Abstract
Retroviral integrase catalyzes the essential step of integrating a double-stranded DNA copy of the viral genome into a host cell chromosome. Mutational studies have revealed that integrase is involved in additional steps of viral replication, but the mechanism for the pleiotropic effect is not well characterized. Since Cys residues generally play crucial roles in protein structure and function, we introduced Cys-to-Ser substitutions at positions 56, 65, and 130 of human immunodeficiency virus type 1 (HIV-1) integrase to determine their effects on integration activity and viral replication. None of the substitutions significantly affected the enzymatic activities in vitro. When introduced into the NL4-3 molecular clone of HIV-1, mutant viruses encoding Cys mutations at positions 56 and 65 of integrase replicated similarly to the wild-type virus in CD4+-T-cell lines, whereas the C130S-containing virus was noninfectious. The entry and postintegration steps of the viral life cycle for all mutant viruses were normal, and all had particle-associated reverse transcriptase (RT) activity. However, early reverse-transcribed DNA products were absent in the lysate of cells infected with the C130S mutant virus, indicating that the mutation abolished the ability of the virus to initiate endogenous reverse transcription. Coimmunoprecipitation using purified integrase and RT showed that the C-terminal domain of wild-type HIV-1 integrase interacted with RT. The interaction between integrase and RT was not affected in the presence of a reducing or alkylating agent, suggesting that the interaction did not involve a disulfide linkage. The C130S substitution within the core region may disrupt the protein recognition interface of the C-terminal domain and abolish its ability to interact with RT. Our results indicate that integrase plays an important role during the reverse-transcription step of the viral life cycle, possibly through physical interactions with RT.
Integration of a double-stranded DNA copy of the viral RNA genome into the host chromosome is essential for retroviral replication (for reviews, see references 7, 12, and 31). The integration reaction is catalyzed by the viral protein integrase (IN), which is encoded by sequences at the 3′ end of the pol gene of the viral genome. IN is initially expressed as part of the Gag-Pol polyprotein precursor, and the subsequent processing events within the virion by the viral protease produce the mature functional protein. After virus entry into a susceptible host cell and reverse transcription, IN removes two nucleotides 3′ to the conserved CA dinucleotide from each end of the linear viral cDNA in a reaction called 3′-end processing. The processed viral cDNA and IN, within a nucleoprotein complex called the preintegration complex (PIC), then move from the cytoplasm into the nucleus of the infected cell. IN inserts the newly processed 3′ ends of the viral DNA into the host genome in a coupled cleavage-ligation reaction called 3′-end joining or strand transfer. Host enzymes are believed to carry out the repair of gaps between viral and host DNAs after strand transfer (6, 14, 60), resulting in the formation of a provirus.
IN comprises three domains. The N-terminal domain contains a highly conserved zinc-binding HHCC motif consisting of two His and two Cys residues (41, 43). Coordination of zinc promotes the multimerization of IN and enhances in vitro activities (9, 36, 62). Mutations of residues of this motif abolish the viral infectivity (18, 57). The central core domain of IN contains a catalytic DD(35)E motif that is conserved in all retroviral INs and retrotransposons, as well as some bacterial transposases (33, 41, 43). The crystal structure of the core domain shows that these residues coordinate a divalent cation that is essential for IN activity (23). The C-terminal domain is the least conserved among retroviral INs and binds DNA nonspecifically (19, 54).
Studies of human immunodeficiency virus type 1 (HIV-1) replication in cell cultures showed that certain mutations of IN have pleiotropic effects. The defects occur at stages of viral replication other than integration, such as reverse transcription, nuclear import, viral protein expression, packaging, and processing, and probably involve different mechanisms (10, 18, 35, 37, 52, 57, 58). The precise determinants in IN that cause each defect have not been well characterized.
Cys residues are often found sparingly in proteins, and when present, they generally play important structural and architectural roles. HIV-1 IN has a total of six Cys residues. Two of these residues are located in the N-terminal domain and are part of the conserved HHCC motif involved in zinc coordination (62). Mutations of the N-terminal Cys residues disrupt IN multimerization, abolish 3′-end processing and strand transfer activities in vitro (17, 53, 62), and inhibit viral replication in cultured cells by blocking reverse transcription (37). Three other Cys residues are located in the core domain at positions 56, 65, and 130, and the remaining one is in the C-terminal domain at position 280. Replacing the Cys at residue position 56, 65, or 280 of HIV-1 IN results in a protein displaying wild-type levels of 3′-end processing and strand transfer activities in vitro (5, 11, 16, 29, 59). However, the effect of C130 substitution on IN activity has not been determined. Although the replication of the C280S-containing virus is indistinguishable from that of the wild type (29), it is not known if the Cys residues at positions 56 and 65 are necessary for virus replication. A virus containing a C130G mutation in IN shows a loss of infectivity (40), but the basis for the defect is not clear.
In this study, we examined the effect of replacing the Cys residues within the core domain of HIV-1 IN on its enzymatic activity, as well as the effect on the replication of viruses harboring such INs. We found that none of the core Cys residues were necessary for IN-catalyzed reactions in vitro. However, mutant viruses encoding a C130S mutation were noninfectious. We determined that this substitution conferred an inability to initiate reverse transcription, indicating that IN plays an important role during reverse transcription. In vitro analysis using coimmunoprecipitation showed that, unlike the wild type and other Cys mutants, IN with the C130S substitution failed to interact with reverse transcriptase (RT). The results suggest that the interaction between IN and RT is functional and critical for viral replication.
MATERIALS AND METHODS
Cells and reagents.
293T cells, a transformed cell line derived from human embryonic kidney cells, were grown in Dulbecco modified Eagle medium containing l-glutamine and 4.5 g of glucose (Gibco)/liter, supplemented with 10% fetal bovine serum (FBS) (HyClone), 100 U of penicillin G (Cellgro)/ml, and 0.1 mg of streptomycin (Sigma)/ml. CEM cells, a human CD4+-T-cell line, were grown in RPMI 1640 medium (Gibco) supplemented with 10% FBS (Omega), 100 U of penicillin G/ml, and 0.1 mg of streptomycin/ml.
Viral p24 levels were measured using an enzyme-linked immunosorbent assay (ELISA) with the HIV-1 p24 antigen assay kit (Coulter). The sera from HIV-1-infected patients were purchased from The Scripps Research Institute. The following reagents were obtained through the AIDS Research and Reference Program: purified recombinant RT heterodimer and the rabbit anti-HIV-1 RT antiserum from Stuart Le Grice, mouse monoclonal antibody against HIV-1 RT (mAb21) from Stephen Hughes, and rabbit anti-HXB2 IN (amino acids 23 to 34) polyclonal antibody from Duane Grandgenett. Purified N-terminally truncated IN (IN51-288) and the core domain (IN51-212) of HIV-1 IN were obtained from Robert Craigie, and the C-terminal domain (IN213-288) of HIV-1 IN was obtained from Alan Engelman.
Construction of Cys mutants of HIV-1 IN.
The wild-type HIV-1 IN gene was derived from the HIV-1 molecular clone NL4-3 (1) and cloned into the E. coli expression vector pT7-7 (47). To facilitate purification, the expression construct contained DNA sequences that encode a seven-His tag followed by amino acid residues constituting a thrombin cleavage site (IVPRGSHM) at the N terminus of the IN gene. Cys residues of HIV-1 IN were replaced with Ser by overlapping PCR in a stepwise fashion (4). The following oligonucleotides (Operon Technologies) were used to introduce the mutations, with nucleotide substitutions shown in boldface type: C56S mutation, HINC56Sss (5′-GTAGACAGTAGCCCAGGAATA-3′) and HINC56Sas (5′-CTGGGCTACTGTCTACTTGT-3′); C65S mutation, HINC65Sss (5′-GCTAGATAGTACACATTTACAAGGAAAAG-3′) and HINC65Sas (5′-TCCTTCTAAATGTGTACTATCTAGCTG-3′); C130S mutation, HINC130Sss (5′-ACAGTTAAGGCCGCCAGTTGGTGG-3′) and HINC130Sas (5′-CCCACCAACTGGCGGCCTT-3′). In the first round of PCR, the 5′ portion of the IN gene was amplified using T7PRO (5′-TAATACGACTCACTATAGGG-3′), which anneals to positions 273 to 292 of the pT7-7/His(T) vector, as the forward primer and the antisense oligonucleotide (with the “as” designation) of the aforementioned primer set as the reverse primer. The 3′ portion of the IN gene was amplified using the sense oligonucleotide (with the “ss” designation) of the primer set as the forward primer and H-IN280Bam (5′-ATTGGATCCTTAATCCTCATCCTGTCTACTTGCCACACTATCATCACC-3′) as the reverse primer. H-IN280Bam contains the C280S mutation (boldface type) and has a BamHI site (underlined) adjacent to the stop codon (italicized) of the IN gene. In the second round of PCR, the amplified 5′ and 3′ portions of IN were mixed, denatured at 90°C for 1 min, annealed at 60°C for 1 min, and chain elongated at 72°C for 5 min. The entire IN sequence with the desired mutation was then amplified using T7PRO and H-IN280Bam as the forward and reverse primers, respectively.
Amplified PCR fragments were digested with NdeI and BamHI and then ligated with pT7-7/His(T) previously digested with NdeI and BamHI. The DNA sequences of all the resulting constructs were confirmed by dideoxy sequencing using the Thermo Sequenase DNA polymerase (Amersham Pharmacia) according to the manufacturer's instructions.
Expression and purification of Cys mutants of HIV-1 IN.
IN expression constructs were transformed into Escherichia coli BL21(DE3). Wild-type and mutant INs were expressed and purified under denaturing conditions using a protocol similar to that described previously (13, 26). Briefly, expression of the recombinant protein was induced by the addition of 1 mM isopropyl-1-thio-β-d-galactopyranoside. The bacterial pellet was lysed by sonication, centrifuged, and suspended in a denaturing buffer containing 20 mM Tris-HCl, pH 8.0, 0.5 M NaCl, and 6 M guanidine-HCl. The supernatant fraction of the bacterial lysate was incubated with Ni-nitrilotriacetic acid agarose (Qiagen). The His-tagged IN was eluted with the denaturing buffer containing imidazole and then renatured during dialysis. The dialysate was concentrated using a stirred cell (Amicon model 8050) with a YM10 ultrafiltration membrane (Millipore), and the protein sample was treated with human thrombin (Sigma) to remove the His tag, leaving the vector-derived tetrapeptide GSHM at the N terminus. The sample was then diluted and loaded onto a high-performance SP-Sepharose column (Amersham Pharmacia). The protein was then eluted from the column with an NaCl linear gradient. The purities of protein fractions were verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie blue staining. The fractions were pooled, dialyzed, and stored at −80°C. Protein concentrations were determined by the Bradford assay (Bio-Rad) according to the manufacturer's instructions using bovine serum albumin as a standard. C-terminally truncated HIV-1 IN (IN1-212) was purified as described previously (26, 47).
In vitro integration assays. The in vitro activities of HIV-1 IN and its mutant variants were determined using established oligonucleotide-based assays (12). The 3′-end processing, strand transfer, and disintegration reactions were carried out at 37°C for 1 h in a 20-μl reaction volume containing 5 nM 32P-labeled substrate, 75 nM purified IN, 20 mM HEPES-Na, pH 7.5, 30 mM NaCl, 10 mM MnCl2, 10 mM dithiothreitol (DTT), and 0.05% IGEPAL (octylphenyl-polyethylene glycol). IGEPAL is a nonionic, nondenaturing detergent. The reaction products were separated on a 15% polyacrylamide-7 M urea gel. The gel was then dried, and the reaction products were analyzed by a PhosphorImager using ImageQuant software (Molecular Dynamics).
Construction of mutant viral clones and preparation of viral stocks.
To prepare viral DNA constructs containing the various mutant INs, the Cys-to-Ser mutations of HIV-1 IN were first introduced into pBSNL-AE, a subclone containing the 2.3-kbp AgeI-EcoRI fragment (nucleotide positions 3486 to 5744) of NL4-3.
(i) Single-Cys mutant subclones.
A C56S mutation in IN was prepared using the QuikChange site-directed mutagenesis kit (Stratagene) with the primer pair C56Sss-KZ (5′-CCATGCATGGACAAGTCGACAGTAGCCCAGGAATATG-3′) and C56Sas-KZ (5′-CATATTCCTGGGCTACTGTCGACTTGTCCATGCATGG-3′). Nucleotide substitutions are indicated in boldface type. To facilitate screening, a new SalI site (underlined) was created by the primer pair in the resulting pBSNL-C56S through a silent mutation of Val at amino acid position 54 of IN. pBSNL-C65S and pBSNL-C130S subclones were constructed by inserting the BstXI-BspMI DNA fragments of the HIV-1 IN expression plasmids pT7-7/IN-C65/280S and pT7-7/IN-C56/130/280S, which encode the C65S and C130S mutations, respectively, in the pBSNL-AE vector previously digested with BstXI and BspMI.
(ii) Double-Cys mutant subclones.
BstXI-EcoRI fragments of pBSNL-C65S and pBSNL-C130S containing the C65S and C130S mutations, respectively, were ligated with BstXI-EcoRI-digested pBSNL-C56S vector containing the C56S mutation, resulting in the double-Cys mutants pBSNL-C56/65S and pBSNL-C56/130S, respectively. pBSNL-C65/130S was constructed by inserting the C130S-containing MscI-EcoRI fragment of pBSNL-C130S into the pBSNL-C65S vector previously digested with MscI and EcoRI.
To construct the mutant viral clones, the AgeI-EcoRI DNA fragment of each mutant subclone was purified and ligated with plasmid NL4-3 previously digested with AgeI and EcoRI. The ligation product was transformed into SURE cells (Stratagene) by electroporation. The presence of mutations was verified by restriction analysis and DNA sequencing.
To prepare virus stocks, 293T cells at 60 to 70% confluence in 75-cm2 flasks were transfected with 20 μg of plasmid DNA containing the wild-type or mutant HIV-1 clone using the calcium phosphate precipitation method (4). The culture media were removed, and the cells were replenished with 15 ml of fresh medium 7 h after the addition of DNA. The culture media were harvested 36 and 60 h posttransfection, pooled, and clarified using low-speed centrifugation (45 × g for 5 min). The supernatant fractions containing the virions were then filtered by gravity drip through cellulose acetate tube top filters (0.45-μm pore size; Corning). The virions were pelleted over a 20% sucrose gradient by ultracentrifugation at 100,000 × g for 2 h at 4°C. Viral titers were determined by ELISA against viral p24 antigen, and the stocks were stored in 0.5-ml aliquots at −80°C.
Replication kinetics of mutant viruses.
Twenty nanograms of p24 equivalent of viral stocks were used to infect 2 × 106 CEM cells in 1 ml of RPMI 1640 containing 5 μg of Polybrene/ml, 100 U of penicillin G/ml, and 0.1 mg of streptomycin/ml at 37°C. At 2 h postinfection, the medium was removed, and the cells were washed twice with 5 ml of phosphate-buffered saline (PBS; 10 mM sodium phosphate, pH 7.4 and 150 mM NaCl) and fed with 6 ml of fresh medium. The cells were monitored for syncytium formation and split 1:3 at 2-day intervals after infection. Aliquots of the culture medium were subjected to p24 analysis to determine the levels of virus production.
Virus attachment and entry assays.
The assays for measuring virus attachment and entry were modified from that described previously (45). CEM cells (106) were incubated in 1 ml of RPMI 1640 at 37°C for 30 min before the addition of 20 ng of p24 equivalent of sucrose cushion-purified viruses. After an additional 2 h at 37°C, the medium was removed, and the virus-exposed cells were washed three times with 1 ml of ice-cold PBS to remove the unattached virus. For measuring virus entry, the same conditions were used, except that after the first PBS wash, the cells were incubated with 0.5 ml of 0.25% trypsin for 5 min at 37°C to remove surface-bound viruses. The protease activity was neutralized by 1 ml of the complete medium containing 10% FBS, and the cells were then washed twice with 1 ml of ice-cold PBS. In both attachment and entry assays, the cell pellets were lysed in PBS containing 1% Triton X-100 and 0.1 mM phenylmethylsulfonyl fluoride. After centrifugation at 10,000 × g for 5 min, the supernatant fraction was collected and diluted twofold with PBS. The p24 concentration of the diluted cell lysate was determined by ELISA.
Western blot analysis of viral proteins.
Sucrose cushion-purified virions were resuspended in a lysis buffer containing 62.5 mM Tris-HCl, pH 6.8, 0.2% SDS, 5% 2-mercaptoethanol, and 10% glycerol. Fifty nanograms of p24 equivalent of viral samples were subjected to SDS-12% PAGE and transferred onto a nitrocellulose membrane (Osmonics). The membrane was blocked with Tris-buffered saline (20 mM Tris-HCl, pH 7.5, and 500 mM NaCl) containing 5% nonfat dry milk for 1.5 h and then incubated with anti-HIV-1 human serum diluted 500-fold in Tris-buffered saline containing 1% nonfat dry milk and 0.05% Tween 20 for 1 h. The blot was then incubated with horseradish peroxidase-conjugated goat anti-human secondary antibody (Zymed) at 1:250,000 dilution for 1 h. A chemiluminescence substrate kit (SuperSignal; Pierce) was used for detection according to the manufacturer's instructions.
Analysis of viral DNA synthesis.
Fifty nanograms of p24 equivalent of viral stock was treated with 5 U of RNase-free DNase I (Amersham Pharmacia) at room temperature for 1 h. An equal amount of viral stock was heat inactivated by incubation at 65°C for 2 h after DNase I treatment. The DNase I-treated virus was used to infect 2 × 106 CEM cells in 1 ml of RPMI 1640 containing 5 μg of Polybrene/ml, 100 U of penicillin G/ml, and 0.1 mg of streptomycin/ml for 1 h at 37°C. The cells were pelleted by centrifugation at 1,000 × g; washed twice with 5 ml of PBS; resuspended in 5 ml of RPMI 1640 with 5% FBS, 5 μg of Polybrene/ml, 100 U of penicillin G/ml, and 0.1 mg of streptomycin/ml; and then incubated in six-well plates at 37°C. At 8 h postinfection, the cells were collected and washed twice with 5 ml of PBS. Total DNA was obtained by using the DNeasy tissue kit (Qiagen) with the optional RNase A treatment step. Amplification of viral DNA using the TaqMan PCR Core Reagent kit (Applied Biosystems) was done essentially as described previously (25). Briefly, 1 μg of each DNA sample in a 15-μl reaction volume was amplified in a 384-well plate format on the ABI Prism 7900 sequence detection system (Applied Biosystems). Reactions were carried out at 50°C for 2 min and 95°C for 5 min, followed by 40 cycles of amplification at 95°C for 15 s and 60°C for 1 min.
PCR primer and probe sequences were obtained from PE-Applied Biosystems and Annovis. The primers SR1 (5′-CAAGTAGTGTGTGCCCGTCTGT-3′), which anneals to nucleotide positions 551 to 571 of NL4-3, and AA55 (5′-CTGCTAGAGATTTTCCACACTGAC-3′), which anneals to nucleotide positions 635 to 612, were used at concentrations of 300 and 150 nM, respectively, to amplify the R-U5 region of the early reverse-transcribed DNA of HIV-1. The fluorescent TaqMan probe ZXF for the amplified R-U5-containing product was modified at the 5′ and 3′ ends with 6-carboxy-fluoresein (FAM) and 6-carboxytetramethyl-rhodamine (TAMRA), respectively (5′-FAM-TGTGACTCTGGTAACTAGAGATCCCTCAGACCC-TAMRA-3′). The probe anneals to nucleotide positions 574 to 606 and was used at a concentration of 200 nM. Another set of primers was used to amplify the human β-globin gene from each DNA sample and served as an internal standard (25, 61). The concentrations of the forward primer BGF1 (5′-CAACCTCAAACAGACACCATG-3′) and the reverse primer BGR1 (5′-TCCACGTTCACCTTGCCC-3′) were 300 and 200 nM, respectively. The fluorescent probe BGX1 for the human β-globin gene (5′-FAM-CTCCTGAGGAGAAGTCTGCCGTTACTGCC-TAMRA-3′) was used at a final concentration of 150 nM.
Plasmid DNA of NL4-3 linearized by EcoRI was amplified in parallel as HIV-1 DNA standards (8 to 16,000 copies/reaction). DNA from uninfected CEM cells was amplified to obtain the standard curve for the β-globin gene (0.8 to 800 ng/reaction).
Virion-associated RT assay.
The HIV-1 RT activities of wild-type and mutant virions were determined essentially as previously described (3). The sucrose cushion-purified virus was resuspended in buffer R (16.7 mM Tris-HCl, pH 7.8, 80 mM KCl, 0.17 mM EDTA, 6.7 mM DTT, 33% glycerol, and 0.3% Triton X-100). Ten microliters of the viral lysate was incubated with a 90-μl reaction mixture containing 0.05 U of poly(rA) · poly(dT)12-18 (Amersham Pharmacia) and 2.5 μCi of [methyl-3H]dTTP (30 Ci/mmol; Amersham Pharmacia) at 37°C for 1 h. The entire sample was then spotted onto a DE81 filter circle (Whatman). The filter was washed four times with 10 ml of 2× SSC (0.03 M sodium citrate, 0.3 M NaCl) and once with 10 ml of 95% ethanol and air dried. The filter was placed in a glass vial containing 5 ml of liquid scintillation fluid (ReadySafe; Beckman), and the amount of radioactivity was determined by scintillation counting (Beckman model LS3801).
Coimmunoprecipitation of RT and IN.
HIV-1 RT heterodimer (p66/p51; 10 pmol) was incubated with 40 pmol of purified recombinant wild-type or mutant HIV-1 INs in 50 μl of buffer B (20 mM Tris-HCl, pH 7.5, 250 mM NaCl, 0.2 mM MgCl2, 5 mg of bovine serum albumin/ml) for 30 min at room temperature. Two microliters of rabbit anti-HIV-1 RT polyclonal antibody diluted with 48 μl of wash buffer (buffer B plus 1% IGEPAL) was added to each sample, and the samples were incubated on ice for 1 h. Ten microliters of immobilized protein A-G beads (UltraLink; Pierce) was then added to each sample, and the mixture was incubated with gentle rocking for 1 h at 4°C. The beads were spun down at 500 × g for 2 min, washed three times with 400 μl of wash buffer, and eluted with 40 μl of buffer containing 62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 0.7 M β-mercaptoethanol (β-ME), and 2% SDS. Seven microliters of eluted proteins was subjected to SDS-10% PAGE and Western blot analysis as described above. RT was probed using the mouse anti-RT monoclonal antibody or the anti-HIV-1 human serum at a 1:500 dilution, whereas IN was probed using the rabbit anti-HIV-1 IN polyclonal antibody or the anti-HIV human serum at a 1:500 dilution. The blot was then incubated with goat anti-mouse, anti-human, or anti-rabbit secondary antibody conjugated with horseradish peroxidase (Zymed) at 1:50,000 dilution for 1 h. Detection was carried out using a chemiluminescence substrate kit (SuperSignal; Pierce) according to the manufacturer's instructions.
RESULTS
Cys residues within the core domain of HIV-1 IN are not essential for its enzymatic activities in vitro.
Wild-type HIV-1 IN has a total of six Cys residues at positions 40, 43, 56, 65, 130, and 280. To examine whether Cys residues at positions 56, 65, and 130 of HIV-1 IN are required for its integration activity in vitro, we constructed mutant HIV-1 INs containing multiple Cys-to-Ser substitutions and purified them as recombinant proteins from E. coli (Fig. 1). One mutant, IN-NR, has all of the Cys residues except C40 and C43 replaced with Ser. The other mutant, IN-C56/65/280S, has Cys residues at positions 56, 65, and 280 replaced with Ser. The in vitro activities of both mutants were compared to those of the wild-type IN (Fig. 2). Both IN-NR and IN-C56/65/280S possessed 3′-end processing, strand transfer, and disintegration activities. Since the mutation of Cys at amino acid positions 56, 65, and 130 did not abolish the catalytic activity of HIV-1 IN, the result indicated that none of the core Cys residues are essential for the integration reaction in vitro. Although IN-NR was catalytically active, its 3′-end processing activity was ∼50% of that of the wild-type IN (Fig. 2A, lane 3 versus lane 2), and the level of strand transfer activity was significantly less than those of the wild type and IN-C56/65/280S (Fig. 2B, lane 3 versus lanes 2 and 4). Considering that the C130S substitution causes instability of HIV-1 IN in a eukaryotic expression system (5), the substitution may produce a folding defect that adversely affects certain functions. However, the retention of enzymatic activities implies that the Cys substitutions did not grossly alter the structure of IN. Also, limited digestion of IN-NR with trypsin or V8 protease produced proteolytic fragments similar to those of wild-type IN (data not shown), again suggesting that the Cys substitutions did not cause extensive changes in IN conformation.
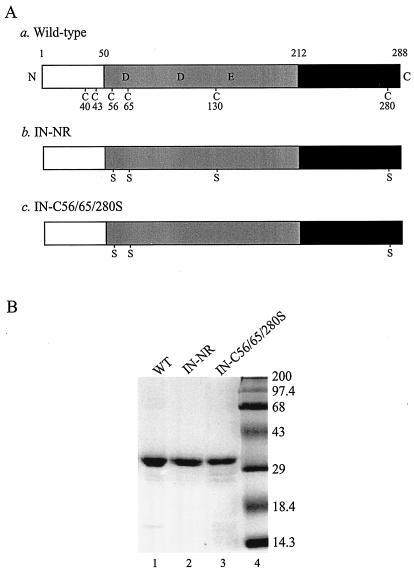
FIG. 1.
Recombinant wild-type and mutant HIV-1 INs. (A) Schematic representation of wild-type HIV-1 IN and Cys mutants used in the in vitro assays. N and C, N and C termini of the protein, respectively. The open boxes represent the N-terminal domain (amino acids 1 to 50) containing the conserved zinc-binding HHCC motif, the shaded boxes represent the central core domain (amino acids 51 to 212) containing the catalytic DD(35)E motif, and the solid boxes represent the C-terminal domain (amino acids 213 to 288). The six Cys residues and their amino acid positions are indicated in the wild-type IN (construct a). Replacements of Cys by Ser are indicated in both IN-NR (construct b) and IN-C56/65/280S (construct c). (B) Coomassie blue-stained SDS-polyacrylamide gel of 100 pmol of purified recombinant wild-type IN (WT; lane 1) and its mutant derivatives containing Cys-to-Ser substitutions at positions 56, 65, 130, and 280 (IN-NR; lane 2) or at positions 56, 65, and 280 (IN-C56/65/280S; lane 3). Molecular mass markers (Gibco) are shown in lane 4 and are labeled in kilodaltons on the right.
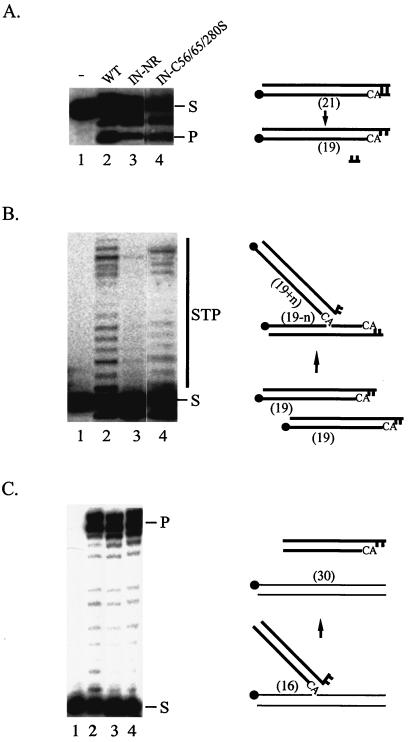
FIG. 2.
Enzymatic activities of wild-type and mutant HIV-1 INs in vitro. (A) 3′-end processing. (B) Strand transfer. (C) Disintegration. In all three panels, lane 1 contained the labeled substrate only, lane 2 had a reaction mixture containing the wild-type IN, and lanes 3 and 4 had reaction mixtures containing the mutant IN-NR and IN-C56/65/280S, respectively. The migration positions of the substrates (S) and products (P, 3′-end processing or disintegration product; STP, strand transfer product) are marked. For each in vitro assay, a model of the reaction substrate and products is at the right of the gel. The solid circles indicate the positions of the 32P label at the 5′ end of the oligonucleotide. The thick lines represent viral sequences, and the thin lines represent target sequences. The numbers in parentheses indicate the lengths in nucleotides of substrates and products.
Mutant viruses encoding the C130S mutation in IN are replication defective.
To determine the role of Cys of HIV-1 IN during viral replication, we constructed several mutant viral clones encoding one or two Cys-to-Ser mutations in the IN gene of the molecular clone NL4-3 (Fig. 3A and B). Virions were prepared by transient transfection of 293T cells with plasmid DNA containing wild-type or mutant HIV-1 clones. The viral titer was determined by measuring the level of viral capsid (CA; p24) protein in the culture medium. The titers of mutant viral clones, ranging from 500 to 1,100 ng of p24/ml, were similar to that of the wild type (800 ng/ml).
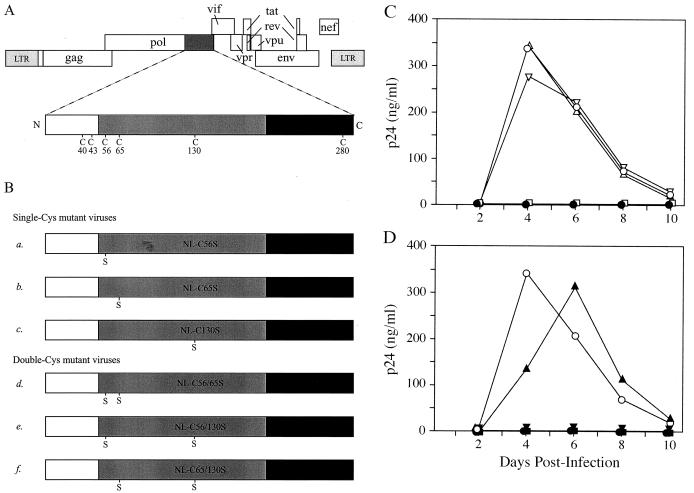
FIG. 3.
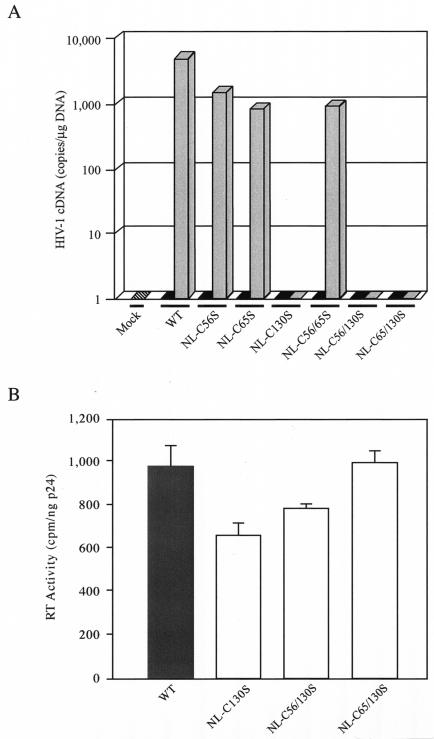
Replication kinetics of mutant HIV-1 viral clones. (A) Genomic organization of HIV-1. The IN-coding sequence, represented by a dark shaded box, is expanded to show the positions of Cys residues. The IN domains are described in the legend to Fig. 1A. (B) HIV-1 clones containing a mutated IN. Viral clones encode either a single Cys-to-Ser mutation at amino acid residue position 56 (construct a), 65 (construct b), or 130 (construct c) or two Cys-to-Ser mutations at positions 56 and 65 (construct d), 56 and 130 (construct e), or 65 and 130 (construct f) of IN. (C) Replication kinetics of wild-type and single-Cys mutant viruses. CEM cells were infected with equal amounts of p24 equivalent of the wild type (○) or single-Cys mutant NL-C56S (Δ), NL-C65S (▿), or NL-C130S (□). (D) Replication kinetics of wild-type and double-Cys mutant viruses. CEM cells were infected with wild-type (○) or double-Cys mutant NL-C56/65S (▴), NL-C56/130S (▾), or NL-C65/130S (▪). In both panels C and D, the culture media were monitored for p24 production at the indicated time points postinfection. Mock infection was carried out with heat-inactivated wild-type virus (•).
Equal amounts (20 ng) of p24 equivalent of wild-type or mutant viral stocks were used to infect CEM cells. The infectivities of mutant viruses were measured by the release of viral particles and the appearance of syncytia in the culture media over 10 days. Cells infected with wild-type NL4-3 or the mutant virus NL-C56S, NL-C65S, or NL-C56/65S developed syncytia 4 days postinfection. However, syncytia were not observed in cells infected with NL-C130S, NL-C56/130S, or NL-C65/130S during the 10-day culture period (data not shown). The mutant viruses NL-C56S, NL-C65S, and NL-C56/65S exhibited wild-type-like replication kinetics, with p24 production peaking 4 to 6 days postinfection (Fig. 3C and D). Cells infected by NL-C130S, NL-C56/130S, or NL-C65/130S failed to generate detectable levels of p24 in the culture media, indicating that these viruses were noninfectious (Fig. 3C and D). Since none of the Cys mutations contained in NL-C56S, NL-C65S, or NL-C56/65S led to altered replication kinetics, the replication defect seen in NL-C130S, NL-C56/130S, and NL-C65/130S mutant viruses is therefore due to the C130S mutation of HIV-1 IN; it is the only mutation absent in the replication-competent mutant viruses and the only mutation shared by all the defective viruses. This finding is consistent with a previous report demonstrating a loss of viral infectivity in an HIV-1 clone, BRU, containing a C130G mutation within IN (40).
Viral attachment and entry, and proteolytic processing and packaging of viral proteins, are normal for all mutant viruses.
We measured the amounts of cell-associated and intracellular p24 in infected cells to examine whether the Cys mutations affected virus attachment and entry. This was particularly pertinent for the C130S-containing viruses, which were unable to replicate. The cell-associated p24 levels, which account for internalized virions as well as those on the membrane surface, from cells infected with either wild-type or various mutant viruses were not significantly different (P < 0.05), ranging from 305.3 ± 45.1 to 422.0 ± 58.0 pg/106 cells (Table 1). The intracellular p24 levels, which were measured after trypsin digestion to remove plasma membrane-attached viruses, were also similar among the wild-type and mutant viruses (Table 1). The result suggested that the Cys mutations of IN have no significant effects on virus attachment and entry.
TABLE 1.
Effects of Cys mutations of IN on CEM cell virus attachment and entry
| Virus | p24 level (pg/106 cells)a
|
|
|---|---|---|
| Without trypsin | With trypsin | |
| Heat inactivatedb | 96.7 ± 5.2 | 3.5 ± 0.7 |
| WTc | 395.3 ± 41.1 | 31.3 ± 4.8 |
| NL-C130S | 305.3 ± 45.1 | 34.5 ± 6.4 |
| NL-C56/130S | 415.3 ± 31.0 | 28.8 ± 3.5 |
| NL-C65/130S | 422.0 ± 58.0 | 29.0 ± 7.0 |
Values are expressed as the mean ± standard error of three or four independent experiments.
Wild-type NL4-3 viruses were heated at 70°C for 20 min.
WT, wild type.
To examine whether the lack of infectivity of C130S mutants of HIV-1 IN was caused by a disruption of the viral protein synthesis, packaging, or polyprotein processing, we examined the composition of the mutant viral particles by Western blotting. The lysates of wild-type and mutant viruses were resolved by SDS-PAGE, blotted onto a nitrocellulose membrane, and probed with anti-HIV-1 human serum (Fig. 4). The Gag precursor proteins Pr55 and Pr41, along with its processed products matrix (MA; p17) and CA (p24), were present in both wild-type and mutant virions. The three processed products of the 160-kDa Gag-Pol precursor, IN (p32) and the p51 and p66 subunits of RT, were also present in all mutant virions. Compared to the level found in the wild-type NL4-3 (Fig. 4, lane 1), the amounts of processed IN present in NL-C130S and NL-C56/130S were lower (Fig. 4, lanes 4 and 6). We do not know if this was due to a lower level of incorporation, an alteration in polyprotein processing or degradation, or the qualitative nature of the Western analysis. However, for NL-C65/130S, which contains the C130S substitution and is replication defective, the levels of IN and RT after normalization with p24 were similar to those of the wild-type virus (Fig. 4, lane 7). The results demonstrated that the mutant viruses did not have a severe defect in viral-protein expression, assembly, release, or maturation.
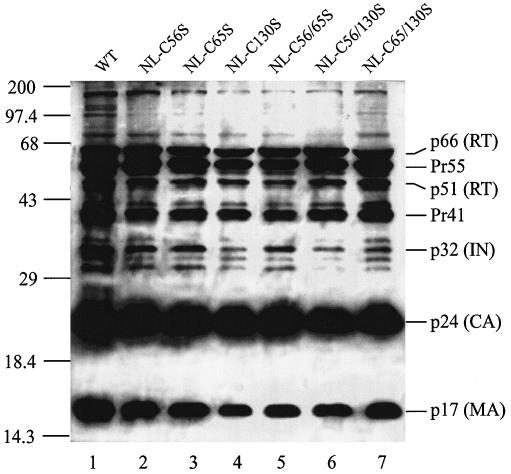
FIG. 4.
Western blot analysis of HIV-1 proteins in viral particles. Each lane contains proteins from 50 ng of p24 equivalent of viral lysates separated on an SDS-12% polyacrylamide gel and probed with anti-HIV-1 human serum. Lane 1, wild-type virus (WT); lanes 2 to 7, various Cys mutant viruses as identified above the lanes. The numbers on the left are molecular masses in kilodaltons, and the labels on the right correspond to positions of viral proteins or their precursors.
Mutant viruses encoding the C130S mutation contain a functional RT but are impaired in viral-DNA synthesis.
Genetic studies have shown that certain mutations in IN affect reverse transcription (18, 35, 37, 52). Since none of the Cys mutants were impaired in entry or in the late stages of viral replication, the abilities of mutant viruses to synthesize viral DNA during infection were examined using the TaqMan real-time PCR method (25). The CD4+ T cells were infected with identical amounts of wild-type or mutant viruses, as determined by the p24 content, and the results were normalized by the total amount of input cellular DNA. For the wild-type virus, 7,000 copies of HIV-1 cDNA per μg of cellular DNA were present 8 h postinfection (Fig. 5A). All the replication-competent mutants, including single- and double-Cys mutants at positions 56 and 65, showed comparable amounts of the early cDNA product, albeit at a level about sevenfold less than that of the wild type. Importantly, for all the C130S-containing mutant viruses, which were replication defective, the early R-U5 reverse-transcription product was completely absent (Fig. 5A). Our results indicated that the loss of infectivity seen in the C130S mutant viruses was due to the defect in viral DNA synthesis before integration.
FIG. 5.
cDNA synthesis and RT activities of wild-type and mutant viruses. (A) Quantitative analysis of viral cDNA synthesis by real-time PCR. The amount of early reverse-transcribed viral DNA present in CEM cells was measured 8 h after infection with the wild-type (WT) or mutant viruses (shaded bars). All viral stocks were treated with RNase-free DNase I to remove potential contamination by plasmid DNA. The culture medium alone was used in the mock infection (hatched bar), and equivalent amounts of the respective heat-inactivated viruses (solid bars) were used as negative controls for infection. For normalization of the DNA input of each sample, the human β-globin gene was amplified under identical PCR conditions. The results are expressed as the number of copies of HIV-1 cDNA detected per microgram of total DNA extracted from infected cells. The values shown represent the averages of two independent experiments. In each experiment, the standards and test samples were run in duplicate. (B) Particle-associated RT activities of wild-type and C130S-containing mutant viruses. After transient transfection of 293T cells, WT (shaded bar) and mutant (open bars) viruses were harvested, concentrated, lysed, and assayed for particle-associated RT activity as described in Materials and Methods. The result for each viral clone is shown as the mean ± standard error of three experiments using independently derived virus stocks.
Certain IN mutations that produced replication-defective virions have reduced levels of particle-associated RT activity (48, 49). To examine whether the lack of viral DNA synthesis in the C130S mutant virion was due to an alteration in the intrinsic activity of RT, the particle-associated activity of RT was assayed. Wild-type and the C130S-containing mutant viruses were lysed in detergent, and an exogenous template-primer and a radiolabeled nucleotide were added to the viral lysate. The activity of RT in the detergent-lysed virions was measured by the retention of the incorporated radioactive nucleotide on the positively charged filter. In three independent experiments, all mutant viruses encoding the C130S mutation exhibited considerable levels of RT activity, ranging from 68 to 100% of that of the wild-type virus (Fig. 5B). Although the intrinsic RT activity of NL-C130S was significantly less (P < 0.05) than that of the wild type, the 32% decrease in activity could not account for the complete absence of early reverse-transcribed products in cells infected with NL-C130S (Fig. 5A). Furthermore, both NL-C56/130S and NL-C65/130S, which also were unable to synthesize viral cDNA (Fig. 5A), had levels of intrinsic RT activity similar to that of the wild-type virus (Fig. 5B). Taken together, the results indicated that the lack of early reverse-transcription products in cells infected with viruses carrying the C130S mutation is not due to a difference between the intrinsic RT activities in the mutant and wild-type viruses.
The C-terminal domain of HIV-1 IN interacts with RT, and C130S substitution of HIV-1 IN disrupts the RT-IN interaction.
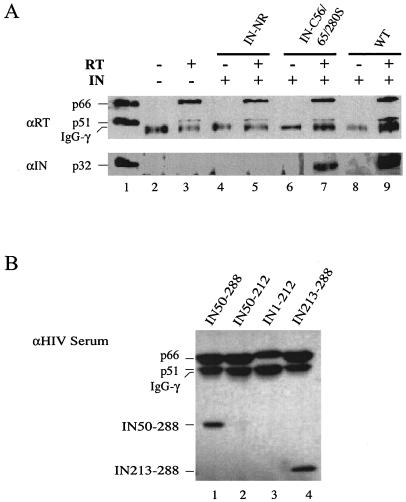
Both RT and IN are parts of the PIC (20, 38), and their enzymatic activities can influence one another (50). Previous studies demonstrated that a physical interaction exists between RT and IN of HIV-1 and murine leukemia virus (MLV) (28, 50, 58). Therefore, we examined the ability of the C130S mutant of HIV-1 IN to interact with RT in vitro (Fig. 6A). In the absence of HIV-1 RT heterodimer, wild-type HIV-1 IN was not immunoprecipitated by the anti-HIV-1 RT polyclonal antibody (Fig. 6A, lane 8), indicating that the anti-HIV-1 RT polyclonal antibody did not cross-react with IN under the assay conditions. Addition of RT to the reaction mixture resulted in coimmunoprecipitation of HIV-1 IN (Fig. 6A, lane 9), confirming the previous findings that RT and IN physically interact (28, 50, 58). However, when the Cys mutant IN-NR (Fig. 6A, lanes 4 and 5) or IN-C56/65/280S (Fig. 6A, lanes 6 and 7) was used instead of the wild-type IN, only IN-C56/65/280S was pulled down by the anti-RT antibody in the presence of RT (Fig. 6A, lane 7). The two IN mutants differ from each other by only one amino acid residue at position 130: IN-C56/65/280S contains Cys, while IN-NR contains Ser (Fig. 1A). The result indicated that the C130S substitution abolishes the in vitro physical interaction between RT and IN.
FIG. 6.
Interaction between HIV-1 IN and RT. (A) Coimmunoprecipitation of RT and IN in vitro. Lane 1 contained the heterodimer purified RT (p66/p51), and IN and served as a positive control for immunoblotting and size markers. Lane 2 was a negative control and contained only the rabbit anti-HIV-1 RT polyclonal antibody. Lane 3 was a complete reaction without IN. For the coimmunoprecipitation experiments, wild-type HIV-1 IN (p32; lanes 8 and 9) or the Cys mutant derivative IN-NR (lanes 4 and 5) or IN-C56/65/280S (lanes 6 and 7) was incubated in the absence (lanes 4, 6, and 8) or presence (lanes 5, 7, and 9) of purified HIV-1 RT. The RT or RT-IN complex was immunoprecipitated with the rabbit anti-HIV-1 RT polyclonal antibody as described in Materials and Methods. The top blot was probed with the mouse anti-RT monoclonal antibody (αRT), and the bottom blot was probed with the rabbit anti-IN polyclonal antibody (αIN). The 50-kDa bands in lanes 2 to 9 correspond to the immunoglobulin G heavy chain (IgG-γ) that cross-reacted with the mouse anti-RT monoclonal antibody used for immunoblotting. (B) Mapping of the IN domain that interacts with RT. Various HIV-1 IN truncation mutants (as labeled above the lanes) were incubated with RT and immunoprecipitated using the rabbit anti-HIV-1 RT polyclonal antibody as described above. The blot was probed with anti-HIV-1 human serum.
A more direct approach than the use of IN-NR to test the role of C130 in interacting with RT is to prepare purified IN containing only the C130S substitution. However, due to extensive degradation, we were unable to obtain a reasonable yield of the mutant protein.
To map the IN domain involved in interacting with RT, we carried out a coimmunoprecipitation experiment using HIV-1 RT heterodimer and various truncation mutant derivatives of HIV-1 IN (Fig. 6B). The ability of the N-terminally truncated mutant (Fig. 6B, lane 1) or the C-terminal domain only (Fig. 6B, lane 4), but not the core domain only (Fig. 6B, lane 2) or the C-terminally truncated mutant (Fig. 6B, lane 3), to coimmunoprecipitate with RT indicated that the C-terminal domain of IN (residues 213 to 288) is involved in interacting with RT.
The RT-IN interaction does not require a disulfide linkage.
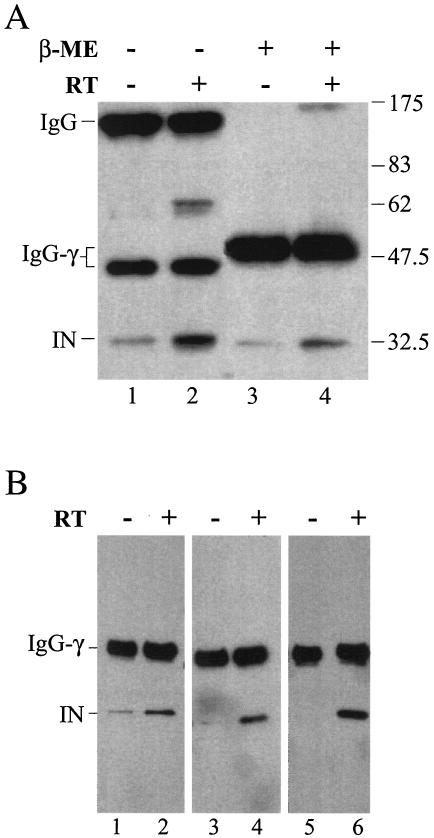
The observed physical interaction between IN and RT may involve the formation of a disulfide linkage between C130 and a free Cys in RT. Such an interaction cannot take place when C130 is replaced with Ser. Immunoprecipitation experiments showed that MLV IN is associated with RT through a disulfide linkage (28). Since C130 is located in the core domain of HIV-1 IN and the interacting domain was mapped within the C terminus, a more plausible explanation is that replacing Cys at position 130 of IN with Ser alters the recognition interface of IN that is critical for interacting with RT. To verify that IN does not interact with RT via disulfide linkages, the IN-RT complex was first immunoprecipitated with anti-RT antibody as described earlier and then treated either with the reducing agent β-ME or left untreated before separation by nonreducing SDS-PAGE and detection by Western blotting using anti-IN antibody (Fig. 7A). As expected, in the presence of β-ME, all the IN that was pulled down by RT was in monomeric form (Fig. 7A, lane 4). As an internal control, the reduction of the disulfide bond by the addition of β-ME was confirmed by the conversion of the 150-kDa rabbit immunoglobulin complex (Fig. 7A, lanes 1 and 2) to the 50-kDa monomeric heavy chain (Fig. 7A, lanes 3 and 4). In the absence of β-ME (Fig. 7A, lane 2), the IN coimmunoprecipitated with RT was mostly monomeric, with ∼15% migrating as a dimer (∼64 kDa). The dimeric IN presumably was linked by an intermolecular disulfide bond via Cys at position 280 (5). However, an immunoreactive product corresponding to the RT-IN complex was not detected when the blot was probed with antibody against IN (Fig. 7A, lane 2) or RT (data not shown). The absence of RT-IN complex under nonreducing conditions suggested that the interaction between IN and RT does not involve disulfide bond formation.
FIG. 7.
RT-IN interaction does not involve disulfide bond formation. (A) Effect of a reducing agent on RT-IN interaction. HIV-1 IN was incubated in the absence (−) (lanes 1 and 3) or presence (+) (lanes 2 and 4) of RT and immunoprecipitated with a rabbit anti-HIV-1 RT polyclonal antibody. The immunoprecipitated complex was left untreated (lanes 1 and 2) or treated with β-ME (lanes 3 and 4), separated by SDS-PAGE, and analyzed by Western blotting using the rabbit anti-IN polyclonal antibody. The 64-kDa band in lane 2 corresponds to IN homodimers (5). The lower level of IN in lane 4 compared to lane 2 is due to a direct effect of β-ME on IN, possibly by altering the reactivity of IN to the anti-IN antibody. IgG-γ, immunoglobulin G heavy chain. (B) Effect of an alkylating agent on RT-IN interaction. Forty picomoles of HIV-1 IN in a final volume of 10 μl was preincubated at room temperature for 15 min in buffer C (20 mM HEPES-Na, pH 6.8, 250 mM NaCl, 0.2 mM MgCl2) with (lanes 1 and 2) or without (lanes 3 to 6) 10 mM DTT (20 mM equivalent of free sulfhydryls). The IN was then treated with 3.6 (2× molar excess; lanes 3 and 4) or 18 (10× molar excess; lanes 1, 2, 5, and 6) mM NEM for 1 h at 4°C. In lanes 3 to 6, the activity of NEM was quenched with 10 mM DTT. The samples were then added to buffer C with (lanes 2, 4, and 6) or without (lanes 1, 3, and 5) RT. Coimmunoprecipitation was carried out with the rabbit anti-HIV-1 RT polyclonal antibody. The immunoprecipitated complex was eluted with the buffer containing β-ME, and the rest of the procedure was as described for panel A. The extent of NEM modification was monitored using electrospray ionization-mass spectrometry. At 2× molar excess of NEM, all INs were modified, and the distribution of INs containing four, five, and six NEM adducts was ∼35, 50, and 10%, respectively (data not shown).The IN with four NEM adducts likely represented Cys modification at positions 56, 65, 130, and 280, since the Cys residues at positions 40 and 43 are bound with zinc and therefore should be much less reactive. The extent of NEM adduct formation at 10× excess was not determined.
To further confirm that IN did not interact with RT via disulfide linkage, wild-type IN was incubated with N-ethylmaleimide (NEM) before being subjected to the coimmunoprecipitation analysis. NEM alkylates reactive Cys residues and renders them incapable of forming disulfide bonds. Treatment of IN with 2- or 10-fold molar excess of NEM did not affect its ability to coimmunoprecipitate with RT (Fig. 7B, lanes 3 to 6). The result is consistent with the observation that the interaction between IN and RT is independent of disulfide linkage formation.
DISCUSSION
We examined the role of Cys residues of HIV-1 IN in the enzymatic activity of the protein in vitro and during replication of the virus in cell cultures. Although none of the four Cys residues located in the core and C-terminal domains are required for the enzymatic activity of IN in vitro, we found that Cys at position 130 is essential during viral replication. Unlike Cys substitutions at positions 56 and 65, mutant viruses encoding the C130S mutation were unable to replicate in T-cell lines. To determine at which step during the viral life cycle the replication block occurred, the mutant viruses were subjected to various biochemical assays. We found that the titers of wild-type and mutant viruses, as measured by the concentration of viral capsid protein, are similar after transient transfection of producer cells. The mutant viruses harvested from the culture medium after transfection contain wild-type levels of viral precursor and mature proteins. These results indicate that the postintegration steps, such as protein expression, packaging, processing, and maturation, are not affected by the Cys mutations. In addition, the early steps of attachment and entry of mutant viruses appear to be normal, as measured by the level of capsid protein in infected cells. The only defect identified during the replication of the C130S-containing virus occurs at reverse transcription, as the early reverse-transcribed DNA product was undetectable in infected cells. However, the defect is not due to the absence or inactivity of RT, since the detergent-lysed virions containing the C130S mutation have intrinsic RT activity comparable to that of the wild type.
Several studies have shown that mutations of IN can produce pleiotropic effects during HIV-1 replication (10, 18, 52, 57). Besides integration, these include early steps in the viral life cycle, such as reverse transcription (35, 37, 52, 58) and nuclear import of PICs (22, 52), as well as the late postintegration steps, such as polyprotein processing, assembly, and maturation (8, 42, 48). The mechanisms by which IN or its mutants exert these multiple effects are likely diverse and are presently poorly understood. Since IN is synthesized and packaged into the immature virion as part of the Gag-Pol polyprotein, many of the alterations in the virion morphology and the late steps of replication are likely attributable to the effects of IN mutations on the Gag-Pol precursor protein (8, 42). However, certain IN mutations, such as H12L, H16V, C43L, S81R, F185A, and Y143G, specifically impair nuclear import or reverse transcription without affecting the Gag-Pol polyprotein and other steps in the life cycle (35, 37, 58). Our results show that the C130S substitution in IN belongs to the latter category. Furthermore, disruption of reverse transcription by IN mutations has also been demonstrated in MLV (34) and the yeast retrovirus-like element Ty3 (39), suggesting that the involvement of IN could be a general property shared by retroviruses and retrotransposons.
How does a mutation in IN specifically interfere with reverse transcription? There is increasing evidence that IN physically interacts with RT. For avian viruses and human T-cell leukemia virus type 1, IN is an integral part of RT and forms a functional heterodimeric or oligomeric complex with the processed RT (24, 51). For HIV-1 and MLV, both RT and IN are present within the same nucleoprotein complex during reverse transcription (20, 21, 30), and direct interaction between the two proteins is supported by biochemical analyses in vitro (28, 50, 58). The specific RT-IN interaction is also confirmed by a pull-down assay with glutathione S-transferase or His tag, and the RT-binding site on IN is mapped to the C-terminal domain (27a), corroborating our results reported here. The finding that IN interacts with RT is consistent with the requirement for IN during reverse transcription. However, the question of whether the RT-IN interaction observed in vitro is biologically significant has not been addressed. Also, it has not been determined if the IN mutations that fail to support reverse transcription in vivo are incapable of interacting with RT in vitro. In this study, we showed that INs containing Cys substitutions at positions 56 and 65, which can support the initiation of reverse transcription, retain the ability to physically interact with RT. Conversely, viruses harboring the C130S mutation have a complete absence of reverse-transcription product, and purified C130S-containing IN fails to interact with RT in vitro. Although the mechanism is still unknown, these results reveal a direct correlation between IN mutations that impair reverse transcription in vivo and the ability of the IN mutant to interact with RT in vitro. Taken together, the data suggest that the RT-IN interaction in vitro is biologically relevant and has an important function during viral replication. Experiments are under way to examine the abilities of other known reverse-transcription-defective IN mutants to coimmunoprecipitate with RT and to determine whether the physical interaction between RT and IN occurs in vivo.
A common mechanism for assembling protein complexes is through the formation of intermolecular disulfide bonds between Cys residues of different protein subunits (for examples, see references 46 and 56). Although the structure of the full-length HIV-1 IN has not been determined, the structures of the truncated variants show that C130 is buried within a hydrophobic cavity (11, 23, 55). Thus, C130 is unlikely to be available to form a disulfide bond with RT. The absence of an RT-IN complex under nonreducing conditions and the inability of NEM to prevent IN from interacting with RT also indicate that the interaction between RT and IN does not involve intermolecular disulfide linkages. Furthermore, the RT-interacting region of IN has been mapped to the C-terminal domain, which does not include the C130 residue. Therefore, we hypothesize that the C130S substitution disrupts the protein recognition interface of the C-terminal domain of IN and abolishes its ability to interact with RT. The presence of a minor structural alteration induced by the Cys substitution is supported by the indirect evidence that the C130S IN mutant has a significantly weak strand transfer activity and an altered physical property that led to its instability during purification (reference 5 and data not shown).
Although RT itself is sufficient to catalyze RNA- and DNA-dependent polymerization in vitro, the reaction in vivo of HIV-1 can be affected by several viral factors, including MA, nucleocapsid, Nef, Tat, and Vif (2, 15, 27, 32, 44). The phenotype of the C130S mutant, together with the activities of other IN mutants described previously (35, 37, 58), provides strong evidence that IN is critical for reverse transcription. However, to conclude unequivocally that the defect caused by the C130S substitution in IN is direct and specific for initiating reverse transcription, additional experiments are needed to test other alternatives, such as the uncoating of viral particles, the assembly and function of the reverse-transcription complex, and the incorporation of the tRNA primer. In addition, we have not formally ruled out the possibility that replacing Cys at residue 130 with Ser may produce a gain-of-function mutation, resulting in the suppression of RT activity. Also, the precise mechanism by which the physical interaction between RT and IN can affect reverse transcription remains to be determined.
In summary, we have demonstrated that although a C130S substitution in HIV-1 IN does not abolish the protein's catalytic activities in vitro, this substitution leads to a block in reverse transcription in vivo. The effects of the various Cys mutants on reverse transcription in infected cells are corroborated by the coimmunoprecipitation experiment in vitro, suggesting that the physical interaction between RT and IN is biologically relevant. Based on these data, we propose that mutation of the Cys residue at position 130 of IN to Ser abolishes the IN-RT interaction, resulting in a defect in the initiation of reverse transcription and the subsequent loss of infectivity of the C130S-containing virus.
Acknowledgments
We thank Ganjam Kalpana and Vinayaka Prasad for communicating unpublished data and careful reading of the manuscript, Stuart Le Grice for the generous supply of purified RT and anti-RT antibody, Jerome Zack for advice on real-time PCR and valuable discussions, Tom Wilkinson for technical assistance and critical review of the manuscript, and the Core Virology Laboratory at the UCLA AIDS Institute for carrying out the ELISA for p24.
This work was supported by National Institutes of Health grant CA68859 to S.A.C. and a Supplementary Grant for Minority Students to C.D.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken, C., and D. Trono. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J. Virol. 69:5048-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldovini, A., and B. D. Walker. 1990. Techniques in HIV research. Stockton Press, New York, N.Y.
- 4.Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1999. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Bischerour, J., H. Leh, E. Deprez, J. C. Brochon, and J. F. Mouscadet. 2003. Disulfide-linked integrase oligomers involving C280 residues are formed in vitro and in vivo but are not essential for human immunodeficiency virus replication. J. Virol. 77:135-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brin, E., J. Yi, A. M. Skalka, and J. Leis. 2000. Modeling the late steps in HIV-1 retroviral integrase-catalyzed DNA integration. J. Biol. Chem. 275:39287-39295. [DOI] [PubMed] [Google Scholar]
- 7.Brown, P. O. 1997. Integration, p. 161-204. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 8.Bukovsky, A., and H. Gottlinger. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 70:6820-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burke, C. J., G. Sanyal, M. W. Bruner, J. A. Ryan, R. L. LaFemina, H. L. Robbins, A. S. Zeft, C. R. Middaugh, and M. G. Cordingley. 1992. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J. Biol. Chem. 267:9639-9644. [PubMed] [Google Scholar]
- 10.Cannon, P. M., W. Wilson, E. Byles, S. M. Kingsman, and A. J. Kingsman. 1994. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J. Virol. 68:4768-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J. C., J. Krucinski, L. J. Miercke, J. S. Finer-Moore, A. H. Tang, A. D. Leavitt, and R. M. Stroud. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. USA 97:8233-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow, S. A. 1997. In vitro assays for activities of retroviral integrase. Methods 12:306-317. [DOI] [PubMed] [Google Scholar]
- 13.Craigie, R., A. B. Hickman, and A. Engelman. 1995. HIV: a practical approach, vol. 2. Oxford University Press, New York, N.Y.
- 14.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 15.Dettenhofer, M., S. Cen, B. A. Carlson, L. Kleiman, and X. F. Yu. 2000. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J. Virol. 74:8938-8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellison, V., J. Gerton, K. A. Vincent, and P. O. Brown. 1995. An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J. Biol. Chem. 270:3320-3326. [DOI] [PubMed] [Google Scholar]
- 17.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelman, A., A. B. Hickman, and R. Craigie. 1994. The core and carboxy-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol. 68:5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of non-dividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldgur, Y., F. Dyda, A. Hickman, T. M. Jenkins, R. Craigie, and D. R. Davies. 1998. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl. Acad. Sci. USA 95:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golomb, M., D. P. Grandgenett, and W. Mason. 1981. Virus-coded DNA endonuclease from avian retrovirus. J. Virol. 38:548-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorry, P. R., G. Bristol, J. A. Zack, K. Ritola, R. Swanstrom, C. J. Birch, J. E. Bell, N. Bannert, K. Crawford, H. Wang, D. Schols, E. De Clercq, K. Kunstman, S. M. Wolinsky, and D. Gabuzda. 2001. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J. Virol. 75:10073-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goulaouic, H., and S. A. Chow. 1996. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J. Virol. 70:37-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrich, D., C. Ulich, L. F. Garcia-Martinez, and R. B. Gaynor. 1997. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 16:1224-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Hehl, E. A., P. Joshi, G. V. Kalpana, and V. R. Prasad. 2004. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J. Virol. 78:5056-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, S. C., D. L. Court, M. Zweig, and J. G. Levin. 1986. Murine leukemia virus pol gene products: analysis with antisera generated against reverse transcriptase and endonuclease fusion proteins expressed in Escherichia coli. J. Virol. 60:267-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenkins, T. M., A. Engelman, R. Ghirlando, and R. Craigie. 1996. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 271:7712-7718. [DOI] [PubMed] [Google Scholar]
- 30.Karageorgos, L., P. Li, and C. Burrell. 1993. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res. Hum. Retrovir. 9:817-823. [DOI] [PubMed] [Google Scholar]
- 31.Katz, R. A., and A. M. Skalka. 1994. The retroviral enzymes. Annu. Rev. Biochem. 63:133-173. [DOI] [PubMed] [Google Scholar]
- 32.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulkosky, J., R. A. Katz, G. Merkel, and A. M. Skalka. 1995. Activities and substrate specificity of the evolutionarily conserved central domain of retroviral integrase. Virology 206:448-456. [DOI] [PubMed] [Google Scholar]
- 34.Lai, L., H. Liu, X. Wu, and J. C. Kappes. 2001. Moloney murine leukemia virus integrase protein augments viral DNA synthesis in infected cells. J. Virol. 75:11365-11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leavitt, A. D., G. Robles, N. Alesandro, and H. E. Varmus. 1996. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J. Virol. 70:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, S. P., J. Xiao, J. R. Knutson, M. S. Lewis, and M. K. Han. 1997. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry 36:173-180. [DOI] [PubMed] [Google Scholar]
- 37.Masuda, T., V. Planelles, P. Krogstad, and I. S. Y. Chen. 1995. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 69:6687-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nymark-McMahon, M. H., N. S. Beliakova-Bethell, J. L. Darlix, S. F. Le Grice, and S. B. Sandmeyer. 2002. Ty3 integrase is required for initiation of reverse transcription. J. Virol. 76:2804-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petit, C., O. Schwartz, and F. Mammano. 1999. Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J. Virol. 73:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polard, P., and M. Chandler. 1995. Bacterial transposases and retroviral integrases. Mol. Microbiol. 15:13-23. [DOI] [PubMed] [Google Scholar]
- 42.Quillent, C., A. M. Borman, S. Paulous, C. Dauguet, and F. Clavel. 1996. Extensive regions of pol are required for efficient human immunodeficiency virus polyprotein processing and particle maturation. Virology 219:29-36. [DOI] [PubMed] [Google Scholar]
- 43.Rice, P., R. Craigie, and D. R. Davies. 1996. Retroviral integrases and their cousins. Curr. Opin. Struct. Biol. 6:76-83. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez-Rodriguez, L., Z. Tsuchihashi, G. M. Fuentes, R. A. Bambara, and P. J. Fay. 1995. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J. Biol. Chem. 270:15005-15011. [DOI] [PubMed] [Google Scholar]
- 45.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 1999. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 18:6771-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Senkevich, T. G., C. L. White, E. V. Koonin, and B. Moss. 2002. Complete pathway for protein disulfide bond formation encoded by poxviruses. Proc. Natl. Acad. Sci. USA 99:6667-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shibagaki, Y., and S. A. Chow. 1997. Central core domain of retroviral integrase is responsible for target site selection. J. Biol. Chem. 272:8361-8369. [DOI] [PubMed] [Google Scholar]
- 48.Shin, C.-G., B. Taddeo, W. A. Haseltine, and C. M. Farnet. 1994. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J. Virol. 68:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taddeo, B., W. A. Haseltine, and C. M. Farnet. 1994. Integrase mutants of human immunodeficiency virus type 1 with a specific defect in integration. J. Virol. 68:8401-8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tasara, T., G. Maga, M. O. Hottiger, and U. Hubscher. 2001. HIV-1 reverse transcriptase and integrase enzymes physically interact and inhibit each other. FEBS Lett. 507:39-44. [DOI] [PubMed] [Google Scholar]
- 51.Trentin, B., N. Rebeyrotte, and R. Z. Mamoun. 1998. Human T-cell leukemia virus type 1 reverse transcriptase (RT) originates from the pro and pol open reading frames and requires the presence of RT-RNase H (RH) and RT-RH-integrase proteins for its activity. J. Virol. 72:6504-6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsurutani, N., M. Kubo, Y. Maeda, T. Ohashi, N. Yamamoto, M. Kannagi, and T. Masuda. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74:4795-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vincent, K. A., V. Ellison, S. A. Chow, and P. O. Brown. 1993. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J. Virol. 67:425-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vink, C., A. A. M. Oude Groeneger, and R. H. A. Plasterk. 1993. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type 1 integrase protein. Nucleic Acids Res. 21:1419-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, J. Y., H. Ling, W. Yang, and R. Craigie. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20:7333-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welker, E., W. J. Wedemeyer, and H. A. Scheraga. 2001. A role for intermolecular disulfide bonds in prion diseases? Proc. Natl. Acad. Sci. USA 98:4334-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiskerchen, M., and M. A. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hehl, G. V. Kalpana, V. Prasad, and J. C. Kappes. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73:2126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yi, J., E. Asante-Appiah, and A. M. Skalka. 1999. Divalent cations stimulate preferential recognition of a viral DNA end by HIV-1 integrase. Biochemistry 38:8458-8468. [DOI] [PubMed] [Google Scholar]
- 60.Yoder, K. E., and F. D. Bushman. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 62.Zheng, R., T. M. Jenkins, and R. Craigie. 1996. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. USA 93:13659-13664. [DOI] [PMC free article] [PubMed] [Google Scholar]