfoot genes (Freeling et al., 2007). Fisher's exact test was used to compare the significance of motif enrichment relative to 11,942,950 nucleotides of Arabidopsis control sequence or 28,035,390 nucleotides of rice control sequence. Intronic sequences and the segment of the genome situated between a gene and its outermost CNS were used as control sequences following removal of CNS and transposable element sequences. Motifs with at least five CNS matches were retained and Fisher's exact test was used to compare motif counts in deep CNSs with counts in control sequences. P values were corrected for multiple testing at a false discovery rate of 5% using the Benjamini-Hochberg method (Benjamini and Hochberg, 1995).
A custom perl script was also used to count motifs appearing more than once in any given CNS. Control data sets were made from random sequences pulled from the Arabidopsis set of control sequences and matched for length and GC content (±5%) with the set of deep CNS sequences. Motif counts were made for 10,000 control data sets, and z-test statistics were used to test for enrichment in the overall number of duplicate motifs in deep CNSs and to test for enrichment of any of the four motifs that had at least 10 duplicate motifs (AAAG, AAAGH, WAAAG, and TGAC) in deep CNSs.
Identifying CNSs with Conserved RNA Secondary Structure
To identify regions with conserved RNA secondary structure, HSPs overlapping intragenic CNSs from Arabidopsis, grape, peach, cacao, and columbine syntenous regions, or rice, sorghum (Sorghum bicolor), and banana syntenous regions, were aligned using Muscle and analyzed on the RNAz Web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAz.cgi) for the potential to form conserved and thermodynamically stable RNA secondary structure (Washietl et al., 2005). The RNAz algorithm incorporates a covariation term that rewards compensatory mutations. Alignments with RNAz-defined RNA class probabilities of at least 0.5 were manually inspected, and structures were classified as likely regions of secondary structure if the structures were phylogenetically conserved in at least three species. Structures based predominantly on base-pairing between G/A- and C/T-rich regions were not retained.
Intragenic CNSs most likely to form secondary structure were further examined for the presence of overlapping reads from a dsRNA library prepared from Arabidopsis unopened flower buds (Zheng et al., 2010). dsRNA reads downloaded from accession SRX026295 were formatted with fastq-dump (http://www.ncbi.nlm.nih.gov/Traces/sra/), 3′ adapter sequences were removed using cutadapt (http://code.google.com/p/cutadapt/), and processed reads were aligned to an indexed TAIR10 version of the Arabidopsis genome using GSNAP (Wu and Nacu, 2010). Aligned reads were compressed into a BAM format using SAMtools (Li et al., 2009) and visualized with the Integrated Genome Browser (Nicol et al., 2009) for reads overlapping RNAz-predicted regions of secondary structure.
Accession Numbers
Genomic sequence data used in this article can be found in the databases listed under the following accession numbers:Arabidopsis thaliana Col-0 v10 (http://www.Arabidopsis.org/); Columbine v1.1 (Aquilegia coerulea Goldsmith; http://www.phytozome.net/aquilegia.php); peach v1 (Prunus persica; http://www.phytozome.net/peach.php) (Verde et al., 2013); grape v2, masked (Vitis vinifera; http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/) (Jaillon et al., 2007); cocoa v1.0 (Theobroma cacao; http://cocoagendb.cirad.fr/) (Argout et al., 2011); Amborella v1.0.27 (Amborella trichopoda; http://www.amborella.org/) (Amborella Genome Project, 2013).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Use of GEvo Panels to Detect Deeply Conserved CNSs.
Supplemental Figure 2. Motifs Overrepresented in Deep CNSs Are Phylogenetically Conserved.
Supplemental Figure 3. ATML and PDF2 Have a 5′ Proximal Experimentally Validated L1-Box and a 3′ UTR CNS with a Potential RNA-Fold.
Supplemental Table 1. Genes Associated with the Deep CNS Sequences Described in Figure 3.
Supplemental Table 2. Genes Associated with Deep CNS Sequences Used to Derive the RNAz-Predicted Consensus RNA Structures Depicted in Figure 4.
Supplemental Table 3. CNSs with Predicted RNA-Folds.
Supplemental Table 4. Experimentally Validated Deep CNSs.
Supplemental Data Set 1. Eudicot Deep CNSs.
Supplemental Data Set 2. CNSs Present in Both Commelinid and Eudicot Lineages.
Supplemental Data Set 3. Eudicot Deep CNS-Associated Genes.
Supplemental Data Set 4. Deep CNS-Associated Genes Are Significantly More Enriched in Trans-Dev GO Terms Compared with α-CNS–Associated Genes.
Supplemental Data Set 5. Genes Associated with Deep CNSs Are Enriched in Transcription and Development GO Terms Regardless of CNS Position.
Supplementary Material
Acknowledgments
We thank The Amborella Genome Project (www.amborella.org), the Aquilegia Genome Sequencing Project, and the U.S. Department of Energy, Joint Genome Initiative (http://www.phytozome.net/aquilegia.php) for the timely release of draft A. trichopoda and columbine genomes. We thank Gina Turco and Sabarinath Subramaniam for computational help and Damon Lisch and the rest of the Freeling lab for comments. This work was funded by the National Science Foundation (IOS1248106 to M.F.).
AUTHOR CONTRIBUTIONS
D.B. designed and performed the research, and wrote the article. M.F. conceived and initiated the project and contributed to writing the article.
Glossary
- CNE
conserved noncoding element
- LIME
long identical multispecies element
- mya
million years ago
- CNS
conserved noncoding sequence
- HSP
high-scoring segment pair
- UTR
untranslated region
- GO
gene ontology
- dsRNA
double-stranded RNA
- ULE
ultraconserved-like element
- WGD
whole-genome duplication
Footnotes
Online version contains Web-only data.
References
- Abe M., Takahashi T., Komeda Y. (2001). Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 26: 487–494. [DOI] [PubMed] [Google Scholar]
- Amborella Genome Project (2013). The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Argout X., et al. (2011). The genome of Theobroma cacao. Nat. Genet. 43: 101–108. [DOI] [PubMed] [Google Scholar]
- Attanasio C., et al. (2008). Assaying the regulatory potential of mammalian conserved non-coding sequences in human cells. Genome Biol. 9: R168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter L., Jironkin A., Hickman R., Moore J., Barrington C., Krusche P., Dyer N.P., Buchanan-Wollaston V., Tiskin A., Beynon J., Denby K., Ott S. (2012). Conserved noncoding sequences highlight shared components of regulatory networks in dicotyledonous plants. Plant Cell 24: 3949–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D. (2004). Ultraconserved elements in the human genome. Science 304: 1321–1325. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bilu Y., Barkai N. (2005). The design of transcription-factor binding sites is affected by combinatorial regulation. Genome Biol. 6: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Bataille A.R., Chen X., Poitras C., Laganière J., Lefèbvre C., Deblois G., Giguère V., Ferretti V., Bergeron D., Coulombe B., Robert F. (2006). Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 16: 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E., Galbiati M., Albertini A., Fornara F., Conti L., Coupland G., Tonelli C. (2011). DOF-binding sites additively contribute to guard cell-specificity of AtMYB60 promoter. BMC Plant Biol. 11: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., et al. (2006). Widespread genome duplications throughout the history of flowering plants. Genome Res. 16: 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Hont A., et al. (2012). The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488: 213–217. [DOI] [PubMed] [Google Scholar]
- Drake J.A., Bird C., Nemesh J., Thomas D.J., Newton-Cheh C., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Hirschhorn J.N. (2006). Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat. Genet. 38: 223–227. [DOI] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund D.M., Cierlik I., Ståldal V., Claes A.R., Vestman D., Chandler J., Sundberg E. (2011). Expression of Arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box-like regulatory element. Plant Physiol. 157: 2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.C., Brown D.D., Brown J.W. (2010). The small nucleolar ribonucleoprotein (snoRNP) database. RNA 16: 664–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Fawcett J.A., Maere S., Van de Peer Y. (2009). Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. USA 106: 5737–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Bi C., Clark B.S., Mady R., Shah P., Kohtz J.D. (2006). The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20: 1470–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Subramaniam S. (2009). Conserved noncoding sequences (CNSs) in higher plants. Curr. Opin. Plant Biol. 12: 126–132. [DOI] [PubMed] [Google Scholar]
- Freeling M., Rapaka L., Lyons E., Pedersen B., Thomas B.C. (2007). G-boxes, bigfoot genes, and environmental response: Characterization of intragenomic conserved noncoding sequences in Arabidopsis. Plant Cell 19: 1441–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Woodhouse M.R., Subramaniam S., Turco G., Lisch D., Schnable J.C. (2012). Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr. Opin. Plant Biol. 15: 131–139. [DOI] [PubMed] [Google Scholar]
- Gandikota M., Birkenbihl R.P., Höhmann S., Cardon G.H., Saedler H., Huijser P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49: 683–693. [DOI] [PubMed] [Google Scholar]
- Gardner P.P., Daub J., Tate J., Moore B.L., Osuch I.H., Griffiths-Jones S., Finn R.D., Nawrocki E.P., Kolbe D.L., Eddy S.R., Bateman A. (2011). Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 39: D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy M., Franco-Zorrilla J.M., Pérez-Pérez J., Oliveros J.C., Lorenzo O., Solano R. (2011). Improved protein-binding microarrays for the identification of DNA-binding specificities of transcription factors. Plant J. 66: 700–711. [DOI] [PubMed] [Google Scholar]
- Gómez-Porras J.L., Riaño-Pachón D.M., Dreyer I., Mayer J.E., Mueller-Roeber B. (2007). Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genomics 8: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Moose S.P. (2003). Conserved noncoding sequences among cultivated cereal genomes identify candidate regulatory sequence elements and patterns of promoter evolution. Plant Cell 15: 1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guturu H., Doxey A.C., Wenger A.M., Bejerano G. (2013). Structure-aided prediction of mammalian transcription factor complexes in conserved non-coding elements. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerry T.E., Gehring W.J. (1996). Intron of the mouse Hoxa-7 gene contains conserved homeodomain binding sites that can function as an enhancer element in Drosophila. Proc. Natl. Acad. Sci. USA 93: 13884–13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmston N., Lenhard B. (2013). Chromatin and epigenetic features of long-range gene regulation. Nucleic Acids Res. 41: 7185–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A., et al. (2013). An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat. Genet. 45: 891–898. [DOI] [PubMed] [Google Scholar]
- Hufton A.L., Mathia S., Braun H., Georgi U., Lehrach H., Vingron M., Poustka A.J., Panopoulou G. (2009). Deeply conserved chordate noncoding sequences preserve genome synteny but do not drive gene duplicate retention. Genome Res. 19: 2036–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- Ito M., Araki S., Matsunaga S., Itoh T., Nishihama R., Machida Y., Doonan J.H., Watanabe A. (2001). G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13: 1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., et al. French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467. [DOI] [PubMed] [Google Scholar]
- Janssen T., Bremer K. (2004). The age of major monocot groups inferred from 800+ rbcL sequences. Bot. J. Linn. Soc. 146: 385–398. [Google Scholar]
- Javelle M., Klein-Cosson C., Vérnoud V., Boltz V., Maher C., Timmermans M., Depège-Fargeix N., Rogowsky P.M. (2011). Genome-wide characterization of the HD-ZIP IV transcription factor family in maize: Preferential expression in the epidermis. Plant Physiol. 157: 790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong M.J., Shih M.C. (2003). Interaction of a GATA factor with cis-acting elements involved in light regulation of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis. Biochem. Biophys. Res. Commun. 300: 555–562. Erratum. Biochem. Biophys. Res. Commun. 333: 1385. [DOI] [PubMed] [Google Scholar]
- Jiang N., Bao Z., Zhang X., Eddy S.R., Wessler S.R. (2004). Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573. [DOI] [PubMed] [Google Scholar]
- Kalyna M., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40: 2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplinsky N.J., Braun D.M., Penterman J., Goff S.A., Freeling M. (2002). Utility and distribution of conserved noncoding sequences in the grasses. Proc. Natl. Acad. Sci. USA 99: 6147–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S., Kern A.D., Bejerano G., Fewell G., Fulton L., Wilson R.K., Salama S.R., Haussler D. (2007). Human genome ultraconserved elements are ultraselected. Science 317: 915. [DOI] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S. (2008). Two different mechanisms control ethylene sensitivity in Arabidopsis via the regulation of EBF2 expression. Plant Signal. Behav. 3: 749–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E.M. (2009). Aquilegia: A new model for plant development, ecology, and evolution. Annu. Rev. Plant Biol. 60: 261–277. [DOI] [PubMed] [Google Scholar]
- Kritsas K., Wuest S.E., Hupalo D., Kern A.D., Wicker T., Grossniklaus U. (2012). Computational analysis and characterization of UCE-like elements (ULEs) in plant genomes. Genome Res. 22: 2455–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E. (2007). Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929. [DOI] [PubMed] [Google Scholar]
- Larsson E., Sundström J.F., Sitbon F., von Arnold S. (2012). Expression of PaNAC01, a Picea abies CUP-SHAPED COTYLEDON orthologue, is regulated by polar auxin transport and associated with differentiation of the shoot apical meristem and formation of separated cotyledons. Ann. Bot. (Lond.) 110: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar G., Goodman H.M. (2000). The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol. Biol. 42: 571–581. [DOI] [PubMed] [Google Scholar]
- Lee A.P., Kerk S.Y., Tan Y.Y., Brenner S., Venkatesh B. (2011). Ancient vertebrate conserved noncoding elements have been evolving rapidly in teleost fishes. Mol. Biol. Evol. 28: 1205–1215. [DOI] [PubMed] [Google Scholar]
- Lee A.P., Yang Y., Brenner S., Venkatesh B. (2007a). TFCONES: A database of vertebrate transcription factor-encoding genes and their associated conserved noncoding elements. BMC Genomics 8: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007b). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licastro D., Gennarino V.A., Petrera F., Sanges R., Banfi S., Stupka E. (2010). Promiscuity of enhancer, coding and non-coding transcription functions in ultraconserved elements. BMC Genomics 11: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifanov A.P., Makeev V.J., Nazina A.G., Papatsenko D.A. (2003). Homotypic regulatory clusters in Drosophila. Genome Res. 13: 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots G.G., Locksley R.M., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A. (2000). Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288: 136–140. [DOI] [PubMed] [Google Scholar]
- Lowe C.B., Kellis M., Siepel A., Raney B.J., Clamp M., Salama S.R., Kingsley D.M., Lindblad-Toh K., Haussler D. (2011). Three periods of regulatory innovation during vertebrate evolution. Science 333: 1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E., Freeling M. (2008). How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 53: 661–673. [DOI] [PubMed] [Google Scholar]
- Moore M.J., Bell C.D., Soltis P.S., Soltis D.E. (2007). Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. USA 104: 19363–19368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello L., Breviario D. (2008). Plant spliceosomal introns: Not only cut and paste. Curr. Genomics 9: 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat F., Van de Peer Y., Salse J. (2012). Decoding plant and animal genome plasticity from differential paleo-evolutionary patterns and processes. Genome Biol. Evol. 4: 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.Z., Grate L., Donohue J.P., Preston C., Nobida N., O'Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Jr (2007). Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 21: 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol J.W., Helt G.A., Blanchard S.G., Jr, Raja A., Loraine A.E. (2009). The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega M.A., Ovcharenko I., Afzal V., Rubin E.M. (2003). Scanning human gene deserts for long-range enhancers. Science 302: 413. [DOI] [PubMed] [Google Scholar]
- Panne D., Maniatis T., Harrison S.C. (2007). An atomic model of the interferon-beta enhanceosome. Cell 129: 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L.A., Loots G.G., Nobrega M.A., Ovcharenko I. (2007). Predicting tissue-specific enhancers in the human genome. Genome Res. 17: 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L.A., et al. (2006). In vivo enhancer analysis of human conserved non-coding sequences. Nature 444: 499–502. [DOI] [PubMed] [Google Scholar]
- Reddy A.S., Ali G.S. (2011). Plant serine/arginine-rich proteins: roles in precursor messenger RNA splicing, plant development, and stress responses. Wiley Interdiscip. Rev RNA 2: 875–889. [DOI] [PubMed] [Google Scholar]
- Reineke A.R., Bornberg-Bauer E., Gu J. (2011). Evolutionary divergence and limits of conserved non-coding sequence detection in plant genomes. Nucleic Acids Res. 39: 6029–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneker J., Lyons E., Conant G.C., Pires J.C., Freeling M., Shyu C.-R., Korkin D. (2012). Long identical multispecies elements in plant and animal genomes. Proc. Natl. Acad. Sci. USA 109: E1183–E1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., et al. (2008). Identification and characterization of new long conserved noncoding sequences in vertebrates. Mamm. Genome 19: 703–712. [DOI] [PubMed] [Google Scholar]
- Sandelin A., Bailey P., Bruce S., Engström P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B. (2004). Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen S., Pheasant M., Makunin I.V., Mattick J.S. (2008). Large-scale appearance of ultraconserved elements in tetrapod genomes and slowdown of the molecular clock. Mol. Biol. Evol. 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Strähle U., Rastegar S. (2008). Conserved non-coding sequences and transcriptional regulation. Brain Res. Bull. 75: 225–230. [DOI] [PubMed] [Google Scholar]
- Subramaniam S., Wang X., Freeling M., Pires J.C. (2013). The fate of Arabidopsis thaliana homeologous CNSs and their motifs in the Paleohexaploid Brassica rapa. Genome Biol. Evol. 5: 646–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Bowers J.E., Wang X., Paterson A.H. (2010). Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc. Natl. Acad. Sci. USA 107: 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Lyons E., Pedersen B., Schnable J.C., Paterson A.H., Freeling M. (2011). Screening synteny blocks in pairwise genome comparisons through integer programming. BMC Bioinformatics 12: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.C., Rapaka L., Lyons E., Pedersen B., Freeling M. (2007). Arabidopsis intragenomic conserved noncoding sequence. Proc. Natl. Acad. Sci. USA 104: 3348–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco G., Schnable J.C., Pedersen B., Freeling M. (2013). Automated conserved non-coding sequence (CNS) discovery reveals differences in gene content and promoter evolution among grasses. Front. Plant Sci. 4: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I., et al. International Peach Genome Initiative (2013). The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45: 487–494. [DOI] [PubMed] [Google Scholar]
- Visel A., Bristow J., Pennacchio L.A. (2007). Enhancer identification through comparative genomics. Semin. Cell Dev. Biol. 18: 140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Prabhakar S., Akiyama J.A., Shoukry M., Lewis K.D., Holt A., Plajzer-Frick I., Afzal V., Rubin E.M., Pennacchio L.A. (2008). Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat. Genet. 40: 158–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter A., Tunc-Ozdemir M., Grove B.C., Green P.J., Shintani D.K., Breaker R.R. (2007). Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell 19: 3437–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker N.S., Stiffler N., Barkan A. (2007). POGs/PlantRBP: A resource for comparative genomics in plants. Nucleic Acids Res. 35: D852–D856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Tian Q., Hou Z., Mucha M., Aukerman M., Olsen O.A. (2007). The Arabidopsis thaliana AT PRP39-1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time. Plant Cell Rep. 26: 1357–1366. [DOI] [PubMed] [Google Scholar]
- Wang J., Lee A.P., Kodzius R., Brenner S., Venkatesh B. (2009). Large number of ultraconserved elements were already present in the jawed vertebrate ancestor. Mol. Biol. Evol. 26: 487–490. [DOI] [PubMed] [Google Scholar]
- Washietl S., Hofacker I.L., Lukasser M., Hüttenhofer A., Stadler P.F. (2005). Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 23: 1383–1390. [DOI] [PubMed] [Google Scholar]
- Wikström N., Savolainen V., Chase M.W. (2001). Evolution of the angiosperms: Calibrating the family tree. Proc. Biol. Sci. 268: 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse M.R., Tang H., Freeling M. (2011). Different gene families in Arabidopsis thaliana transposed in different epochs and at different frequencies throughout the rosids. Plant Cell 23: 4241–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse M.R., Schnable J.C., Pedersen B.S., Lyons E., Lisch D., Subramaniam S., Freeling M. (2010). Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biol. 8: e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A., Elgar G. (2008). Organization of conserved elements near key developmental regulators in vertebrate genomes. Adv. Genet. 61: 307–338. [DOI] [PubMed] [Google Scholar]
- Woolfe A., et al. (2005). Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.D., Nacu S. (2010). Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yu J., Li D., Zhang Z., Liu F., Zhou X., Wang T., Ling Y., Su Z. (2010). PMRD: Plant microRNA database. Nucleic Acids Res. 38: D806–D813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Ryvkin P., Li F., Dragomir I., Valladares O., Yang J., Cao K., Wang L.-S., Gregory B.D. (2010). Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 6: e1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Perry S.E. (2005). Control of expression and autoregulation of AGL15, a member of the MADS-box family. Plant J. 41: 583–594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.