Abstract
The SecA2 proteins are a special class of transport-associated ATPases that are related to the SecA component of the general Sec system, and are found in an increasingly large number of Gram-positive bacterial species. The SecA2 substrates are typically linked to the cell wall, but may be lipid-linked, peptidoglycan-linked, or non-covalently associated S-layer proteins. These substrates can have a significant impact on virulence of pathogenic organisms, but may also aid colonization by commensals. The SecA2 orthologues range from being highly similar to their SecA paralogues, to being distinctly different in apparent structure and function. Two broad classes of SecA2 are evident. One transports multiple substrates, and may interact with the general Sec system, or with an as yet unidentified transmembrane channel. The second type transports a single substrate, and is a component of the accessory Sec system, which includes the SecY paralogue SecY2 along with the accessory Sec proteins Asp1-3. Recent studies indicate that the latter three proteins may have a unique role in coordinating post-translational modification of the substrate with transport by SecA2. Comparative functional and phylogenetic analyses suggest that each SecA2 may be uniquely adapted for a specific type of substrate.
1. Introduction and Overview
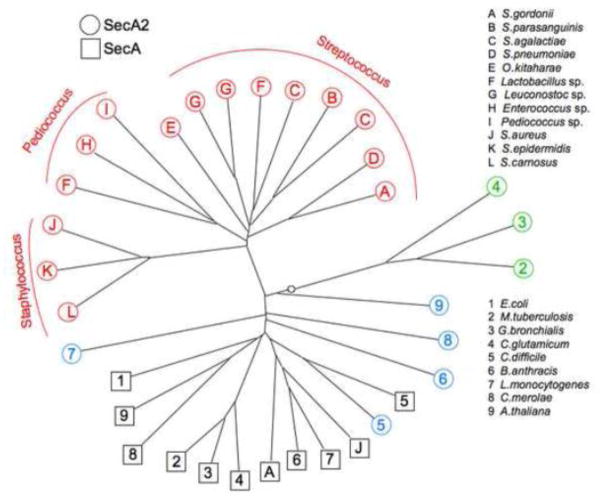
The genomes of many Gram-positive bacterial species encode two homologues of the SecA transport-associated ATPase. The first examples of a second SecA, or SecA2 (a SecA homologue different from the core component of the general Sec system), were noted in Mycobacterium tuberculosis, Listeria monocytogenes and Streptococcus gordonii a little more than a decade ago [1–3]. Until very recently, it was thought that only a small group of pathogenic, Gram-positive bacteria expressed two distinctly different SecA homologues. However, as large-scale microbiome projects have made the genome sequences of more understudied organisms available, it has become apparent that a much larger group of bacteria, including commensal and food-grade organisms, also encode SecA2 proteins. The precise number of bacterial species that express SecA2 is difficult to assess, in part because there are as yet few defining characteristics for the SecA2 orthologues (interspecies homologues). Thus the SecA2 proteins are often not identified in the databases as such, but rather may be incorrectly annotated as SecA. Although the SecA2 proteins of mycobacteria, listeria and streptococci are all similar to SecA proteins of the general Sec system, it has become apparent that these SecA2s are not closely related to each other, either phylogenetically (Fig. 1) or functionally. The SecA2 proteins have typically been classified into two categories: those that have a corresponding SecY2 and thus belonging to a SecA2/Y2 (or accessory Sec) system, and those that do not. As we will discuss, there is increasing evidence for a much larger spectrum of SecA2 types.
Figure 1. Phylogenetic relatedness of the SecA and SecA2 proteins.
Phylogenetic analysis of the SecA and SecA2 proteins from selected Gram-positive bacterial species was performed using DARWIN [96]. Species are presented more than once if there are strains that encode significantly different SecA2 orthologues. The unrooted tree shows three main branches of SecA2 divergence: 1) SecA2s of the accessory Sec system (red), 2) SecA2s of the Actinobacteria species (green), and 3) other SecA2s (cyan) and SecA proteins of the general Sec system (black), with E. coli SecA included as a point of reference. SecA2 homologues from Arabidopsis thaliana and the unicellular red alga Cyanidioschyzon merolae are included for comparison.
2. General characteristics of the SecA2 proteins
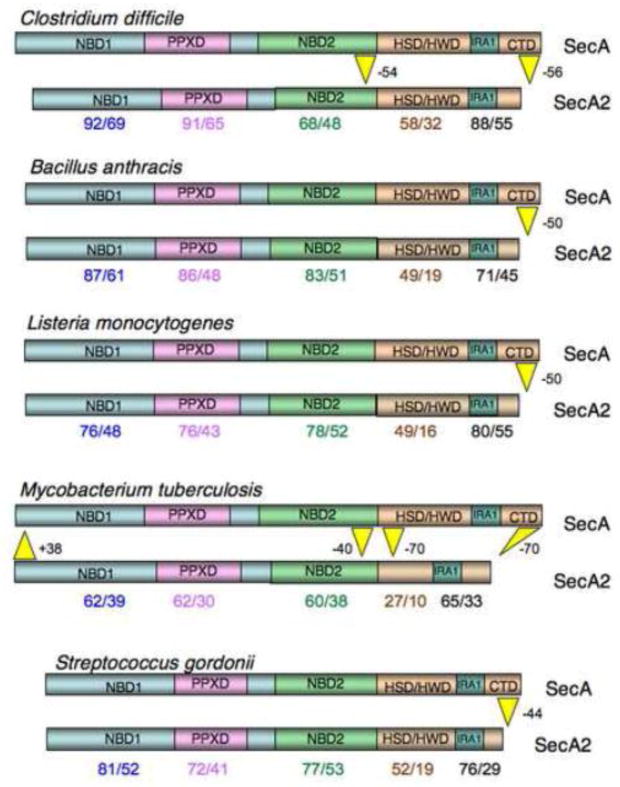
The SecA family of ATPases is characterized by four major domains: two nucleotide binding folds (NBD1 and NBD2), a preprotein binding domain (PPXD) located within NBD1, and a C-terminal region that encompasses several functional or structural domains (the HWD/HSD, IRA1 and CTD; Fig. 2). The HWD/HSD provides a platform to which NBD1 and NBDs are anchored, and interacts with both SecY and preproteins during transport [4–8]. The IRA1 and CTD domains have a significant impact on ATP binding and hydrolysis by the NBD, and preprotein binding by the PPXD, respectively [5, 9]. While it is clear that the ability to hydrolyze ATP is essential for the function of SecA, and that this activity is affected by preprotein binding, the precise means by which SecA couples the energy of ATP hydrolysis with movement of the preprotein through the SecYEG channel is still under intense debate [10–12].
Figure 2. Comparative domain organization among SecA2 proteins.
SecA2 domains were identified by alignment with E. coli SecA. Domain boundaries are based on those determined by Papanikolau et al [4]. NBD, nucleotide binding domain; PPXD, preprotein cross-linking domain; HWD, helical wing domain; HSD, helical scaffold domain; IRA, intramolecular regulator of ATPase activity; CTD, C-terminal domain. IRA1 is a functional domain that overlaps with the two-helix finger structural domain [6, 97]. Numbers shown below the SecA2 proteins indicate the percent similarity/identity with the corresponding domain of the SecA paralogue.
Like SecA, the SecA2 proteins have two NBDs that flank a PPXD. However, they are all shorter than their SecA paralogues (i.e. SecA from the same organism, and sometimes referred to as SecA1 in organisms that have a SecA2), and may contain deletions within one or more of the domains. In contrast to SecA, which is involved in the transmembrane transport of a number of secreted and cell wall proteins, SecA2 is typically involved in the transport of a very limited number of substrates. With a few exceptions, SecA2 is not essential for viability, but instead has a major impact on virulence or colonization. In general, this is because one or more of the SecA2 substrates contributes to survival or the occupation of a specific niche in vivo. So what is the role of SecA2 in transport? The answer may depend on the substrate.
3. Transport by multi-substrate SecA2 proteins
One broad set of SecA2 proteins lack a corresponding SecY2 partner, and facilitate transport in what are sometimes called “SecA2 only” systems. This type of SecA2-mediated transport has been recently reviewed [13, 14], so we will only briefly summarize the composite findings, and present some additional observations and new perspectives. Although the means by which they interact with a transmembrane channel may not be the same in all cases, it is apparent that they all are more selective than SecA, with regard to the substrates they transport. The SecA2-dependent substrates are often, but not always, encoded in the same locus as SecA2. Of note, the substrates for SecA2-dependent transport identified thus far appear to be involved in oxidative stress response, cell wall metabolism or structural modification of the cell wall. These substrates are not readily distinguishable from general Sec system substrates, and are at least partially transportable by SecA/YEG, although the secA2 phenotype may be more apparent under certain growth conditions or in vivo.
3.1 Mycobacteria and Corynebacteria (Actinobacterial type)
The first SecA2 homologue was noted upon completion of the M. tuberculosis genome in 2000 [2]. It has subsequently become apparent that there are closely related (80–90% identity) SecA2 homologues in all other mycobacterial species. There are also very similar (50–60% identity) SecA2 orthologues in other actinobacterial genera such as Gordinia and Corynebacteria (Table I and Fig. 1), although these differ somewhat from each other and from the mycobaterial SecA2s with regard to deletions or additions within the SecA2 domains. The apparent phylogenetic and structural differences between the Mycobacteria and Corynebacteria SecA2s may reflect a functional difference, since the SecA2 of Corynebacterium glutamicum was found to be essential for viability [15], whereas the mycobacterial SecA2s are not.
Table I.
Gram-positive bacterial species that encode SecA2.a
| Actinobacteria | Listeria | Bacillusb | Streptococcusc | Staphylococcus | Pediococcusd |
|---|---|---|---|---|---|
| Mycobacteria | Listeria | Bacillus | Streptococcus | Staphylococcus | Pediococcus |
| M. tuberculosis | L.monocytogenes | B. anthracis | S. pneumoniae | S. aureu | P.pentosacceus |
| M. smegmatis | L. inocua | B. cereus | S. mitis | S. epidermidis | P. acidilactici |
| M. leprae | L. seeligeri | B. thuringiensis | S. oralis | S. warneri | P.lolii |
| M. bovis | L. marthii | B. smithii | S. sanguinis | S. carnosus | P.claussenii |
| M. marinum | L. vanovii | B.methanolicus | S. salivarius | S.intermedius | |
| M.canetti | L. welshimeri | S.suis | Enterococcus | ||
| M.avium | L. grayi | S.iniae | E.saccharolyticus | ||
| M.abscessus | S.porcinus | E.raffinosus | |||
| S.australis | E.avium | ||||
| Gordonia | S.vestibularis | E.sulfureus | |||
| G.otidis | S.cristatus | E.pallens | |||
| G.rhizosphera | |||||
| G.bronchialis | Lactobacillus | Lactobacillus | |||
| G.alkanivorans | L. salivarius | L.paracasei | |||
| G.hirsuta | L. johnsonii | L.suebicus | |||
| G.effusa | L.murinus | L.fructivorans | |||
| G.sputi | L.gastricus | L.rhamnosus | |||
| L.zeae | |||||
| Corynebacterium | Leuconostoc | ||||
| C. glutamicum | L.gasicomitatum | ||||
| C. diptheriae | L.gelidium | ||||
| C.durum | L.kimchii |
grouped according to the major and minor branch types in Fig. 1 (not all-inclusive)
absent from B. subtilis
absent from S. pyogenes and S. mutans
absent from E. faecalis and E. faecium
The most extensively characterized SecA2 proteins are those of Mycobacterium tuberculosis and Mycobacterium smegmatis. Mutational analysis identified two substrates for the M. tuberculosis SecA2, KatG and the Fe-dependent SodA [16]. Both of theses substrates lack a canonical Sec signal peptide. For M. smegmatis, two lipoproteins that are predicted to be involved in sugar uptake have been documented as SecA2 substrates [17]. Although both proteins have a traditional lipoprotein signal peptide, recent evidence indicates that the mature regions confer a dependence on SecA2 for export [18]. That is, rapid folding of the mature region (versus a specific sequence in the mature region) may render the preprotein more dependent on SecA2 for transport.
Consistent with a role as a transport motor, the ATPase activity of mycobacterial SecA2 has been documented by both mutational analyses and by in vitro studies using the purified protein [19]. However, this SecA2 shares just 29% identity (54% similarity) with its SecA paralogue. Most of the similarity between SecA and SecA2 resides within the NBD and PPXD domains (Fig. 2), although SecA2 has a ~35aa deletion between the ATP-binding motifs V and VI of NBD2. This deletion is likely to be functionally relevant, since alterations in NBD2 can affect the rate of ATP hydrolysis by E. coli SecA [20–23]. Of note, the affinity of M. tuberculosis SecA2 for ATP is ten-fold higher, and the resting or “basal” rate of hydrolysis is five-fold lower, than that of SecA [19]. These differences thus suggest that preprotein binding and ATP hydrolysis by SecA2 may be differently coordinated, as compared with SecA.
The mycobacterial SecA2 also has a 38 amino acid extension of NBD1, but conversely has a 70 amino acid CTD truncation. Perhaps more significantly, this SecA2 has a large deletion in the HWD/HSD. Since the HSD provides a major platform for interacting with SecY [6–8], the omission within this region could be a means to prevent SecA2 from interacting directly with the SecYEG translocon. Functional studies support this possibility [24]. That is, SecA2 is primarily cytosolic, whereas SecA is both cytosolic and membrane-associated, although the expression levels of the two proteins are similar. In addition, SecA depletion studies indicate that SecA2 cannot function independently of SecA. The combined data thus support the proposed role for SecA2, in which it interacts with the SecA/YEG translocase via interaction with SecA, rather than interacting directly with SecYEG.
3.2 Listeria
The SecA2 of L. monocytogenes was fortuitously identified because mutations in secA2 can have a dramatic effect on colony and cellular morphology [3]. SecA2 mutants display a rough colony phenotype, elongated cells and more extensive chaining. This is at least in part due to impaired export of two enzymes involved in cell wall metabolism, the p60 (CwhA) autolysin and the NamA (MurA) hydrolase. p60 is encoded in the same locus as SecA2 (Fig. 3) whereas NamA is encoded elsewhere in the genome. Each of these substrates has a canonical signal peptide. Deletion of secA2 markedly reduces (but does not abolish) the export of these two proteins. Accordingly, SecA2 is not essential for viability but instead has a significant effect on virulence [25]. More than a dozen additional SecA2 substrates have been reported in L. monocytogenes [25–27]. One of these, an Mn-containing superoxide dismutase (MnSod), lacks an apparent N-terminal signal peptide, so it is unclear what features affect recognition by SecA2. However, MnSod was shown to undergo cytosolic phosphorylation, which renders it inactive, yet only the active, non-phosphorylated form is secreted [27]. Although there is a correlation between the MnSod phosphorylation level and a dependence on SecA2 for secretion, it has not been determined whether SecA2 recognizes the phosphorylated or dephosphorylated form of the preprotein.
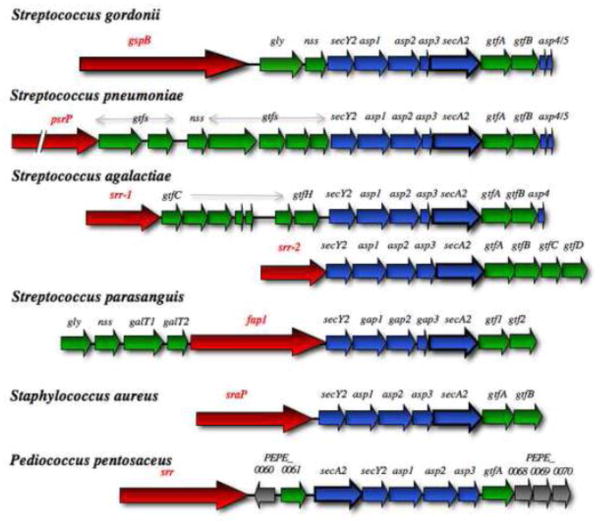
Figure 3. Genetic loci encoding the multi-substrate SecA2 proteins.
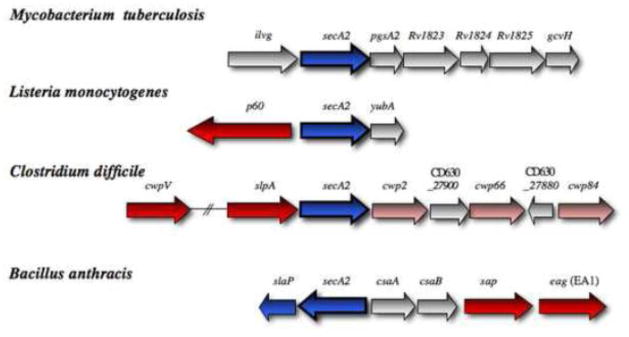
The chromosomal regions flanking secA2 (blue) in Mycobacterium tuberculosis, Listeria monocytogenes, Clostridium difficile and Bacillus anthracis are shown. A component required for SecA2 transport in B. anthracis is also in blue. Genes encoding defined SecA2 substrates are shown in red, and presumed substrates are indicated in pink. Substrates encoded elsewhere in the chromosome are not shown.
The Listeria SecA2 proteins share 40% identity (70% similarity) with their SecA paralogues, with dissimilarities most prevalent in the C-terminal regions. Although they lack a C-terminal CTD, there are otherwise no deletions within SecA2, as compared with the SecA paralogue (Fig. 2). The composite findings thus far do not illuminate whether SecA2 is likely to interact with SecYEG or with a different transmembrane channel. However, a recent report has indicated that the cell division protein DivIVa affects substrate localization to the septal region, whereas SecA2 localizes to the same region independently of DivIVA [28]. The authors suggest that SecA2 may selectively facilitate export of the cell wall hydrolases near the cell septum. In addition to p60, the SecA2 locus encodes YubA, a putative membrane permease that is predicted to have seven transmembrane segments, although a possible role for YubA in transport has not been addressed. A secA2 locus with a similar gene arrangement is present in non-pathogenic listeria species (Table I), and SecA2 mediates NamA export in at least one of these species (L. innocua) [29].
3.3 Bacillus
As was the case in Listeria, the SecA2 of Bacillus anthracis was identified and characterized because of its impact on cellular morphology, septation and chain length [30]. The B. anthracis SecA2 is encoded within a cluster of genes that are responsible for the formation of the S-layer, a paracrystalline lattice of proteins on the bacterial cell surface (Fig. 3). A similar genetic locus is present in several other Bacillus species (Table I), but is apparently absent from B. subtilis. The B. anthracis SecA2 is not essential for viability, and deletion of secA2 results in a decrease, but does not abolish, secretion of Sap and EA1, two major components of the S-layer. The two SecA2 substrates have nearly identical 29 amino acid canonical signal peptides. Sap, but not EA1, is encoded in the secA2 locus.
Regarding how the B. anthracis SecA2 may function, it lacks a C-terminal CTD but otherwise has no deletions compared with SecA (Fig. 2). It shows 75% similarity (45% identity) to its SecA paralogue. The dissimilarities are scattered throughout the protein, but most of the differences are concentrated in the HSD/HWD. A secY2 is present in the genome, but deletion does not result in decreased Sap or EA1 secretion, indicating that SecA2 does not interact with SecY2 to transport these substrates. Conversely, SecA2 may work in conjunction with the Slayer assembly protein SlaP, which is encoded just downstream of SecA2. Deletion of slaP results in a phenotype identical to that resulting from deletion of secA2. Topological predictions suggest that SlaP should be cytoplasmic, but cell fractionation studies point to a peripheral membrane location that is independent of SecA2 and the S-layer proteins. Co-purification experiments indicate that the soluble form of SlaP is not tightly associated with any other protein. Other evidence suggests that SecA2 and SlaP affect the uniform distribution of Sap in the cell envelope. That is, in the absence of SecA2 or SlaP, Sap appears in patches deposited throughout the envelope. The even distribution of Sap and EA1 in the envelope appears to be essential for the localization of the cell wall protein BslO at the septal region, where it catalyzes the separation of daughter cells [30, 31]. While it is possible that SecA2/SlaP simply expedite transport through SecYEG, Nguyen-Mau et al proposed a more intriguing hypothesis in which SecA2/SlaP may confine the export of Sap and EA1 to discrete subcellular locations, perhaps near folding catalysts.
3.4 Clostridium difficile
The only clostridial SecA2 characterized to date is that of C. difficile [32]. As in Bacillus species, secA2 resides within an S-layer gene cluster. However, the C. difficile SecA2 is essential for viability. This may be because one of the SecA2-dependent substrates, SlpA (the main component of the S-layer), is essential for viability. The SecA2-dependent substrates were identified through the combined use of dominant negative variants of SecA2 (NBD mutations) and anti-sense RNA expression (suppression of SecA versus SecA2 expression). In addition to SlpA, an S-layer protein encoded near the secA2 locus (CwpV) was found to be SecA2-dependent. SlpA and CwpV both have traditional Sec-type signal peptides. Additional putative substrates include Cwp2, Cwp66 and Cwp84, which are also encoded in the secA2 locus. Cwp84 is a cysteine protease that is responsible for processing SlpA after it is translocated but prior to its incorporation into the S-layer [33].
Unlike most of the other SecA2 orthologues, the C. difficile SecA2 is very closely related to its SecA paralogue (77% over-all similarity, 54% identity). Most of the differences are found in the HSD/HWD, but there is a 54 aa deletion in NBD2 between Motif V and Motif VI, and a 54 aa C-terminal truncation (Fig. 2). As in mycobacteria, SecA2 is cytosolic, whereas SecA localizes to the membrane [32]. The data suggest that C. difficile SecA2 can function independently of SecA, and thus may interact directly with SecYEG. There do not appear to be SecA2 orthologues in other clostridial species, including the medically relevant C. tetani, C. botulinum, or C. perfringens. Thus, the question of why C. difficile employs a SecA2 for Slayer protein transport is intriguing.
3.6 Possible roles in transport
The over-all similarity of the multi-substrate SecA2 proteins to SecA, rather than some other type of chaperone or ATPase, for example, suggests that the coupling of preprotein binding with ATP hydrolysis is important for function. However, nearly all of the multi-substrate SecA2s have a deletion in the NBD2 domain, suggesting they may bind or hydrolyze ATP differently than SecA, and thus may be differently regulated. Indeed, in vitro experiments with the mycobacterial SecA and SecA2 proteins have demonstrated such differences. If the acquisition of SecA2, a specialized motor protein, provides even a small growth advantage (via a slightly more efficient use of ATP), it is likely to persist in the population. A better understanding of SecA2 function, and whether there are significant differences from the SecA paralogues, will help to determine whether they can be selectively targeted for antimicrobial development.
The composite studies and structural comparisons also suggest that the means by which the motor activity of SecA2 is coupled to active transport may differ from that of SecA, and may further vary amongst the SecA2 orthologues. A direct interaction with SecYEG is most likely for the C. difficile SecA2, which is highly similar to SecA. However, other SecA2s differ significantly from their SecA paralogues in the HSD/HWD domain, which is a region known to provide extensive contacts between SecA and SecY during transport [6–8]. It is therefore less likely that the SecA2s interact directly with the SecYEG translocon, or at least not in the same way that SecA does. Instead, a different mode of interaction with SecYEG could be facilitated through another protein docked at SecYEG. In mycobacteria, SecA2 appears to interact with SecYEG via SecA. In this case, SecA2 might have a SecB-like chaperone or targeting role. Of probable functional significance, all SecA2s have a deletion in the CTD (except the C. glutamicum SecA2, which has a CTD insertion). In SecA, this domain is thought to help discriminate between true transport substrates (bound to SecB) and hydrophobic regions of cytoplasmic proteins [5].
Another possibility that cannot be excluded is that SecA2 may interact with an entirely different transmembrane channel. The identification of interacting partners for SecA2, and a more thorough assessment of the role of other secA2 locus components in post-translational modification or transport, will shed light on these possibilities. In addition, since they are related to the newly-discovered SecA2 proteins in plastids of unicellular algae and higher plants [34–36] (Fig. 1), additional studies of these bacterial motor proteins may inform, and be informed by, related studies in eukaryotes [37].
In cases where SecA2 interacts with SecYEG or SecA/YEG, SecA2 may provide “priority boarding” or an express lane to the translocon. Each SecA2 may have different criteria for what merits priority, and the net effect would be to shift the balance of preproteins that are transported. However, the combined findings thus far do not clearly illuminate whether the SecA2 proteins may differ in how they select substrates for transport, whether there are unique features of the preprotein that may be recognized by SecA2, or whether the SecA2 substrates are simply the most abundant preproteins. Targeted mutations within the SecA2 substrates along with an analysis of the effect on SecA2-dependent transport should help to resolve this issue.
4. The accessory Sec system
Many species of streptococci and staphylococci express a specialized transporter known as the accessory Sec (or aSec) system. Along with SecA2, the aSec system invariably includes SecY2 (a paralogue of SecY) and the accessory Sec proteins Asp1-3. The Asps are unlike any other characterized proteins, and appear to be unique to the accessory Sec system. Unlike the general Sec system, which exports a variety of proteins, each aSec system is dedicated to the transport of a large, extensively glycosylated cell wall-anchored protein. These unique substrates, known as serine-rich repeat (SRR) glycoproteins, undergo extensive glycosylation intracellularly, prior to being transported to the bacterial cell surface.
The aSec system has been most extensively studied in S. gordonii and Streptococcus parasanguinis, two opportunistic pathogens normally present among the oral microbiota, but frequently associated with cardiovascular infections. Of historical note, the aSec system was first identified in a screen of S. gordonii mutants for decreased binding to human platelets [1]. The aSec system has since been identified in nine distinct genera and at least 32 different species to date (Table I), and there is a striking conservation of gene organization within the chromosomal loci that encompass this system (Fig. 4). Along with the transport components, the locus also encodes the transport substrate, and two or more glycosyl transferases that are responsible for the post-translational modification of the preprotein with carbohydrate moieties. It is not entirely clear why a dedicated system is necessary for the export of the SRR glycoproteins. One previous notion has been that the aSec system is essential for these unusual substrates because SecA or SecYEG cannot accommodate glycosylated proteins. However, several lines of evidence indicate that this is not entirely true, and suggest a more complex role for the aSec system in SRR glycoprotein expression.
Figure 4. Gene organization within the accessory Sec loci.
Comparison of the aSec locus in selected Gram-positive bacterial species. Genes encoding the aSec system components are colored blue, and the export susbstrate is shown in red. Glycosyl transferases and other enzymes involved solely in carbohydrate modifications are shown in green. Genes of unknown function are shown in grey, while genes encoding potential regulatory proteins are depicted in black. Two different variations found in Streptococcus agalactiae strains are shown. The Staphylococcus locus is representative of most Staphylococcal species. A slightly different arrangement, in which secA2 is immediately upstream of secY2 and the asp123 genes, is found in Pediococcus, Enterococcus and some Lactobacillus species.
4.1 substrates of the aSec system
The SRR glycoproteins comprise a unique family of adhesins that bind a wide range of ligands, and have a significant impact on biofilm formation and virulence in a diversity of infections. For example, GspB and Hsa (the SRR adhesins of S. gordonii strains M99 and DL1, respectively) bind sialylated carbohydrates on both the salivary mucin MG2 (MUC7), and on the platelet membrane protein GPIb [38–43]. This latter interaction appears to be a major factor in the pathogenesis of endocarditis [44, 45]. Fap1 expression by S. parasanguinis enhances adherence to saliva and subsequent biofilm formation [46, 47]. The SRR adhesins of Streptococcus pneumoniae, Streptococcus agalactiae, and Staphylococcus aureus have been linked to pneumonia, meningitis and neonatal sepsis, and endocarditis, respectively [48–54]. Although the ligands for the SRR glycoproteins are not known for all species, it is clear that they encompass both carbohydrates (e.g. sialyl T-antigen for GspB), as well as proteins (e.g. human keratin 4 and fibrinogen for the Srr1 adhesin of S. agalactiae) [55–57]. This diversity of ligands most likely reflects specific targets for microbial adhesion in different biologic niches.
Although the binding region of these adhesins can differ significantly, the overall domain organization is conserved, with an atypical N-terminal signal peptide followed by a short SRR domain, a ligand binding domain, a long SRR domain, and a C-terminal LPXTG cell wall anchoring motif (Fig. 5). Carbohydrate moieties are rapidly added to the SRR domains through the combined action of two or more intracellular enzymes. Studies in S. gordonii and S. parasanguinis have indicated that a heterodimeric complex of GtfA/B (Gtf1/2) catalyzes the proximal linkage of N-acetyl glucosamine (GlcNAc) to the polypeptide backbone [58, 59]. The GlcNAc is presumed to be O-linked to serine residues in the SRR domains, but the precise linkage has not yet been verified, nor has any consensus motif for glycosylation been identified. Other glycosyl transferases may add additional glycan moieties following the initial GlcNAc deposition. The precise structure of the glycan on the mature adhesins is not known, but rough estimates for GspB indicate that approximately 100 monosaccharide residues may be added per each polypeptide backbone [60]. The extensive glycosylation often has a notable affect on solubility, stability, and resistance to proteolysis. In addition, the glycan modifications can have a dramatic impact on the binding properties of the adhesins [53, 61–63].
Figure 5. SRR glycoprotein domain organization and conserved signal peptide features.
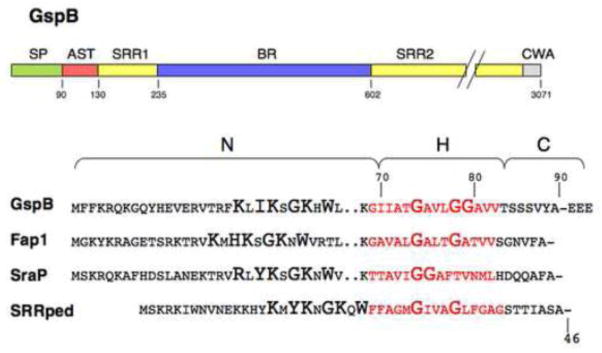
Upper diagram: The domains identified in GspB are characteristic of the SRR glycoprotein family. SP, signal peptide; AST, accessory Sec transport domain; SRR1 and SRR2, serine-rich repeat regions 1 and 2, respectively; BR, ligand binding region; CWA, cell wall anchoring domain (including an LPxTG motif).
Lower diagram: Amino-terminal sequence of the SRR glycoproteins from S. gordonii (GspB), S. parasangunis (Fap1), S. aureus (SraP), and P. acidilactici (SRRpp).
One reason the aSec systems went undetected for so long may be due to the nature of the substrates, which can be extremely difficult to detect and characterize. With the apparent masses ranging from 200 kDa to much greater than 400 kDa, the SRR glycoproteins are highly refractory to staining with most conventional reagents, and differently glycosylated forms may not be recognized by the same antibodies or lectins. These complications should not be overlooked, since they can make it difficult to distinguish primary effects on glycosylation and transport from secondary effects on these processes (see sections 4.2.2 and 4.3.3).
4.2 Preprotein recognition and trafficking to the aSec system
4.2.1 The SRR signal peptides
To better define the mechanisms of transport, truncated versions of the SRR glycoproteins have been used as model substrates. These have been highly valuable analytical tools since they lack a C-terminal cell wall anchoring domain and therefore are secreted freely into the culture medium, making it easier to quantify changes in export. Moreover, sufficient truncation of the extensive SRR2 region renders the preprotein stable even when not glycosylated, and thus allows examination of export independently of glycosylation. Such studies have revealed some key requirements for transport by the aSec versus canonical Sec system. Surprisingly, glycosylation of the substrate is not necessary for recognition by the aSec system. Instead, a 90 amino acid N-terminal signal sequence is one key portion of the preprotein that is essential for transport [64, 65]. The atypical signal peptide has a tri-partite structure characteristic of general Sec system signal peptides, although the N region is approximately three times longer (Fig. 5). The extended N region invariably includes a “KxYKxGKxW motif”. The functional relevance of this motif is not yet known, but it is predicted to form an amphipathic helix that may aid in targeting of the preprotein to anionic phospholipid patches in the membrane. A targeted deletion within the N region confirmed that it is important for transport, but does not affect the transport route [66].
In S. gordonii, the hydrophobic core of the GspB signal peptide (the H region), rather than the extended N region, was found to be a primary determinant of trafficking to the aSec system [66]. In particular, three glycine residues in the H region have a major impact on this process. Results suggest that the glycines decrease the propensity to form an α-helix, but may also reduce the H region hydrophobicity (both properties are known to affect recognition and transport by the general Sec system). Replacement of one or two glycines with residues that increase the propensity for α-helix formation decreases the stringency of trafficking, such that the preproteins (i.e. truncated variants) are more readily transported by the general Sec system. Replacing all three glycine residues completely abrogates aSec transport, and re-directs the protein to the Sec system. Conversely, substitution of a glycine with proline only partially interferes with aSec transport, and simultaneously prevents Sec transport. Thus, very subtle changes in the hydrophobic core of the signal peptide have a strong influence on the trafficking of the preprotein, and the three glycine residues therefore contribute to a novel type of “Sec avoidance” mechanism.
Nearly all of the SRR glycoproteins identified to date have a similar ~90 residue amino-terminal sequence. One exception is found in the aSec substrates of pediococci, where the signal peptide has a KxYKxGKxW motif that is immediately followed by the H region (i.e. the signal peptide lacks the intervening portion of the N region; Fig. 5). Another apparent, but not likely valid, exception is in the signal peptides of pneumococcal aSec substrates such as PsrP. The annotated sequences indicate a translational start site at the first methionine codon, which corresponds to codon 38 or 42 of other SRR glycoprotein signal sequences. However, the actual open reading frame is likely to begin at an atypical ATA codon further upstream, although this has not been verified experimentally.
The H regions of the signal peptides display two apparent varieties. In the streptococcal SRR proteins, the H region typically includes three glycine residues arranged in a GxxGG motif, and has a low hydrophobicity. As described above, these combined features may correspond to a unique type of Sec avoidance motif. The staphylococcal substrates have a central pair of glycines, and a slightly higher hydrophobicity in the signal peptide H region (Fig. 5), and may lack a Sec avoidance mechanism. Although this has not been thoroughly assessed, in S. aureus some transport of the aSec substrate SraP (also known as SasA) is evident in secA2 deletion strains [67, 68], which suggests that the substrate may be less stringently trafficked, such that it can be transported by either the Sec or aSec route (Fig. 6C). A recent report indicates that SecY2 expression is up-regulated during the early exponential phase of growth [69]. Thus, trafficking of the SRR glycoprotein in S. aureus could be influenced by the relative expression levels of the two transporters.
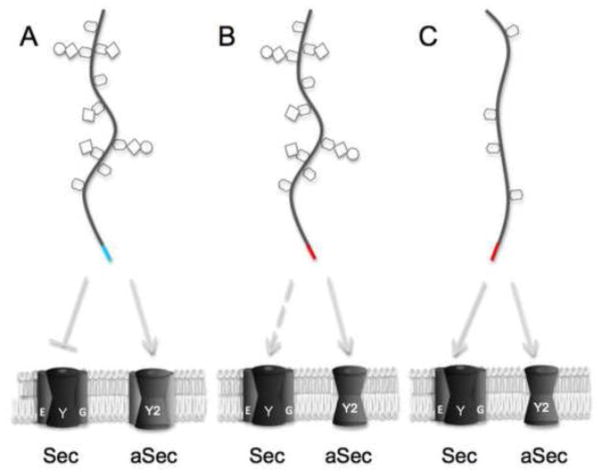
Figure 6. Impact of signal peptides and post-translational modifications on trafficking through the general Sec or aSec system.
A signal sequence with a relatively strong Sec avoidance motif is indicated in cyan, and signal sequences that can support transport by the general Sec system are indicated in red. A: In most streptococci, the aSec locus encodes four or more putative glycosyl transferases, and the SRR glycoprotein signal peptide is not optimal for general Sec transport. B: GspB variants with one or more substitutions of critical glycine residues in the signal peptide H region, or wild-type Fap1, may be inefficiently transported by the general Sec system (dashed line indicates transport of partially or incorrectly glycosylated substrates). C: In staphylococci, the aSec locus includes just the core GtfA/B glycosyl transferase, and the SRR glycoprotein signal peptide can facilitate transport via either the Sec or aSec pathway.
4.2.2 Impact of glycosylation on trafficking
Although the carbohydrate moieties are not necessary for recognition by the aSec system, glycosylation of the SRR domains can influence trafficking by impeding general Sec transport. However, the extent to which glycosylation affects trafficking may depend on the nature of the signal peptide. For example, in S. gordonii and S. agalactiae, where the signal peptide of the SRR glycoprotein has an apparently strong Sec avoidance motif, little or no preprotein is transported by the general Sec system as long as all of the glycosyl transferases are intact (Fig. 6A). In S. parasanguinis, the Fap1 signal peptide has one less glycine in the H region (Fig. 5), and can readily support transport of a nonglycosylated substrate (e.g. when gtf1 is deleted) by the general Sec pathway [65]. Accordingly, multiple reports have indicated that, even when the entire gtf cluster is intact, a partially glycosylated Fap1 precursor is transported by the general Sec system (perhaps inefficiently) when aSec transport is impaired (Fig. 6B, and see section 4.3.3). In S. aureus, which lacks additional glycosyl transferases and expresses just the core GtfA/B complex, there is also evidence that at least some SRR glycoprotein can be transported by SecA/YEG [67, 68] (Fig. 6C). Thus, in cases where the signal peptide can facilitate transport by the general Sec pathway, partially glycosylated preproteins may be transported by SecA/YEG upon deletion of any aSec component.
4.2.3 The AST domain
In addition to the signal peptide, the aSec substrate must have a specific segment (the accessory Sec transport or AST domain) at the amino terminus of the mature region. Studies in S. gordonii and S. parasanguinis have demonstrated that deletion of this region abolishes aSec transport [65, 70]. Even single amino acid substitutions within the AST domain can dramatically impair aSec transport. The signal peptide and AST domain together are sufficient for the transport of a heterologous substrate (specifically, a slow-folding variant of the MalE maltose binding protein) by the aSec system. Curiously, the sequence of the AST domain is not highly conserved, and diverges much more rapidly than does the signal peptide. Extensive mutational analysis of the AST domain of GspB, in combination with “pre-gated” variants of the S. gordonii SecY2 (see section 4.4 below), indicated that the AST domain affects both targeting to the translocon and opening of the channel. These earlier results therefore suggested that the AST domain might interact with both SecA2 and SecY2.
Subsequent in vivo photo-cross-linking experiments were undertaken to capture any aSec components that interact with the preprotein during transport, and only SecA2 was found to contact the AST domain [71]. This result implies that targeting and gating may both be driven by AST domain interactions with SecA2. Whereas SecA2 can bind the end of the AST domain adjacent to the signal peptide independently of other aSec components, full engagement of the AST domain, as well as transport of the preprotein by SecA2, requires one or more of the Asps (see section 4.5 below). The AST engagement by SecA2 is reminiscent of the high affinity interactions that occur between preprotein mature regions and the SecA/YEG translocon [72]. However, the requirement of a specific segment in the preprotein, along with the involvement of the Asps, is a unique feature of aSec transport that may ensure the passage of just a single substrate via this channel.
4.3 the accessory Sec translocase
4.3.1 SecA2
The SecA2 proteins belonging to the aSec system have several common features. They all have a 45 aa truncation of the CTD, as compared with their SecA paralogues, and typically have a proline residue at the C-terminus. They have 70% similarity (35 to 40% identity) to SecA, and most of the similarity is limited to the nucleotide binding motifs of NBD1 and NBD2 (Fig. 2). Mutations of conserved NBD1 residues known to impair nucleotide binding and transport by E. coli SecA abolish transport mediated by S. gordonii SecA2, whereas mutations in NBD2 have a less dramatic impact [73]. Compared with SecA, the S. gordonii SecA2 has a lower basal rate of ATP hydrolysis, and requires higher magnesium concentrations for activity. The composite data indicate that the streptococcal SecA2 may be more tightly regulated than SecA, and support the possibility that one or more of the Asps may be required to stimulate ATP binding or hydrolysis, as discussed below (section 4.3.3).
In spite of the similarities amongst the aSec motor proteins, the SecA2 orthologues diverge quite rapidly and are not readily interchangeable. For example, the SecA2 proteins from Streptococcus sanguinis and S. pneumoniae show 91% and 79% similarity (81% and 61% identity), respectively, to that of S. gordonii. Although the S. sanguinis SecA2 can partially complement a ΔsecA2 mutation in S. gordonii, the S. pneumoniae SecA2 is unable to transport GspB [71]. Thus, at least some of the divergence in the SecA2 proteins must reflect differing inter- versus intra-molecular interactions. It is possible that the SecA2 differences may parallel those of the preprotein substrates, but the driving force behind such rapid divergence remains to be determined.
4.3.2 The SecY2 translocon
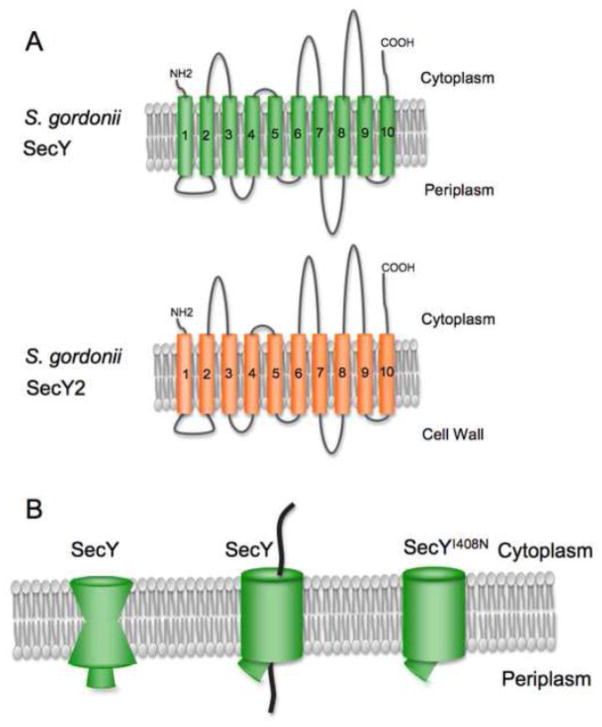
SecY2 is presumed to form the transmembrane channel through which the SRR glycoproteins exit the cytoplasm. The predicted topology of SecY2 is nearly identical to that of SecY (Fig. 7A). However, the SecY2 orthologues have very low primary sequence similarity to the SecY paralogues (20% identity, 60% similarity), and only a limited number of the SecY residues involved in protein-protein interactions are conserved in SecY2. Importantly, none of the C5 cytosolic loop residues that contact SecA (summarized in Lycklama et al [11]) are present in SecY2, which is indicative of a lack of cross-talk between the general Sec and aSec systems. The inactive SecY channel has an hourglass shape (Fig. 7B), and two features help to seal the closed channel: a ring of hydrophobic residues near the central pore (the “pore ring” residues), and a “plug” on the periplasmic or external side comprised of residues in TM2 [74–76]. Upon intercalation of a signal peptide between two of the transmembrane segments, the SecY channel widens, and the plug is displaced [77, 78]. Compared with SecY, the putative pore ring residues of SecY2 include slightly bulkier residues, such as leucine or methionine, versus isoleucines. As yet, there is nothing obviously different about the SecY2 proteins that might account for the ability to transport a glycosylated protein.
Figure 7. SecY versus SecY2 topology and structural features.
A: The predicted topology of SecY2 is very similar to that of SecY. B: The inactive SecY channel is impermeable to small molecules, due to the constriction by a ring of hydrophobic residues near the center of the channel along with a plug formed by residues of TM2. In the active channel, the central pore widens and the plug is displaced. Some prl mutations in E. coli SecY result in a partially or more readily opened channel.
In the general Sec system, two small proteins combine with SecY to form the translocon. SecE is essential for transport and helps stabilize the open form of the channel [78, 79]. SecG enhances the translocation efficiency and was recently shown to contact SecA during transport [7]. In some streptococcal species, the aSec system includes one or two additional small proteins (Asp4 and Asp5) that may be structural components of the transmembrane channel [80]. Asp4 and Asp5 have one or two predicted transmembrane segments, respectively, and thus display some topological similarity to SecE and SecG. It is suspected that they may interact with SecY2, but this has not been verified experimentally. Functional studies in S. gordonii indicate that Asp4 is partially dispensable for the export of truncated or non-glycosylated GspB variants via the aSec route [81]. This is consistent with a role of Asp4 in stabilizing the open state of the transmembrane channel, rather than a role in the initiation of translocation. In S. aureus, there is evidence for interaction between SecY2 and SecG [82]. In other organisms that lack Asp4 and Asp5, it is possible that the SRR signal peptide could facilitate or stabilize the open SecY2 channel, and therefore preclude the need for these Asps.
Several lines of experimental evidence suggest that SecA2 interacts directly with SecY2 to facilitate SRR glycoprotein transport. First, deletion of secY2 results in the same phenotype as deletion of secA2 [1, 67]. Second, certain alterations in SecY2 mimic the effect of prl (protein localization) mutations in SecY and compensate for defects in the GspB signal peptide and some AST mutations [70]. For example, the SecY2 substitution I382N mirrors the I408N (prlA4) mutation in SecY [83], a substitution of one of the pore ring residues that destabilizes the closed channel and is thought to override a gating mechanism that is usually facilitated by signal peptide intercalation between the second and seventh transmembrane segments of SecY [77] (Fig. 7B). It is therefore highly likely that SecY2 is a structural component of the SecA2 translocase.
4.3.3 additional accessory Sec system components
Although Asps1-3 are invariable components of the aSec system, their precise roles in SRR glycoprotein expression are just beginning to be understood. In S. gordonii these proteins clearly have a direct role specifically related to aSec transport, and are essential for this process even when glycosylation of the preprotein is blocked by deletion of the core glycosyl transferase gene gtfA [81]. Since the Asps have no primary sequence similarity to any proteins of known function, a variety of genetic and biochemical techniques have been used to assess how and where they function. Asp2 and Asp3 have been shown to have a range of interactions. Both proteins directly bind the SRR regions of the GspB preprotein [84]. Rather than binding to specific motifs, Asps2 and 3 appear to recognize the unstructured or non-folded sections of the preprotein. Although these Asps bind GspB directly, they do not appear to function as conventional chaperones, since they are not required for GspB stability or targeting to SecA2. As described above, evidence indicates that they are important for the full engagement of the AST domain with SecA2, and subsequent GspB transport. Experiments also indicate that Asp2 and Asp3 both directly bind SecA2 [81]. Asp2, in particular, appears to have a high affinity for SecA2, since it localizes at the inner membrane in the same punctate pattern as SecA2, when co-expressed with this protein in E. coli [85]. Thus, either or both Asps may directly alter the conformation of SecA2 to facilitate the full engagement of the preprotein AST domain, or to stimulate ATP binding or hydrolysis.
Subcellular localization studies of Asp1, Asp2 and Asp3 indicate that all three of these proteins fractionate to both the membrane and cytosol compartments, and their location is at least partially affected by other aSec components. Thus, all three of the Asps may transit between the cytosol and the cytoplasmic membrane. There is also evidence that Asps1-3 form a complex that is soluble and cytosolic, but will partially localize to the membrane when co-expressed with SecA2 [85]. What is as yet unresolved is whether these proteins transit individually or collectively between the two compartments.
The Asp homologues of S. parasangunis have been designated as glycosylation-associated proteins Gap1-3, in part because they are thought to have a primary affect on glycosylation rather than transport, and in part because a portion of Gap1 resembles some glycosyl transferases [86]. In addition, results of earlier studies suggested different roles for SecA2 (i.e. a clear role in transport) versus SecY2, Gap1 and Gap3 (an impact on glycosylation) [87–89]. However, later reports indicate that deletion of gap1, gap2 or gap3 results in a phenotype identical to that of a secY2 deletion, which is the loss of correctly glycosylated or “mature” Fap1 on the cell surface, and a loss of binding to saliva-coated hydroxylapatite [90, 91]. Rather than abolishing transport, deletion of any of these components results in the expression of a partially glycosylated Fap1 precursor on the cell surface, but possibly in reduced amounts. This is most likely due to the absence of a strong Sec avoidance motif in the Fap1 signal sequence, and less stringent trafficking of the glycosylated preprotein (see section 4.2.2). Thus, if the Gaps primarily affect glycosylation and not transport, it is unclear why the Fap1 precursor is not efficiently transported by the aSec system, as happens upon deletion of other glycosyl transferases such as Gtf1 or GalT2 [88, 92]. As with the Asps, a number of interactions amongst the Gap proteins, and between the Gaps and SecA2, have been characterized [90, 93, 94]. However, unlike the Asps, the stability of the Gaps is interdependent. That is, Gap1 and Gap2 affect the stability of Gap3, and are thus thought to serve as chaperones for Gap3. The importance of these interactions towards either the glycosylation or transport of Fap1 as yet has not been defined.
4.3.4. Asp1-3 (Gap1-3) as bi-functional components of the aSec system
Based on the combined results of studies in S. parasanguinis, S. gordonii and S. salivarius, Zhou and Wu [95] suggested that the processes of transport and glycosyation are coupled, and proposed a two-step model for the glycosylation of Fap1 in which SecA2, SecY2, Gap1 and Gap3 convert a partially glycosylated precursor to a mature glycoform. Recent studies on the function of Asp2 in S. gordonii demonstrated that this component of the aSec system does indeed affect the carbohydrate content of GspB [63]. Importantly, the specific effect on glycosylation was not apparent upon deletion of asp2, but only by making targeted substitutions in the protein. Tertiary structure predictions indicate that a region of Asp2 resembles the catalytic region of esterases and hydrolases. Alignment of the Asp2 sequence with that of the esterase catalytic domains led to the identification of a Ser-Glu-His catalytic triad. Somewhat surprisingly, mutation of these residues did not impair transport, but did result in changes in the glycan composition of GspB, as indicated by decreased electrophoretic mobility, altered affinity for several lectins, and increased incorporation of GlcNAc. Moreover, the altered glycoform of GspB was unable to facilitate binding of S. gordonii to human platelets. Thus, Asp2 is clearly involved in both the transport and glycosylation processes. However, no catalytic activity was evident when assessing the isolated, purified protein. It was therefore postulated that the Asp2 catalytic activity might be dependent upon the formation of a multimeric Asp complex.
5. New model and key unanswered questions
Based on the most recent composite data, we propose a new model for aSec transport, and a new mechanistic role for the Asps in SRR glycoprotein biogenesis (Figure 8). In this new model, the Asps are bifunctional proteins involved in both transport and glycosylation. The Asps have individual roles in transport that are not dependent on the glycosylation process. In addition, however, they have a collective and direct role in glycosylation that is dependent on transport via SecA2/Y2. More specifically, one or more of the Asps facilitate the full engagement of the preprotein AST domain by SecA2 and the initiation of transport. A complex of the Asps may then modify the glycan composition of the SRR glycoprotein as it undergoes transport by SecA2/Y2. By functioning as the nexus of these two processes, the Asps serve as a control point for the correct biogenesis of the SRR proteins. Glycosylation is essential for stability, but must be done with high precision for the SRR protein to have optimal binding properties. Thus, the specialized aSec transporter may have evolved not merely to accommodate a glycoprotein, but to couple the modulation of glycan composition with transport. Determining exactly how this coupling occurs is fundamental to understanding SRR glycoprotein expression. This model is more consistent with data that indicate at least some of the post-translational modifications are compatible with SecA/YEG transport (as is the case for S. aureus and S. parasanguinis), but the precise glycan composition of the substrate is affected by the Asps associated with the SecA2/SecY2 translocon. The model also raises an interesting question concerning the role of SecA2 proteins in general: do all SecA2 proteins facilitate the coupling of post-translational (or post-translocational) modifications with transport? In cases where they are not essential for viability, further study of SecA2 transporters may provide significant insights and understanding of general Sec system transport in Gram-positive bacteria.
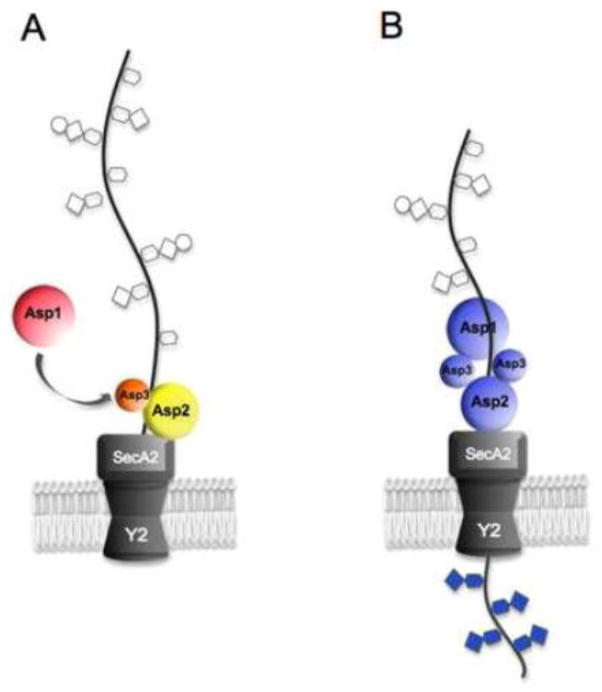
Figure 8. Model for SRR glycoprotein biogenesis.
A: A partially glycosylated preprotein arrives at the translocon, and one or more of the Asps facilitate an interaction between SecA2 and the preprotein AST domain that trigger opening of the SecY2 channel. B: An Asp123 complex further modifies the glycan composition as translocation of the SRR glycoprotein proceeds through SecA2/Y2. The Asp123 modifications could include trimming, replacement, or addition of carbohydrate moieties. SecA2 and SecY2 are thought not to have a direct enzymatic role in glycan modification, but instead facilitate the interaction between the Asp complex and the substrates.
Highlights.
SecA2 proteins comprise a selective class of transport-associated ATPases.
SecA2 is found in an increasingly large number of Gram-positive bacterial species.
One type of SecA2 transports multiple substrates, and may interact with the general Sec system.
A second type transports a single substrate, and is a component of the accessory Sec system.
The accessory Sec system couples post-translational modifications with transport.
Acknowledgments
This work was supported by the Department of Veterans Affairs and the VA Merit Review program; the Northern California Institute for Research and Education; grant R01-AI41513from the NIH (P.M.S.); and a Fellowship Award from the American Heart Association (AHA), Western Affiliate (YY).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bensing BA, Sullam PM. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol Microbiol. 2002;44:1081–94. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 2.Braunstein M, Brown AM, Kurtz S, Jacobs WR., Jr Two nonredundant SecA homologues function in mycobacteria. J Bacteriol. 2001;183:6979–90. doi: 10.1128/JB.183.24.6979-6990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenz LL, Portnoy DA. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol Microbiol. 2002;45:1043–56. doi: 10.1046/j.1365-2958.2002.03072.x. [DOI] [PubMed] [Google Scholar]
- 4.Papanikolau Y, Papadovasilaki M, Ravelli RB, McCarthy AA, Cusack S, Economou A, Petratos K. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J Mol Biol. 2007;366:1545–57. doi: 10.1016/j.jmb.2006.12.049. [DOI] [PubMed] [Google Scholar]
- 5.Gelis I, Bonvin AM, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–69. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–43. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das S, Oliver DB. Mapping of the SecA.SecY and SecA.SecG interfaces by site-directed in vivo photocross-linking. J Biol Chem. 2011;286:12371–80. doi: 10.1074/jbc.M110.182931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper DB, Smith VF, Crane JM, Roth HC, Lilly AA, Randall LL. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol. 2008;382:74–87. doi: 10.1016/j.jmb.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vrontou E, Karamanou S, Baud C, Sianidis G, Economou A. Global co-ordination of protein translocation by the SecA IRA1 switch. J Biol Chem. 2004;279:22490–7. doi: 10.1074/jbc.M401008200. [DOI] [PubMed] [Google Scholar]
- 10.Chatzi KE, Sardis MF, Karamanou S, Economou A. Breaking on through to the other side: protein export through the bacterial Sec system. Biochem J. 2013;449:25–37. doi: 10.1042/BJ20121227. [DOI] [PubMed] [Google Scholar]
- 11.Lycklama ANJA, Driessen AJ. The bacterial Sec-translocase: structure and mechanism. Philos Trans R Soc Lond B Biol Sci. 2012;367:1016–28. doi: 10.1098/rstb.2011.0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park E, Rapoport TA. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- 13.Feltcher ME, Braunstein M. Emerging themes in SecA2-mediated protein export. Nat Rev Microbiol. 2012;10:779–89. doi: 10.1038/nrmicro2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freudl R. Leaving home ain’t easy: protein export systems in Gram-positive bacteria. Res Microbiol. 2013;164:664–74. doi: 10.1016/j.resmic.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Caspers M, Freudl R. Corynebacterium glutamicum possesses two secA homologous genes that are essential for viability. Arch Microbiol. 2008;189:605–10. doi: 10.1007/s00203-008-0351-0. [DOI] [PubMed] [Google Scholar]
- 16.Braunstein M, Espinosa BJ, Chan J, Belisle JT, Jacobs WR., Jr SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:453–64. doi: 10.1046/j.1365-2958.2003.03438.x. [DOI] [PubMed] [Google Scholar]
- 17.Gibbons HS, Wolschendorf F, Abshire M, Niederweis M, Braunstein M. Identification of two Mycobacterium smegmatis lipoproteins exported by a SecA2-dependent pathway. J Bacteriol. 2007;189:5090–100. doi: 10.1128/JB.00163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feltcher ME, Gibbons HS, Ligon LS, Braunstein M. Protein export by the mycobacterial SecA2 system is determined by the preprotein mature domain. J Bacteriol. 2013;195:672–81. doi: 10.1128/JB.02032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou JM, D’Lima NG, Rigel NW, Gibbons HS, McCann JR, Braunstein M, Teschke CM. ATPase activity of Mycobacterium tuberculosis SecA1 and SecA2 proteins and its importance for SecA2 function in macrophages. J Bacteriol. 2008;190:4880–7. doi: 10.1128/JB.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sianidis G, Karamanou S, Vrontou E, Boulias K, Repanas K, Kyrpides N, Politou AS, Economou A. Cross-talk between catalytic and regulatory elements in a DEAD motor domain is essential for SecA function. Embo J. 2001;20:961–70. doi: 10.1093/emboj/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–26. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 22.Karamanou S, Gouridis G, Papanikou E, Sianidis G, Gelis I, Keramisanou D, Vrontou E, Kalodimos CG, Economou A. Preprotein-controlled catalysis in the helicase motor of SecA. Embo J. 2007;26:2904–14. doi: 10.1038/sj.emboj.7601721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karamanou S, Vrontou E, Sianidis G, Baud C, Roos T, Kuhn A, Politou AS, Economou A. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol. 1999;34:1133–45. doi: 10.1046/j.1365-2958.1999.01686.x. [DOI] [PubMed] [Google Scholar]
- 24.Rigel NW, Gibbons HS, McCann JR, McDonough JA, Kurtz S, Braunstein M. The accessory SecA2 system of mycobacteria requires ATP binding and the canonical SecA1. J Biol Chem. 2009 doi: 10.1074/jbc.M900325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenz LL, Mohammadi S, Geissler A, Portnoy DA. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc Natl Acad Sci U S A. 2003;100:12432–7. doi: 10.1073/pnas.2133653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renier S, Chambon C, Viala D, Chagnot C, Hebraud M, Desvaux M. Exoproteomic analysis of the SecA2-dependent secretion in Listeria monocytogenes. EGD-e J Proteomics. 2013;80C:183–195. doi: 10.1016/j.jprot.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 27.Archambaud C, Nahori MA, Pizarro-Cerda J, Cossart P, Dussurget O. Control of Listeria superoxide dismutase by phosphorylation. J Biol Chem. 2006;281:31812–22. doi: 10.1074/jbc.M606249200. [DOI] [PubMed] [Google Scholar]
- 28.Halbedel S, Hahn B, Daniel RA, Flieger A. DivIVA affects secretion of virulence-related autolysins in Listeria monocytogenes. Mol Microbiol. 2012;83:821–39. doi: 10.1111/j.1365-2958.2012.07969.x. [DOI] [PubMed] [Google Scholar]
- 29.Mishra KK, Mendonca M, Aroonnual A, Burkholder KM, Bhunia AK. Genetic organization and molecular characterization of secA2 locus in Listeria species. Gene. 2011;489:76–85. doi: 10.1016/j.gene.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen-Mau SM, Oh SY, Kern VJ, Missiakas DM, Schneewind O. Secretion genes as determinants of Bacillus anthracis chain length. J Bacteriol. 2012;194:3841–50. doi: 10.1128/JB.00384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kern VJ, Kern JW, Theriot JA, Schneewind O, Missiakas D. Surface-layer (Slayer) proteins sap and EA1 govern the binding of the S-layer-associated protein BslO at the cell septa of Bacillus anthracis. J Bacteriol. 2012;194:3833–40. doi: 10.1128/JB.00402-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fagan RP, Fairweather NF. Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem. 2011;286:27483–93. doi: 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang TH, de la Riva L, Fagan RP, Storck EM, Heal WP, Janoir C, Fairweather NF, Tate EW. Chemical probes of surface layer biogenesis in Clostridium difficile ACS. Chem Biol. 2010;5:279–85. doi: 10.1021/cb9002859. [DOI] [PubMed] [Google Scholar]
- 34.Koyama Y, Kaneko Y, Matsuoka S, Matsumoto K, Hara H, Ohta N. Expression and localization of two SecA homologs in the unicellular red alga Cyanidioschyzon merolae. Biosci Biotechnol Biochem. 2012;76:417–22. doi: 10.1271/bbb.110833. [DOI] [PubMed] [Google Scholar]
- 35.Koyama Y, Takimoto K, Kojima A, Asai K, Matsuoka S, Mitsui T, Matsumoto K, Hara H, Ohta N. Characterization of the nuclear- and plastid-encoded secA-homologous genes in the unicellular red alga Cyanidioschyzon merolae. Biosci Biotechnol Biochem. 2011;75:2073–8. doi: 10.1271/bbb.110338. [DOI] [PubMed] [Google Scholar]
- 36.Skalitzky CA, Martin JR, Harwood JH, Beirne JJ, Adamczyk BJ, Heck GR, Cline K, Fernandez DE. Plastids contain a second sec translocase system with essential functions. Plant Physiol. 2011;155:354–69. doi: 10.1104/pp.110.166546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Celedon JM, Cline K. Intra-plastid protein trafficking: how plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim Biophys Acta. 2013;1833:341–51. doi: 10.1016/j.bbamcr.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bensing BA, Lopez JA, Sullam PM. The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane. glycoprotein. Ibalpha Infect Immun. 2004;72:6528–37. doi: 10.1128/IAI.72.11.6528-6537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi Y, Konishi K, Cisar JO, Yoshikawa M. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect Immun. 2002;70:1209–18. doi: 10.1128/IAI.70.3.1209-1218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi Y, Sandberg AL, Ruhl S, Muller J, Cisar JO. A specific cell surface antigen of Streptococcus gordonii is associated with bacterial hemagglutination and adhesion to alpha2-3-linked sialic acid-containing receptors. Infect Immun. 1997;65:5042–51. doi: 10.1128/iai.65.12.5042-5051.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi Y, Yajima A, Cisar JO, Konishi K. Functional analysis of the Streptococcus gordonii DL1 sialic acid-binding adhesin and its essential role in bacterial binding to platelets. Infect Immun. 2004;72:3876–82. doi: 10.1128/IAI.72.7.3876-3882.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takamatsu D, Bensing BA, Cheng H, Jarvis GA, Siboo IR, Lopez JA, Griffiss JM, Sullam PM. Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein. Ibalpha Mol Microbiol. 2005;58:380–92. doi: 10.1111/j.1365-2958.2005.04830.x. [DOI] [PubMed] [Google Scholar]
- 43.Takamatsu D, Bensing BA, Prakobphol A, Fisher SJ, Sullam PM. Binding of the streptococcal surface glycoproteins GspB and Hsa to human salivary proteins. Infect Immun. 2006;74:1933–40. doi: 10.1128/IAI.74.3.1933-1940.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi Y, Takashima E, Shimazu K, Yagishita H, Aoba T, Konishi K. Contribution of sialic acid-binding adhesin to pathogenesis of experimental endocarditis caused by Streptococcus gordonii DL1. Infect Immun. 2006;74:740–3. doi: 10.1128/IAI.74.1.740-743.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiong YQ, Bensing BA, Bayer AS, Chambers HF, Sullam PM. Role of the serine-rich surface glycoprotein GspB of Streptococcus gordonii in the pathogenesis of infective endocarditis. Microb Pathog. 2008;45:297–301. doi: 10.1016/j.micpath.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H, Mintz KP, Ladha M, Fives-Taylor PM. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol Microbiol. 1998;28:487–500. doi: 10.1046/j.1365-2958.1998.00805.x. [DOI] [PubMed] [Google Scholar]
- 47.Froeliger EH, Fives-Taylor P. Streptococcus parasanguis fimbria-associated adhesin fap1 is required for biofilm formation. Infect Immun. 2001;69:2512–9. doi: 10.1128/IAI.69.4.2512-2519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obert C, Sublett J, Kaushal D, Hinojosa E, Barton T, Tuomanen EI, Orihuela CJ. Identification of a Candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect Immun. 2006;74:4766–77. doi: 10.1128/IAI.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rose L, Shivshankar P, Hinojosa E, Rodriguez A, Sanchez CJ, Orihuela CJ. Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J Infect Dis. 2008;198:375–83. doi: 10.1086/589775. [DOI] [PubMed] [Google Scholar]
- 50.Seifert KN, Adderson EE, Whiting AA, Bohnsack JF, Crowley PJ, Brady LJ. A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae. Microbiology. 2006;152:1029–40. doi: 10.1099/mic.0.28516-0. [DOI] [PubMed] [Google Scholar]
- 51.Siboo IR, Chambers HF, Sullam PM. Role of SraP, a Serine-Rich Surface Protein of Staphylococcus aureus, in binding to human platelets. Infect Immun. 2005;73:2273–80. doi: 10.1128/IAI.73.4.2273-2280.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Sorge NM, Quach D, Gurney MA, Sullam PM, Nizet V, Doran KS. The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J Infect Dis. 2009;199:1479–87. doi: 10.1086/598217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mistou MY, Dramsi S, Brega S, Poyart C, Trieu-Cuot P. Molecular dissection of the secA2 locus of group B Streptococcus reveals that glycosylation of the Srr1 LPXTG protein is required for full virulence. J Bacteriol. 2009;191:4195–206. doi: 10.1128/JB.01673-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seo HS, Xiong YQ, Sullam PM. Role of the serine-rich surface glycoprotein Srr1 of Streptococcus agalactiae in the pathogenesis of infective endocarditis. PLoS One. 2013;8:e64204. doi: 10.1371/journal.pone.0064204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seo HS, Mu R, Kim BJ, Doran KS, Sullam PM. Binding of glycoprotein Srr1 of Streptococcus agalactiae to fibrinogen promotes attachment to brain endothelium and the development of meningitis. PLoS Pathog. 2012;8:e1002947. doi: 10.1371/journal.ppat.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shivshankar P, Sanchez C, Rose LF, Orihuela CJ. The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells. Mol Microbiol. 2009;73:663–79. doi: 10.1111/j.1365-2958.2009.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samen U, Eikmanns BJ, Reinscheid DJ, Borges F. The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect Immun. 2007;75:5405–14. doi: 10.1128/IAI.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takamatsu D, Bensing BA, Sullam PM. Four proteins encoded in the gspB-secY2A2 operon of Streptococcus gordonii mediate the intracellular glycosylation of the platelet-binding protein. GspB J Bacteriol. 2004;186:7100–11. doi: 10.1128/JB.186.21.7100-7111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bu S, Li Y, Zhou M, Azadin P, Zeng M, Fives-Taylor P, Wu H. Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin. J Bacteriol. 2008;190:1256–66. doi: 10.1128/JB.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bensing BA, Gibson BW, Sullam PM. The Streptococcus gordonii platelet binding protein GspB undergoes glycosylation independently of export. J Bacteriol. 2004;186:638–45. doi: 10.1128/JB.186.3.638-645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou M, Zhu F, Dong S, Pritchard D, Wu H. A novel glucosyltransferase is required for glycosylation of a serine-rich adhesin and biofilm formation by Streptococcus parasanguinis. J Biol Chem. 2010 doi: 10.1074/jbc.M109.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takamatsu D, Bensing BA, Sullam PM. Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein. GspB Mol Microbiol. 2004;52:189–203. doi: 10.1111/j.1365-2958.2004.03978.x. [DOI] [PubMed] [Google Scholar]
- 63.Seepersaud R, Bensing BA, Yen YT, Sullam PM. The accessory Sec protein Asp2 modulates GlcNAc deposition onto the serine-rich repeat glycoprotein. GspB J Bacteriol. 2012;194:5564–75. doi: 10.1128/JB.01000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bensing BA, Takamatsu D, Sullam PM. Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol Microbiol. 2005;58:1468–81. doi: 10.1111/j.1365-2958.2005.04919.x. [DOI] [PubMed] [Google Scholar]
- 65.Chen Q, Sun B, Wu H, Peng Z, Fives-Taylor PM. Differential roles of individual domains in selection of secretion route of a Streptococcus parasanguinis serine-rich adhesin, Fap1. J Bacteriol. 2007;189:7610–7. doi: 10.1128/JB.00748-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bensing BA, Siboo IR, Sullam PM. Glycine residues in the hydrophobic core of the GspB signal sequence route export toward the accessory Sec pathway. J Bacteriol. 2007;189:3846–54. doi: 10.1128/JB.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siboo IR, Chaffin DO, Rubens CE, Sullam PM. Characterization of the accessory Sec system of Staphylococcus aureus. J Bacteriol. 2008;190:6188–96. doi: 10.1128/JB.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeDent A, Bae T, Missiakas DM, Schneewind O. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. Embo J. 2008;27:2656–68. doi: 10.1038/emboj.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Y, Dong J, Wu N, Gao Y, Zhang X, Mu C, Shao N, Fan M, Yang G. The production of extracellular proteins is regulated by ribonuclease III via two different pathways in Staphylococcus aureus. PLoS One. 2011;6:e20554. doi: 10.1371/journal.pone.0020554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bensing BA, Sullam PM. Transport of preproteins by the accessory Sec system requires a specific domain adjacent to the signal peptide. J Bacteriol. 2010;192:4223–32. doi: 10.1128/JB.00373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bensing BA, Yen YT, Seepersaud R, Sullam PM. A Specific interaction between SecA2 and a region of the preprotein adjacent to the signal peptide occurs during transport via the accessory Sec system. J Biol Chem. 2012;287:24438–47. doi: 10.1074/jbc.M112.378059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gouridis G, Karamanou S, Gelis I, Kalodimos CG, Economou A. Signal peptides are allosteric activators of the protein translocase. Nature. 2009;462:363–7. doi: 10.1038/nature08559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bensing BA, Sullam PM. Characterization of Streptococcus gordonii SecA2 as a paralogue of SecA. J Bacteriol. 2009;191:3482–91. doi: 10.1128/JB.00365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 75.Saparov SM, Erlandson K, Cannon K, Schaletzky J, Schulman S, Rapoport TA, Pohl P. Determining the conductance of the SecY protein translocation channel for small molecules. Mol Cell. 2007;26:501–9. doi: 10.1016/j.molcel.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 76.Gumbart J, Schulten K. The roles of pore ring and plug in the SecY protein-conducting channel. J Gen Physiol. 2008;132:709–19. doi: 10.1085/jgp.200810062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li W, Schulman S, Boyd D, Erlandson K, Beckwith J, Rapoport TA. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol Cell. 2007;26:511–21. doi: 10.1016/j.molcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 78.Tam PC, Maillard AP, Chan KK, Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. Embo J. 2005;24:3380–8. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith MA, Clemons WM, Jr, DeMars CJ, Flower AM. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J Bacteriol. 2005;187:6454–65. doi: 10.1128/JB.187.18.6454-6465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takamatsu D, Bensing BA, Sullam PM. Two additional components of the accessory Sec system mediating export of the Streptococcus gordonii platelet-binding protein. GspB J Bacteriol. 2005;187:3878–83. doi: 10.1128/JB.187.11.3878-3883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seepersaud R, Bensing BA, Yen YT, Sullam PM. Asp3 mediates multiple protein-protein interactions within the accessory Sec system of Streptococcus gordonii. Mol Microbiol. 2010;78:490–505. doi: 10.1111/j.1365-2958.2010.07346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sibbald MJ, Winter T, van der Kooi-Pol MM, Buist G, Tsompanidou E, Bosma T, Schafer T, Ohlsen K, Hecker M, Antelmann H, Engelmann S, van Dijl JM. Synthetic effects of secG and secY2 mutations on exoproteome biogenesis in Staphylococcus aureus. J Bacteriol. 2010;192:3788–800. doi: 10.1128/JB.01452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Flower AM, Doebele RC, Silhavy TJ. PrlA and PrlG suppressors reduce the requirement for signal sequence recognition. J Bacteriol. 1994;176:5607–14. doi: 10.1128/jb.176.18.5607-5614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yen YT, Seepersaud R, Bensing BA, Sullam PM. Asp2 and Asp3 interact directly with GspB, the export substrate of the Streptococcus gordonii accessory Sec System. J Bacteriol. 2011;193:3165–74. doi: 10.1128/JB.00057-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yen YT, Cameron TA, Bensing BA, Seepersaud R, Zambryski PC, Sullam PM. Differential localization of the streptococcal accessory sec components and implications for substrate export. J Bacteriol. 2013;195:682–95. doi: 10.1128/JB.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li Y, Chen Y, Huang X, Zhou M, Wu R, Dong S, Pritchard DG, Fives-Taylor P, Wu H. A conserved domain of previously unknown function in Gap1 mediates protein-protein interaction and is required for biogenesis of a serine-rich streptococcal adhesin. Mol Microbiol. 2008;70:1094–104. doi: 10.1111/j.1365-2958.2008.06456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Q, Wu H, Fives-Taylor PM. Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguis FW213. Mol Microbiol. 2004;53:843–56. doi: 10.1111/j.1365-2958.2004.04116.x. [DOI] [PubMed] [Google Scholar]
- 88.Wu H, Bu S, Newell P, Chen Q, Fives-Taylor P. Two gene determinants are differentially involved in the biogenesis of Fap1 precursors in Streptococcus parasanguis. J Bacteriol. 2007;189:1390–8. doi: 10.1128/JB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peng Z, Wu H, Ruiz T, Chen Q, Zhou M, Sun B, Fives-Taylor P. Role of gap3 in Fap1 glycosylation, stability, in vitro adhesion, and fimbrial and biofilm formation of Streptococcus parasanguinis. Oral Microbiol Immunol. 2008;23:70–8. doi: 10.1111/j.1399-302X.2007.00401.x. [DOI] [PubMed] [Google Scholar]
- 90.Echlin H, Zhu F, Li Y, Peng Z, Ruiz T, Bedwell GJ, Prevelige PE, Jr, Wu H. Gap2 promotes the formation of a stable protein complex required for mature Fap1 biogenesis. J Bacteriol. 2013;195:2166–76. doi: 10.1128/JB.02255-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou M, Peng Z, Fives-Taylor P, Wu H. A conserved C-terminal 13-amino-acid motif of Gap1 is required for Gap1 function and necessary for the biogenesis of a serine-rich glycoprotein of Streptococcus parasanguinis. Infect Immun. 2008;76:5624–31. doi: 10.1128/IAI.00534-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu H, Zeng M, Fives-Taylor P. The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect Immun. 2007;75:2181–8. doi: 10.1128/IAI.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou M, Zhang H, Zhu F, Wu H. Canonical SecA associates with an accessory secretory protein complex involved in biogenesis of a streptococcal serine-rich repeat glycoprotein. J Bacteriol. 2011;193:6560–6. doi: 10.1128/JB.05668-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou M, Zhu F, Li Y, Zhang H, Wu H. Gap1 functions as a molecular chaperone to stabilize its interactive partner Gap3 during biogenesis of serine-rich repeat bacterial adhesin. Mol Microbiol. 2012;83:866–78. doi: 10.1111/j.1365-2958.2012.07970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou M, Wu H. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology. 2009;155:317–27. doi: 10.1099/mic.0.025221-0. [DOI] [PubMed] [Google Scholar]
- 96.Gonnet GH, Hallett MT, Korostensky C, Bernardin L. Darwin v. 2.0: an interpreted computer language for the biosciences. Bioinformatics. 2000;16:101–3. doi: 10.1093/bioinformatics/16.2.101. [DOI] [PubMed] [Google Scholar]
- 97.Erlandson KJ, Miller SB, Nam Y, Osborne AR, Zimmer J, Rapoport TA. A role for the two-helix finger of the SecA ATPase in protein translocation. Nature. 2008;455:984–7. doi: 10.1038/nature07439. [DOI] [PMC free article] [PubMed] [Google Scholar]