Abstract
Given the recent scale-up of antiretroviral therapy (ART) in sub-Saharan Africa, we sought to determine how often and at what levels do drug-resistant mutant variants exist in ART-naïve HIV subtype C infected individuals. Samples from 10 ART-naïve Zambian individuals were subjected to ultra-deep pyrosequencing (UDPS) to characterize the frequency of low-abundance drug resistance mutations in the pol gene. Low-abundance clinically relevant variants were detected for nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs) and protease inhibitors (PIs) in eight of the ten subjects. Intermediate to high-level resistance was predicted for the majority of NRTIs. Mutations conferring resistance to most first-line and some second-line therapy drugs were also observed. UDPS detected a number of additional major resistant mutations suggesting that these individuals may have an increased risk of virological failure after initiating ART. Moreover, the effectiveness of first-line and even some second-line ART may be compromised in this setting.
Keywords: Antiretroviral therapy, drug naïve, HIV-1, low abundance variants, subtype C, ultra-deep pyrosequencing, Zambia
INTRODUCTION
As access to antiretroviral drugs is expanding in sub-Saharan Africa and second-line treatments are often not readily available in the setting, it is imperative that adequate assessment on the prevalence and transmission of drug resistance mutations is performed to ensure optimization of treatment efficacy [1]. Antiretroviral therapy (ART) has been successful in suppressing the human immunodeficiency virus (HIV), however it fails to completely eliminate HIV from infected individuals and often elicits drug-resistant variants. These mutations can persist in chronically HIV-infected, ART-naïve individuals, and the prevalence of these resistance variants is likely to increase overtime [2]. In addition the presence of these mutations has been demonstrated to have a negative impact on the antiretroviral therapeutic response and shortens the time to the first virological failure [3]. Recently it has been demonstrated by ultra-deep pyrosequencing (UDPS), that resistance variants as low as 1% are clinically relevant and can have a profound impact on treatment outcome even in ART-naïve individuals that have recently initiated therapy [4, 5]. These mutations found in less than 20% are not detectable by standard genotyping sequencing techniques [4]. Furthermore, the low-abundance resistant variants can out-compete wild-type virus under the selection pressures of antiretrovirals, ultimately lead to virologic failure [4, 6, 7].
Currently, little is known about the prevalence of low-abundance drug-resistant variants that exist in treatment-naïve patients especially in resource-limited settings such as sub-Saharan Africa. While subtype B predominates in the U.S., over 50% of worldwide infections are subtype C, with approximately 98% of HIV positive individuals in southern Africa [8]. One such country in sub-Saharan Africa where we have been investigating is Zambia, where subtype C is the predominant circulating HIV strain [9–12]. HIV treatment in Zambia became available to the public sector in 2002 and was expanded in 2004 [13].
Previously, we demonstrated an increasing trend in the number of minor, borderline and partial resistance mutations, as well as major resistance mutations after the availability free ART in Zambia [11]. Here, we studied a subset of those patients by UDPS to assess the frequency of low-level drug-resistant variants. To our knowledge this is the first time that HIV-1 subtype C infected patients from a sub-Saharan African setting have been directly evaluated for low-abundance drug-resistant viral variants using UDPS in comparison to conventional sequencing. The present findings demonstrate that there is a high prevalence of primary low-abundance drug-resistant variants against the current available treatment regimen in drug-naïve individuals, which were not detected by conventional technologies and may in turn be of clinical importance.
MATERIALS AND METHODS
Selection of Study Participants and Sample Preparation
Frozen HIV-1 proviral DNA samples from Zambian adults recently infected with HIV subtype C were randomly selected from two cohorts of archived specimens. All subjects were ART-naïve at the time of sample collection. A written informed consent was obtained from all individuals. The study was conducted in accordance with the principles of the Helsinki Declaration and approved by the Zambian Ministry of Health, the Research and Ethics Committee of the University Teaching Hospital in Lusaka, Zambia and the Institutional Review Board of the University of Nebraska, Lincoln. The first cohort comprised of four randomly selected Zambian women whose peripheral blood mononuclear cells (PBMCs) DNA was available for the study. These samples were obtained in Lusaka, Zambia between 1998 and 2002, before ART was available in Zambia to the public sector. The second set was from two males and four females, collected in 2005 from a surveillance of HIV subtyping and genotyping, after ART was freely available in Zambia. DNA extraction from these PBMC specimens was previously described by Gonzalez et al. 2010 [11]. In order to determine the HIV copy number per μg of DNA in each sample, a real time PCR amplification of the HIV LTR region was performed following the conditions previously reported by Yun et al. 2002 [14].
PCR Amplification for Amplicon Library Preparation and UDPS
In order to determine the frequency of low-abundance ART resistance mutations within the viral population of each study participant, UDPS was performed on barcoded overlapping amplicons querying positions of HIV drug - resistance mutations in the protease (PR) and reverse transcriptase (RT)-coding regions. The first step in the amplicon library preparation was to generate a fragment 1686 bp amplicon containing the PR and the RT genes from the DNA samples using the primers reported by Zhang et al. 2004 [15] and the FastStart High Fidelity PCR System (Roche, Indianapolis, IN). For each sample, an average of 815 HIV DNA copies was amplified to generate these amplicons. The amplicon library was generated using eleven pairs of 6n barcoded primers adapted from Hoffman et al. 2009 [16]. These overlapping fragments were amplified using the FastStart High Fidelity PCR System. The positive PCR products were purified using the E.Z.N.A. Gel Extraction Kit (Omega Bio-Tech, Norcross, GA) and quantitated by PicoGreen fluorescence (Invitrogen, Carlsbad, CA). After pooling the amplicons in equimolar concentrations, the samples were processed and sequenced on a Genome Sequencer FLX (Roche/454 Life Sciences, Branford, CT) at the University of Nebraska Lincoln's Applied Genomics and Ecology core facility.
UDPS Sequence Analysis
The initial sequence reaction yielded 42,099 sequence reads that passed quality filtering. To ensure high quality reads and to reduce the typical sequencing errors from pyrosequencing the following quality control strategy was used. All reads that had ambiguous bases (N) or whose lengths lay outside the main distribution, as well as inexact matches to the primer or 6-bp barcoding sequence were discarded. In addition reads with low quality scores (<20) were excluded. The quality control procedure was implemented using an in-house Perl script with both forward and reverse primers removed. An additional analysis was performed to exclude sequence reads that were suspected to have resulted from G-to-A hypermutations [17].
For each patient a direct clonal sequence served as a reference template in this study. Each sequence read was mapped onto the direct PCR sequence using the Smith-Waterman algorithm with the following parameters for the alignment; gap opening (−4), gap division (4), match (+1), transition divisor (2) and transversion (−2). Drug-resistant mutations were identified using the 2009 surveillance drug resistant mutation (SDRM) list obtained from Stanford University. Drug resistance was predicted by using the Stanford Genotypic Resistance Interpretation Algorithm (version 6.0.8) available at http://hivdb.stanford.edu/pages/algs/HIVdb.html.
To measure the accuracy of UDPS, an analysis based on four pNL43 clonal sequences performed on the same plates with the clinical samples was carried out. The mean error rate was estimated by comparing each UDPS sequencing read to the control sequence. The overall mean mismatch error rate was 0.195%. To distinguish sequence errors from authentic minor variants we adopted an exclusionary cutoff of 0.2% because of the a priori interest in mutations such as those at known drug resistance positions. However, in order to eliminate the possibility of artifacts, only mutations with frequencies greater than 1% were included in the analyses.
RESULTS
Patient Characteristics
Ultra-deep pyrosequencing (UDPS) was applied to characterize the frequency of low-abundance drug resistant variants in clinical samples obtained from 10 HIV-1 subtype C infected patients. All selected patients were adults, HIV-1 positive and ART-naïve. Patients were strategically chosen from a number of different towns in order to represent the Zambian population. An average of 4093.5 HIV copies/ μg of DNA were isolated from each sample, and a mean of 815 copies of HIV DNA were used to obtain the 1686 bp amplicon containing the PR and the RT genes.
After a correction step was applied, UDPS generated on average 3961 (3192 – 4858) reads per sample with a mean length of 211 bases. For each patient sample an average of 98.88% of the GS FLX nucleotides were mapped onto each reference template previously derived yielding an average coverage of approximately 600 sequence reads per base.
Comparison of UDPS and Standard Direct Sequencing Methods
The number of participants with a specific HIV ART-resistance major mutation detected by standard direct sequencing and UDPS was analyzed and compared (Table 1). Eight ART-resistance mutations were detected by UDPS, three for PIs, three for NRTIs and two for NNRTIs, and only one ART-resistance mutation, K65R (NRTI), was observed in more than one study participant. Only two mutations, M184V (NRTI) and K103N (NNRTI) were detected by both UDPS and standard direct sequencing. These two mutations were confined to detection by direct sequencing in a single patient (KA02). These results indicate that UDPS identified all DRMs at levels ≥ 20% that were detected by standard sequencing.
Table 1.
HIV ART Resistance Mutations Detected by Direct and Ultra-Deep Sequencing
| Resistance Mutations | Sequencing Method* | ||
|---|---|---|---|
| Direct Only | Ultra-Deep Only | Direct and Ultra-Deep | |
| PI | |||
| D30N | 0 | 1 | 0 |
| M46I | 0 | 1 | 0 |
| G73S | 0 | 1 | 0 |
| NRTI | |||
| K65R | 0 | 6 | 0 |
| M184V | 1 | 1 | 1 |
| K219R | 0 | 1 | 0 |
| NNRTI | |||
| K103N | 1 | 1 | 1 |
| G190A | 0 | 1 | 0 |
Number of participants with a specific HIV ART resistance major mutation detected by a specific sequencing method.
Overall Rates of Drug-Resistance Mutations
For all subjects low-abundance drug resistant mutations were detected using the 2009 SDRM list. ART mutations detected by UDPS were categorized as having only low abundance variants (prevalence level of <20%) and those with >20% prevalence level. Low-abundant drug resistance mutations constituted 85% off all resistance mutations found (Fig. 1). Only 15% of resistance mutations found in all subjects were detected in a frequency >20%.
Fig. (1).
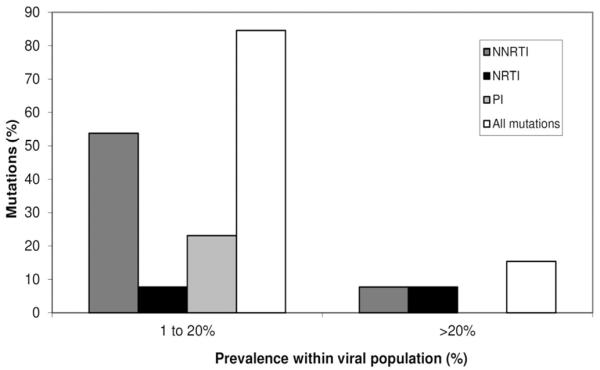
Proportion of resistance mutations within the viral population of subtype C infected ART-naïve Zambian individuals. Proportion of mutations detected by UDPS with an abundance of 1 to 20% and > 20% of the virus population. Each bar indicates percentages of NNRTIs (dark gray), NRTIs (black), PIs (gray) and overall mutations (light gray) that comprise each prevalence range category.
Specific Drug Resistance Mutations Per Class
Of the 10 subjects analyzed only one had a NNRTI mutations. These mutations were K103N and G190A, and were present in 71% and 5% of the sequence reads respectively from a single patient (KA02) (Table 2).
Table 2.
Frequency of RT Inhibitor Resistance Mutations Detected by Ultra-Deep Sequencing
| Reverse Transcriptase | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed Prevalence for Each Resistance Mutation (%) within the Viral Population of Each Study Participant | |||||||||||
| Mutation | Class | MO09 | 924 | LUK06 | MO03 | LV04 | 1690 | 1931 | KA02 | CH02 | 1994 |
| K65R | NRTI | 0 | 1.85 | 0 | 0 | 1.37 | 1.19 | 1.42 | 1.14 | 1.32 | 0 |
| M184V | NRTI | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 70.23 | 0 | 0 |
| K219R | NRTI | 1.53 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K103N | NNRTI | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 70.81 | 0 | 0 |
| G190A | NNRTI | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.04 | 0 | 0 |
Similarly, only three NRTI mutations were detected in these patients One of these NRTI mutations, K65R, which was detected in 6 of the 10 patients (Table 2). Another primary major mutation that can confer NRTI resistance detected in low frequencies was K219R.
Similar to the NNRTI mutations the majority of major PI mutations were observed at a prevalence rate of 1 to 20% (Fig. 1). Only three mutations were observed at frequencies greater than 1%, two of which, D30N and M46I, were found in the same patient LUK06 (Table 3).
Table 3.
Frequency of PR Inhibitor Resistance Mutations Detected by Ultra-Deep Sequencing
| Protease | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed Prevalence for Each Resistance Mutation (%) within the Viral Population of Each Study Participant | |||||||||||
| Mutation | Class | MO09 | 924 | LUK06 | MO03 | LV04 | 1690 | 1931 | KA02 | CH02 | 1994 |
| D30N | PI | 0 | 0 | 3.71 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| M46I | PI | 0 | 0 | 7.83 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| G73S | PI | 0 | 5.96 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Predicted Genotype Resistance Using the Stanford Interpretation Algorithm
The resistance mutations associated with NRTIs from all patients was analyzed using the Stanford algorithm (Table 4). While patients MO09 and KA02 had potential low-level resistance and low-level resistance to D4T respectively, subjects CH02, 1690,924, LV04 and 1931 had intermediate resistance. Intermediate resistance to 3TC, ABC, DDI, FTC and TDF was observed in patients CH02, 1690,924, LV04 and 1931. On the other hand, high level resistance to 3TC, ABC and FTC was observed in patient KA02. In addition, a potential low-level resistance to AZT was observed in patient MO09. As for NNRTIs, high level resistance to EFV and NVP and partial low-level resistance to ETR and RPV were observed in subject KA02 (Table 5). Resistance to the new generation NNRTI ETR was attributed to the presence of G190A, which is one of the 13 mutations earmarked in the DUET study trials with a response to ETR (Katlama C, et al. (2007) 4th IAS Conference on HIV Pathogenesis, Treatment and Prevention http://www.ias2007.org).
Table 4.
Genotypic Resistance to NRTIs in ART-Naïve Individuals as Determined by the Stanford Interpretation Algorithm
| Subject ID | Resistance Mutations | Predicted Susceptibility | ||||||
|---|---|---|---|---|---|---|---|---|
| 3TC | ABC | AZT | D4T | DDI | FTC | TDF | ||
| MO09 | K219R | S | S | PLLR | PLLR | S | S | S |
| 1994 | n | S | S | S | S | S | S | S |
| CH02 | K65R | I | I | S | I | I | I | I |
| 1690 | K65R | I | I | S | I | I | I | I |
| LUK06 | n | S | S | S | S | S | S | S |
| MO03 | n | S | S | S | S | S | S | S |
| KA02 | K65R, M184V | R | R | S | LLR | I | R | I |
| 924 | K65R | I | I | S | I | I | I | I |
| LV04 | K65R | I | I | S | I | I | I | I |
| 1931 | K65R | I | I | S | I | I | I | I |
List of abreviations: 3TC, lamivudine; ABC, abacavir; AZT, zidovudine; D4T, stavudine; DDI, didanosine; FTC, emtricitabine; TDF, tenofovir; I, intermediate resistance; LLR, low-level resistance; PLLR, potential low-level resistance; R, high-level resistance; S, susceptible; n, no resistance mutations.
Table 5.
Genotypic Resistance to NNRTIs in ART-Naïve Individuals as Determined by the Stanford Interpretation Algorithm
| Subject ID | Resistance Mutations | Predicted Susceptibility | |||
|---|---|---|---|---|---|
| EFV | ETR | NVP | RPV | ||
| MO09 | n | S | S | S | S |
| 1994 | n | S | S | S | S |
| CH02 | n | S | S | S | S |
| 1690 | n | S | S | S | S |
| LUK06 | n | S | S | S | S |
| MO03 | n | S | S | S | S |
| KA02 | K103N, G190A | R | PLLR | R | PLLR |
| 924 | n | S | S | S | S |
| LV04 | n | S | S | S | S |
| 1931 | n | S | S | S | S |
List of abreviations: EFV, efavirenz; ETR, etravirine; NVP, nevirapine; I, intermediate resistance; RPV, rilpivirine; LLR, low-level resistance; PLLR, potential low-level resistance; R, high-level resistance; S, susceptible; n, no resistance mutations.
Among PIs, patient LUK06 had HIV high level resistance to NFV partial low-level resistance to ATV, FPV, IDV and LPV (Table 6). Meanwhile, patient 924 harbored partial level resistance to ATV, NFV and SQV.
Table 6.
Genotypic Resistance to Pis in ART-Naïve Individuals as Determined by the Stanford Interpretation Algorithm
| Subject ID | Resistance Mutations | Predicted Susceptibility | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ATV/r | DRV/r | FPV/r | IDV/r | LPV/r | NFV | SQV/r | TPV/r | ||
| MO09 | n | S | S | S | S | S | S | S | S |
| 1994 | n | S | S | S | S | S | S | S | S |
| CH02 | n | S | S | S | S | S | S | S | S |
| 1690 | n | S | S | S | S | S | S | S | S |
| LUK06 | D30N, M46I | PLLR | S | PLLR | PLLR | PLLR | R | S | S |
| MO03 | n | S | S | S | S | S | S | S | S |
| KA02 | n | S | S | S | S | S | S | S | S |
| 924 | G73S | PLLR | S | S | S | S | PLLR | PLLR | S |
| LV04 | n | S | S | S | S | S | S | S | S |
| 1931 | n | S | S | S | S | S | S | S | S |
List of abreviations: ATV/r, atazanavir/ritonavir; DRV/r, darunavir/ritonavir; FPV/r, fosamprenavir/ritonavir; IDV/r, indinavir/ritonavir; LPV/r, lopinavir/ritonavir; NFV, nelfinavir; SQV/r, saquinavir/ritonavir; TPV/r, tipranavir/ritonavir; I, intermediate resistance; LLR, low-level resistance; PLLR, potential low-level resistance; R, high-level resistance; S, susceptible; n, no resistance mutations.
DISCUSSION
In this study we have used UDPS technology to detect minor virus variants in HIV-1 pol from 10 drug-naive individuals from Zambia infected with HIV-1 subtype C. In recent years, accumulating evidence has suggested that drug-resistant viral variants present at low frequency in the population are clinically important, as they may have a profound impact on treatment outcomes such as virologic failure to NNRTI-based therapy [4, 6, 18, 19]. The detected frequency of these clinically relevant minority variants could be as low as 1% by UDPS [4, 5], or 0.07% by allele specific PCR [18]. Conversely, not every study can associate minor resistant variants with therapy failure [20–22]. Since 2004, first-line ART in Zambia has comprised of a NRTI backbone, AZT or d4T plus lamivudine, in combination with a non-NRTI such as EFV or NVP. In July 2007, the Zambian Ministry of Health introduced TDF as part of first-line therapy, making it the first African country to use it as a nucleotide/NRTI backbone on a wide scale Mwinga A. (2007) HIV/AIDS Implementer's meeting http://hivimplem enters.com/2007/). Early indications describing the clinical outcomes among patients prescribed TDF have demonstrated a favorable benefit with similar clinical and programmatic outcomes as AZT and d4T [23]. Results from our analysis suggest that the viral quasispecies within these therapy-naïve individuals harbor low-abundance resistance mutations that may compromise the first line therapy, including TDF, currently available to patients in Zambia. Intermediate to high-level resistance was predicted for the majority of NRTIs. Intermediate resistance to TDF was observed in 6 of the 10 individuals and attributed to the presence of K65R. This polymorphism was detected at an average of 1.36% of the viral population of each these participants (Table 2). However, one individual harbored M184V in combination with K65R, which increases the level of susceptibility to TDF [24–26]. This K65R mutation is one of the most important HIV-1 drug-resistance mutations that confer decreased susceptibility to all clinically used NRTIs except zidovudine. Recent clinical studies had shown the preferential emergence of K65R in patients infected with subtype C from a range of different treatment regimens in sub-Saharan Africa and elsewhere [27–29]. Additionally, low abundance of K65R have been detected by UDPS in ART-naïve non-B subtypes and correlated with virological failure [5]. The predominance of K65R in subtype C can be explained by a template pausing molecular mechanism that facilitates the selection of K65R in the KKK motif at codons 64, 65, 66 of the RT [30–32], and has been suggested that this characteristic of the KKK motif can lead to PCR induced K65R [33]. However, a recent UDPS study were controls of different percentages of K65R mutation were included and the theoretical and observed values of K65R plasmid dilutions were similar (100%:94%, 5%:2.30% and 1%:0.90%), indicates that the frequencies of K65R obtained by UDPS are reasonably accurate [34]. The presence of TDF resistance in the population should be a concern for the current ART regimen.
Generally, second-line treatment is limited to boosted protease inhibitor based regimes for patients failing first-line therapy. ETR offers a higher genetic barrier to the development of viral resistance than the first-generation NNRTIs [35]. Yet, to date little is known about the drug's potential effectiveness in sub-Saharan Africa although on a positive note over 90% of patients previously exposed in southern Africa treatment programs would remain susceptible to ETR [36]. Recently, UDPS of treatment-naïve and NNRTI experienced patients found that ETR may be effective in treatment-naive patients with the most commonly occurring NNRTI-resistant mutation K103N [37]. Interestingly, when we assessed the potential of ETR resistance in our subjects, we observed a potential low-level resistance attributed to G190A in only one out of the ten subjects. Therefore, a majority of the ART naive patients in Zambia may still have a good optimum response to ETR.
Overall, the knowledge about the prevalence of drug-resistance mutations among both ART-naïve and treated individuals in Zambia is limited [9–12]. A recent study that assessed the mutational patterns and factors associated with baseline drug-resistant HIV present at the initiation of baseline ART shows that resistance mutations may compromise the response to standard first-line ART in 6% of patients in Lusaka, Zambia [10]. In addition, three of the NRTI and one of the NNRTI mutations detected in our cohort were also observed in a cohort of Zambian children during 24 months of first-line ART [9]. By using UDPS, in addition to the reported mutations from these studies, we have also detected additional potential resistance variants, probably due to the detection limit of direct sequencing employed by these studies. It is possible that many of the resistance mutants detected after initiation of treatment in the treated children were likely to exist prior to treatment initiation but were undetectable by conventional sequencing. Therefore, if an analysis of low-abundance drug-resistance mutations is available prior to the initiation of treatment, an optimal ART combination can be used from the beginning. It is likely that the UDPS technology will continue to be improved and will become economically feasible to monitor drug-resistant variants even in the developing country settings.
There are several limitations to the present investigation. First, this was a preliminary study examining the feasibility of using UDPS in limited resource settings such as sub-Saharan Africa. Therefore, our sample size is limited. However this analysis gives us an insight into the level of primary antiretroviral resistance at a population level in Zambia. Secondly, a widely recognized problem associated with UDPS is the possibility of under or oversampling of the viral quasispecies pool due to a variable number of genome copy numbers amplified from each subject. Given this variability there is always likelihood for resampling the same template due to a low copy number. In turn this may distort the true frequencies of variants within a giving population. Nevertheless it is unlikely that our frequencies are over inflated by target resampling as there is a higher likelihood of resampling the wild type variant as it is the predominant form within the population. Thus, we may be actually underestimating the significance of the low-level drug-resistant mutations in the population. The 454/Roche method similar to any other sequencing technique involving PCR steps may induce base pair substitutions leading to drug resistance mutations that are from PCR error [33]. Despite this challenge the detection of a large proportion of the mutations found in this study is in agreement with previous reports demonstrating that most of these mutations are authentic and currently affecting the first-line treatment of patients in Zambia.
Unfortunately, in this study it was not possible to analyze viral RNA due to the difficulties associated with the transport and storage of plasma. Therefore, resistance mutations were analyzed from PBMCs. While proviral DNA provides a good account of archived resistance mutations it may not represent those variants involved in active replication. However, previous studies using UDPS have revealed that proviral DNA may hide segregated variants [38, 39] that represent a minor subpopulation of the viral quasispecies and may be a source of replication competent virus. Although it is not possible to evaluate the level of proviral variants being expressed we can assume that some of them will generate viral progeny or even recombine with replicating virus as previously shown [38]. As a result archived provirus variants may preferential replicate at a given time point and undergo selective expansion in the presence of ART, possibly affecting the therapeutic response. In fact it has been now shown that the rates of detection of ART mutations in plasma RNA are similar to those observed in proviral DNA from PBMC samples [40].
CONCLUSIONS
In conclusion, we have demonstrated that UDPS can provide new insights on the burden of drug-resistance in treatment-naïve subjects in developing countries. With this technology we were able to identify and quantify an array of preexisting HIV-1 resistance variants that were not detected by conventional sequencing approaches. Given the increasing prevalence of subtype C infections worldwide it is vital that minor drug-resistance variants are identified to ensure optimization of treatment regimens.
ACKNOWLEDGEMENTS
We thank all the study participants and the personnel at the University Teaching Hospital and Voluntary Counseling and Testing centers in Zambia for their collaboration. This study was supported by Public Health Service grants CA75903, P01AI48240, NIGMS COBRE grant P30 GM103509, D43 TW001492 and T32 AI060547 to CW; SG was an NIH Ruth L. Kirschstein Fellow and CG was a Fogarty Fellow.
Footnotes
AUTHORS' CONTRIBUTIONS Conceived and designed the experiments: CW, SG and CG. Contributed with reagents/materials/analysis tools: CW and DT. Performed the experiments: SG and CG. Analyzed and interpreted the data: DT, SG, and CG. Drafted the article: SG, DT and CW. All authors read and approved the final manuscript.
CONFLICT OF INTEREST No financial or non-financial conflict of interests exist.
PATIENT CONSENT Declared none.
HUMAN/ANIMAL RIGHTS Declared none.
REFERENCES
- [1].Buckton AJ. New methods for the surveillance of HIV drug resistance in the resource poor world. Curr Opin Infect Dis. 2008;21:653–8. doi: 10.1097/QCO.0b013e3283186d1a. [DOI] [PubMed] [Google Scholar]
- [2].Novak RM, Chen L, MacArthur RD, et al. Prevalence of antiretroviral drug resistance mutations in chronically HIV-infected, treatment-naive patients: implications for routine resistance screening before initiation of antiretroviral therapy. Clin Infect Dis. 2005;40:468–74. doi: 10.1086/427212. [DOI] [PubMed] [Google Scholar]
- [3].Little SJ, Holte S, Routy JP, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347:385–94. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- [4].Simen BB, Simons JF, Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199:693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- [5].Lataillade M, Chiarella J, Yang R, et al. Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naive subjects in the CASTLE study. PLoS One. 2010;5:e10952. doi: 10.1371/journal.pone.0010952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Le T, Chiarella J, Simen BB, et al. Low-abundance HIV drug-resistant viral variants in treatment-experienced persons correlate with historical antiretroviral use. PLoS One. 2009;4:e6079. doi: 10.1371/journal.pone.0006079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hedskog C, Mild M, Jernberg J, et al. Dynamics of HIV-1 quasispecies during antiviral treatment dissected using ultra-deep pyrosequencing. PLoS One. 2010;5:e11345. doi: 10.1371/journal.pone.0011345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20(16):W13–23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- [9].Gupta RK, Ford D, Mulenga V, et al. Drug resistance in human immunodeficiency virus type-1 infected Zambian children using adult fixed dose combination stavudine, lamivudine, and nevirapine. Pediatr Infect Dis J. 2010;29:e57–62. doi: 10.1097/INF.0b013e3181e47609. [DOI] [PubMed] [Google Scholar]
- [10].Hamers RL, Siwale M, Wallis CL, et al. HIV-1 drug resistance mutations are present in six percent of persons initiating antiretroviral therapy in Lusaka, Zambia. J Acquir Immune Defic Syndr. 2010;55:95–101. doi: 10.1097/QAI.0b013e3181e544e0. [DOI] [PubMed] [Google Scholar]
- [11].Gonzalez S, Gondwe C, Tully DC, et al. Short communication: antiretroviral therapy resistance mutations present in the HIV type 1 subtype C pol and env regions from therapy-naive patients in Zambia. AIDS Res Hum Retroviruses. 2010;26:795–803. doi: 10.1089/aid.2009.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Handema R, Terunuma H, Kasolo F, et al. Prevalence of drug-resistance-associated mutations in antiretroviral drug-naive Zambians infected with subtype C HIV-1. AIDS Res Hum Retroviruses. 2003;19:151–60. doi: 10.1089/088922203762688667. [DOI] [PubMed] [Google Scholar]
- [13].Stringer JS, Zulu I, Levy J, et al. Rapid scale-up of antiretroviral therapy at primary care sites in Zambia: feasibility and early outcomes. Jama. 2006;296:782–93. doi: 10.1001/jama.296.7.782. [DOI] [PubMed] [Google Scholar]
- [14].Yun Z, Fredriksson E, Sonnerborg A. Quantification of human immunodeficiency virus type 1 proviral DNA by the TaqMan real-time PCR assay. J Clin Microbiol. 2002;40:3883–4. doi: 10.1128/JCM.40.10.3883-3884.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang H, Zhou Y, Alcock C, et al. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J Virol. 2004;78:1718–29. doi: 10.1128/JVI.78.4.1718-1729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hoffmann C, Minkah N, Leipzig J, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gifford RJ, Rhee SY, Eriksson N, et al. Sequence editing by Apolipoprotein B RNA-editing catalytic component-B and epidemiological surveillance of transmitted HIV-1 drug resistance. AIDS. 2008;22:717–25. doi: 10.1097/QAD.0b013e3282f5e07a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Metzner KJ, Giulieri SG, Knoepfel SA, et al. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin Infect Dis. 2009;48:239–47. doi: 10.1086/595703. [DOI] [PubMed] [Google Scholar]
- [19].Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008;5:e158. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Peuchant O, Thiebaut R, Capdepont S, et al. Transmission of HIV-1 minority-resistant variants and response to first-line antiretroviral therapy. AIDS. 2008;22:1417–23. doi: 10.1097/QAD.0b013e3283034953. [DOI] [PubMed] [Google Scholar]
- [21].Messiaen P, Verhofstede C, Vandenbroucke I, et al. Ultra-deep sequencing of HIV-1 reverse transcriptase before start of an NNRTI-based regimen in treatment-naive patients. Virology. 2012;426:7–11. doi: 10.1016/j.virol.2012.01.002. [DOI] [PubMed] [Google Scholar]
- [22].Jakobsen MR, Tolstrup M, Sogaard OS, et al. Transmission of HIV-1 drug-resistant variants: prevalence and effect on treatment outcome. Clin Infect Dis. 2010;50:566–73. doi: 10.1086/650001. [DOI] [PubMed] [Google Scholar]
- [23].Chi BH, Mwango A, Giganti M, et al. Early clinical and programmatic outcomes with tenofovir-based antiretroviral therapy in Zambia. J Acquir Immune Defic Syndr. 2010;54:63–70. doi: 10.1097/QAI.0b013e3181c6c65c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wolf K, Walter H, Beerenwinkel N, et al. Tenofovir resistance and resensitization. Antimicrob Agents Chemother. 2003;47:3478–84. doi: 10.1128/AAC.47.11.3478-3484.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Naeger LK, Margot NA, Miller MD. Increased drug susceptibility of HIV-1 reverse transcriptase mutants containing M184V and zidovudine-associated mutations: analysis of enzyme processivity, chain-terminator removal and viral replication. Antivir Ther. 2001;6:115–26. [PubMed] [Google Scholar]
- [26].Whitcomb JM, Parkin NT, Chappey C, Hellmann NS, Petropoulos CJ. Broad nucleoside reverse-transcriptase inhibitor cross-resistance in human immunodeficiency virus type 1 clinical isolates. J Infect Dis. 2003;188:992–1000. doi: 10.1086/378281. [DOI] [PubMed] [Google Scholar]
- [27].Doualla-Bell F, Avalos A, Brenner B, et al. High prevalence of the K65R mutation in human immunodeficiency virus type 1 subtype C isolates from infected patients in Botswana treated with didanosine-based regimens. Antimicrob Agents Chemother. 2006;50:4182–5. doi: 10.1128/AAC.00714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Toni TA, Brenner BG, Asahchop EL, Ntemgwa M, Moisi D, Wainberg MA. Development of an allele-specific PCR for detection of the K65R resistance mutation in patients infected with subtype C human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2010;54:907–11. doi: 10.1128/AAC.01080-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Orrell C, Walensky RP, Losina E, Pitt J, Freedberg KA, Wood R. HIV type-1 clade C resistance genotypes in treatment-naive patients and after first virological failure in a large community antiretroviral therapy programme. Antivir Ther. 2009;14:523–31. [PMC free article] [PubMed] [Google Scholar]
- [30].Coutsinos D, Invernizzi CF, Moisi D, et al. A template-dependent dislocation mechanism potentiates K65R reverse transcriptase mutation development in subtype C variants of HIV-1. PLoS One. 2011;6:e20208. doi: 10.1371/journal.pone.0020208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Coutsinos D, Invernizzi CF, Xu H, Brenner BG, Wainberg MA. Factors affecting template usage in the development of K65R resistance in subtype C variants of HIV type-1. Antivir Chem Chemother. 2010;20:117–31. doi: 10.3851/IMP1443. [DOI] [PubMed] [Google Scholar]
- [32].Coutsinos D, Invernizzi CF, Xu H, et al. Template usage is responsible for the preferential acquisition of the K65R reverse transcriptase mutation in subtype C variants of human immunodeficiency virus type 1. J Virol. 2009;83:2029–33. doi: 10.1128/JVI.01349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Varghese V, Wang E, Babrzadeh F, et al. Nucleic acid template and the risk of a PCR-Induced HIV-1 drug resistance mutation. PLoS One. 2010;5:e10992. doi: 10.1371/journal.pone.0010992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Recordon-Pinson P, Papuchon J, Reigadas S, Deshpande A, Fleury H. K65R in Subtype C HIV-1 Isolates from Patients Failing on a First-Line Regimen Including d4T or AZT: Comparison of Sanger and UDP Sequencing Data. PLoS One. 2012;7:e36549. doi: 10.1371/journal.pone.0036549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jayaweera DT, Espinoza L, Castro J. Etravirine: the renaissance of non-nucleoside reverse transcriptase inhibitors. Expert Opin Pharmacother. 2008;9:3083–94. doi: 10.1517/14656560802489569. [DOI] [PubMed] [Google Scholar]
- [36].Stevens WS, Wallis CL, Sanne I, Venter F. Will etravirine work in patients failing nonnucleoside reverse transcriptase inhibitor-based treatment in southern Africa? J Acquir Immune Defic Syndr. 2009;52:655–6. doi: 10.1097/QAI.0b013e3181ba1b00. [DOI] [PubMed] [Google Scholar]
- [37].Varghese V, Shahriar R, Rhee SY, et al. Minority variants associated with transmitted and acquired HIV-1 nonnucleoside reverse transcriptase inhibitor resistance: implications for the use of second-generation nonnucleoside reverse transcriptase inhibitors. J Acquir Immune Defic Syndr. 2009;52:309–15. doi: 10.1097/QAI.0b013e3181bca669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rozera G, Abbate I, Bruselles A, et al. Archived HIV-1 minority variants detected by ultra-deep pyrosequencing in provirus may be fully replication competent. AIDS. 2009;23:2541–3. doi: 10.1097/QAD.0b013e32832fd2e1. [DOI] [PubMed] [Google Scholar]
- [39].Rozera G, Abbate I, Bruselles A, et al. Massively parallel pyrosequencing highlights minority variants in the HIV-1 env quasispecies deriving from lymphomonocyte sub-populations. Retrovirology. 2009;6:15. doi: 10.1186/1742-4690-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Soto-Ramirez LE, Rodriguez-Diaz R, Duran AS, et al. Antiretroviral resistance among HIV type 1-infected women first exposed to antiretrovirals during pregnancy: plasma versus PBMCs. AIDS Res Hum Retroviruses. 2008;24:797–804. doi: 10.1089/aid.2007.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]


