Abstract
Transforming growth factors β (TGF-βs) inhibit growth of epithelial cells and induce differentiation changes, such as epithelial-mesenchymal transition (EMT). On the other hand, bone morphogenetic proteins (BMPs) weakly affect epithelial cell growth and do not induce EMT. Smad4 transmits signals from both TGF-β and BMP pathways. Stimulation of Smad4-deficient epithelial cells with TGF-β1 or BMP-7 in the absence or presence of exogenous Smad4, followed by cDNA microarray analysis, revealed 173 mostly Smad4-dependent, TGF-β-, or BMP-responsive genes. Among 25 genes coregulated by both factors, inhibitors of differentiation Id2 and Id3 showed long-term repression by TGF-β and sustained induction by BMP. The opposing regulation of Id genes is critical for proliferative and differentiation responses. Hence, ectopic Id2 or Id3 expression renders epithelial cells refractory to growth inhibition and EMT induced by TGF-β, phenocopying the BMP response. Knockdown of endogenous Id2 or Id3 sensitizes epithelial cells to BMP, leading to robust growth inhibition and induction of transdifferentiation. Thus, Id genes sense Smad signals and create a permissive or refractory nuclear environment that defines decisions of cell fate and proliferation.
Developmental choices between cell proliferation and differentiation are regulated by morphogenetic, growth, and transcription factors (1). Transforming growth factor β (TGF-β) represents a diverse family of secreted polypeptides that influence cell proliferation and differentiation during development and tissue homeostasis (34). Epithelial cell growth and differentiation are inhibited by TGF-β, which can also induce de-differentiation or epithelial-mesenchymal transition (EMT) (8). In contrast, bone morphogenetic proteins (BMPs) either do not affect or at high doses weakly inhibit epithelial cell growth while they stimulate mitogenesis on mesenchymal or neuronal cells (2, 30). Carcinoma cells may exhibit weak antiproliferative responses to BMPs (9, 10, 19).
TGF-βs and BMPs signal via the Smad pathway, in which Smad4 is a unique common effector (33). Smad4 is frequently mutated in human carcinomas, leading to loss of growth inhibition by TGF-β (32). Smad4-deficient (knockout or carcinoma) cells revealed an important role for Smad4 in growth inhibition and suggested alternative signaling pathways operating in parallel to Smads (7, 37, 38). The genes that control cell proliferation and differentiation by TGF-β are under intense investigation. Recent microarray screens identified many target genes, such as (i) genes of the epithelial cytostatic program of TGF-β1 (4, 14); (ii) genes of the mesenchymal extracellular matrix program of TGF-β1 (41); and (iii) genes of various epithelial, mesenchymal, or endothelial cell differentiation programs of TGF-β1, BMP-2/4, activin-A, or specific BMP receptors (3, 6, 26, 42, 45). In the third class, the genes encoding inhibitors of DNA binding/differentiation (Id genes) play prominent roles (25, 29).
Id proteins (Id1 to Id4) antagonize basic helix-loop-helix (bHLH) transcription factors and mediate mitogenic signals, inhibit differentiation, and play critical roles in animal development and cancer (29, 36, 44). Id1 is a well-established BMP target and regulates differentiation switches in osteoblasts, fibroblasts, epithelial cells, and endothelial cells (3, 5, 25, 40). Recently, TGF-β1-mediated repression of epithelial Id1 expression was suggested to be important for the cytostatic program of TGF-β (14). Among the other Id genes, Id2 and Id3 expression is induced by BMPs, but the physiological role for such regulation remains unknown (25). Thus, Id genes are regulated by TGF-β members in a cell-type-specific manner and play diverse and yet poorly understood physiological roles as targets of these pathways.
We applied transcriptomic analysis to measure the distinct and common gene targets of two major TGF-β members, TGF-β1 and BMP-7. Using mammary epithelial cells either lacking or expressing Smad4, we mostly identified Smad4-dependent targets, and among these, BMP-7 regulated a larger number of genes than TGF-β1. Among the coregulated genes, Id2 and Id3 exhibited exceptionally robust regulation. We establish the physiological role for such regulation by demonstrating that Id2 and Id3 define the epithelial context that allows proper growth and differentiation responses to specific TGF-β superfamily factors.
MATERIALS AND METHODS
Cells, viruses, proteins, and nucleic acids.
Human HaCaT keratinocytes, mouse mammary epithelial NMuMG, and human mammary carcinoma MDA-MB-468 cells have been described elsewhere (18). NMe cells, a clonal epithelial derivative of NMuMG, were from K. Vershueren, Leuven, Belgium, and α-TN4 murine lens epithelial cells were from J. Piatigorsky, Bethesda, Md. Primary normal human mammary epithelial cells and normal human epithelial keratinocytes were from Cambrex-Clonetics. Adenoviruses expressing LacZ, Flag-tagged wild-type Smad4, and its C-terminal deletion mutant (Smad4 515-ter) were from K. Miyazono, Tokyo, Japan; that expressing Flag-tagged wild-type Id3 was from P. ten Dijke, Amsterdam, The Netherlands, after permission from T. Taga, Kunamoto, Japan; myc-tagged wild-type Id2 was from F. D. Miller, Montréal, Canada; and the CAGA9-MLP-Luciferase promoter-reporter from S. Dooley, Aachen, Germany, and green fluorescent protein (GFP) were based on the Adeasy system. Adenoviruses were amplified and titrated in 293 cells as described previously (28). Recombinant mature TGF-β1 was from PeproTech EC Ltd. or N. Ferrara (Genentech, Inc.), and recombinant mature BMP-7 was from K. Sampath (Curis, Inc.). The human Id2-specific siRNA, 5′-CACGGATATCAGCATCCTG-3′ (sense strand), corresponds to bp 321 to 339 relative to the A (bp 1) of the start codon (accession no. M97796), and differs from the mouse Id2 cDNA by 1 bp. The human Id3-specific siRNA, 5′-TCCTACAGCGCGTCATCGA-3′ (sense strand), corresponds to bp 206 to 224 (accession no. X69111). For knockout of mouse mRNAs a set of two different siRNAs was used for each Id member. Accordingly, the two mouse Id2-specific siRNAs were: 5′-GCACGTCATCGATTACATC-3′ and 5′-TGTCGAATGATAGCAAAGT-3′ (both sense strands), which correspond to bp 267 to 285 and bp 443 to 461, respectively, of the mouse Id2 cDNA (accession no. BC053699). The two mouse Id3-specific siRNAs were, 5′-GCCTCTTGGACGACATGAA-3′ and 5′-GCTCACTCCGGAACTTGTG-3′ (both sense strands), which correspond to bp 173 to 191 and bp 366 to 384, respectively, of the mouse Id3 cDNA (accession no. NM_008321). siRNA against the luciferase reporter vector pGL2 (accession no. X65324) served as a control. All siRNAs were from Dharmacon Research, Inc. The human Id2 promoter-reporter construct pGId2-2750 was from A. Iavarone, New York, N.Y., and the mouse Id3 promoter construct p-389Id3CAT was from R. W. Lim, Columbia, Mo.
Transfections, infections, and gene reporter assays.
Transient transfections with promoter-reporter plasmids or siRNAs using Lipofectamine and Lipofectamine 2000 (Invitrogen) were according to manufacturer's protocols and as described previously (18). For luciferase reporter assays the enhanced luciferase assay kit was used according to the manufacturer's protocol (BD Pharmingen Inc.). Adenoviral transient infections using the multiplicity of infection (MOI) specified in the figures were performed as described previously (18).
cDNA microarray analysis.
Total RNA was isolated from adenovirus-infected MDA-MB-468 cells after mock treatment or stimulation with TGF-β1 or BMP-7 using the RNeasy kit (Qiagen). RNA sample (50 μg) pairs from mock-treated and factor-treated cells were labeled by reverse transcriptase with dCTP-Cy3 and dCTP-Cy5, respectively, and in the inverse order (dye swap). Equal amounts of labeled cDNA probes per pair were hybridized in triplicate to cDNA microarray chips (Hver1.2.1) from the Sanger/LICR/CRUK Consortium (for details and hybridization protocols, see http://www.sanger.ac.uk/Projects/Microarrays/), containing 9,932 spotted elements corresponding to 6,000 unique cDNAs. Chips were scanned in a Perkin-Elmer/GSI Lumonics ScanArray 4000 scanner, and spot intensity was analyzed by QuantArray (GSI Lumonics). Statistical analysis of the triplicate data sets was performed using GeneSpring 5.1 (Silicon Genetics). Raw data were normalized by nonlinear Lowess normalization. Regulated genes were selected based on average ratio values of ≥1.7 (up-regulation) and ≤0.59 (down-regulation) and had to be expressed in all three replicates (intensity of the gene spot exceeded the background intensity plus 3 standard deviations based on the intensity of all spots). In each replicate, the ratio was >1.2 (up-regulation) or <0.83 (down-regulation). These values were based on the standard deviation of the ratio for all expressed genes. Finally, regulated genes had to score significantly in a t test (P for ratios within triplicates, <0.05). Description of the genes was based on the Hver1.2.1_NCBI31 annotation list from the Sanger Center. All files containing the raw data of the microarray analysis have been deposited to ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) at the European Bioinformatics Institute, Hinxton, United Kingdom, and are publicly available under accession no. E-MEXP-78".
RT-PCR.
Total DNA-free cellular RNA was extracted with the RNeasy kit (Qiagen). Reverse transcription (RT)-PCRs were performed as described previously (18). Specific primers were designed according to sequences available in the databanks or published by others (Table 1). Glyceraldehyde-3′-phosphate dehydrogenase (GAPDH) served as control for equivalent cDNA synthesis. RT controls (no SuperSript II in reaction) and PCR controls (H2O instead of cDNA) were performed in every reaction set and never yielded amplified DNA fragments (unpublished results).
TABLE 1.
Oligonucleotide primers used for RT-PCR
| Gene | Primer sequence | Strand | Product size (bp) | Temp (°C) | PCR cycle | Reference |
|---|---|---|---|---|---|---|
| Human Id2 | 5′-ACGACCCGATGAGCCTGCTA-3′ | + | 213 | 55 | 30 | 22 |
| 5′-TCCTGGAGCGCTGGTTCTG-3′ | − | |||||
| Human Id3 | 5′-TGAGCTTGCTGGACGAC-3′ | + | 571 | 51 | 24 | 22 |
| 5′-CCTTGGCATAGTTTGGAGAG-3′ | − | |||||
| Mouse Id2 | 5′-GCATCCCCCAGAACAAGAAGGT-3′ | + | 451 | 57 | 28 | NM_010496 |
| 5′-CCAGGCCGGAGAACAAGACAC-3′ | − | |||||
| Mouse Id3 | 5′-GGTGCGCGGCTGCTACGAG-3′ | + | 555 | 63 | 28 | NM_008321 |
| 5′-CAGGCCACCCAAGTTCAGTCC-3′ | − | |||||
| GATA2 | 5′-CCCCATCCCCACCTACCCCTCCTA-3′ | + | 504 | 62 | 31 | M77810 |
| 5′-TCCGCCCCTTTCTTGCTCTTCTTG-3′ | − | |||||
| OVOL1 | 5′-GCGTGCGGCCCTACAAGTG-3′ | + | 277 | 62 | 31 | NM_004561 |
| 5′-TGCAGCAGGGAAGTGACAGTG-3′ | − | |||||
| p21 | 5′-CTGCCCAAGCTCTACCTTCC-3′ | + | 123 | 57 | 30 | 31 |
| 5′-CAGGTCCACATGGTCTTCCT-3′ | − | |||||
| c-myc | 5′-ACCCGGACGACGAGACCTTCATCA-3′ | + | 683 | 63 | 26 | 18 |
| 5′-GGGGCTGGTGCATTTTCGGTTGTT-3′ | − | |||||
| Fibronectin | 5′-TGGAACTTCTACCAGTGCGAC-3′ | + | 450 | 59 | 33 | 13 |
| 5′-TGTCTTCCCATCATCGTAACAC-3′ | − | |||||
| Smad6 | 5′-GACCAGTACAAGCCACTGGA-3′ | + | 628 | 54 | 37 | 39 |
| 5′-ATCTGTGGTTGTTGAGTAGG-3′ | − | |||||
| Smad7 | 5′-CAGATTCCCAACTTCTTCTG-3′ | + | 533 | 53 | 40 | 39 |
| 5′-GTTGAAGATGACCTCCAGCC-3′ | − | |||||
| GAPDH | 5′-ATCACTGCCACCCAGAAGAC-3′ | + | 443 | 57 | 30 | 39 |
| 5′-ATGAGGTCCACCACCCTGTT-3′ | − |
Immunoblot-immunofluorescence analysis.
Immunoblotting of total protein extracts was carried out as described previously (18). Densitometric protein analysis was performed using the AIDA software (version 3.10.039; FujiFilm). The specific protein bands from the immunoblots were normalized to the constant internal control (β-tubulin), and the values were plotted in bar graphs. For immunofluorescence, cells were infected with adenoviruses and then treated with TGF-β1 or BMP-7 as indicated in the figure legends; rabbit polyclonal anti-Id2 (C-20), anti-Id3 (C-20), anti-myc (9E10), and anti-Smad4 (H-552), were from Santa Cruz Biotechnology, Inc.; mouse monoclonal anti-Flag (M2 and M5), anti-smooth muscle actin (anti-SMA) (A2547), antivimentin (clone V9, V6630), and anti-β-tubulin (T8535) were from Sigma-Aldrich; mouse monoclonal anti-E-cadherin (C20820) was from BD Transduction Laboratories; rat monoclonal anti-ZO-1 (MAB1520) was from Chemicon International; and rabbit polyclonal anti-Smad2 (MLP) was produced in-house. All photomicrographs were obtained by a Zeiss Axioplan 2 microscope with a Hammamatsu C4742-95 digital camera, using the Zeiss Plan-neofluar 40×/0.75 objective lens and photographing at ambient temperature in the absence of immersion oil. Image memory content was reduced and brightness and contrast were adjusted using Adobe Photoshop after image acquisition with the camera's QED software.
Thymidine incorporation assays.
After a 24-h adenoviral infection in the presence of 3% fetal bovine serum (Sigma-Aldrich), MDA-MB-468 cells were treated with vehicle, TGF-β1, or BMP-7 for 18 h, during the last 2 h of which [3H]thymidine (1 μCi/ml; NEN) was added. For Id2/3 studies in MDA-MB-468, HaCaT, or NMe cells, cultures were stimulated 72 h after infection in the absence of serum or 24 h after siRNA transfection. Thus, thymidine incorporation was analyzed between 42 and 90 h postinfection or posttransfection and 18 h post-factor addition, when monolayers were 50 to 70% confluent. For dose-response experiments, the same protocol was used except that three different doses of TGF-β1 were used. Assays were performed as previously described (18). Data are plotted as averages with standard errors of triplicates for each condition per independent experiment. Independent experiments were repeated at least twice. Statistical analysis was performed by two-tailed paired Student's t test (significance, P ≤ 0.02).
RESULTS
Restoration of Smad4 function and TGF-β/BMP signaling.
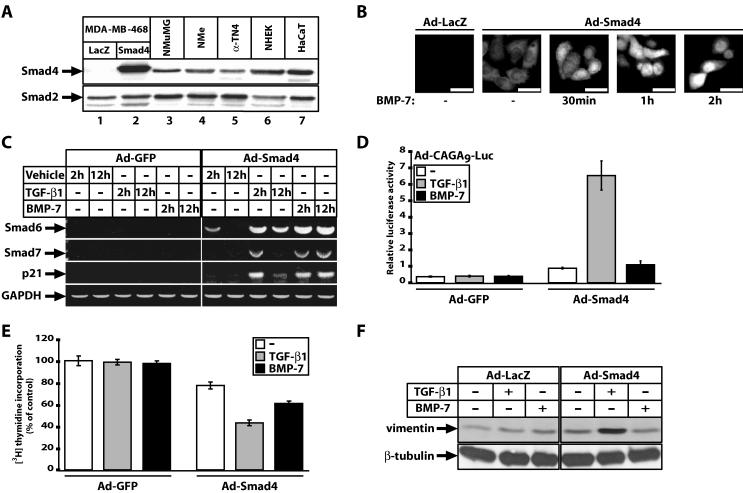
Restoration of TGF-β1 signaling by stable transfection of wild-type Smad4 into MDA-MB-468 cells has been previously described (7). However, it was important for us to additionally validate the adenoviral method of gene delivery, the BMP-7 response, and additional physiological responses than those previously characterized. The MOI of Ad-Smad4 chosen to infect MDA-MB-468 monolayers was a compromise between protein expression levels and efficiency of infection. Under standardized conditions, ectopic Smad4 levels were two- to threefold higher than those of several primary and established normal epithelial cell lines that express endogenous levels of Smad4 (Fig. 1A). As a negative control, we infected cells with adenoviruses expressing LacZ or GFP. Immunofluorescence experiments verified that the reconstituted Smad4 localized diffusely in both cytoplasm and nuclei of infected cells, and upon stimulation with either TGF-β1 or BMP-7 for 30 min to 2 h it rapidly accumulated in the nucleus, as expected (Fig. 1B and unpublished results).
FIG. 1.
Smad4 reconstitutes multiple physiological responses to TGF-β1 and BMP-7 in MDA-MB-468 cells. (A) Relative Smad4 protein expression levels in MDA-MB-468 cells infected with control Ad-LacZ (lane 1) and specific Ad-Smad4 (lane 2) adenoviruses, compared to endogenous levels of Smad4 from five epithelial cell lines (lanes 3 to 7) using the same anti-Smad4 antibody. Endogenous Smad2 expression levels serve as controls. (B) Proper nuclear translocation of infected Smad4 protein in response to BMP-7 (300 ng/ml) for the indicated times in adenovirus-infected cells with control Ad-LacZ and specific Ad-Smad4 viruses and processed with anti-Flag antibody against the Smad4 epitope tag. Bars in all photomicrographs represent 10 μm. (C) Proper regulation of endogenous Smad6, Smad7, and p21 mRNA levels in response to growth factor dilution buffer (vehicle), TGF-β1 (0.5 ng/ml) or BMP-7 (300 ng/ml) for the indicated time periods, after restoration of Smad4 levels, as revealed by semiquantitative RT-PCR assays, using GAPDH as control. (D) Responsiveness of a Smad3/Smad4-specific synthetic promoter (CAGA9)-luciferase reporter to control vehicle (white bars), TGF-β1 (5 ng/ml) (grey bars), or BMP-7 (300 ng/ml) (black bars) for 24 h, after adenoviral infection of the reporter construct together with control Ad-GFP or specific Ad-Smad4 adenovirus. Average values with standard errors (error bars) from triplicate determinations are shown. (E) Relative levels of [3H]thymidine incorporation into nuclei of cells infected with control Ad-GFP or specific Ad-Smad4 adenovirus prior to serum starvation and stimulation with vehicle (-, white bars), TGF-β1 (0.5 ng/ml) (grey bars), and BMP-7 (300 ng/ml) (black bars). The data are expressed as percentages relative to the control vehicle condition in the presence of Ad-GFP and represent averages with standard errors (error bars) derived from triplicate measurements. (F) Regulation of the fibroblastic marker vimentin indicates epithelial cells undergoing fibroblastoid differentiation after infection with control Ad-LacZ or specific Ad-Smad4 adenovirus and stimulation with vehicle (−), TGF-β1 (5 ng/ml), or BMP-7 (300 ng/ml). The β-tubulin levels serve as control.
The reconstituted Smad4 also restored proper immediate-early gene responses of the inhibitory Smads, Smad6 and Smad7, or the cell cycle inhibitor p21Cip1, to TGF-β1 and BMP-7 (Fig. 1C). Smad6 and Smad7 represent genes activated by Smads as part of a negative feedback loop as their protein products bind to TGF-β/BMP receptor complexes and inhibit Smad phosphorylation and oligomerization with Smad4, or they induce receptor ubiquitination and degradation (33). The cell cycle inhibitor p21Cip1 is known to participate in the growth-inhibitory response of epithelial cells and is also regulated by the Smad pathway (38).
A specific synthetic promoter (CAGA9) that potently responds to Smad3 and Smad4 due to their direct binding to its multimerized Smad binding elements, showed a clear and robust response only after Smad4 reconstitution and stimulation with TGF-β1 (Fig. 1D). BMP-7 fails to induce this promoter-reporter as previously demonstrated (28). We also measured more global cellular responses to TGF-β1 and BMP-7, such as proliferation and differentiation of the MDA-MB-468 cells in the absence and presence of reconstituted Smad4 (Fig. 1E and F). As expected, only upon reconstitution of Smad4 did TGF-β1 and BMP-7 lead to a reproducible inhibition of proliferation (Fig. 1E). As predicted for epithelial tumor cells, the TGF-β1 response was stronger than the BMP-7 response. Using the protein vimentin as a marker of EMT, we could also detect that only upon Smad4 reconstitution and stimulation with TGF-β1 the vimentin levels increased significantly in the cells, indicating a more fibroblastoid differentiation state (Fig. 1F). BMP-7 was rather inert with respect to this response despite the reconstitution of Smad4, data reproducing results obtained in another mammary epithelial cell line, NMuMG, which exhibits more overt EMT (28). We conclude that Smad4 reintroduction into MDA-MB-468 mammary carcinoma cells restores multiple physiological responses to TGF-β1 and BMP-7. We decided that it was meaningful to characterize novel gene targets of the two signaling pathways in this cell system.
Transcriptomic analysis of TGF-β1 and BMP-7-responsive genes.
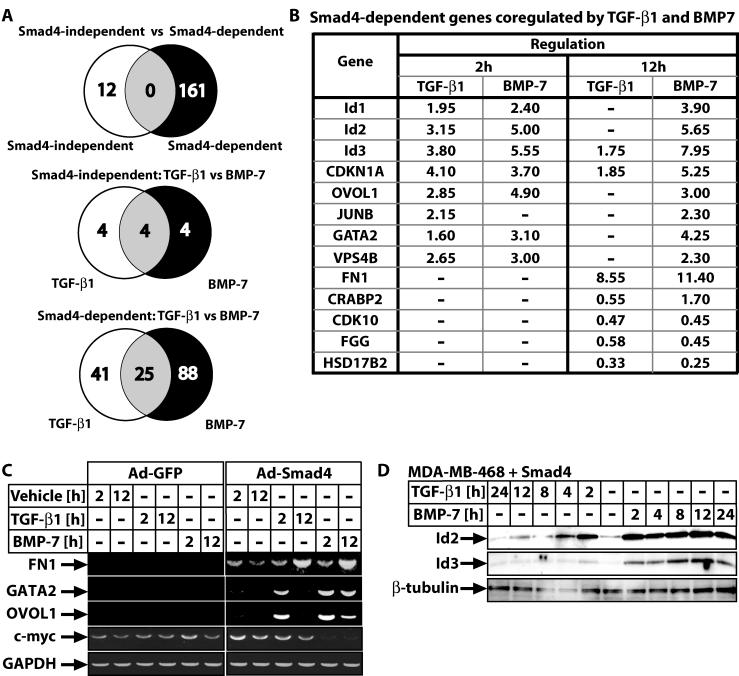
To monitor expression of large numbers of genes in MDA-MB-468 cells we performed cDNA microarray analysis using human arrays (Hver1.2.1; Sanger Center) with 9,932 elements corresponding to 6,000 unique cDNAs. To distinguish between the Smad4-independent and Smad4-dependent gene responses we used mock GFP-infected or Smad4-infected cells in the absence or presence of stimulation with TGF-β1 or BMP-7 for 2 h (early response) and 12 h (late response) (Fig. 2A). The screen identified 173 highly regulated genes, of which 39 currently lack functional annotation and the rest 134 have known function and were clustered in 6 functional groups (Fig. 2). Among these, 46% exhibited positive regulation and 11% showed negative regulation, whereas the rest 43% did not change significantly at the early time point. In contrast, at the late time point, 70% showed induction, 23% showed repression and 7% remained unaltered. Many of the obtained genes have not been previously linked to TGF-β superfamily signals.
FIG. 2.
Gene clusters of highly regulated genes in MDA-MB-468 cells. (A) Experimental design of the microarray experiment defines the Smad4-dependent and -independent gene response. (B) Clusters of TGF-β1- and BMP-7-responsive genes in the absence of Smad4. (C) Clusters of Smad4-responsive genes in the absence of growth factor stimulation. (D) Clusters of TGF-β1-responsive genes in the presence of Smad4. (E) Clusters of BMP-7-responsive genes in the presence of Smad4. (B to E) Gene expression data are shown based on a color and brightness scale (with a scale of 0 to 10 arbitrary units) (E), for the control (−) and 2- and 12-h time points, or the absence (−) or presence (+) of Smad4, followed by the corresponding gene number and name as defined in the human genome by Ensembl. “Unknown” stands for genes with fully unknown annotation, whereas “EST” stands for nonannotated genes identified at least once as expressed sequence tags. Unclassified genes are those that failed to group to a clear functional group.
The Smad4-independent group of genes was dramatically small (12 genes) for both ligands tested (Fig. 3A). Most (83%) of the Smad4-independent gene targets represent cDNAs without current annotation (Fig. 2B). Among these genes, four were regulated by TGF-β1, four were regulated by BMP-7, and four were regulated by both (Fig. 3A). Reconstitution of Smad4 in the absence of ligand stimulation also led to a small but significant group of gene responses (16 induced genes in total), which were mostly (63%) part of the TGF-β1 or BMP-7 response (Fig. 2C). This attests to the presence of autocrine TGF-β/BMP signaling in the carcinoma cells. The genes of this group represent those most sensitive and highly regulated by the two ligands studied and included six unique genes, half of which belong to the metallothionein family, a fact of currently unknown biological importance. Finally, the most informative profiles were those obtained after stimulation of wild-type Smad4-reconstituted cells with either ligand (Fig. 2D and E). We identified 66 TGF-β1 gene targets and 114 BMP-7 targets (Fig. 3A), which included previously characterized genes such as p21Cip1, JunB, c-myc, and those encoding fibronectin and tissue inhibitor of metalloproteases 3. The BMP-7 genomic program listed 1.7-fold more members than the TGF-β1 program, and this enhanced response was evident in all six functional categories. A significant number of unique genes to each ligand were identified, 41 TGF-β1-specific and 88 BMP-7-specific genes (Fig. 3A). The difference between the two profiles must be understood at the level of individual genes or differential modes of regulation of the same genes. The TGF-β1 genomic response is more rapid (74% of genes regulated at the early time point), whereas the BMP-7 response is more delayed and sustained (only 55% of genes regulated at the early time point).
FIG. 3.
Summary and validation of highly regulated Smad4-dependent genes in MDA-MB-468 cells. (A) Venn diagrams of gene expression itemized in three ways. The two gene groups are colored white or black and their intersection grey. (B) Top list of genes coregulated by TGF-β1 and BMP-7 in a Smad4-dependent manner. Gene names use Ensembl nomenclature and values represent average change of mRNA levels estimated by microarray analysis (−, not statistically significant changes). (C) Semiquantitative RT-PCR for four selected genes and control GAPDH in cells infected with control Ad-GFP or specific Ad-Smad4 adenovirus and treated for the times shown with vehicle, TGF-β1 (0.5 ng/ml), or BMP-7 (300 ng/ml). (D) Immunoblot analysis of endogenous Id2 and Id3 proteins in MDA-MB-468 cells reconstituted with wild-type Smad4 after transient adenoviral infection. Time course stimulation for the indicated time points with TGF-β1 (0.5 ng/ml) and BMP-7 (300 ng/ml). The specific protein bands and the β-tubulin control are shown with arrows.
The Smad4-dependent response included 25 genes regulated by both ligands, and among these genes are the most sensitive targets, cell cycle regulator and transcription/differentiation factor genes (Fig. 3A and B, which shows the top 13 coregulated genes). To validate the rigidity of the microarray data we confirmed the regulation of nine genes at the mRNA and protein level (see Fig. 1C for p21 mRNA; Fig. 3C for fibronectin 1 [fn1], gata2, ovol1, c-myc mRNAs; Fig. 3D for Id2 and Id3 protein; and unpublished results). Overall, we observed close concordance between microarray data and RT-PCR or protein data, with the microarray analysis underestimating the amplitude of gene response.
We also performed an additional specificity control in order to verify that the gene expression measured was indeed due to the wild-type Smad4 transgene and not due to a random effect of the specific adenovirus during the infection process. For this, we infected cells with a Smad4 mutant, Smad4(515-ter), derived from human cancer cells and whose nonsense mutation at residue 515 leads to a short C-terminal truncation and complete loss of function (24). The gene expression profile generated by reconstitution of this mutant into MDA-MB-468 cells is essentially identical to that of mock-GFP reconstitution. No specific gene regulation could be measured by this mutant after stimulation with TGF-β1 or BMP-7 (unpublished results). In the absence of ligand stimulation, 19 genes regulated only by the presence of Smad4(515-ter) were found. While 16 of these genes were unique, only 3 overlapped with genes regulated by wild-type Smad4. This experiment confirmed the essential role of Smad4 in TGF-β signal transduction and suggested that mutations accumulating in human cancers primarily function by blocking the normal flow of these pathways. However, such cancer mutants can induce a limited number of novel and TGF-β-independent transcriptional events, at least under conditions of ectopic expression.
Id2 and Id3 genes are differentially regulated by TGF-β1 and BMP-7.
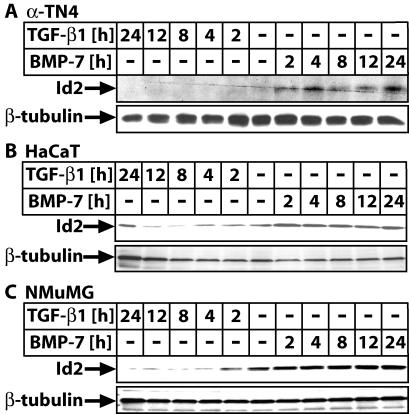
Among the most highly coregulated genes were Id2 and Id3 (Fig. 3B), which have not been previously studied extensively in epithelial cells responding to TGF-β or BMP, and thus it was of interest to understand why these signaling pathways targeted Id genes. Detailed mRNA and protein expression analysis in several epithelial cell types allowed us to test the generality of mode and kinetics of Id2 and Id3 regulation (Fig. 3D and 4; also see Fig. S1 and S2A in the supplemental material). Several noteworthy characteristics were observed. First, the TGF-β1 response was mostly bimodal, with an early mRNA induction phase and a late phase of downregulation and eventually repression below the background levels, while the BMP-7 response was continuously sustained and positive. Second, the early mRNA induction phase by TGF-β1 was primarily observed in primary human mammary epithelial cells (see Fig. S1A in the supplemental material), normal human epithelial keratinocytes (see Fig. S1B in the supplemental material), or immortalized normal lens epithelial cells α-TN4 (see Fig. S1C in the supplemental material). The same profile of mRNA and protein kinetics was observed in MDA-MB-468 cells in which we performed the microarray screen (Fig. 3B and D). In immortalized normal HaCaT (see Fig. S1D in the supplemental material) and immortalized normal NMuMG (see Fig. S2A in the supplemental material) cells, this transient induction phase was not as easily detectable or possibly shorter than our earliest 2-h time point. The latter is in agreement with similar studies recently reported by others (14, 21, 35). However, the bimodal kinetics of Id2 and Id3 mRNA regulation by TGF-β1 was not directly translated at the protein level in most cell types (Fig. 4), with the exception of the MDA-MB-468 cells (Fig. 3D), in which mRNA and protein levels showed faithful concordance. Thus, it is rather difficult at this point to reconcile the physiological significance of this early and transient regulation of Id2/3 mRNA levels. The BMP-7 response is more universally conserved between epithelial and mesenchymal cells since the profiles shown here resemble those recently reported for the Id1 gene in osteoblasts (15, 17, 23).
FIG. 4.
Epithelial Id2 exhibits sustained induction by BMP-7 and repression by TGF-β1. (A to C) Immunoblot analysis of Id2 and control β-tubulin in α-TN4 (A), HaCaT (B), and NMuMG (C) cells treated for the times shown with vehicle (−), TGF-β1 (5 ng/ml), or BMP-7 (300 ng/ml).
In order to test whether the early response of these genes was direct or whether it required protein synthesis, we inhibited ribosomal activity using cycloheximide prior to cell stimulation with TGF-β1 or BMP-7 for a short period of 0 to 4 h (see Fig. S2 in the supplemental material). Repression of Id2/3 expression by TGF-β1 and induction by BMP-7 could occur even in the presence of cycloheximide, albeit not as potently as under control conditions. This is primarily due to the strong positive effect cycloheximide has on Id2/3 mRNA expression in the absence of stimulation with growth factors. Finally, we tested the responsiveness of two promoter constructs, a 2.8-kbp human Id2 (20) and a 389-bp mouse Id3 (43). We observed proper regulation of the Id2 promoter by the two ligands in several of the above cell lines, while we failed to record regulation of the short Id3 promoter (unpublished results; also see Fig. S2B in the supplemental material). These results are in agreement with the involvement of direct signaling effectors of TGF-β/BMP, the Smads, together with indirect cooperating transcription factors in Id gene regulation (14, 17, 35).
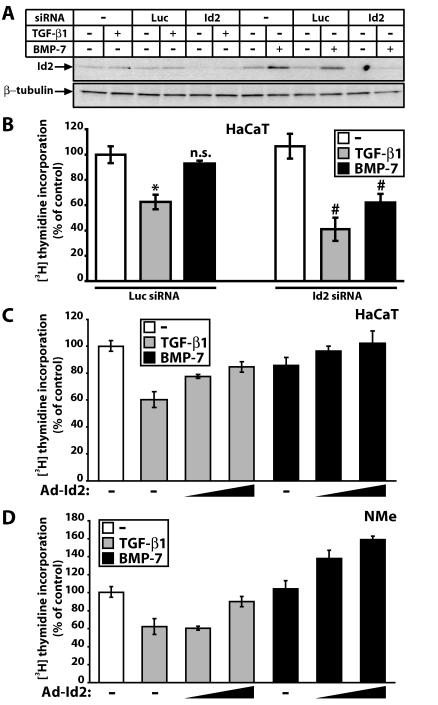
Id2 regulates the growth-inhibitory response of epithelial cells to TGF-β and BMP.
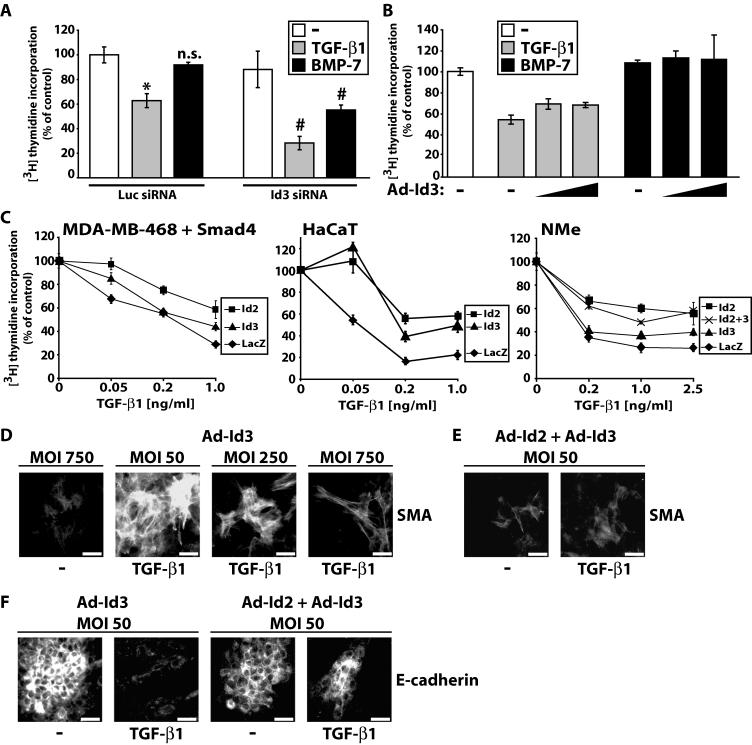
To elucidate the functional role for the observed Id2 and Id3 gene regulation in epithelial cells, we focused on regulation of cell proliferation and the Id2 protein, as the role of Id2 in regulation of the cell cycle has been analyzed more extensively (29, 36, 44). We studied HaCaT and mouse mammary (NMuMG/NMe) cells, two widely used cell models whose growth is inhibited by TGF-β. Using an Id2-specific siRNA we blocked basal expression and induction of Id2 by BMP-7 (Fig. 5A). TGF-β1 suppresses HaCaT S-phase entry (measured by thymidine incorporation), whereas BMP-7 has minimal effects (Fig. 5B), as described previously (19). Surprisingly, knockdown of Id2 enhanced HaCaT growth inhibition by TGF-β1, demonstrating that sustained repression of the Id2 gene is physiologically relevant for cell cycle control (Fig. 5B). Knockdown of Id2 also led to significant growth inhibition by BMP-7, suggesting that one reason why BMP-7 fails to arrest epithelial cell growth is the sustained Id2 induction.
FIG. 5.
Id2 is a critical mediator of epithelial cell proliferation responses to TGF-β1 and BMP-7. (A) Immunoblot analysis of Id2 and control β-tubulin in HaCaT cells shows knockdown of constitutive and BMP-7-inducible Id2 expression by specific (Id2) but not by control (Luc) siRNA. Transfected cells were treated for 4 h with vehicle (−), TGF-β1 (1 ng/ml), or BMP-7 (300 ng/ml). (B) Relative levels of [3H]thymidine incorporation into HaCaT nuclei after control (Luc) or specific (Id2) siRNA transfection, serum starvation, and stimulation with vehicle (−) (white bars), TGF-β1 (1 ng/ml) (grey bars), and BMP-7 (300 ng/ml) (black bars). Data are expressed as percentages relative to vehicle (−) in transfectants of control (Luc) siRNA, and represent averages of triplicates with standard errors (error bars). Statistical significance of the difference between vehicle (−) and TGF-β1 at P ≤ 0.02 (*), nonsignificance for the difference between vehicle (−) and BMP-7 at P ≤ 0.1 (n.s.), and significance of the difference between Id2 and Luc siRNA for each growth factor treatment at P ≤ 0.02 (#) is indicated. (C and D) Relative [3H]thymidine incorporation into HaCaT (C) or NMe (D) nuclei infected with control Ad-LacZ (−) or specific Ad-Id2 adenoviruses (triangles show increasing MOI [20 or 100 for HaCaT and 250 or 750 for NMe]), prior to serum starvation and stimulation with vehicle (-, white bars), TGF-β1 (1 ng/ml) (grey bars), and BMP-7 (300 ng/ml) (black bars). Data are expressed as percentages relative to control vehicle in the presence of the respective adenovirus.
To confirm these results we ectopically expressed Id2 and predicted that high constitutive Id2 levels should block the strong growth-inhibitory effect of TGF-β1 and neutralize the weak BMP-7 effect. Indeed, dose-dependent increase of Id2 levels (see Fig. S3A in the supplemental material) attenuated HaCaT and NMe growth inhibition by TGF-β1 and completely neutralized the weak BMP-7 effect (Fig. 5C and D). We conclude that Id2 defines regulation of epithelial cell growth by TGF-β or BMP: sustained Id2 repression by TGF-β allows growth inhibition whereas sustained Id2 induction prohibits BMP-7 from acting as a potent antiproliferative factor.
Id2 regulates EMT induced by TGF-β.
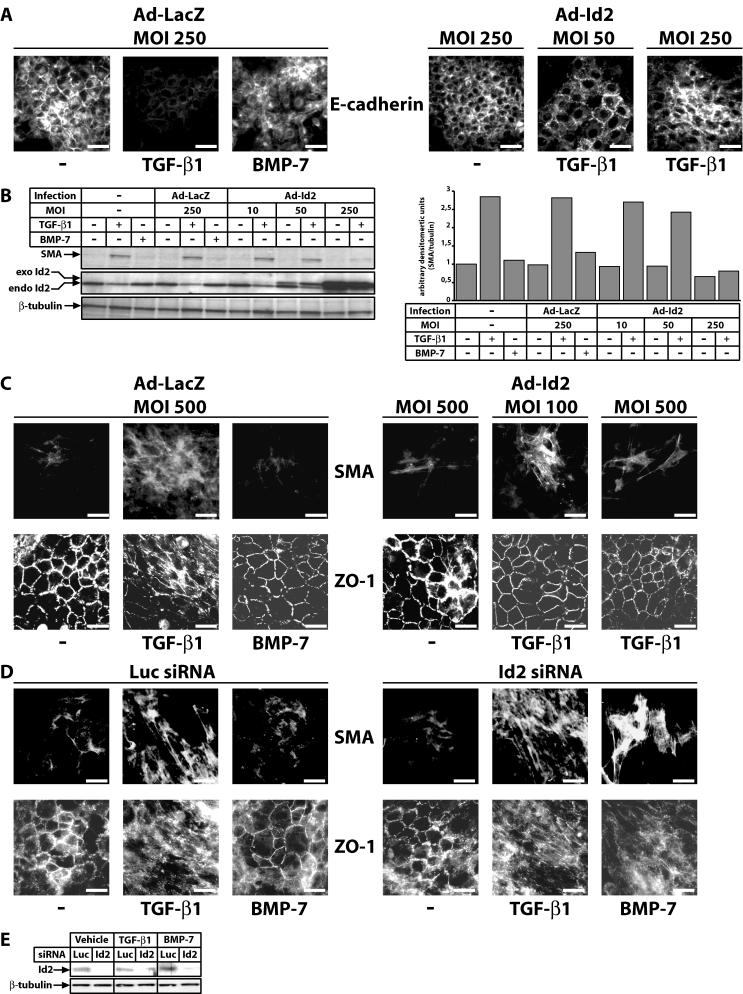
Since Id proteins inhibit differentiation of many cell types (29, 36, 44), and based on the role of Id2 in regulating cell growth by TGF-β and BMP (shown above), we tested whether Id2 could be implicated in regulation of differentiation by TGF-β and BMP. We focused on EMT due to its pivotal role in cancer progression and metastasis (11), in normal mammary (NMuMG/NMe) and lens (α-TN4) epithelial cells. Both cell types transdifferentiate to a fibroblastoid or myofibroblastic phenotype in response to TGF-β1 only but not to BMP-7 (Fig. 6A to C). Thus, infection of NMe cells (or NMuMG cells; unpublished results) with increasing MOI of Id2 adenovirus blocked E-cadherin downregulation by TGF-β1 (for Id2 protein levels, see Fig. 6A and S3A in the supplemental material). Induction of anti-SMA, a myofibroblast marker, during EMT of NMe cells (Fig. 6B), strongly correlated with Id2 downregulation by TGF-β1. Ectopic Id2 expression bypasses the endogenous Id2 downregulation by TGF-β1 and thus dose-dependently blocks SMA induction. Similarly, in α-TN4 cells in which TGF-β1 induces SMA and delocalizes the epithelial tight junction marker ZO-1 (α-TN4 cells do not express detectable E-cadherin protein levels), ectopic Id2 expression dose-dependently blocked the TGF-β1 response (Fig. 6C). We conclude that Id2 overexpression blocks EMT to the myofibroblastic phenotype. Conversely, Id2 knockdown in α-TN4 cells not only enhanced EMT induced by TGF-β1 but, more importantly, permitted EMT by BMP-7 (Fig. 6D and E). Thus, Id2 plays parallel roles in the determination of both the growth and transdifferentiation response of epithelial cells to TGF-β superfamily ligands.
FIG. 6.
Id2 controls the EMT response to TGF-β1 and BMP-7. (A) Epithelial marker E-cadherin immunofluorescence in NMe cells infected with the indicated MOI of control Ad-LacZ or specific Ad-Id2 adenoviruses prior to 48-h stimulation with vehicle (−), TGF-β1 (5 ng/ml), or BMP-7 (300 ng/ml). (B) Myoepithelial marker anti-SMA immunoblot in NMe cells left intact (−) or infected with the indicated MOI of control Ad-LacZ or specific Ad-Id2 adenoviruses prior to 48-h stimulation with vehicle (−), TGF-β1 (5 ng/ml), or BMP-7 (300 ng/ml). Endogenous (endo) and exogenous (exo) Id2 and control β-tubulin proteins are shown. On the right, a bar graph presents densitometric analysis of the SMA protein band of the immunoblot on the left, after normalization to the corresponding β-tubulin levels. (C) Myoepithelial marker SMA and epithelial tight junction marker ZO-1 immunofluorescence in α-TN4 cells infected with the indicated MOI of control Ad-LacZ or specific Ad-Id2 adenoviruses prior to 48-h stimulation with vehicle (−), TGF-β1 (10 ng/ml), or BMP-7 (300 ng/ml). (D) SMA and ZO-1 immunofluorescence in α-TN4 cells transfected with control (Luc) or specific (Id2) siRNA prior to 48-h stimulation with vehicle (−), TGF-β1 (10 ng/ml), or BMP-7 (300 ng/ml). (A, C, and D) Bars represent 10 μm. (E) Endogenous Id2 and control β-tubulin immunoblot in α-TN4 cells transfected with control (Luc) or specific (Id2) siRNA prior to 48-h stimulation with vehicle, TGF-β1 (10 ng/ml), or BMP-7 (300 ng/ml).
Id3 is a less-sensitive sensor of cell growth and transdifferentiation responses to TGF-β and BMP.
Knockdown of endogenous Id3 expression in HaCaT cells (see Fig. S3B in the supplemental material) enhanced growth inhibition by TGF-β1 and BMP-7 (Fig. 7A), almost as potently as the equivalent Id2 knockdown (Fig. 5B). This proves that the knockdown of Id3 achieved in these experiments is functional. In contrast, ectopic expression of Id3 (see Fig. S3C in the supplemental material) weakly blocked and neutralized the growth-inhibitory response to TGF-β1 and BMP-7, respectively (Fig. 7B), which is a less-efficient effect compared to ectopic Id2 expression (Fig. 5C and D). To provide more compelling evidence for the differential effect of Id2 and Id3 on epithelial growth inhibition in response to TGF-β1, we performed dose-response assays using increasing doses of TGF-β1 (Fig. 7C). In all three cell lines tested, ectopic Id2 could fully block the effect of 0.05-ng/ml TGF-β1 (unpublished results for NMe cells). In MDA-MB-468 cells reconstituted with wild-type Smad4, Id2 decreased the effect of TGF-β1 on cell growth at all doses tested whereas Id3 scored significantly only at the highest ligand dose. In HaCaT cells, ectopic expression of Id2 or Id3 reduced significantly the growth-inhibitory effect of TGF-β1 at all doses tested, however, Id2 was more efficient at doses higher than 0.05 ng/ml. Finally, in NMe cells the difference between Id2 and Id3 was even more exacerbated, and the combination of Id2 plus Id3 overexpression gave the same response curve as Id2 alone. The latter observation suggests that the two Id members mediate their cell cycle related effects through common mechanisms (presumably regulation of common gene targets). If each Id member was acting independently, one would have expected at minimum additive effects after coexpression of Id2 and Id3 on cell growth, which was never seen.
FIG. 7.
Id3 is a weaker but essential mediator of epithelial proliferative and EMT responses to TGF-β1 and BMP-7. (A) Relative levels of [3H]thymidine incorporation into HaCaT nuclei transfected with control (Luc) or specific (Id3) siRNA prior to serum starvation and stimulation with vehicle (−) (white bars), TGF-β1 (1 ng/ml) (grey bars), and BMP-7 (300 ng/ml) (black bars). The data are expressed as in Fig. 5B. (B) Relative levels of [3H]thymidine incorporation into HaCaT nuclei infected with control Ad-LacZ (−) or specific Ad-Id3 adenoviruses (triangles show increasing MOI: 20 and 500), prior to serum starvation and stimulation with vehicle (−) (white bar), TGF-β1 (1 ng/ml) (grey bars), and BMP-7 (300 ng/ml) (black bars). The data are expressed as in Fig. 5C. (C) Dose-response graphs of relative levels of [3H]thymidine incorporation into MDA-MB-468, HaCaT, and NMe nuclei in response to increasing doses of TGF-β1. MDA-MB-468 cells were reconstituted with wild-type Smad4 using transient adenoviral infection prior to the experiment. All cells were coinfected with control Ad-LacZ or specific Ad-Id2 or Ad-Id3 adenoviruses or both together. The MOI of each adenovirus was 80 for MDA-MB-468, 500 for HaCaT, and 750 for NMe cells. (D and E) SMA immunofluorescence in α-TN4 cells infected with the indicated MOI of Ad-Id3 adenovirus (D) or low MOI combination of Ad-Id2 and Ad-Id3 (E) prior to 48-h stimulation with vehicle (−) or TGF-β1 (10 ng/ml). (F) E-cadherin immunofluorescence in NMe cells infected with the indicated low MOI of Ad-Id3 or combination of Ad-Id2 and Ad-Id3 prior to 48-h stimulation with vehicle (−) or TGF-β1 (5 ng/ml). (D to F) Bars represent 10 μm.
In parallel EMT assays of α-TN4 and NMe cells, Id3 overexpression (see Fig. S3D in the supplemental material) blocked EMT in response to TGF-β1 (Fig. 7D and F), but not as efficiently as ectopic Id2 expression (Fig. 6A to C). Infection of α-TN4 or NMe cells with low doses of single Id2 or Id3 adenoviruses had no significant effects; however, when combined, they completely blocked EMT (Fig. 7E and F). We conclude that Id3 plays similar but not as potent roles in determining the response of epithelial cells to TGF-β1 and BMP-7. However, concerted regulation of both Id2 and Id3 genes seems to be the critical target of TGF-β superfamily signaling.
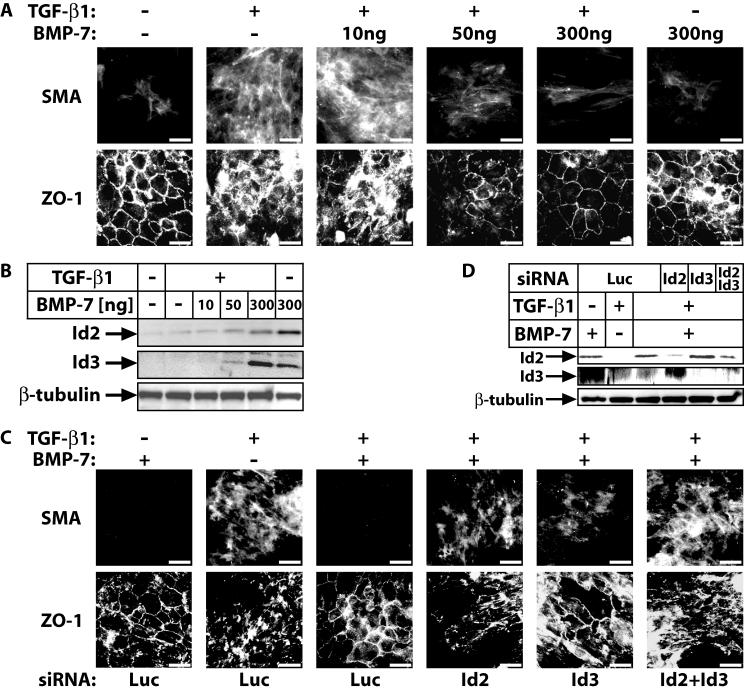
To verify this model bypassing the use of transfection or viral infection, we simultaneously treated cells with TGF-β1 and BMP-7 (Fig. 8). Since BMP-7 induces sustained Id2 and Id3 levels, we reasoned that combining a constant dose of TGF-β1 with increasing doses of BMP-7 might allow us to observe antagonism of the EMT response. Indeed, experiments in α-TN4 cells established that myofibroblast transdifferentiation in response to TGF-β1 can be fully blocked by coincubation of cells with BMP-7 (50 ng/ml) (Fig. 8A). The neutralizing effect of BMP-7 against TGF-β1 was as potent as that of ectopic Id2 and Id3 expression in these cells (Fig. 6C and 7D) and reflected the mode of endogenous Id2/3 protein expression, whereby, increasing BMP-7 doses overcame the negative regulation by TGF-β1 (Fig. 8B). In order to prove that Id2 and Id3 were the critical players in these costimulation experiments with both TGF-β1 and BMP-7, we repeated the experiment with α-TN4 cells in which endogenous Id2 or Id3 or both were knocked down via RNA interference (Fig. 8C and D). Immunofluorescence for the SMA and ZO-1 markers confirmed that when endogenous Id2 or Id3 levels were reduced, the antagonistic effect of BMP-7 against TGF-β1 was essentially neutralized. A stronger effect was observed when both Id2 and Id3 were knocked down, and in these experiments we also observed that Id2 knockdown had a more potent effect compared to Id3 knockdown. Thus, Id2 and Id3 represent critical nuclear effectors that mediate antagonism between TGF-β and BMP pathways.
FIG. 8.
BMP-7 antagonizes TGF-β1-induced lens myoepithelial transdifferentiation. (A) ZO-1 and SMA immunofluorescence in α-TN4 cells stimulated with vehicle (−), TGF-β1 alone (10 ng/ml), TGF-β1 (10 ng/ml) together with increasing doses (10 to 300 ng/ml) of BMP-7 or BMP-7 alone (300 ng/ml) for 48 h. (B) Immunoblot analysis of endogenous Id2 and Id3 and control β-tubulin levels in α-TN4 cells stimulated with vehicle (−), with TGF-β1 (10 ng/ml) alone or together with increasing doses (10 to 300 ng/ml) of BMP-7, or with BMP-7 alone (300 ng/ml) for 48 h. (C) SMA and ZO-1 immunofluorescence in α-TN4 cells stimulated for 48 h with BMP-7 (300 ng/ml), TGF-β1 (10 ng/ml), or BMP-7 (300 ng/ml) plus TGF-β1 (10 ng/ml), after transient transfection with the indicated siRNAs. (A and C) Bars represent 10 μm. (D) Immunoblot analysis of endogenous Id2 and Id3 and control β-tubulin levels in α-TN4 cells stimulated with the same conditions and after transient transfection with the same siRNA combinations as in panel C.
DISCUSSION
Transcriptomic analysis of two major signaling pathways of the TGF-β superfamily unexpectedly showed few, poorly annotated Smad4-independent genes (Fig. 2 and 3). Previously, fibroblasts from Smad4 knockout mice gave evidence for a more significant Smad4-independent cell response (37). However, the physiological effects of TGF-β superfamily members differ dramatically between mesenchymal and epithelial cells, making it difficult to compare studies on such disparate cell types. Our findings agree with independent epithelial cell studies using mutant TGF-β receptors unable to activate the Smad pathway (13) and are in favor of the central role of Smad4 in all TGF-β superfamily pathways (33). The common genomic program regulated by TGF-β1 and BMP-7 is rather short and includes genes implicated in cell proliferation control (p21Cip1, Id1 to -3, c-myc; Fig. 3B). In addition, we identified several novel gene targets of both pathways in epithelial cells, such as ovol1, gata2, Crabp2, vps4b, cdk10, fgg, and hsd17b2. These genes may be of importance for understanding distinct functions of the two signaling pathways and attract our future focus. Our microarray screen revealed 129 genes regulated specifically by one of the two growth factors (Fig. 3A). It is possible that among them are critical genes that define the final and specific effect of TGF-β1 or BMP-7 on epithelial cells. However, as we have shown for Id2 and Id3, differential regulation of common gene targets by TGF-β and BMP can explain distinct physiological effects of these two pathways.
We focused on Id2 and Id3 because of their robust gene regulation (Fig. 3B), and due to the limited understanding of their role in TGF-β physiology, while Id1 is the best-analyzed Id member in terms of its regulation by BMP and TGF-β, it remains unclear how it contributes to cell proliferation control by these factors (14, 21, 25, 29). Induction of Id1 to -3 by BMP-2 in breast cancer cells or by TGF-β1 in fibroblasts was recently shown (3, 5). Studies of lymphocyte differentiation provide convincing evidence for Id2 and Id3 roles in TGF-β physiology (12, 16). Using a panel of normal epithelial cells we establish here a general profile of Id2 and Id3 regulation (Fig. 3D and 4; also see Fig. S1 and S2A in the supplemental material). This includes the MDA-MB-468 mammary carcinoma cell where the initial microarray screen was performed, whereby Id2 and Id3 scored as positively regulated genes at the early time point of TGF-β stimulation (Fig. 3B). Detailed immunoblot analysis and time course experiments showed that this cell line, like more normal epithelial cells, also responds to TGF-β after Smad4 reconstitution, by eventually downregulating Id2 and Id3 levels at late time points. On the other hand, we still do not understand why TGF-β can transiently induce Id2 or Id3 mRNA but not protein expression in certain cell types (see Fig. S1 in the supplemental material) and both Id2/Id3 mRNA and protein levels in MDA-MB-468 carcinoma cells (Fig. 3B and D). Our focus on the functional role of Id2 and Id3 regulation downstream of TGF-β and BMP signaling led to the finding that these genes define specificity of the two pathways in epithelial cell biology.
Since knockdown of Id2 (Fig. 5B) and Id3 (Fig. 7A) enhances growth inhibition by TGF-β1 and BMP-7, we consider these proteins to be important components controlling cell proliferation by TGF-β members. It would be interesting to test whether loss of Id1 function could have similar effects on TGF-β/BMP-mediated epithelial growth inhibition. Throughout this study we observed that Id2 was always more efficient to regulate cell growth in response to TGF-β members, when compared to Id3 (Fig. 5 and 7). This cannot be due to the expression levels of the two proteins as the same result was reached when high ectopic levels of each Id member were expressed in various epithelial cell models. Finally, when both Id2 and Id3 were ectopically expressed, we never observed additive or synergistic effects compared to experimental perturbation of each Id member separately (Fig. 7). These observations suggest that Id2 and Id3 have common targets in epithelial cells, e.g., common bHLH proteins which regulate the same critical target genes (see below). If each Id protein had distinct targets that were cooperatively contributing to the control of cell proliferation, then we should have observed at least additive if not synergistic effects. It is therefore possible that the higher efficiency by which Id2 blocks TGF-β-mediated physiological effects reflects a posttranslational modification which is directly induced by the TGF-β signal. Such modification could either enhance the inhibitory activity of Id2 or inversely dampen the activity of Id3. Alternatively, a more likely scenario is the possibility that Id2 modulates E-protein transcriptional activity more efficiently compared to Id3, either because of higher affinity for partner bHLH members or because of more efficient recruitment of cofactors that help mediate a complete repression of E-protein activity. Both alternative scenarios are worth investigating in the future.
Two prominent models explain the effects of Id members on cell cycle regulation (29, 36, 44). Id proteins inhibit the transcriptional activity of E-proteins such as the products of the E2A gene, E12 and E47, which positively regulate the cell cycle regulatory genes p15Ink4b, p16Ink4a, and p21Cip1. Alternatively, Id2 can reverse the cell cycle-inhibitory action of pocket proteins of the retinoblastoma family. Id3 cannot interact with pocket proteins, and thus, the retinoblastoma model cannot be applied to this protein. Based on our data, we suggest that TGF-β, which leads to sustained downregulation of Id2 and Id3 in epithelial cells, can block the cell cycle by the synergistic effects resulting from relief of E-protein activity on as yet unidentified gene targets and from direct transcriptional induction of the cell cycle inhibitor genes p15Ink4b and p21Cip1. It is worth noting here that both of these genes are induced by the TGF-β/Smad pathway directly (34). On the other hand, BMP induces sustained Id2 and Id3 expression and thus provides constant repressor function against E- and pocket proteins. BMP also induces p21Cip1 despite its positive effect on Id expression, and downregulates c-myc, both events favoring a halt of the cell cycle. Thus, induction of antagonistic factors by BMP may lead to almost neutral functional outcomes of cell cycle regulation in epithelial cells, which can explain why BMPs have weak and variable effects on epithelial cell growth. Considering the combined effects of TGF-β and BMP signaling on Id protein expression, we favor the model whereby Id2/Id3 repress bHLH factor activity on as yet unrecognized gene targets, distinct from cell cycle inhibitors such as p21Cip1, whose products must be directly linked to cell cycle control. We are using cDNA microarray screens to address this important question.
The established roles of Ids as differentiation inhibitors led us to investigate such a role in response to TGF-β and BMP (Fig. 6 and 7). We focused on a mammary and a lens EMT cell model because EMT plays important roles in tumor progression (11), and this is an epithelial response to TGF-β but not to BMP (28). Our evidence supports a model whereby Id2/3 regulation renders TGF-β capable of eliciting EMT, whereas it waives such an instructive role from BMP. For the first time, we demonstrate that BMP induces EMT under conditions of Id2 or Id3 knockout (Fig. 6 and 8). In agreement with this model BMP-7 could neutralize the EMT response to TGF-β1, since BMP-7 could override the downregulation of Id2/3 by TGF-β1 (Fig. 8). Thus, during tumor progression, misregulation of epithelial Id2/3 expression could possibly allow even BMP pathways to cause EMT and enhance tumor invasiveness.
Similar to the epithelial cell growth results, the EMT data of both ectopic expression and endogenous knockdown (Fig. 7 and 8) revealed that Id2 had higher potential in blocking TGF-β-induced EMT. However, in contrast to their effects on cell growth inhibition, combinations of Id2 and Id3 ectopic expression (Fig. 7E and F) or, alternatively, combinations of Id2 and Id3 knockdown (Fig. 8C) demonstrated that these two proteins acted in a manner which was more than additive and possibly synergistic. This strongly implies that there exist distinct targets of Id2 and Id3 which act in a combined manner to regulate the process of EMT. We currently do not know which are the targets of Id proteins that mediate EMT. One possibility is that E-proteins such as E12 and E47, in addition to their roles in cell cycle control, may also link Id-mediated repression to EMT. This hypothesis is compatible with the known role of E2A gene products on regulation of the E-cadherin gene, a hallmark of epithelial differentiation (27). However, as E-cadherin does not play a causal role in the process of EMT, there must be additional genes involved in such a process whose regulation is mediated by bHLH proteins and thus link to the action of Id2 and Id3 as described for the first time here.
We demonstrate that Id2 and Id3 are important for the concerted regulation of cell proliferation and transdifferentiation downstream of TGF-β pathways. We conclude that in epithelial tissues where multiple TGF-β ligands are active, the physiological outcome of responding epithelial cells will depend on regulation of sensitive gene targets such as Id2 and Id3. Id genes provide the first molecular targets of TGF-β pathways that explain biological specificity with respect to regulation of cell growth and differentiation.
Supplementary Material
Acknowledgments
The microarray consortium is funded by the Wellcome Trust, Cancer Research UK and the Ludwig Institute of Cancer Research. This research was funded in part by grants from the Human Frontier Science Program (A.M.), a postdoctoral fellowship from the French Association pour la Recherche sur le Cancer (U.V.), and graduate student fellowships from the University of Uppsala (M.K. and R.B.).
We thank the staff of the Sanger Center Microarray Facility (http://www.sanger.ac.uk/Projects/Microarrays/) for supplying arrays, laboratory protocols, technical advice (David Vetrie, Cordelia Langford, Adam Whittaker, and Neil Sutton), and Quantarray/GeneSpring data files and databases relating to array elements (Kate Rice, Rob Andrews, Adam Butler, and Harish Chudasama). We acknowledge the MRC HGMP Resource Centre (Hinxton, United Kingdom) for the human I.M.A.G.E. cDNA clone collection. cDNA clone resequencing was performed by Team 56 at the Sanger Institute. We thank S. White and E. Hunter for advice on cDNA microarray analysis. We thank S. Dooley, N. Ferrara, N. Fusenig, F. W. Miller, K. Miyazono, J. Piatigorski, K. Sampath, T. Taga, P. ten Dijke, G. Valdimarsdottir, K. Verschueren, and B. Vogelstein for reagents.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Barolo, S., and J. W. Posakony. 2002. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16:1167-1181. [DOI] [PubMed] [Google Scholar]
- 2.Botchkarev, V. A. 2003. Bone morphogenetic proteins and their antagonists in skin and hair follicle biology. J. Investig. Dermatol. 120:36-47. [DOI] [PubMed] [Google Scholar]
- 3.Chambers, R. C., P. Leoni, N. Kaminski, G. J. Laurent, and R. A. Heller. 2003. Global expression profiling of fibroblast responses to transforming growth factor-β1 reveals the induction of inhibitor of differentiation-1 and provides evidence of smooth muscle cell phenotypic switching. Am. J. Pathol. 162:533-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C. R., Y. Kang, and J. Massagué. 2001. Defective repression of c-myc in breast cancer cells: a loss at the core of the transforming growth factor β growth arrest program. Proc. Natl. Acad. Sci. USA 98:992-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clement, J. H., N. Marr, A. Meissner, M. Schwalbe, W. Sebald, K. O. Kliche, K. Hoffken, and S. Wolfl. 2000. Bone morphogenetic protein 2 (BMP-2) induces sequential changes of Id gene expression in the breast cancer cell line MCF-7. J. Cancer Res. Clin. Oncol. 126:271-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jong, D. S., E. J. van Zoelen, S. Bauerschmidt, W. Olijve, and W. T. Steegenga. 2002. Microarray analysis of bone morphogenetic protein, transforming growth factor β, and activin early response genes during osteoblastic cell differentiation. J. Bone Miner. Res. 17:2119-2129. [DOI] [PubMed] [Google Scholar]
- 7.de Winter, J. P., B. A. Roelen, P. ten Dijke, B. van der Burg, and A. J. van den Eijnden-van Raaij. 1997. DPC4 (SMAD4) mediates transforming growth factor-β1 (TGF-β1) induced growth inhibition and transcriptional response in breast tumour cells. Oncogene 14:1891-1899. [DOI] [PubMed] [Google Scholar]
- 8.Derynck, R., and Y. E. Zhang. 2003. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425:577-584. [DOI] [PubMed] [Google Scholar]
- 9.Franzén, A., and N. E. Heldin. 2001. BMP-7-induced cell cycle arrest of anaplastic thyroid carcinoma cells via p21CIP1 and p27KIP1. Biochem. Biophys. Res. Commun. 285:773-781. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh-Choudhury, N., K. Woodruff, W. Qi, A. Celeste, S. L. Abboud, and G. Ghosh-Choudhury. 2000. Bone morphogenetic protein-2 blocks MDA MB 231 human breast cancer cell proliferation by inhibiting cyclin-dependent kinase-mediated retinoblastoma protein phosphorylation. Biochem. Biophys. Res. Commun. 272:705-711. [DOI] [PubMed] [Google Scholar]
- 11.Grünert, S., M. Jechlinger, and H. Beug. 2003. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat. Rev. Mol. Cell Biol. 4:657-665. [DOI] [PubMed] [Google Scholar]
- 12.Hacker, C., R. D. Kirsch, X. S. Ju, T. Hieronymus, T. C. Gust, C. Kuhl, T. Jorgas, S. M. Kurz, S. Rose-John, Y. Yokota, and M. Zenke. 2003. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat. Immunol. 4:380-386. [DOI] [PubMed] [Google Scholar]
- 13.Itoh, S., M. Thorikay, M. Kowanetz, A. Moustakas, F. Itoh, C.-H. Heldin, and P. ten Dijke. 2003. Elucidation of Smad requirement in transforming growth factor-β type I receptor-induced responses. J. Biol. Chem. 278:3751-3761. [DOI] [PubMed] [Google Scholar]
- 14.Kang, Y., C. R. Chen, and J. Massagué. 2003. A self-enabling TGFβ response coupled to stress signaling. Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell 11:915-926. [DOI] [PubMed] [Google Scholar]
- 15.Katagiri, T., M. Imada, T. Yanai, T. Suda, N. Takahashi, and R. Kamijo. 2002. Identification of a BMP-responsive element in Id1, the gene for inhibition of myogenesis. Genes Cells 7:949-960. [DOI] [PubMed] [Google Scholar]
- 16.Kee, B. L., R. R. Rivera, and C. Murre. 2001. Id3 inhibits B lymphocyte progenitor growth and survival in response to TGF-β. Nat. Immunol. 2:242-247. [DOI] [PubMed] [Google Scholar]
- 17.Korchynskyi, O., and P. ten Dijke. 2002. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277:4883-4891. [DOI] [PubMed] [Google Scholar]
- 18.Kurisaki, K., A. Kurisaki, U. Valcourt, A. A. Terentiev, K. Pardali, P. ten Dijke, C.-H. Heldin, J. Ericsson, and A. Moustakas. 2003. Nuclear factor YY1 inhibits transforming growth factor β- and bone morphogenetic protein-induced cell differentiation. Mol. Cell. Biol. 23:4494-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lange, D., U. Persson, U. Wollina, P. ten Dijke, E. Castelli, C.-H. Heldin, and K. Funa. 1999. Expression of TGF-β related Smad proteins in human epithelial skin tumors. Int. J. Oncol. 14:1049-1056. [DOI] [PubMed] [Google Scholar]
- 20.Lasorella, A., M. Noseda, M. Beyna, Y. Yokota, and A. Iavarone. 2000. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature 407:592-598. [DOI] [PubMed] [Google Scholar]
- 21.Ling, M. T., X. Wang, S. W. Tsao, and Y. C. Wong. 2002. Down-regulation of Id-1 expression is associated with TGF-β1-induced growth arrest in prostate epithelial cells. Biochim. Biophys. Acta 1570:145-152. [DOI] [PubMed] [Google Scholar]
- 22.Locklin, R. M., B. L. Riggs, K. C. Hicok, H. F. Horton, M. C. Byrne, and S. Khosla. 2001. Assessment of gene regulation by bone morphogenetic protein 2 in human marrow stromal cells using gene array technology. J. Bone Miner. Res. 16:2192-2204. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Rovira, T., E. Chalaux, J. Massagué, J. L. Rosa, and F. Ventura. 2002. Direct binding of Smad1 and Smad4 to two distinct motifs mediates bone morphogenetic protein-specific transcriptional activation of Id1 gene. J. Biol. Chem. 277:3176-3185. [DOI] [PubMed] [Google Scholar]
- 24.Maurice, D., C. E. Pierreux, M. Howell, R. E. Wilentz, M. J. Owen, and C. S. Hill. 2001. Loss of Smad4 function in pancreatic tumors: C-terminal truncation leads to decreased stability. J. Biol. Chem. 276:43175-43181. [DOI] [PubMed] [Google Scholar]
- 25.Miyazono, K., and K. Miyazawa. 2002. Id: a target of BMP signaling. Sci. STKE. 2002:PE40. [DOI] [PubMed] [Google Scholar]
- 26.Ota, T., M. Fujii, T. Sugizaki, M. Ishii, K. Miyazawa, H. Aburatani, and K. Miyazono. 2002. Targets of transcriptional regulation by two distinct type I receptors for transforming growth factor-β in human umbilical vein endothelial cells. J. Cell. Physiol. 193:299-318. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Moreno, M. A., A. Locascio, I. Rodrigo, G. Dhondt, F. Portillo, M. A. Nieto, and A. Cano. 2001. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J. Biol. Chem. 276:27424-27431. [DOI] [PubMed] [Google Scholar]
- 28.Piek, E., A. Moustakas, A. Kurisaki, C.-H. Heldin, and P. ten Dijke. 1999. TGF-β type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J. Cell Sci. 112:4557-4568. [DOI] [PubMed] [Google Scholar]
- 29.Ruzinova, M. B., and R. Benezra. 2003. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 13:410-418. [DOI] [PubMed] [Google Scholar]
- 30.Sakou, T. 1998. Bone morphogenetic proteins: from basic studies to clinical approaches. Bone 22:591-603. [DOI] [PubMed] [Google Scholar]
- 31.Schonherr, E., B. Levkau, L. Schaefer, H. Kresse, and K. Walsh. 2001. Decorin-mediated signal transduction in endothelial cells. Involvement of Akt/protein kinase B in up-regulation of p21WAF1/CIP1 but not p27KIP1. J. Biol. Chem. 276:40687-40692. [DOI] [PubMed] [Google Scholar]
- 32.Schutte, M. 1999. DPC4/SMAD4 gene alterations in human cancer, and their functional implications. Ann. Oncol. 10:56-59. [PubMed] [Google Scholar]
- 33.Shi, Y., and J. Massagué. 2003. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113:685-700. [DOI] [PubMed] [Google Scholar]
- 34.Siegel, P. M., and J. Massagué. 2003. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat. Rev. Cancer. 3:807-820. [DOI] [PubMed] [Google Scholar]
- 35.Siegel, P. M., W. Shu, and J. Massagué. 2003. Mad Upregulation and Id2 Repression Accompany Transforming Growth Factor (TGF)-β-mediated Epithelial Cell Growth Suppression. J. Biol. Chem. 278:35444-35450. [DOI] [PubMed] [Google Scholar]
- 36.Sikder, H. A., M. K. Devlin, S. Dunlap, B. Ryu, and R. M. Alani. 2003. Id proteins in cell growth and tumorigenesis. Cancer Cell 3:525-530. [DOI] [PubMed] [Google Scholar]
- 37.Sirard, C., S. Kim, C. Mirtsos, P. Tadich, P. A. Hoodless, A. Itie, R. Maxson, J. L. Wrana, and T. W. Mak. 2000. Targeted disruption in murine cells reveals variable requirement for Smad4 in transforming growth factor β-related signaling. J. Biol. Chem. 275:2063-2070. [DOI] [PubMed] [Google Scholar]
- 38.ten Dijke, P., M.-J. Goumans, F. Itoh, and S. Itoh. 2002. Regulation of cell proliferation by Smad proteins. J. Cell. Physiol. 191:1-16. [DOI] [PubMed] [Google Scholar]
- 39.Valcourt, U., J. Gouttenoire, A. Moustakas, D. Herbage, and F. Mallein-Gerin. 2002. Functions of transforming growth factor-β family type I receptors and Smad proteins in the hypertrophic maturation and osteoblastic differentiation of chondrocytes. J. Biol. Chem. 277:33545-33558. [DOI] [PubMed] [Google Scholar]
- 40.Valdimarsdottir, G., M.-J. Goumans, A. Rosendahl, M. Brugman, S. Itoh, F. Lebrin, P. Sideras, and P. ten Dijke. 2002. Stimulation of Id1 expression by bone morphogenetic protein is sufficient and necessary for bone morphogenetic protein-induced activation of endothelial cells. Circulation 106:2263-2270. [DOI] [PubMed] [Google Scholar]
- 41.Verrecchia, F., M. L. Chu, and A. Mauviel. 2001. Identification of novel TGF-β /Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J. Biol. Chem. 276:17058-17062. [DOI] [PubMed] [Google Scholar]
- 42.Xu, R. H., X. Chen, D. S. Li, R. Li, G. C. Addicks, C. Glennon, T. P. Zwaka, and J. A. Thomson. 2002. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20:1261-1264. [DOI] [PubMed] [Google Scholar]
- 43.Yeh, K., and R. W. Lim. 2000. Genomic organization and promoter analysis of the murine Id3 gene. Gene 254:163-171. [DOI] [PubMed] [Google Scholar]
- 44.Yokota, Y., and S. Mori. 2002. Role of Id family proteins in growth control. J. Cell. Physiol. 190:21-28. [DOI] [PubMed] [Google Scholar]
- 45.Zavadil, J., M. Bitzer, D. Liang, Y. C. Yang, A. Massimi, S. Kneitz, E. Piek, and E. P. Böttinger. 2001. Genetic programs of epithelial cell plasticity directed by transforming growth factor-β. Proc. Natl. Acad. Sci. USA 98:6686-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.