SUMMARY
Multimeric, ring-shaped molecular motors rely on the coordinated action of their subunits to perform crucial biological functions. During these tasks, motors often change their operation in response to regulatory signals. Here, we investigate a viral packaging machine as it fills the capsid with DNA and encounters increasing internal pressure. We find that the motor rotates the DNA during packaging and that the rotation per basepair increases with filling. This change accompanies a reduction in the motor’s step size. We propose that these adjustments preserve motor coordination by allowing one subunit to make periodic, specific, and regulatory contacts with the DNA. At high filling, we also observe the down-regulation of the ATP-binding rate and the emergence of long-lived pauses, suggesting a throttling-down mechanism employed by the motor near the completion of packaging. This study illustrates how a biological motor adjusts its operation in response to changing conditions, while remaining highly coordinated.
INTRODUCTION
Ring-shaped oligomeric molecular motors hydrolyze nucleoside triphosphates to drive the directed movement of substrates during essential cellular processes such as DNA translocation, protein unfolding, and proton gradient generation (Lyubimov et al., 2011; Singleton et al., 2007). To perform processive mechanical work, these motors must coordinate the activity of individual subunits that constitute the ring (Iino and Noji, 2013). Moreover, as they proceed through their biological tasks, these molecular machines often need to respond to environmental changes, such as fluctuating chemical concentrations (Berg, 2003), varying mechanical loads (Sen et al., 2013), and ancillary ligand interactions (Ilves et al., 2010). How ring motors modulate their mechanochemical cycle and intersubunit coordination in response to these factors is not fully understood.
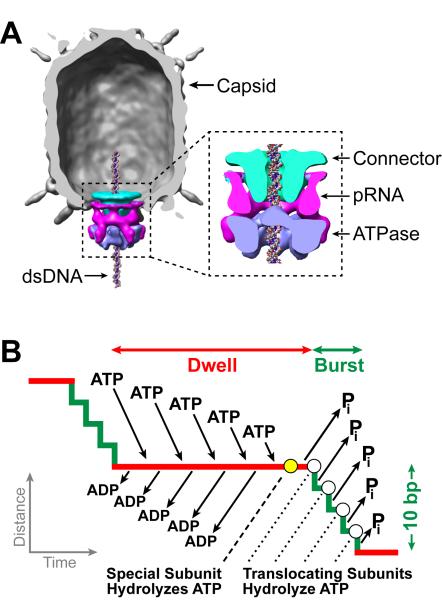
Double-stranded DNA (dsDNA) viruses, including tailed bacteriophages and eukaryotic herpes and adenoviruses, employ a ring-shaped packaging motor to pump the viral genome into a pre-formed protein capsid during viral self-assembly (Casjens, 2011; Hetherington et al., 2012; Rao and Feiss, 2008). To compact the stiff, highly-charged dsDNA to near-crystalline densities inside the capsid, the packaging motor needs to perform large amounts of mechanical work (Evilevitch et al., 2003; Gelbart and Knobler, 2009; Smith et al., 2001). The packaging motor of the Bacillus subtilis bacteriophage φ29 is a model system for investigating viral packaging and, more generally, for characterizing the operation of ring motors (Morais, 2012). This motor packages a 19.3-kilo-basepair (kbp) genome into a capsid that is 40 nm in diameter and 50 nm in height, and can exert forces beyond 60 pN (Smith et al., 2001). The motor complex consists of three coaxial rings through which the DNA is threaded into the capsid (Cao et al.; Morais et al., 2008): a dodecameric connector, a pentameric prohead-RNA (pRNA), and a homo-pentameric ATPase [gene product (gp) 16] that generates the driving force for genome packaging (Figure 1A). Our earlier single-molecule studies on the φ29 motor (Chemla et al., 2005; Chistol et al., 2012; Moffitt et al., 2009) have revealed a unique packaging mechanism in which each packaging cycle is composed of a dwell phase and a burst phase (Figure 1B). During the dwell phase, all five gp16 subunits release ADP and load ATP in an interlaced fashion. During the burst phase, sequential ATP hydrolysis and inorganic phosphate (Pi) release by four gp16 subunits result in the translocation of 10 bp of DNA in four 2.5-bp steps. Our previous data also suggest that, during the dwell phase, one of the ATPase subunits makes load-bearing, regulatory electrostatic contacts with adjacent pairs of backbone phosphates on one DNA strand that are spaced every 10 bp between consecutive cycles (Aathavan et al., 2009).
Figure 1. Overview of the φ29 Packaging Motor.
(A) Cryo-EM reconstruction of the φ29 capsid (gray), connector (cyan), pRNA (magenta), and gp16 ATPase (blue). Reproduced from (Morais et al., 2008), with permission from Elsevier.
(B) Mechanochemical model of the dwell-burst packaging cycle at low capsid filling.
Several key aspects regarding the operation of the φ29 packaging motor have not been elucidated. First, it has been long proposed that the DNA within the viral capsid organizes as a spool that may require the DNA to rotate relative to the capsid and/or the motor to relieve torsional strain (Earnshaw and Casjens, 1980; Hendrix, 1978). In addition, the difference between the 10-bp burst size of the motor and the 10.4-bp helical pitch of the B-form DNA (Wang, 1979) also suggests that the DNA may need to rotate relative to the motor in order to re-form the crucial electrostatic contacts after each dwell-burst cycle. Although these fundamental considerations have motivated great curiosity in simultaneously measuring DNA translocation and rotation, such information has not been experimentally accessible. Second, it was shown that packaging gradually slows down as the capsid fills with DNA (Smith et al., 2001), but the mechanism responsible for this phenomenon was not well understood. The reduction in packaging velocity has been explained by a model in which an internal pressure as high as ~100 atm builds up inside the capsid, slowing down and eventually stalling the motor toward the end of packaging (Rickgauer et al., 2008; Smith et al., 2001). However, this model relied on the untested assumption that the effect of DNA confinement on the motor dynamics is equivalent to that of an opposing force applied externally with optical tweezers. Moreover, it remained unclear how the details of motor operation and intersubunit coordination are modified by the increasing capsid filling.
Here we present experiments designed to characterize the rotation and translocation of the DNA being packaged by the motor throughout the entire range of capsid filling. Our data show that the motor indeed rotates the DNA during packaging. Moreover, we find that as the capsid fills and the packaging velocity drops, the motor simultaneously adjusts its step size and the amount of DNA rotation per cycle in order to retain its mechanism of intersubunit coordination and motor-DNA interaction. Finally, we uncover an intricate throttle control mechanism in which the motor modifies multiple parameters of its mechanochemical cycle to complete genome packaging.
RESULTS
The φ29 Motor Rotates DNA during Packaging
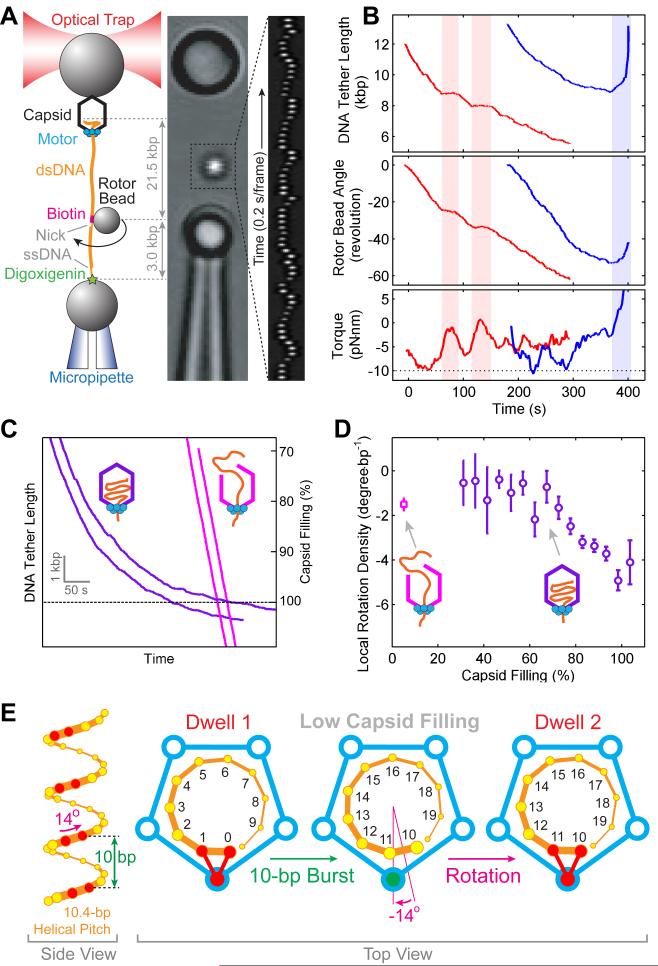
To determine whether the motor rotates DNA, we modified the standard two-bead optical tweezers packaging assay by introducing a third “rotor bead” (Bryant et al., 2003) (Figure 2A). This three-bead assay makes it possible to monitor changes in the angle of the DNA around its axis while simultaneously observing DNA translocation into the viral capsid. The tether was held under a constant tension of 10 pN so that the dsDNA was nearly completely extended and, thus, any twist applied to it during packaging was converted fully to rotation of the rotor bead (Strick et al., 1999). As the tether shortened due to DNA packaging, the rotor bead rotated about the DNA helical axis in a left-handed direction, denoted here as a negative angle (Figures 2A and 2B, and Movie S1). The rotation is ATP-dependent and correlated with packaging because pauses in translocation occur concomitantly with pauses in rotation (Figure 2B, red shades). The observation of DNA rotation during packaging disproves a recent conjecture that rejects the existence of such rotation in φ29 (Schwartz et al., 2013). Because of the large hydrodynamic drag of the rotor bead, its rotation angle lags behind the true rotation angle of the DNA. Since the torsional rigidity of the DNA is known, it is possible to derive the amount of rotation imposed on the DNA by the motor from the observed bead rotation (Figures S1A and S1B). We also calculated the torque stored in the DNA during packaging (Figure 2B, bottom panel). Remarkably, the stored torque can briefly reach −10 pN·nm, a value at which underwound DNA denatures (Bryant et al., 2003). Thus, besides being able to generate forces sufficient to overstretch dsDNA (Smith et al., 1996), the φ29 packaging motor is also able to overcome any torque load this motor may encounter in vivo.
Figure 2. The φ29 Motor Rotates DNA during Packaging.
(A) Left: Experimental geometry of the rotation assay. The packaging complex is tethered between two beads. Biotin-streptavidin linkages torsionally couple the rotor bead to the optically-trapped bead via dsDNA. A nick and a single-stranded DNA region ensure that the rotor bead is torsionally decoupled from the micropipette-bound bead. Middle: Micrograph of the experimental geometry. Right: Kymograph of the rotor bead position during packaging.
(B) Sample traces displaying DNA tether length (top), rotor bead angle (middle), and torque stored in the DNA (bottom) during packaging. Pauses in translocation are concomitant with pauses in rotation (shaded light red). Slipping (shaded light blue) causes a reversal in the rotation direction.
(C) Sample packaging traces with intact proheads (purple) and trepanated proheads (magenta).
(D) Local DNA rotation density versus capsid filling. The data point obtained with trepanated proheads – corresponding to very low capsid filling conditions – is shown as a magenta square. Error bars represent SEM.
(E) The geometric basis for DNA rotation at low capsid filling. Left: B-form DNA backbone diagram (only the 5′–3′ strand is shown). The φ29 motor forms specific contacts with pairs of phosphates (red) every 10 bases. Right: Top view of the motor (blue) and the DNA (orange). The same subunit contacts the DNA backbone phosphates in consecutive dwells. After a 10-bp burst, a 14° clockwise DNA rotation is needed to bring the DNA and the motor into perfect register.
We then measured the DNA rotation angle per unit length packaged (termed as rotation density, ρ) as a function of capsid filling. This analysis revealed that ρ increases in magnitude from ~ −1.5 to ~ −5 °/bp as packaging progresses from 50% to 100% filling (Figures 2C and 2D, purple), indicating that the magnitude of rotation is influenced by the amount of DNA confined inside the capsid. The alternative explanation that the change in rotation density is a sequence-dependent effect can be shown to be invalid (Figure S1C).
In order to decouple the motor’s intrinsic ability to rotate DNA from any twist imposed by the chiral arrangement of the DNA inside the capsid, it is necessary to measure rotation in the absence of intra-capsid DNA organization. The φ29 motor was occasionally observed to translocate amounts of DNA well in excess of the capsid capacity and without decelerating (Figure 2C, magenta traces), a phenomenon also seen in bacteriophage λ (Fuller et al., 2007). We surmise that these events correspond to ruptured capsids and we refer to them as “trepanated” proheads. We found that this rupture can be induced by repeatedly freezing and thawing the φ29 capsids prior to motor assembly. Motors on trepanated proheads were observed to rotate the DNA during packaging in the same direction as motors on intact proheads (Figure S1D). A constant rotation density of −1.5±0.2 °/bp was measured for trepanated proheads (Figure 2D, magenta square), in agreement with the ρ values from intact proheads at low filling (Figure 2D, purple circles). Importantly, this result indicates that the motor does actively rotate the DNA as part of its packaging mechanism, independent of any genome organization inside the capsid.
Furthermore, the measurement of the rotation density with trepanated proheads provides a clue to the coordination of the motor-DNA complex. After a 10-bp translocation burst, the nucleic acid backbone has only wound by 346°, i.e., 14° less than the amount needed to get the motor and the DNA back in register (Figure 2E). Therefore, a −14° rigid-body rotation would realign the DNA with the motor after every 10 bp packaged, yielding a rotation density of −1.4 °/bp, in agreement with the measured value of −1.5±0.2 °/bp. Given the previous evidence for contacts between the motor and a pair of adjacent backbone phosphates spaced every 10 bp along the DNA (Aathavan et al., 2009), our measurement is consistent with a model in which the motor rotates the DNA to align the (n+10)th phosphate with the same ATPase subunit that contacted the nth phosphate during the previous cycle (Figure 2E). Alternative scenarios where the DNA-contacting subunit changes around the ring from one cycle to the next predict rotation densities incompatible with the experimental data (Figures S1E and S1F).
Motor Coordination Is Preserved at High Filling
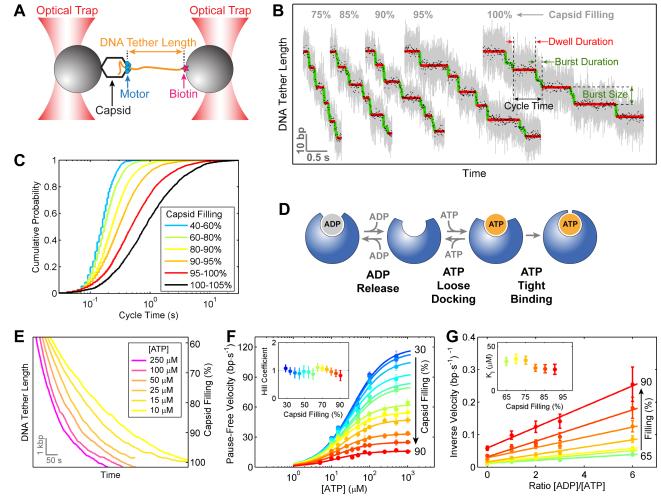
To understand what causes both the previously reported decrease in packaging velocity and the newly found change in DNA rotation density at high capsid filling (Figures 2C and 2D), we used dual-trap optical tweezers to monitor DNA translocation at various filling levels with basepair-scale resolution (Figure 3A). Previous experiments showed that at low capsid filling all five ring subunits must load ATP before any of them initiates hydrolysis, resulting in the dwell-burst partitioning of the packaging cycle (Chistol et al., 2012; Moffitt et al., 2009) (Figure 1B). Here we found that the biphasic dwell-burst structure is retained at high filling (Figure 3B), i.e., ATP hydrolysis events remain segregated from nucleotide binding events throughout the entire packaging process. However, the amount of time that the motor spends on each packaging cycle increases significantly, with the median cycle time rising from 0.15 s at 40% filling to 0.66 s at 100% filling (Figures 3B and 3C).
Figure 3. Motor Coordination Is Preserved at High Capsid Filling.
(A) Experimental geometry of the high-resolution packaging assay.
(B) Sample traces displaying individual packaging cycles at various levels of capsid filling. Raw 2500-Hz data are shown in gray and downsampled 100-Hz data in black. Stepwise fit to the data highlights dwells and bursts in red and green, respectively.
(C) Cumulative probability distribution of the packaging cycle times. Each color corresponds to a certain filling level.
(D) The nucleotide exchange scheme for each ATPase subunit.
(E) Sample packaging traces at low external loads (7–10 pN) and various ATP concentrations.
(F) Mean packaging velocity (pauses and slips removed) versus [ATP]. Each color represents a different filling level. The data are fit to the Hill equation, v = Vmax·[ATP]n/(KMn + [ATP]n). Inset: The Hill coefficient n from the fit.
(G) Inverse of the mean packaging velocity versus the ratio of [ADP] to [ATP] at different filling levels. [ATP] is fixed at 250 μM. The data are fit to a competitive-inhibition model, . Inset: The dissociation constant for ADP, KI, from the fit.
In panels F and G, error bars represent 95% confidence intervals (CI) estimated via bootstrapping.
See also Figure S2.
Our earlier studies have shown that the gp16 subunits within the ring release ADP and bind ATP in an interlaced fashion (Chistol et al., 2012) (Figure 1B), and that each ATP binding event is composed of two stages: a reversible loose-docking transition followed by an irreversible tight-binding transition (Moffitt et al., 2009) (Figure 3D). The two-stage ATP loading scheme gives rise to an apparent lack of ATP binding cooperativity between subunits (Hill coefficient n = 1) (Chemla et al., 2005; Moffitt et al., 2009). To test whether there is a filling-dependent change in this ATP loading sequence, we monitored DNA packaging at various ATP concentrations (Figure 3E). The average packaging velocity as a function of [ATP] at any filling level is well fit by the general Hill equation, v = Vmax·[ATP]n/(KMn + [ATP]n), with a near-unity Hill coefficient (Figure 3F). This result indicates that capsid filling does not affect the motor’s ATP-loading scheme in which the subunits bind ATP sequentially and binding events are separated by irreversible tight-binding transitions.
In the interlaced ADP-release/ATP-loading scheme (Figures 1B and 3D), there is at most one empty nucleotide-binding pocket in the ATPase ring at any given time. Thus, when ADP is added to the packaging buffer as an inhibitor, the inverse of the packaging velocity, 1/v, should be linearly proportional to the concentration of ADP (Chemla et al., 2005; Chistol et al., 2012). To test whether capsid filling affects the ADP-ATP exchange mechanism, we monitored packaging at a fixed concentration of ATP (250 μM) and various concentrations of ADP (0 to 1500 μM) (Figure S2). We observed a linear relationship between 1/v and [ADP] at all filling levels (Figure 3G), suggesting that the highly coordinated nucleotide exchange sequence is also retained at high capsid filling.
Capsid Filling Slows ATP Tight Binding
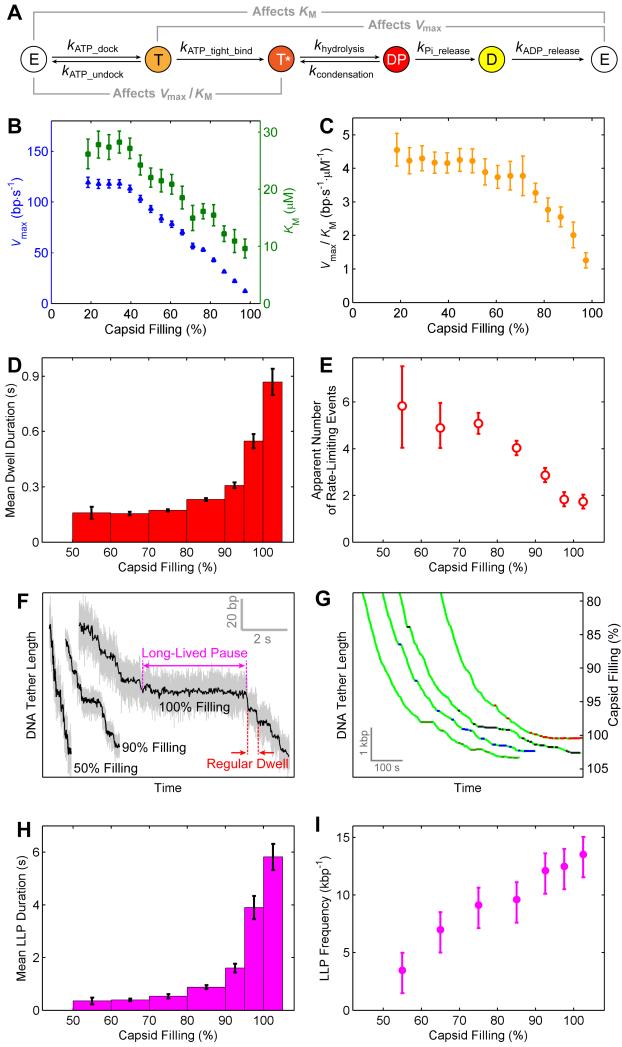
The results above indicate that the changes in the φ29 motor’s operation at high filling (i.e., slower translocation velocity and greater rotation density) are not caused by disruptions of the overall motor coordination; rather, they are most likely caused by modulations of certain kinetic transitions within the same mechanochemical framework (Figure 1B). To dissect which chemical rates (Figure 4A) are affected by capsid filling, we plotted the maximum velocity Vmax and the Michaelis constant KM as a function of filling (Figure 4B). Both Vmax and KM decrease as capsid filling increases. Their ratio Vmax/KM, which describes the effective ATP-binding rate constant (v ≈ (Vmax/KM)·[ATP] at limiting [ATP]) (Chemla et al., 2005; Visscher et al., 1999), also drops with filling (Figure 4C). The dependence of these kinetic parameters on capsid filling provides information about the identity of the chemical transition perturbed by DNA accumulation inside the capsid (Figure S3). In particular, the observed drop in Vmax/KM with capsid filling reflects a reduction of the effective ATP-binding rate. The value of Vmax/KM depends on the rates of all kinetic transitions reversibly connected to ATP binding, namely ATP docking (kATP_dock), undocking (kATP_undock), and tight binding (kATP_tight_bind) (Figure 4A) (Keller and Bustamante, 2000). Thus, the Vmax/KM drop could be caused by a decrease of kATP_dock, an increase of kATP_undock, or a reduction of kATP_tight_bind (Figures S3A–S3C), whereas changes in other chemical rates (hydrolysis, condensation, Pi release, ADP release) have no effect on this ratio (Figure S3D). Moreover, changes in docking or undocking rate alone only affect KM but not Vmax, whereas a decrease in the tight-binding rate causes Vmax, KM, and Vmax/KM to simultaneously drop (Figure S3A), as found experimentally (Figures 4B and 4C). Therefore, this analysis suggests that DNA filling of the capsid slows down ATP tight binding, a transition during which the ATPase commits to hydrolysis by making stronger contacts with the nucleotide (Chemla et al., 2005; Oster and Wang, 2000).
Figure 4. High Capsid Filling Slows ATP Tight Binding and Induces Long-Lived Pauses.
(A) The chemical cycle of each φ29 ATPase subunit.
(B) Vmax (blue) and KM (green) versus capsid filling.
(C) Vmax/KM versus capsid filling.
(D) Mean dwell duration versus capsid filling at saturating [ATP].
(E) Apparent number of rate-limiting kinetic events during the dwell versus capsid filling.
(F) Sample packaging traces at various filling levels highlighting regular dwells and long-lived pauses (LLPs).
(G) Sample traces highlighting the locations of LLPs. For clarity only LLPs longer than 5 s are shown. LLPs from different packaging complexes are colored green, blue, black, and red.
(H) Mean LLP duration versus capsid filling at saturating [ATP].
(I) Frequency of LLP occurrence versus capsid filling at saturating [ATP].
Error bars represent 95% CI.
See also Figures S3 and S4.
The above analysis suggests that the decrease of kATP_tight_bind is the predominant effect of capsid filling on the φ29 motor’s kinetic cycle. Thus, ATP-tight-binding events most likely become rate-limiting for the overall cycle at high filling. This notion is corroborated by the observation of a 6-fold increase in the mean dwell duration from 50% to 100% capsid filling (Figure 4D). In contrast, we recently showed that, at low filling and saturating [ATP], ADP release events are rate-limiting for the overall cycle (Chistol et al., 2012). Interestingly, we found that the apparent number of rate-limiting transitions, which was computed from the distribution of dwell durations (Extended Experimental Procedures), decreases from ~5 at 50% filling to ~2 at 100% filling (Figure 4E). This finding indicates that not all five ATPase subunits have the same ATP-tight-binding rate (or else the apparent number of rate-limiting transitions should remain 5), reflecting functional differences between the subunits of this homomeric ring ATPase. An apparent number of rate-limiting transitions of two could be caused by multiple combinations of ATP-tight-binding rates in the five subunits (Moffitt et al., 2010). In the simplest possible scenario, four subunits have equal tight-binding rates while the remaining subunit has a ~8× lower rate.
High Filling Induces Long-Lived Pauses
In addition to the increase in the duration of regular dwells, at high capsid filling a series of long-lived pauses (LLPs) punctuate processive DNA translocation at random locations along single-molecule packaging trajectories (Figures 4F and 4G). Both the average duration of LLPs and the frequency of their occurrence increase with filling (Figures 4H and 4I). Analysis of high-resolution packaging traces revealed that LLPs predominantly occur during the dwell phase of the mechanochemical cycle (Figures S4A and S4B). The probability of observing a regular dwell of such long duration is vanishingly small (Figures S4C and S4D). Thus LLPs represent kinetic processes distinct from dwells in regular packaging cycles. At high filling, these translocation-inactive processes contribute significantly to the reduction in the net packaging velocity (see Discussion).
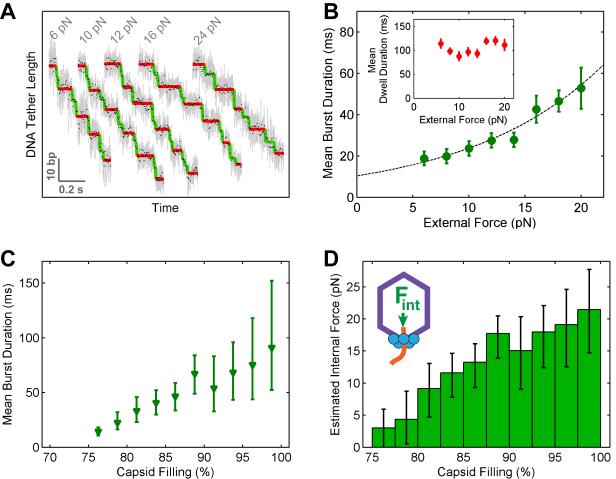
Burst Is Lengthened due to an Internal Force Up to 20 pN
It was previously proposed that the reduced packaging velocity at high capsid filling was caused by an internal force resisting packaging due to DNA confinement, Fint, whose effect was equivalent to the slowing down of the motor by an opposing force applied externally, Fext (Rickgauer et al., 2008; Smith et al., 2001). Because of the assumed equivalence between Fint and Fext, it was thought that the magnitude of the former could be inferred from the observed velocity dependence on capsid filling using the Fext-velocity calibration. In this way, it was estimated that Fint reaches 60 pN at 100% capsid filling (Smith et al., 2001), a value later revised to 110 pN (Rickgauer et al., 2008). The capability of resolving individual dwell-burst cycles enables us to dissect the effect of external force and that of capsid filling on motor operation in greater detail. We found that external force increases only the duration of the DNA-translocating burst phase, but has little effect on the duration of the stationary dwell phase (Figures 5A and 5B). In contrast, capsid filling prolongs both dwell and burst durations (Figures 4D and 5C). The distinct effects of external force and capsid filling on the dwell-burst cycle suggest that the observed slowdown at high filling is due only partially to the internal resisting force exerted by the confined DNA, and thus that the magnitude of the internal force must be reassessed.
Figure 5. Capsid Filling Modulates the Burst Duration.
(A) Sample packaging traces collected at low capsid filling (15–30%) and various external loads. Raw 2500-Hz data are shown in gray and downsampled 100-Hz data in black. Stepwise fit to the data highlights dwells and bursts in red and green, respectively.
(B) Mean burst duration versus external force at low capsid filling (15–30%). The data are fit to an Arrhenius-type equation, τburst(F) = τburst (0)·eFΔx/kT (dashed curve, Δx = 0.33±0.08 nm from the fit). Inset: Mean dwell duration versus external force.
(C) Mean burst duration versus capsid filling.
(D) The magnitude of the internal force as a function of capsid filling, obtained by applying the τburst-F curve (panel B) to the τburst-filling dependence (panel C).
Error bars represent 95% CI.
To revise the Fint estimate, we assumed that the burst duration depends only on the opposing force acting on the motor through DNA, either externally or internally. We can then use the Fext-burst duration calibration (Figure 5B) to estimate Fint responsible for lengthening the burst during capsid filling (Figure 5C). This revised analysis yields an internal force of only 20±7 pN at 100% filling (Figure 5D). Notably, this value is 3–5 times lower than those obtained from prior single-molecule experiments and is in better agreement with predictions from analytical modeling and numerical simulation studies (Evilevitch et al., 2004; Kindt et al., 2001; Purohit et al., 2005; Spakowitz and Wang, 2005).
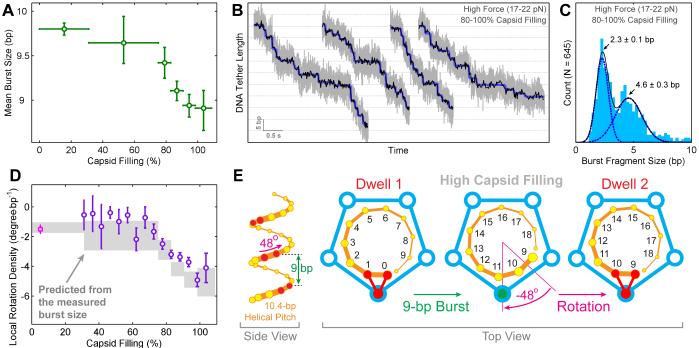
Smaller Burst Size Accounts for Larger Rotation Density at High Filling
Significantly, we found that the mean burst size, i.e., the average amount of DNA being translocated in each cycle, gradually decreases from 9.8±0.1 bp at <30% filling to 8.9±0.3 bp at 100% filling (Figure 6A). Our analysis indicates that the observed non-integer-basepair average burst size can arise from sub-populations of integer-basepair burst size (mainly 10-bp and 9-bp) whose relative proportions change with the amount of capsid filling (Figure S5A). Accordingly, the smooth decrease in the mean burst size is most likely due to shorter bursts becoming more frequent at higher filling. These shorter bursts appear to be composed of shorter steps rather than fewer steps, since the spatiotemporal structure of stepping within a burst does not change as the capsid fills (Figures S5B and S5C). To directly observe the steps that constitute a burst, we followed high-filling packaging against high external loads (~20 pN). As expected, high force slowed down DNA translocation during the burst and consequently we were able to detect steps of ~2.3 bp and integer multiples thereof (Figures 6B and 6C). This result strongly suggests that the 9-bp bursts observed at high filling and low force are still composed of four steps.
Figure 6. Capsid Filling Modulates the Step Size of the Motor without Affecting Subunit Coordination.
(A) Mean burst size versus capsid filling.
(B) Sample packaging traces at high capsid filling and high external loads revealing fragmented bursts. Raw 2500-Hz data are shown in gray, downsampled 150-Hz data in black, and stepwise fits in blue.
(C) Histogram of burst fragment sizes at high force and high filling. The distribution is well fit by two Gaussians that correspond to individual 2.3-bp steps and 4.6-bp burst fragments consisting of two consecutive 2.3-bp steps.
(D) DNA rotation densities inferred from the observed burst sizes (gray) and the measured rotation density values (purple and magenta) as a function of capsid filling.
(E) Illustration of how a smaller burst size results in a larger amount of DNA rotation (compare to Figure 2E).
Error bars represent 95% CI.
See also Figure S5.
To our knowledge, this is the first example of a molecular motor changing its step size during the course of its biological task. More importantly, this observation rationalizes the aforementioned change in DNA rotation density with capsid filling (Figure 2D). To explain the observed DNA rotation at low filling, we propose a model in which the motor rotates the DNA by −14° after each 10-bp burst in order to realign the same DNA-contacting subunit with two adjacent DNA backbone phosphates (Figure 2E). If this model is correct, a decrease in burst size from 10 bp to 9 bp should increase the DNA rotation from −14° to −48° per cycle (Figures 6D and 6E), corresponding to rotation densities of −1.4 °/bp and −5.3 °/bp respectively. The rotation density inferred from the decreasing mean burst size match well with the measured rotation density throughout the whole range of capsid filling (Figure 6D). These results suggest that, as the motor’s step size changes in response to the increasing capsid filling, the amount of DNA rotation by the motor changes accordingly. These concerted changes ensure that the same subunit interacts with the DNA backbone phosphates in successive cycles separated by an integer number of base pairs. Thus, the roles and coordination of all subunits – the one that contacts the DNA during the dwell and the four that do not – remain unchanged across all filling levels.
DISCUSSION
The φ29 Packaging Motor Employs a Throttle Control Mechanism
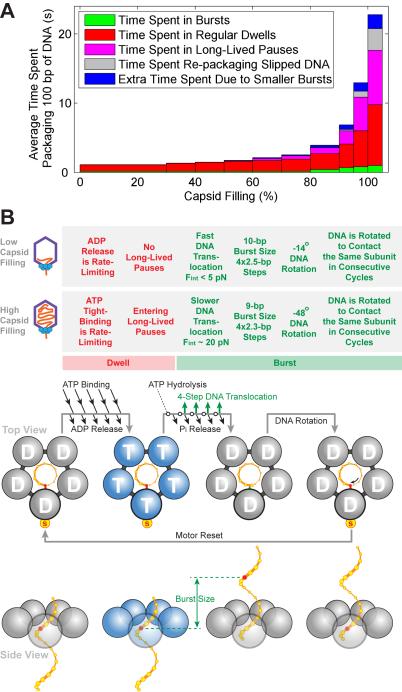
Viral packaging requires the motor to overcome major energetic barriers due to the compaction of DNA inside the capsid. The high-resolution data presented here both quantified those barriers and revealed how the motor adapts to them. It was known that the motor slows at high filling; here we show that the motor throttles down by modifying multiple aspects of its mechanochemical cycle: lengthening of the dwell phase, emergence of LLPs, lengthening of the burst phase, reduction in the burst size, and increased frequency of slipping (Figures 7A and 7B). Among these modifications, the lengthening of the dwell phase and the occurrence of LLPs are the major contributors to the drastic reduction in the packaging velocity at high filling (Figure 7A, red and purple). These diverse effects are allosteric in origin, possibly caused by the internal pressure changing the conformation of the capsid and/or connector, which in turn could cause the ATPase to alter its behavior. On the other hand, the internal force that resists packaging by pushing against the motor’s DNA-translocating motif has the same effect as tension applied externally to the DNA via optical tweezers. Namely, the internal force slows only the burst phase of the cycle – when DNA translocation occurs – thus having only a minor effect on the overall velocity (Figure 7A, green).
Figure 7. The φ29 Motor Adjusts Its Operation in Response to Increasing Capsid Filling.
(A) Average time needed to package 100 bp of DNA at different filling levels. Each color denotes a distinct mechanism that contributes to the slowing down of the motor as the capsid fills.
(B) Characteristics of packaging at low capsid filling and high capsid filling. The φ29 packaging cycle consists of a dwell phase (red) during which five ATPs are loaded sequentially, and a burst phase (green) during which four subunits translocate DNA. In each cycle, the motor rotates the DNA to form specific electrostatic contacts with the DNA backbone. These contacts anchor the motor onto the DNA during the subsequent dwell and determine the identity of the special subunit. Although here rotation is depicted to occur at the end of the burst, alternative scenarios where rotation occurs elsewhere during the cycle cannot be strictly ruled out. Several mechanisms modulate the motor operation at high filling while preserving the overall motor coordination: (1) The ATP-tight-binding rate is down-regulated, prolonging the dwell; (2) The motor enters the LLP state; (3) The duration of the force-sensitive burst phase is prolonged, most likely due to the rising internal force; (4) The step size of the motor decreases, which results in smaller bursts; (5) The amount of DNA rotation per cycle increases.
The precise measurement of the force-sensitive burst duration presented here leads to a new estimate of the internal force as a function of capsid filling. At 100% filling, the internal force is on the order of 20 pN, which corresponds to an internal pressure of ~20 atm. Internal pressure of this magnitude is at least partially counterbalanced by the cytoplasmic pressure in vivo and thus it may be insufficient to drive ejection of the entire viral genome into the host cell (Sao-Jose et al., 2007; Van Valen et al., 2012). Several other mechanisms have been proposed for the completion of viral genome delivery, such as hydrodynamic drag or cellular processes that pull the viral DNA into the host (Gonzalez-Huici et al., 2004; Molineux and Panja, 2013).
As the internal pressure builds during packaging, the frequency of long-lived pauses increases dramatically from essentially zero at low filling to ~1 LLP per 10 cycles at 100% filling (Figure 4I). What causes LLPs? Since large external loads that act on the ATPase channel along the DNA do not induce LLPs (Moffitt et al., 2009), it is unlikely that LLPs are caused by the direct application of an internal force on the ATPase central pore through the DNA. Instead, LLPs may reflect an allosteric communication between the tightly packed capsid and the packaging motor, which induces the motor to adopt a stalled conformation. We speculate that LLPs may allow the encapsidated DNA to relax and thus to avoid being trapped in an unfavorable configuration. Moreover, LLP-like processes may play a role in the “head-full” mechanism employed by some bacteriophages to halt translocation and trigger the endonuclease activity of their terminases when the capsid is full (Casjens and Weigele, 2005; Droge and Tavares, 2005; Lander et al., 2006).
Adjustable Step Size and DNA Rotation Maintain Robust Motor Coordination
Our recent study suggested that the φ29 homo-pentameric ATPase ring contains a special, non-translocating subunit that regulates DNA translocation by the other four subunits (Chistol et al., 2012). The symmetry-breaking mechanism that is responsible for such division of labor in a homomeric ATPase ring, however, has remained mysterious. The new data presented here provide insight into this mechanism: the asymmetric motor-DNA interaction during the dwell phase may represent the physical basis for the functional asymmetry observed during the burst phase. The subunit that anchors the motor onto the DNA backbone at the end of every burst is likely the one that adopts the non-translocating identity. In this picture, it is the DNA substrate that serves as the coordinating agent that breaks the symmetry of the ring.
Furthermore, we found that the motor adapts its burst size and the amount of substrate rotation to ensure that the same subunit makes specific electrostatic contacts with the DNA backbone phosphates every cycle. Remarkably, this coordination mechanism is maintained throughout the entire packaging process even as the packaging velocity drops by two orders of magnitude. While the identity of the DNA-contacting subunit may change during rare events such as slipping, the DNA rotation measurements suggest that during normal operation the motor tends to conserve the identity of the special, substrate-contacting subunit (Figures 2E and 6E).
Implications for Other Molecular Motors
In our model for the φ29 motor, the amount of substrate rotation is set by the geometry of the dsDNA and the variable burst size of the motor. Other ring translocases that operate on helical filament substrates may also recognize a periodic structural element in the substrate every cycle and thus generate substrate rotation. The magnitude of such rotation would depend on the step size and intersubunit coordination of the motor (Saleh et al., 2005). The mechanism described here, in which specific interactions between the motor and the polymeric substrate determine subunit coordination, may be a general feature of a broader class of ring ATPases.
A growing number of molecular motors are now being studied in mechanistic detail through in vitro biophysical methods. However, these experiments often remove the motor from its native biological context, thus becoming blind to the effects of crucial regulatory factors that modulate the activity of the motor. In this work, we examined a viral packaging motor at different stages of its biological task, and discovered the multiple and specific ways in which its mechanisms are modified in response to external signals. The strategy employed by this motor as documented here – utilizing a combination of flexibility and mechanistic invariance to allow robust activity across a range of changing conditions – may provide a blueprint for how other molecular motors can be efficiently regulated during the course of their in vivo tasks. Moreover, as demonstrated by our work, studies on ring ATPases that simultaneously track substrate translocation and rotation – that is, track the full three-dimensional trajectory of the substrate – should provide a more complete picture of the physical principles and biological designs underlying the operation of this family of molecular machines.
EXPERIMENTAL PROCEDURES
Preparation of Viral Components
Fiberless proheads, pRNA-free proheads, ATPase gp16, φ29 DNA-gp3 (gp3 is the terminal protein), and 120-base pRNA were produced and purified as described previously (Zhao et al., 2008). Trepanated proheads were generated by freezing and thawing 12.5 μl pRNA-free proheads comprising capsid and connector (166 nM) in 1× TMS buffer (50 mM Tris-HCl, pH 7.8, 10 mM MgCl2, and 100 mM NaCl) for 10 cycles at −20 °C and room temperature, respectively, and subsequently reconstituting the pRNA ring onto the prohead with fresh pRNA (1.6 mM). After this procedure, ~25% of the proheads appeared trepanated and showed nearly wild-type levels of tether formation efficiency and tether strength against external loads.
Preparation of DNA Constructs
In the rotation assay, the DNA substrate is a composite comprising viral DNA-gp3 to initiate packaging, a spacer DNA to allow the observation of full-genome-length packaging, a segment labeled with biotin, a spacer DNA with a nick to serve as a free swivel, and a single-stranded DNA (ssDNA) tail labeled with multiple digoxigenins (Figure 2A). The DNA construct is a product of three ligations. A PacI-linearized 13-kbp phagemid was extended by terminal transferase (New England Biolabs) in the standard reaction buffer plus 0.25 mM CoCl2 in the presence of digoxigenin-labeled (Roche) and unlabeled nucleotides (New England Biolabs), producing a 3′ ssDNA tail containing a few digoxigenins. That molecule was then digested with BamHI to yield two 6.5-kbp fragments and ligated to a dephosphorylated ~100-bp patch of biotinylated DNA generated by PCR as described previously (Bryant et al., 2003). After subsequent re-phosphorylation, the patch was ligated via an EcoRI site to another linearized, dephosphorylated, and bisected 11.6-kbp plasmid, which was in turn re-phosphorylated and ligated to the DNA-gp3 through the SpeI site.
In the high-resolution translocation assay, to generate DNA substrates shorter than 15 kbp, DNA-gp3 was digested with restriction enzymes (ClaI, XbaI, BstEII, or NcoI, New England Biolabs) and the overhang was filled in with biotinylated nucleotides (Invitrogen) using Klenow exo- (New England Biolabs). To generate DNA substrates longer than 15 kbp, NcoI-digested DNA-gp3 was ligated to a 2-kbp, 4-kbp, or 6-kbp biotinylated PCR product.
DNA Rotation Assay
Packaging complexes were assembled by mixing DNA, proheads, and gp16 as described previously (Smith et al., 2001). Assembled complexes were mixed with 250 μM ATP (Sigma-Aldrich) for 30 s to initiate packaging and then 250 μM ATPγS (Sigma-Aldrich) was added to stall the reaction. In the optical tweezers chamber filled with a stall buffer containing 0.5× TMS, 100 μM ATPγS, and 100 μM ATP, a 2.88-μm anti-capsid-antibody-coated polystyrene bead (Spherotech), previously incubated with the stalled complexes, was held next to a 2.1-μm anti-digoxigenin-coated polystyrene bead. The resulting tether was flow-extended, and a 0.88-μm streptavidin-coated polystyrene bead was optically manipulated to bind the internal biotinylated patch. After rotor bead attachment, the trap was released and returned to the anti-capsid bead. To restart packaging in the optical tweezers, buffer was exchanged for a packaging buffer containing 0.5× TMS and 500 μM (saturating) ATP. During packaging, the tether was held at a constant tension (10 pN) and tether extension was monitored as described before (Smith et al., 2003) while the rotor bead was tracked with bright-field video microscopy (Movie S1).
High-Resolution DNA Translocation Assay
The high-resolution translocation assay was performed using a dual-trap optical tweezers instrument (Moffitt et al., 2006). Stalled packaging complexes were assembled as described in the previous section. Tethers were formed between a 0.90-μm streptavidin-coated bead and a 0.88-μm anti-capsid-antibody-coated bead held in separate optical traps. The DNA tether’s contour length was calculated from the tension and extension using the worm-like-chain model of DNA elasticity (Bustamante et al., 1994). Trap stiffness and detector response were calibrated by fitting a modified Lorentzian to the fluctuation power spectrum of a trapped bead (Tolic-Norrelykke et al., 2004). DNA packaging experiments were performed in a semi-passive force feedback mode in which the DNA tether tension was kept in a small force range (Moffitt et al., 2009). The packaging solution contained 0.5× TMS, saturating ATP (unless specified otherwise), and an oxygen scavenging system (100 μg ml−1 glucose oxidase, 20 μg ml−1 catalase, and 5 mg ml−1 dextrose; Sigma-Aldrich).
Data Analysis
Detailed data analysis procedures are provided in the Extended Experimental Procedures.
The DNA rotation at the motor was derived from the motion of the rotor bead. The hydrodynamic drag torque Tdrag acting on the rotor bead as it rotates can be calculated from the fluid’s viscosity η, the rotor bead’s radius r, and its angular velocity ω: Tdrag = −14πηr3ω (Happel and Brenner, 1965). During packaging, hydrodynamic drag causes the angle of the rotor bead θbead to lag by an amount Δθlag behind the angle of the rotating DNA. This difference is absorbed by the DNA between the motor and the rotor bead (with a length L), which behaves as a torsional spring and generates an elastic torque Telastic = (C/L)Δθlag that drives the bead rotation. Here, C = 460 pN·nm2 is the torsional modulus of DNA (Bryant et al., 2003). Because at steady state the drag torque must be equal and opposite to the elastic torque, it is possible to estimate the angular lag Δθlag from 14πηr3ω = (C/L)Δθlag, and the actual DNA rotation angle from θDNA = θbead + Δθlag (Figure S1A).
To calculate the packaging velocity, raw data recorded at 2500 Hz were downsampled to 100 Hz and divided into slip-free and pause-free segments spanning 1–2 pN of force change. The packaging velocity was calculated by measuring the slope of a linear fit to the data. Packaging segments with clear motor stepping, evaluated by their pairwise distance distributions as described previously (Chistol et al., 2012), were used for further dwell-burst analysis. Dwells were detected using a modified Schwarz Information Criterion (mSIC) algorithm (Chistol et al., 2012). To measure the burst duration, the SIC algorithm was run in its original form (Kalafut and Visscher, 2008). Mini-dwells were identified during the burst phase, reflecting the finite duration of individual translocation events. The duration of a given burst was then computed as the sum of the durations of mini-dwells between two validated regular dwells. To distinguish LLPs from regular dwells, the left 90th percentile of the measured dwell duration distribution was fit to a Gamma distribution (Figures S4C and S4D). The long events unaccounted for by the Gamma distribution were designated as LLPs. The Gamma distribution fit also yielded the apparent number of rate-limiting events during the regular dwell.
Supplementary Material
Research Highlights.
The φ29 packaging motor rotates DNA during translocation.
High capsid filling slows ATP binding and induces motor pausing.
Concerted changes in step size and DNA rotation preserve motor coordination.
The internal force resisting DNA packaging reaches 20 pN at 100% capsid filling.
ACKNOWLEDGMENTS
We thank T. Lionberger, N. Liu, J. Moffitt, and M. Sen for critical readings of the manuscript; Z. Bryant, A. Lee, C. Lee, J. Moffitt, M. Nöllmann, and D. Reid for help with preliminary experiments and analysis; J. Kittleson for plasmids; and L. Comolli, A. Edelstein, Y. Mejia, M. Morais, and S. Smith for discussions. This work is supported in part by the U.S. National Institutes of Health under grants R01-GM071552 (C.B.) and R01-GM059604 (S.G.), the U.S. Department of Energy under contract number DEAC02-05CH11231 (C.B.), and the Howard Hughes Medical Institute (C.B.). S.L. acknowledges support from the NIH Pathway to Independence Award K99-GM107365.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes five figures, one movie, Extended Experimental Procedures, and Supplemental References.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–673. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- Bryant Z, Stone MD, Gore J, Smith SB, Cozzarelli NR, Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- Cao S, Saha M, Zhao W, Jardine PJ, Zhang W, Grimes S, Morais MC. Insights into the structure and assembly of the bacteriophage φ29 dsDNA packaging motor. J Virol. doi: 10.1128/JVI.03203-13. published online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Weigele P. DNA packaging by bacteriophage P22. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Landes Bioscience; Georgetown, TX: 2005. pp. 80–88. [Google Scholar]
- Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Chistol G, Liu S, Hetherington CL, Moffitt JR, Grimes S, Jardine PJ, Bustamante C. High degree of coordination and division of labor among subunits in a homomeric ring ATPase. Cell. 2012;151:1017–1028. doi: 10.1016/j.cell.2012.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droge A, Tavares P. Bacteriophage SPP1 DNA packaging. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Landes Bioscience; Georgetown, TX: 2005. pp. 89–101. [Google Scholar]
- Earnshaw WC, Casjens SR. DNA packaging by the double-stranded DNA bacteriophages. Cell. 1980;21:319–331. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- Evilevitch A, Castelnovo M, Knobler CM, Gelbart WM. Measuring the force ejecting DNA from phage. J Phys Chem B. 2004;108:6838–6843. [Google Scholar]
- Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Osmotic pressure inhibition of DNA ejection from phage. Proc Natl Acad Sci USA. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DE. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart WM, Knobler CM. Pressurized viruses. Science. 2009;323:1682–1683. doi: 10.1126/science.1170645. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Huici V, Salas M, Hermoso JM. The push-pull mechanism of bacteriophage phi29 DNA injection. Mol Microbiol. 2004;52:529–540. doi: 10.1111/j.1365-2958.2004.03993.x. [DOI] [PubMed] [Google Scholar]
- Happel J, Brenner H. Low Reynolds number hydrodynamics, with special applications to particulate media. Prentice-Hall; Englewood Cliffs, NJ: 1965. [Google Scholar]
- Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci USA. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington CL, Moffitt JR, Jardine PJ, Bustamante C. Viral DNA packaging motors. In: Goldman YE, Ostap EM, editors. Molecular Motors and Motility. Elsevier; Oxford, UK: 2012. pp. 420–446. [Google Scholar]
- Iino R, Noji H. Intersubunit coordination and cooperativity in ring-shaped NTPases. Curr Opin Struct Biol. 2013;23:229–234. doi: 10.1016/j.sbi.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalafut B, Visscher K. An objective, model-independent method for detection of non-uniform steps in noisy signals. Comput Phys Commun. 2008;179:716–723. [Google Scholar]
- Keller D, Bustamante C. The mechanochemistry of molecular motors. Biophys J. 2000;78:541–556. doi: 10.1016/S0006-3495(00)76615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt J, Tzlil S, Ben-Shaul A, Gelbart WM. DNA packaging and ejection forces in bacteriophage. Proc Natl Acad Sci USA. 2001;98:13671–13674. doi: 10.1073/pnas.241486298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol. 2011;21:240–248. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Bustamante C. Mechanistic constraints from the substrate concentration dependence of enzymatic fluctuations. Proc Natl Acad Sci USA. 2010;107:15739–15744. doi: 10.1073/pnas.1006997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc Natl Acad Sci USA. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineux IJ, Panja D. Popping the cork: mechanisms of phage genome ejection. Nat Rev Microbiol. 2013;11:194–204. doi: 10.1038/nrmicro2988. [DOI] [PubMed] [Google Scholar]
- Morais MC. The dsDNA packaging motor in bacteriophage phi29. In: Rossmann MG, Rao VB, editors. Viral Molecular Machines. Springer; New York: 2012. pp. 511–547. [Google Scholar]
- Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson DL, Rossmann MG. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure. 2008;16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster G, Wang H. Reverse engineering a protein: the mechanochemistry of ATP synthase. Biochim Biophys Acta. 2000;1458:482–510. doi: 10.1016/s0005-2728(00)00096-7. [DOI] [PubMed] [Google Scholar]
- Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondev J, Phillips R. Forces during bacteriophage DNA packaging and ejection. Biophys J. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- Rickgauer JP, Fuller DN, Grimes S, Jardine PJ, Anderson DL, Smith DE. Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage phi29. Biophys J. 2008;94:159–167. doi: 10.1529/biophysj.107.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh OA, Bigot S, Barre FX, Allemand JF. Analysis of DNA supercoil induction by FtsK indicates translocation without groove-tracking. Nat Struct Mol Biol. 2005;12:436–440. doi: 10.1038/nsmb926. [DOI] [PubMed] [Google Scholar]
- Sao-Jose C, de Frutos M, Raspaud E, Santos MA, Tavares P. Pressure built by DNA packing inside virions: enough to drive DNA ejection in vitro, largely insufficient for delivery into the bacterial cytoplasm. J Mol Biol. 2007;374:346–355. doi: 10.1016/j.jmb.2007.09.045. [DOI] [PubMed] [Google Scholar]
- Schwartz C, De Donatis GM, Zhang H, Fang H, Guo P. Revolution rather than rotation of AAA+ hexameric phi29 nanomotor for viral dsDNA packaging without coiling. Virology. 2013;443:28–39. doi: 10.1016/j.virol.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen M, Maillard RA, Nyquist K, Rodriguez-Aliaga P, Presse S, Martin A, Bustamante C. The ClpXP protease unfolds substrates using a constant rate of pulling but different gears. Cell. 2013;155:636–646. doi: 10.1016/j.cell.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- Smith SB, Cui Y, Bustamante C. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 2003;361:134–162. doi: 10.1016/s0076-6879(03)61009-8. [DOI] [PubMed] [Google Scholar]
- Spakowitz AJ, Wang ZG. DNA packaging in bacteriophage: is twist important? Biophys J. 2005;88:3912–3923. doi: 10.1529/biophysj.104.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick T, Allemand JF, Bensimon D, Lavery R, Croquette V. Phase coexistence in a single DNA molecule. Physica A. 1999;263:392–404. [Google Scholar]
- Tolic-Norrelykke IM, Berg-Sorensen K, Flyvbjerg H. MatLab program for precision calibration of optical tweezers. Comput Phys Commun. 2004;159:225–240. [Google Scholar]
- Van Valen D, Wu D, Chen YJ, Tuson H, Wiggins P, Phillips R. A single-molecule Hershey-Chase experiment. Curr Biol. 2012;22:1339–1343. doi: 10.1016/j.cub.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher K, Schnitzer MJ, Block SM. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- Wang JC. Helical repeat of DNA in solution. Proc Natl Acad Sci USA. 1979;76:200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Morais MC, Anderson DL, Jardine PJ, Grimes S. Role of the CCA bulge of prohead RNA of bacteriophage phi29 in DNA packaging. J Mol Biol. 2008;383:520–528. doi: 10.1016/j.jmb.2008.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.