Abstract
Aldolase C is a brain-specific glycolytic isozyme whose complete repertoire of functions are obscure. This lack of knowledge can be addressed using molecular tools that discriminate the protein from the homologous, ubiquitous paralog aldolase A. The anti-aldolase C antibodies currently available are polyclonal and not highly specific. We obtained the novel monoclonal antibody 9F against human aldolase C, characterized its isoform specificity and tested its performance. First, we investigated the specificity of 9F for aldolase C. Then, using bioinformatic tools coupled to molecular cloning and chemical synthesis approaches, we produced truncated human aldolase C fragments, and assessed 9F binding to these fragments by western blot and ELISA assays. This strategy revealed that residues 85–102 harbor the epitope-containing region recognized by 9F. The efficiency of 9F was demonstrated also for immunoprecipitation assays. Finally, surface plasmon resonance revealed that the protein has a high affinity toward the epitope-containing peptide. Taken together, our findings show that epitope recognition is sequence-driven and is independent of the three-dimensional structure. In conclusion, given its specific molecular interaction, 9F is a novel and powerful tool to investigate aldolase C’s functions in the brain.
Keywords: aldolase C, monoclonal antibodies, epitope mapping, ELISA, western blot, SPR
Introduction
Vertebrates express three tissue-specific isoforms of aldolase, namely aldolase A, B and C.1 Aldolase A is ubiquitously expressed and mutations in this gene have been associated to hemolytic anemia and myopathy.1-5 Aldolase B is mainly expressed in the liver, where it is involved in the utilization of exogenous fructose and mutations in its gene cause an autosomal recessive disease known as hereditary fructose intolerance (HFI).6-10 The aldolase C protein is the brain-specific isoform of the glycolytic enzyme fructose 1–6 bisphosphate aldolase. It catalyzes the reversible cleavage of fructose-1,6-bisphosphate (Fru 1,6-P2) to dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (G3P).11Aldolase C is selectively expressed in the central nervous system (CNS) and in tissue of neuronal origin.12,13
In the CNS, the aldolase C protein is always co-expressed with aldolase A but, unlike the latter, it shows a peculiar stripe-like distribution pattern in different areas of the human, mouse and rat brain.14-16 The glycolytic role of aldolase C is not considered sufficient to explain this peculiar expression in well-delimited regions of the CNS nor to justify its co-expression with aldolase A, since the two isoforms catalyze the same glycolytic reaction with comparable, although different, kinetics.17,18These observations, considered together with the high conservation of the aldolase C gene during evolution,19-24 prompted the hypothesis that aldolase C exerts additional and specific physiological functions within the CNS beyond its role in glycolysis.15 Although various observations support this hypothesis,15,25-28 no molecular mechanism has been so far identified. Moreover, no gene variants have been found for the aldolase C isoform and its putative moonlighting functions in the CNS are still unclear.
Understanding the isozyme-specific functions of aldolase C is hampered not only by the co-expression of aldolase A and C in the CNS, but also by the lack of tools able to efficiently discriminate between the various homologous aldolase isoforms. Indeed, elucidation of specific protein functions largely depends on the possibility of specifically targeting and studying the molecule of interest in its biological context. Antibodies are now widely used tools in practically every area of molecular investigation. However, few anti-aldolase C antibodies are commercially available and all are polyclonal. The quality of results obtained with monoclonal antibodies (mAbs) is generally more consistent than results obtained with polyclonal antibodies because mAbs are generally highly specific and thus they decrease background noise and cross-reactivity. Consequently, their use is to be preferred when working with proteins like aldolase C that have paralogs.
The aim of this study was to provide a new specific anti-aldolase C mAb that can serve as a tool in studies designed to investigate the function of aldolase C in tissues and cells co-expressing aldolase C and A. We have obtained a novel anti-human aldolase C mAb, characterized the target sequence on the protein, and validated its efficiency in multiple molecular applications.
Results
mAb preparation and assessment of specificity for the aldolase C isozyme
The human cDNA encoding the full-length aldolase C protein was cloned in the pET16b+ vector to be expressed in fusion with a His7-tag at the N-terminus. The resulting recombinant product was used first as an immunogen to produce mAbs and, subsequently, as a test antigen to verify the immunoreactivity of the initial mAb clones obtained from the immunization procedure. One clone (namely, 9F) was selected because of its stronger immunoreactive signal (data not shown). We characterized its specificity for aldolase C, mapped the targeted epitope and tested its performance in multiple molecular applications.
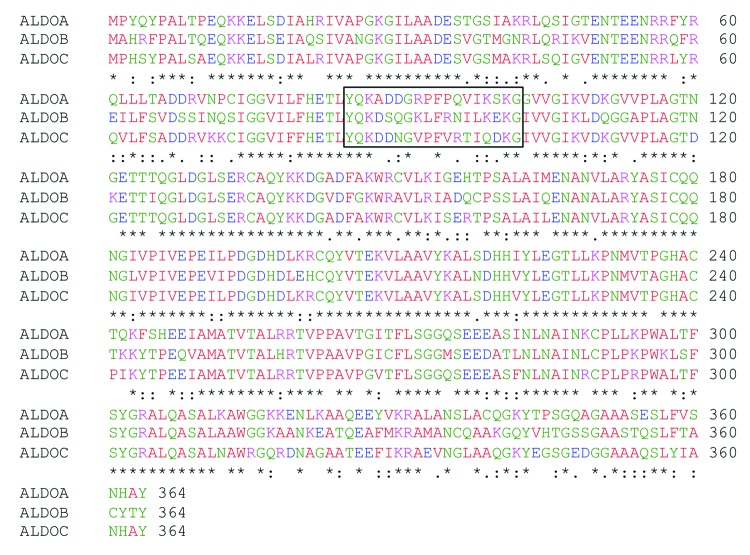
Aldolase isoforms share a very high percentage of sequence identity and similarity (Fig. 1). Indeed, although in vertebrates the three isozymes are encoded by three different genes,3,12,29 phylogenetic studies indicate that they diverged from a common ancestral gene.20 Accordingly, there is a sequence identity higher than 80% between the human aldolase A and C proteins.30 Therefore, we investigated whether the 9F mAb cross-reacts with aldolase A or B, and tested its specificity for aldolase C by western blot on total protein extracts from mouse brain, kidney, liver and heart tissues (Fig. 2). Importantly, a specific positive signal, corresponding to the molecular mass of aldolase (39kDa), was observed only in the brain tissue extract. Moreover, no cross-reactivity occurred with either of the other two aldolase isoforms in liver, kidney and heart tissues where aldolases A and B, but not C, are expressed (Fig. 2). This result demonstrated the specificity of the 9F mAb for the aldolase C isozyme.
Figure 1. Sequence alignment between the three human aldolase isozymes. Comparison of human aldolase A, B and C amino acid sequences. Sequence 85–102 is boxed. Asterisks (*) represent identical matches in the alignment; colons (:), conserved substitutions; periods (.), semi-conserved substitution. The alignment was performed using the “ClustalW2-Multiple Sequence Alignment” software (www.ebi.ac.uk/Tools/msa/clustalw2).” Red, green, blue and violet correspond to neutral, polar, negative and positive amino acids, respectively.
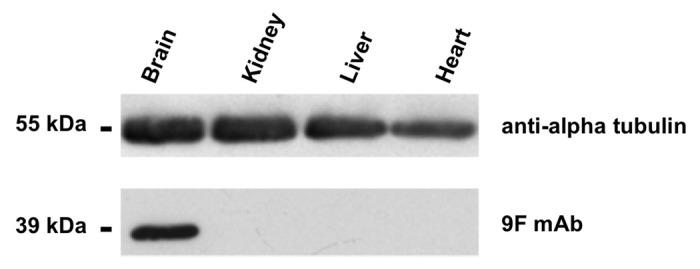
Figure 2. western blot with the anti-aldolase C 9F mAb. Enhanced chemiluminescence detection of aldolase C protein expression in heart, liver, kidney and brain tissue extracts (mouse). Proteins were separated by 12% SDS-PAGE and probed with the anti-aldolase C 9F mAb. Alpha-tubulin immunoblot was used as loading control.
Domain mapping using a panel of overlapping aldolase C fragments
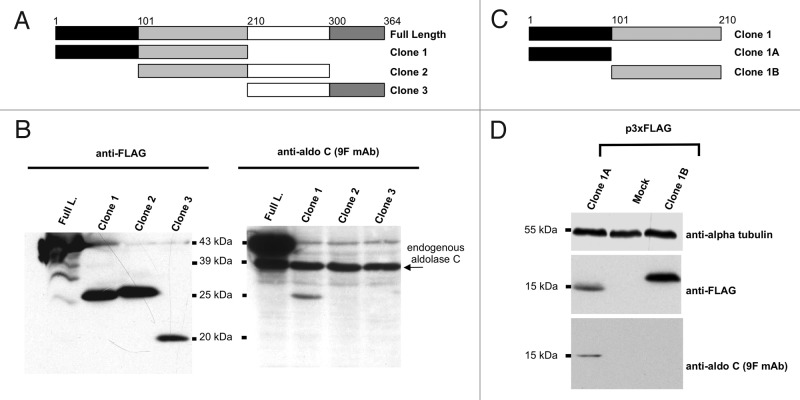
To identify the position of the domain targeted by the 9F mAb, we amplified by PCR three partially overlapping portions of the human aldolase C protein and cloned them into the p3xFLAG-CMV7.1 vector with N-terminal 3xFLAG tags (Fig. 3A; Table S1).The resulting plasmid constructs, namely Clone 1, Clone 2 and Clone 3, as well as the plasmid encoding the human full-length aldolase C protein, were transfected in Neuro2a cells (neuronal mammalian cell line) and the total protein extracts were analyzed by western blotting. The probing of the membrane with anti-Flag antibody showed that all aldolase C fragments were expressed (Fig. 3B, left panel). Moreover, as shown in Figure 3B, right panel, the 9F mAb detected three proteins of different molecular weights. The 39-kDa band was present in all of the four tested lanes, as expected because of the expression of endogenous aldolase C in Neuro2a cells. On the contrary, among the 3xFLAG-tagged aldolase C products, only the exogenous full-length (43kDa; Figure 3B, right panel, lane 1) and Clone 1 proteins (26kDa; Figure 3B, right panel, lane 2) were recognized. This experiment demonstrated that the domain targeted by 9F was contained in the Clone 1 protein region (spanning from aa 1 to aa 210 of the human aldolase C protein); it also revealed that the domain was specifically localized within the 1–100 aa region, as proven by the absence of ECL signal from Clone 2 that overlaps Clone 1 from residue 101 to residue 210. To verify this finding, we split Clone 1 into two additional sub-clones, namely Clone 1A (including amino acids from 1 to 100) and Clone 1B (including amino acids from 101 to 210) (Fig. 3C; Table S1). Immunoblot assays confirmed the domain localization within the region of the aldolase C protein spanning residues 1–100. Indeed, the 9FmAb detected Clone 1A (15kDa) but not Clone 1B (16kDa) (Fig. 3D).
Figure 3. Mapping of the aldolase C region recognized by 9F mAb. (A) and (C) Scheme of the regions of the human aldolase C protein, cloned in the 3xFLAG CMV 7.1 vector. The corresponding clone names are reported on the right. The amino acid limits of each sub-region are indicated by numbers and colors. (B) western blot analysis of total protein lysates from Neuro2a cells alternatively transfected with the 3xFLAG tagged full-length (Full. L.), clone 1, clone 2 or clone 3 fusion products. Proteins were separated by 15% SDS-PAGE and probed with an anti-FLAG antibody (left panel) and with the anti-aldolase C 9F mAb (right panel). (D) western blot analysis of total protein lysates from Neuro2a cells alternatively transfected with the 3xFLAG tagged clone 1A and clone 1B fusion products or mock-transfected. Proteins were separated by 15% SDS-PAGE and probed with an anti-FLAG antibody (upper panel) and with the anti-aldolase C 9F mAb (lower panel). Alpha-tubulin served as loading control.
Bioinformatic prediction of putative epitope-containing peptides
Considering the high specificity demonstrated by the 9F mAb for aldolase C, we hypothesized that its antigenic determinant should include an aldolase C isozyme-specific amino acid stretch. To define the possible antigenic determinant located within the first 100 residues, we used the ElliPro software (http://tools.immuneepitope.org/tools/ElliPro)31 that predicts and visualizes putative antibody epitopes on a given protein 3D structure, according to surface exposure and solvent accessibility of residues.32 Using as input the X-ray structure of the aldolase C protein (PDB accession code: 1XFB),33 ElliPro predicted 12 putative epitope-containing peptides distributed along the entire molecular surface. We focused on the four sequences localized within the first 100 amino acids (Table 1, epitopes No. 2, 3, 5 and 12). Starting from the ElliPro prediction, we defined four 16-mer/18-mer peptide sequences to be tested as the putative epitope-containing regions. It should be noted that the Ellipro-predicted peptides were sometimes extended to include flanking aldolase C-specific-residues, or deprived/extended of terminal residues because of technical issues related to peptide synthesis (compare with shaded sequences in Table 1). These four peptides were produced both in a biotinylated form to conduct ELISA assays, and as fusion proteins with GFP to perform immunoblot experiments.
Table 1. Predicted linear epitope(s) in the aldolase C protein and the corresponding synthesized four peptides.
| No. | Start | End | Peptide sequence | No. of residues | Score |
|---|---|---|---|---|---|
| 1 | 114 | 122 | VPLAGTDGE | 9 | 0.851 |
| 2 | 3 | 18 | HSYPALSAEQKKELSD | 16 | 0.832 |
| 2 | 17 | PHSYPALSAEQKKELS§ | 16 | ||
| 3 | 85 | 93 | YQKDDNGVP | 9 | 0.8 |
| 85 | 102 | YQKDDNGVPFVRTIQDKG§ | 18 | 85 | |
| 4 | 312 | 327 | NAWRGQRDNAGAATEE | 16 | 0.792 |
| 5 | 41 | 58 | AKRLSQIGVENTEENRRL | 18 | 0.751 |
| 41 | 58 | AKRLSQIGVENTEENRRL§ | 18 | ||
| 6 | 340 | 344 | QGKYE | 5 | 0.75 |
| 7 | 235 | 251 | TPGHACPIKYTPEEIAM | 17 | 0.739 |
| 8 | 153 | 166 | KISERTPSALAILE | 14 | 0.716 |
| 9 | 289 | 296 | RCPLPRPW | 8 | 0.704 |
| 10 | 193 | 204 | PDGDHDLKRCQY | 12 | 0.668 |
| 11 | 126 | 133 | QGLDGLSE | 8 | 0.631 |
| 12 | 65 | 73 | SADDRVKKC | 9 | 0.561 |
| 60 | 75 | RQVLFSADDRVKKCIG§ | 16 |
§ Synthesized peptides are shaded
Epitope mapping refinement by ELISA and western blot analysis
To refine the mapping of the 9F mAb-related epitope, we first performed an enzyme-linked immunosorbent assay (ELISA) by spotting the four different biotinylated aldolase C peptides in streptavidin-coated plates. Four mAb dilutions (1:5,000;1:10,000; 1:20,000 and 1:40,000) were tested. A specific peptide-antibody interaction occurred only in the wells containing peptide 85–102. The ELISA data were analyzed and plotted on a graph, relating the measured absorbance values to the corresponding mAb dilution factors (Fig. 4). This result allowed us to narrow down the position of the epitope targeted by the mAb to the small peptide region of the human aldolase C protein spanning residues 85–102.
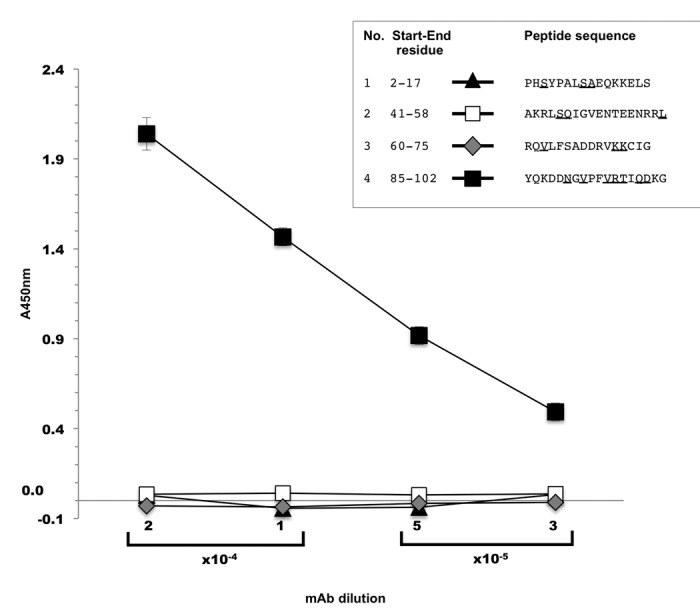
Figure 4. ELISA screening of biotinylated aldolase C peptides. Four different mAb dilution factors were tested in the experiment (1:5,000; 1:10,000; 1:20,000; 1:40,000). X axis, mAb dilution factors; Y axis, absorbance values measured at 450 nm. The marker styles of the four test peptides are indicated in the figure inset together with their amino acid sequence. Underlined letters indicate aldolase C isoenzyme sequence-specific residues as compared with human aldolase A and B. Values are the mean ± SD of duplicate samples and are representative of three independent experiments.
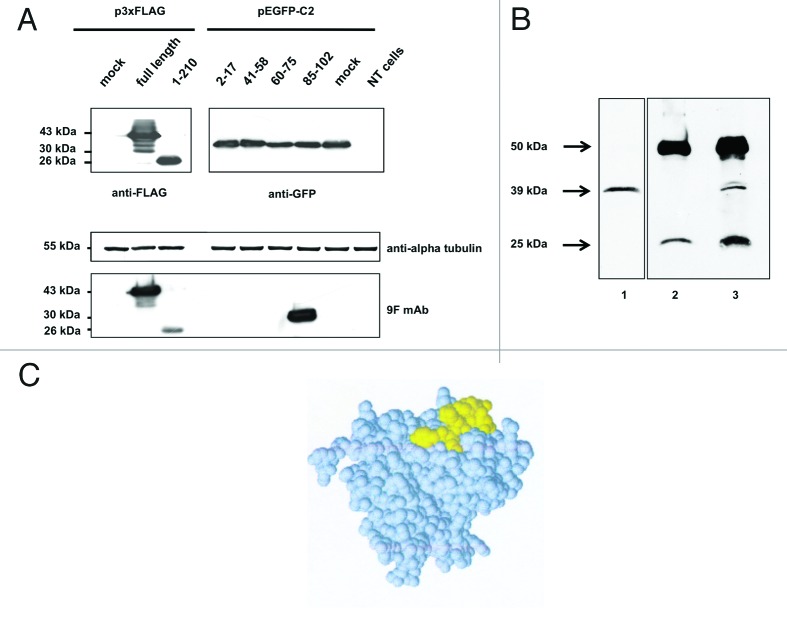
To verify this finding, we cloned the cDNA sequences encoding the four selected peptides of interest in the pEGFP-C2 vector, with the aim of expressing them with a GFP tag fused to their N-terminus (Table S2). We transfected the four pEGFP-C2 clones in Neuro2a cells and assayed the resulting total protein extracts by immunoblotting. Probing of the membrane with an anti-GFP antibody revealed comparable expression levels of all clones (Fig. 5A, anti-GFP immunoblot). Using the anti-aldolase C 9F mAb, ECL detection occurred only in the lane corresponding to the transfection of the GFP-aldolase C aa 85–102 fusion protein (Fig. 5A, anti-aldolase C 9F mAb immunoblot). This assay confirmed that the epitope targeted by the 9F mAb was located within residues 85–102 of the human aldolase C protein.
Figure 5. western blot analysis of (A) aldolase C peptides expressed in fusion with a GFP tag at the N-terminus. Total protein extracts obtained from Neuro2a cells alternatively transfected with the four pEGFP-C2 recombinant clones (see Table S2 for details), mock transfected or not transfected were separated by 12% SDS-PAGE and probed using the anti-aldolase C 9F mAb. To have a complete overview of the epitope-mapping, the 3xFLAG-tagged full-length aldolase C and clone 1 fusion proteins were included in the immunoblot panel. Alpha-tubulin served as loading control; anti-FLAG and anti-GFP antibodies were used to verify the transfections. (B) Aldolase C immunoprecipitation with the 9F mAb. Mouse brain lysate (1mg) was immunoprecipitated with the anti-aldolase C 9F mAb (lane 3, IP) after pre-clearing with mouse IgGs (lane 2, IgG). 1/10 of the total immunoprecipitated fractions were analyzed by western blot using the same 9F mAb. Twenty μg of total protein extract were included in the panel as input control (lane 1, Input). (C) Epitope-containing peptide 85–102 mapped on the protein subunit structure. Space filling representation of the aldolase C 3D structure (PDB code 1XFB). Peptide 85–102, which is highly exposed on the protein surface, is shown in yellow.
Evaluation of anti-aldolase C mAb efficiency in the immunoprecipitation application
After having mapped the epitope targeted by the mAb and demonstrated its efficiency in both immunoblotting and ELISA applications, we tested its efficiency in immunoprecipitation protocols. Because of the specific expression of the aldolase C protein in the CNS, we used total protein extract from adult mouse brain as input sample. As shown in Figure 5B, the experiment revealed that the mAb was efficient also in the immunoprecipitation application. Indeed, the immunoreactive aldolase C signal (39kDa) was observed in the IP fraction (lane 3, IP), but not in the negative control lane (lane 2, IgG). This experiment demonstrated that the 9F mAb immunoprecipitated the aldolase C isozyme and revealed that it targeted the protein also in its endogenous native state.
Aldolase C-NFL RNA co-immunoprecipitation with the 9F mAb
The list of enzymes that have been found to participate in additional cellular processes is constantly expanding. Several well-known metabolic enzymes have been shown to bind RNA.34 An unexpected ability to interact with RNA has been described also for aldolase C: it was found to bind to the 3′UTR of the light neurofilament (NFL) mRNA, known to regulate the transcript stability in neuronal cells.25,28 This provides evidence that aldolase C is involved in processes other than glycolysis, although the functional relevance of the interaction needs to be clarified.
To explore the potential of the 9F mAb as a tool to investigate further the RNA-binding properties of aldolase C, we performed a protein-RNA co-immunoprecipitation assay. As a cellular model, we used HEK293 cells because of their negligible expression of endogenous aldolase C (data not shown). Total protein extracts from HEK293 cells, transfected with the 3xFLAG-tagged full-length aldolase C or mock-transfected (p3xFLAG) were incubated with equal amounts of NFL RNA encompassing the mapped aldolase C-binding site.25 The IP fractions were isolated using the 9F mAb or the control anti-GFP antibody. To verify NFL co-immunoprecipitation, the RNA was extracted and retro-transcribed using an appropriate NFL antisense oligonucleotide. When the 9F mAb was used in the IP step, real-time PCR experiments revealed a 2fold increase in NFL mRNA immunoprecipitation in the 3xFLAG-aldolase C transfected sample vs. the co-immunoprecipitation with anti-GFP antibody (Fig. 6, full-length). On the contrary, no enrichment in NFL mRNA levels was observed in co-immunoprecipitation experiments performed with mock-transfected extracts (Fig. 6, mock).
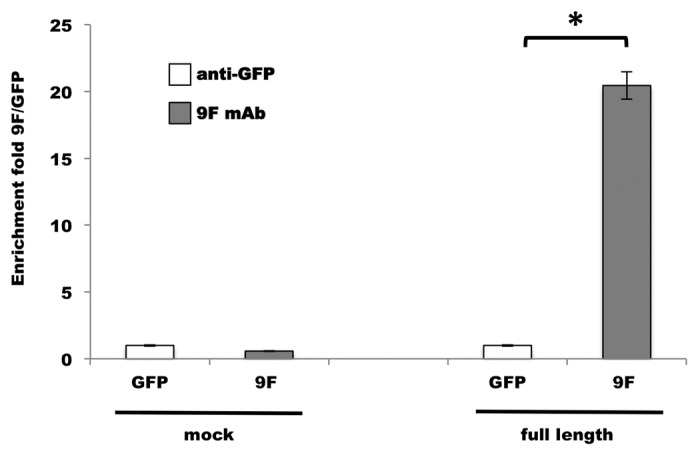
Figure 6. Aldolase C-NFL RNA co-immunoprecipitation with the 9F mAb. The quantity of NFL RNA immunoprecipitated by 9F mAb (gray bars) from 3xFLAG-tagged aldolase C-transfected cells (full-length) was analyzed by qPCR. The fold enrichment is referred to the anti-GFP immunoprecipitation (white bars) performed as a control experiment. The co-immunoprecipitation from HEK293 cells transfected with the p3xFLAG empty vector (mock) was used as negative control. Results are means ± SD from three independent experiments in duplicate. * P < 0.05 vs. anti-GFP.
Determination of the mAb-epitope affinity constant
To gain information about the folded/unfolded state of the tested ligands, CD spectra were recorded for the four synthesized peptides shaded in Table 1 (and Figure 4) and for the His7-tagged full-length aldolase C (Fig. S1). The spectra clearly showed that all the peptides lack a secondary structure, being completely disordered, whereas the full-length protein adopts a typical α/β fold. Indeed, the CD profile of aldolase C is virtually identical to those reported for aldolase A2 and aldolase B,35 which is consistent with their high structural similarity.
To obtain quantitative insights into the affinity of the 9F mAb toward human aldolase C and the 85–102 peptide, we used surface plasmon resonance (SPR) technology.36 Human aldolase C was covalently immobilized on a sensor chip CM5 whereas the four biotinylated aldolase C peptides were immobilized with a capturing approach on sensor chips SA (streptavidin), designed to bind biotinylated molecules. A series of concentrations of the mAb solution (analyte) were allowed to flow on all the immobilized molecules. A ligand-analyte molecular interaction was detected only when the immobilized molecules were the full-length His7-tagged aldolase C and peptide 85–102 (Fig. S2), in full agreement with outcomes from the all other techniques used in this study. Experimental data were fitted using a 1:1 binding model, to calculate the equilibrium dissociation constants of the mAb-epitope complexes. Kd values are (4.31*10−7 ± 1.1*10−8SD) M and (1.98*10−7 ± 1.0*10−8SD) M for the affinity of the mAb toward the peptide 85–102 and the full-length enzyme, respectively. It is noteworthy that these Kd values represent apparent affinities, because bivalent binding of 9F cannot be excluded. Nevertheless, these results confirm the high specificity and affinity of the 9F mAb toward the aldolase C sequence 85–102, and provide valuable insights into the interaction. Indeed, Kd values indicate that: 1) the binding is quite strong; 2) the enzyme and the isolated peptide exhibit a comparable affinity toward the mAb; and 3) the recognition is not guided by the 3D-structure, but only by the sequence.
Discussion
To overcome the difficulty of probing the functions of the aldolase C protein because of its co-expression with aldolase A, we produced and characterized a novel specific anti-aldolase C mAb 9F.
Using diverse methodologies, we localized the epitope targeted by 9F to a region of the aldolase C protein consisting of the 18 amino acids between residues 85–102 of the enzyme sequence. SPR, which gives a quantitative measure of the affinity of 9F toward aldolase C, revealed Kd values within 10−7M order of magnitude, thereby demonstrating the strength of the interaction. Furthermore, the full-length enzyme and the isolated peptide have a similar affinity toward 9F, thereby supporting the absence of any other epitope region within the aldolase C sequence. Importantly, the CD data demonstrated that the epitope recognition is guided by the sequence and not by the 3D structure because 9F binds with a similar affinity to the unfolded peptide and to the folded enzyme.
Although the structural features responsible for the specific functions of the three different aldolase isoforms are largely unknown, they are believed to be determined by the isozyme-specific residues that are mainly localized on the protein surface.37
The peptide region 85–102 of the human aldolase C protein is localized on the protein surface and is characterized by the presence of four highly exposed aldolase C isozyme sequence-specific residues: N90, V92, R96 and D100. Indeed, these residues are not conserved in the corresponding amino acid positions of the human aldolase A or aldolase B enzymes. Moreover, their ElliPro protrusion index values > 0.7 (N90:0.947; V92:0.827; R96:0.789; D100:0.772), as well as the solvent-accessible surface area (N90:36.6Å2; V92:15.4Å2; R96:39.3Å2; D100:29.6Å2) consistently indicate that these residues are highly exposed to the solvent on the protein surface (Fig. 5C). Therefore, fragment 85–102 is prone to recognition by other molecules and eventually to interact with them. These observations suggest that the four amino acids mentioned above play a crucial immunogenic role in determining the anti-aldolase C specificity of the 9F mAb.
Furthermore, the presence of the four aldolase C isozyme sequence-specific residues (N90, V92, R96 and D100) in the epitope-containing region 85–102 (Fig. 5C) also suggests the intriguing possibility that this portion of the protein might be involved in determining aldolase C isozyme-specific functions, acquired through evolution.37 Studies of this specific protein region could help to verify this aspect of aldolase C physiology. Given that the epitope-containing region 85–102 does not include any of the residues that are known to affect catalytic activity, we surmise that the new mAb does not interfere with enzymatic activity.
We show that the 9F anti-aldolase C mAb is efficient in multiple molecular applications. It is noteworthy that, although 9F was raised against the human protein, our immunoblot experiments performed on Neuro2a and PC12 cell lysates, as well as on mouse tissue protein extracts, demonstrated that it is also able to recognize the aldolase C enzyme of mouse and rat origin (data not shown for PC12 immunoblots). Therefore, this antibody has the additional advantage of specifically targeting multiple aldolase C orthologs without cross-reacting with the aldolase A and B paralogs. Moreover, the aldolase C-NFL RNA co-immunoprecipitation results lay the groundwork for further investigations on unconventional functions of the enzyme. In fact, the 9F mAb might help to map the still unknown RNA-binding domain of the protein or to identify additional RNAs possibly interacting with aldolase C.
In conclusion, we generated and characterized a novel, anti-aldolase C mAb whose specificity and broad range of applications make it suitable as a powerful tool for research on this emblematic brain-specific isozyme whose moonlighting physiological roles in the CNS still remains obscure.
Materials and Methods
Preparation of mAbs
ProMab Biotechnologies, Inc. was commissioned to generate the anti-aldolase C mAb. It was produced in mouse using, as immunogen, the full-length human aldolase C protein expressed in fusion with a His7-tag at the N-terminus. To this aim, the human cDNA encoding the full-length aldolase C protein was cloned in the pET16b+ vector and expressed and affinity purified in E. coli BL21DE3 cells. The recombinant product was also used as test antigen to verify the immunoreactivity of the mAb clones. The clone showing the strongest immunoreactive signal was selected within the whole set: its identity code was 9F1B5 and is herein referred to as anti-aldolase C 9F mAb. The method used to prepare the panel of screened antibodies is based on mouse hybridoma development and is under patent (ProMab Biotechnologies Inc.).
Cell culture and plasmids transfection
The cell lines used for transfection experiments were Neuro2a (mouse neuroblastoma, from CEINGE Cell Bank, Naples, Italy) and HEK293 (human embryonic kidney). Cells were maintained, sub-cultured and stored according to the instructions of Deutsche Sammlung von. Mikroorganismen und Zellkulturen GmbH. Medium composition: 87% Dulbecco’s modified Eagle’s medium (Sigma), 10% FBS (HyClone), 2% ultra-glutamine (Lonza), 1% non-essential amino acids (Gibco). One day before transfection, cells were plated in growth medium (without antibiotics) to obtain a 70–90% confluence at the time of transfection. Cells were transfected with lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions and incubated at 37 °C in a CO2 incubator for 48h prior to testing for transgene expression. The culture medium was always changed 6h after transfection. All clones transfected in this study are listed in Tables S1 and S2 together with the expression plasmids and the primer pairs used in the cloning procedure.
Protein extraction from cells and tissues
Cells and tissues were washed twice with cold PBS and lysed for 30 min on ice in lysis buffer (50mM TRIS-HCl, pH 7.5, 150mM NaCl, 0.1% Triton X-100, 0.5 mM PMSF). Before lysis, tissues were dissociated to obtain cells in a dounce homogenizer with a tight-fitting glass pestle. Lysates were clarified by centrifugation at 16000xg for 30min at 4 °C in an Eppendorf microcentrifuge and the total protein concentration was determined with classical Bradford method.38
Western blotting
Protein extracts were resolved on SDS-PAGE gels and transferred onto nitrocellulose membranes (GE Healthcare, RPN1520D) at 200mA for 90min in 1X Transfer Buffer (25mM Tris, 192mM glycine, 10–20% methanol). After transfer, membranes were stained with Ponceau solution to verify the efficiency of the blotting process, and then incubated for 1h in blocking solution (5% non-fat dry milk [Biorad, 170–6404] in PBS pH 7.6 containing 0.1% Tween-20 [T-PBS]) to prevent non-specific binding. Membranes were then incubated for 1h at room temperature (RT) or overnight (4 °C) with specific primary antibodies (9F mAb, 1:1000; α-FLAG, 1:1000; α-GFP, 1:1000) diluted in blocking solution. After three washes with T-PBS, membranes were incubated 45min at RT with a horseradish peroxidase-conjugated secondary antibody (anti-mouse 1:5000; anti-rabbit 1:5000; GE Healthcare). A mouse anti-α-tubulin primary antibody (1:2000) was always used as loading control. Immunoblots were developed by enhanced chemiluminescence (ECL) using Amersham ECL Western Blotting Detection Reagents (GE Healthcare, RPN2106).
ELISA assay of biotinylated peptides
For ELISA assays, aldolase C peptides of interest were synthesized with a biotin coupled to their N-terminus, and immobilized on a solid phase by streptavidin capture. Peptide quality was controlled after chemical synthesis by mass spectrometry (ESI ion trap amaZonSL, Bruker Daltonics, operated in positive ion mode), and resulted in a highly pure single peak. In this experiment, each well of a Nunc-Immuno MaxiSorb plate was coated with 100μl of a streptavidin solution (5μg/ml in 10mM Na2CO3 pH 9.6) by leaving the plate exposed to the air for 12–16 h at 37 °C to allow the solution to evaporate. The next day, non-specific absorption was blocked by dispensing 200μl of blocking buffer (10mg/ml BSA in 1X PBS, 0.05% Tween-20) and incubating the plate 1 h at room temperature (RT) on a shaker. After three washes with wash buffer (1X PBS, 0.05% Tween-20), 100μl of the appropriate peptide solution (1μM peptide in wash buffer) were added to the corresponding well and incubated 1h at RT on a shaker. Subsequently, the peptide solution was removed and the washing procedure was repeated as previously specified. The anti-aldolase C 9F mAb was diluted in 1X PBS, 0.5% BSA, 0.05% Tween-20 (1:5000; 1:10000; 1:20000; 1:40000 starting from the 1.8mg/ml stock solution) and 100μl of these dilutions were added to each well and incubated at RT on a shaker for 1h. After three washes, 100μl/well of a horseradish peroxidase-conjugated anti-mouse IgG solution (1:2000 secondary antibody dilution in 1X PBS, 0.5% BSA, 0.05% Tween-20) where added to each well and incubated 45min at RT on a shaker. Five final washes were performed using the wash buffer. Then 100μl of a 3,3′,5,5′ tetramethylbenzidine substrate solution (TMB, Sigma, T4444) were added to each well and the enzymatic reaction was stopped using 1N sulfuric acid. The results were spectrophotometrically analyzed at 450nm using an appropriate plate reader. The peptides used in this study were chemically synthesized. Each peptide was synthesized including a GSGSG spacer sequence between the N-terminal biotin and the aldolase C peptide.
Results from replicate experiments were analyzed by descriptive statistics with Microsoft Office Excel 2007. Results are shown as mean and standard deviation.
Aldolase C immunoprecipitation using the anti-aldolase C 9F mAb
To test the 9F mAb in immunoprecipitation (IP) assays, the input sample consisted of adult mouse brain total protein extract (1mg) and was pre-cleared by incubation (1h at 4 °C on a rotating wheel) with 2μg of mouse control IgG (Santa Cruz Biotechnology, sc-2025). The solution was then incubated with 50µl of slurry Protein A/G plus-agarose beads (Santa Cruz Biotechnology, sc-2003) for 1h at 4 °C on a rotating wheel and subsequently centrifuged at 4000xg for 5min at 4 °C. The resulting supernatant was incubated with 10μg of the anti-aldolase C 9F mAb for 1h on a rotating wheel at 4 °C. Then, 50µl of fresh Protein A/G plus-agarose beads were added to the test tube and incubated for 2h at 4 °C. After extensive washing of the pellet beads, the resulting immunoprecipitated complex was eluted incubating the resin at 99 °C for 5min in 40µl of 2X protein sample buffer. Immunoprecipitation lysis buffer was constituted by 10% glycerol, 50mM TRIS-HCl pH 8, 150mM NaCl, 0.1–1% NP40, 1X Complete Roche Protease inhibitors, 0.5mM PMSF. For western blot analysis 1/10 of the total immunoprecipitated fraction was assayed. The 9F mAb was used also in the immunoblot analysis of the immunoprecipitation result.
Aldolase C-NFL RNA co-immunoprecipitation assay
To generate the RNA target for co-immunoprecipitation experiment, the region of interest of the mouse 3′ UTR light neurofilament mRNA (NFL RNA), spanning from nucleotide -75 upstream of stop codon to nucleotide +261 downstream of stop codon, was in vitro transcribed. The sequence of interest was amplified from mouse genomic cDNA by using an appropriate primer set that allowed the incorporation of the T7 promoter sequence in the PCR product (mouse_NFL-75s_T7_pro, 5′-aagtccTAATACGACTCACTATAGGGccaaagaatctgaagaggaagaga-3′; mouse_NFL+261as, 5′-CACATTGCCATAGATCCTGAACT-3). The purified PCR product (1000ng) was mixed with 0.5 µl of T7 RNA polymerase enzyme (50 U/µl, Stratagene), 6 µl of 5x Transcription Buffer (Stratagene), 3 µl DTT 100 mM, 3 µl of rNTP 15 mM, 0.5 µl of RNase inhibitor (40 U/µl, Ambion) in a final reaction volume of 30 µl, and the sample was incubated for 2 h at 37 °C. After transcription, 1 µl of DNase (Roche Diagnostics) was added in the reaction tube and incubated 15 min at 37 °C to remove any residual DNA. The transcribed RNA was purified with the TriFast reagent (Euroclone). HEK293 cells, transfected with the 3xFLAG-tagged full-length aldolase C expressing vector or mock transfected, were lysed in HEGN buffer (20 mM Hepes pH 7.7, 150 mM NaCl, 0.5 mM EDTA, 10% glycerol, 0.1% Triton X-100, 1 mM DDT, and 2x Protease Inhibitors Cocktail [Roche Diagnostics]).
Total cell extract (100 μg) was incubated with 0.2 ng of the mouse NFL RNA and subsequently split in three aliquots to be used for the input reference sample, for the control immunoprecipitation with anti-GFP antibody (5μg, Santa Cruz Biotechnology) and for aldolase C immunoprecipitation with mAb 9F (5 μg). Additional HEGN buffer was added to the test tubes (500 μl final volume) that were incubated 2 h at 4 °C on a rotating wheel. Subsequently, 30 μl of A/G plus agarose-beads (Santa Cruz Biotechnology) were added to each sample and incubated for additional 2 h at 4 °C on a rotating wheel. Beads were then washed three times with HEGN buffer supplemented with 0.05% sodium deoxycholate and the RNA was finally extracted from the immunoprecipitated fraction and from the input reference sample. The RNA was retro-transcribed using the mouse NFL+261 as primer and the resulting cDNA was diluted 1:40 in sterile ultrapure water and used to evaluate the NFL mRNA enrichment fold by qPCR with the mouse primer pair NFL-75s (without T7 promoter) and NFL +91as (5′-ATTTGTATAGGATCTGGAACTCAACTG-3). To calculate the fold enrichment, all data were initially normalized to the respective inputs. Then, the enrichment was calculated by subtraction of control anti-GFP co-IP ΔCt from 9F mAb co-IP ΔCt, using the 2^-ΔΔCT method.38
Circular dichroism
Circular dichroism (CD) spectra were recorded with a Jasco J-815 spectropolarimeter equipped with a Peltier type temperature control system (Model PTC-423S). CD measurements (200–250 nm) were performed in MES 15 mM pH 6.0 KCl, 70 mM, at 20 °C, by using a 0.1 cm optical path length cell. CD spectra were recorded on the His-tagged full-length aldolase C (13 µM) and the isolated biotinylated peptide(s) (200 µM). CD spectra, recorded with a time constant of 4s, a 2 nm band width, and a scan rate of 20nm min−1, were signal-averaged over at least three scans. The baseline was corrected by subtracting the buffer spectrum.
Surface plasmon resonance
All SPR experiments were conducted on a Biacore T200 system (GE Healthcare), at 25 °C. Binding assays were performed between the 9F mAb and i) the His-tagged human aldolase C and ii) the 4 biotinylated peptides. First, human aldolase C was covalently immobilized on a CM5 sensor chip (research grade), with a final immobilization level of 3000 response units (RU), by the standard amine coupling procedure, using HBS-EP buffer (HEPES 10 mM, NaCl 150 mM, EDTA 3 mM, 0.005% Surfactant P20, pH 7.4) as running buffer. Immobilization was performed through activation of the sensor chip with 60μL of N-hydroxysuccinimide and N-ethyl-N-(dimethylaminopropyl)-carbodiimide at 10 μLmin−1, for 30s, followed by a 30 μL injection of the enzyme diluted in 10 mM phosphate buffer pH 6.5. Unreacted activated groups were blocked by a 60 μL injection of 1M ethanolamine at 10 μL min−1. Subsequently, a solution of 9F mAb was injected as analyte at various concentrations (from 50 nM to 50 μM), using HBS-EP as running buffer.
Second, the 4 biotinylated peptides, (50µM in HBS-EP buffer), were immobilized through a capturing approach for a contact time of 120s on a sensor chip SA, based on the non-covalent strong coupling of biotinylated probes to the streptavidin. The final immobilization level of the 4 peptides was in the interval 400–500 RU. The 9F mAb was injected, as analyte, at various concentrations (from 2nM to 1μM) using HBS-EP buffer. For all SPR experiments, the chip was regenerated by removing bound analytes using the mobile phase MgCl2 0.25 M, injected for 30s at 30 μL min−1, after the antibody injection. Data were corrected using a blank sensor chip as control. Moreover, binding experiments were repeated in triplicate. The SPR sensorgrams, for each interacting pairs, were analyzed by curve-fitting using a 1:1 binding model of the Biacore T200 BIA evaluation 4.1 software. Dissociation equilibrium constants were calculated from the kinetic association and dissociation constants for the binding of 9F mAb with human aldolase C and with the 85–102 peptide.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Jean Ann Gilder (Scientific Communication srl, Naples, Italy) for revising and editing the text; and Vittorio Lucignano, CEINGE-Biotecnologie Avanzate for technical assistance. We also thank Sara Spaziani for helpful assistance in the immunoprecipitation assay. The authors declare that there is no conflict of interests regarding the publication of this article.
This work was supported by grant RF-2010–2318372 from the Ministry of Health (to F.S.), grant PON01_02589 (MICROMAP) - 2012 and Grant PON02_00677 (BIOGENE) potenziamento lab.8 – 2012 from the Ministry of University and Research (both to F.S.) and grant POR Campania FSE 2007/2013 (Campus-Bioframe) from the Regione Campania (to F.S.).
Glossary
Abbreviations:
- CD
circular dichroism
- CNS
central nervous system
- DHAP
dihydroxyacetone phosphate
- ECL
enhanced chemiluminescence
- Fru 1,6-P2
fructose-1,6-bisphosphate
- G3P
glyceraldehyde 3-phosphate
- HFI
hereditary fructose intolerance
- IP
immunoprecipitation
- mAbs
monoclonal antibodies
- NFL
light neurofilament
- SA
streptavidin
- SPR
surface plasmon resonance
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/28191
References
- 1.Beernink PT, Tolan DR. Subunit interface mutants of rabbit muscle aldolase form active dimers. Protein Sci. 1994;3:1383–91. doi: 10.1002/pro.5560030904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esposito G, Vitagliano L, Cevenini A, Amelio T, Zagari A, Salvatore F. Unraveling the structural and functional features of an aldolase A mutant involved in the hemolytic anemia and severe rhabdomyolysis reported in a child. Blood. 2005;105:905–6. doi: 10.1182/blood-2004-09-3558. [DOI] [PubMed] [Google Scholar]
- 3.Izzo P, Costanzo P, Lupo A, Rippa E, Paolella G, Salvatore F. Human aldolase A gene. Structural organization and tissue-specific expression by multiple promoters and alternate mRNA processing. Eur J Biochem. 1988;174:569–78. doi: 10.1111/j.1432-1033.1988.tb14136.x. [DOI] [PubMed] [Google Scholar]
- 4.Kreuder J, Borkhardt A, Repp R, Pekrun A, Göttsche B, Gottschalk U, Reichmann H, Schachenmayr W, Schlegel K, Lampert F. Brief report: inherited metabolic myopathy and hemolysis due to a mutation in aldolase A. N Engl J Med. 1996;334:1100–4. doi: 10.1056/NEJM199604253341705. [DOI] [PubMed] [Google Scholar]
- 5.Yao DC, Tolan DR, Murray MF, Harris DJ, Darras BT, Geva A, Neufeld EJ. Hemolytic anemia and severe rhabdomyolysis caused by compound heterozygous mutations of the gene for erythrocyte/muscle isozyme of aldolase, ALDOA(Arg303X/Cys338Tyr) Blood. 2004;103:2401–3. doi: 10.1182/blood-2003-09-3160. [DOI] [PubMed] [Google Scholar]
- 6.Cox TM. Aldolase B and fructose intolerance. FASEB J. 1994;8:62–71. doi: 10.1096/fasebj.8.1.8299892. [DOI] [PubMed] [Google Scholar]
- 7.Esposito G, Vitagliano L, Santamaria R, Viola A, Zagari A, Salvatore F. Structural and functional analysis of aldolase B mutants related to hereditary fructose intolerance. FEBS Lett. 2002;531:152–6. doi: 10.1016/S0014-5793(02)03451-8. [DOI] [PubMed] [Google Scholar]
- 8.Lameire N, Mussche M, Baele G, Kint J, Ringoir S. Hereditary fructose intolerance: a difficult diagnosis in the adult. Am J Med. 1978;65:416–23. doi: 10.1016/0002-9343(78)90767-2. [DOI] [PubMed] [Google Scholar]
- 9.Odièvre M, Gentil C, Gautier M, Alagille D. Hereditary fructose intolerance in childhood. Diagnosis, management, and course in 55 patients. Am J Dis Child. 1978;132:605–8. doi: 10.1001/archpedi.1978.02120310069014. [DOI] [PubMed] [Google Scholar]
- 10.Santamaria R, Esposito G, Vitagliano L, Race V, Paglionico I, Zancan L, Zagari A, Salvatore F. Functional and molecular modelling studies of two hereditary fructose intolerance-causing mutations at arginine 303 in human liver aldolase. Biochem J. 2000;350:823–8. doi: 10.1042/0264-6021:3500823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salvatore F, Izzo P, Paolella G. Aldolase gene and protein families: structure, expression and pathophysiology. Horiz Biochem Biophys. 1986;8:611–65. [PubMed] [Google Scholar]
- 12.Rocchi M, Vitale E, Covone A, Romeo G, Santamaria R, Buono P, Paolella G, Salvatore F. Assignment of human aldolase C gene to chromosome 17, region cen----q21.1. Hum Genet. 1989;82:279–82. doi: 10.1007/BF00291170. [DOI] [PubMed] [Google Scholar]
- 13.Willson VJ, Thompson RJ. Human brain aldolase C4 isoenzyme: purification, radioimmunoassay, and distribution in human tissues. Ann Clin Biochem. 1980;17:114–21. doi: 10.1177/000456328001700303. [DOI] [PubMed] [Google Scholar]
- 14.Ahn AH, Dziennis S, Hawkes R, Herrup K. The cloning of zebrin II reveals its identity with aldolase C. Development. 1994;120:2081–90. doi: 10.1242/dev.120.8.2081. [DOI] [PubMed] [Google Scholar]
- 15.Buono P, D’Armiento FP, Terzi G, Alfieri A, Salvatore F. Differential distribution of aldolase A and C in the human central nervous system. J Neurocytol. 2001;30:957–65. doi: 10.1023/A:1021828421792. [DOI] [PubMed] [Google Scholar]
- 16.Hawkes R, Herrup K. Aldolase C/zebrin II and the regionalization of the cerebellum. J Mol Neurosci. 1995;6:147–58. doi: 10.1007/BF02736761. [DOI] [PubMed] [Google Scholar]
- 17.Penhoet EE, Kochman M, Rutter WJ. Molecular and catalytic properties of aldolase C. Biochemistry. 1969;8:4396–402. doi: 10.1021/bi00839a026. [DOI] [PubMed] [Google Scholar]
- 18.Penhoet EE, Rutter WJ. Catalytic and immunochemical properties of homomeric and heteromeric combinations of aldolase subunits. J Biol Chem. 1971;246:318–23. [PubMed] [Google Scholar]
- 19.Berardini TZ, Drygas-Williams M, Callard GV, Tolan DR. Identification of neuronal isozyme specific residues by comparison of goldfish aldolase C to other aldolases. Comp Biochem Physiol A Physiol 1997; 117:471-6. [DOI] [PubMed]
- 20.Kukita A, Mukai T, Miyata T, Hori K. The structure of brain-specific rat aldolase C mRNA and the evolution of aldolase isozyme genes. Eur J Biochem. 1988;171:471–8. doi: 10.1111/j.1432-1033.1988.tb13813.x. [DOI] [PubMed] [Google Scholar]
- 21.Lannoo MJ, Hawkes R. A search for primitive Purkinje cells: zebrin II expression in sea lampreys (Petromyzon marinus) Neurosci Lett. 1997;237:53–5. doi: 10.1016/S0304-3940(97)00802-1. [DOI] [PubMed] [Google Scholar]
- 22.Paolella G, Buono P, Mancini FP, Izzo P, Salvatore F. Structure and expression of mouse aldolase genes. Brain-specific aldolase C amino acid sequence is closely related to aldolase A. Eur J Biochem. 1986;156:229–35. doi: 10.1111/j.1432-1033.1986.tb09572.x. [DOI] [PubMed] [Google Scholar]
- 23.Shiokawa K, Kajita E, Hara H, Yatsuki H, Hori K. A developmental biological study of aldolase gene expression in Xenopus laevis. Cell Res. 2002;12:85–96. doi: 10.1038/sj.cr.7290114. [DOI] [PubMed] [Google Scholar]
- 24.Skala H, Vibert M, Lamas E, Maire P, Schweighoffer F, Kahn A. Molecular cloning and expression of rat aldolase C messenger RNA during development and hepatocarcinogenesis. Eur J Biochem. 1987;163:513–8. doi: 10.1111/j.1432-1033.1987.tb10898.x. [DOI] [PubMed] [Google Scholar]
- 25.Cañete-Soler R, Reddy KS, Tolan DR, Zhai J. Aldolases a and C are ribonucleolytic components of a neuronal complex that regulates the stability of the light-neurofilament mRNA. J Neurosci. 2005;25:4353–64. doi: 10.1523/JNEUROSCI.0885-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slemmer JE, Haasdijk ED, Engel DC, Plesnila N, Weber JT. Aldolase C-positive cerebellar Purkinje cells are resistant to delayed death after cerebral trauma and AMPA-mediated excitotoxicity. Eur J Neurosci. 2007;26:649–56. doi: 10.1111/j.1460-9568.2007.05708.x. [DOI] [PubMed] [Google Scholar]
- 27.Staugaitis SM, Zerlin M, Hawkes R, Levine JM, Goldman JE. Aldolase C/zebrin II expression in the neonatal rat forebrain reveals cellular heterogeneity within the subventricular zone and early astrocyte differentiation. J Neurosci. 2001;21:6195–205. doi: 10.1523/JNEUROSCI.21-16-06195.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefanizzi I, Cañete-Soler R. Coregulation of light neurofilament mRNA by poly(A)-binding protein and aldolase C: implications for neurodegeneration. Brain Res. 2007;1139:15–28. doi: 10.1016/j.brainres.2006.12.092. [DOI] [PubMed] [Google Scholar]
- 29.Tolan DR, Penhoet EE. Characterization of the human aldolase B gene. Mol Biol Med. 1986;3:245–64. [PubMed] [Google Scholar]
- 30.Rottmann WH, Deselms KR, Niclas J, Camerato T, Holman PS, Green CJ, Tolan DR. The complete amino acid sequence of the human aldolase C isozyme derived from genomic clones. Biochimie. 1987;69:137–45. doi: 10.1016/0300-9084(87)90246-X. [DOI] [PubMed] [Google Scholar]
- 31.Ponomarenko J, Bui HH, Li W, Fusseder N, Bourne PE, Sette A, Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novotný J, Handschumacher M, Haber E, Bruccoleri RE, Carlson WB, Fanning DW, Smith JA, Rose GD. Antigenic determinants in proteins coincide with surface regions accessible to large probes (antibody domains) Proc Natl Acad Sci U S A. 1986;83:226–30. doi: 10.1073/pnas.83.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arakaki TL, Pezza JA, Cronin MA, Hopkins CE, Zimmer DB, Tolan DR, Allen KN. Structure of human brain fructose 1,6-(bis)phosphate aldolase: linking isozyme structure with function. Protein Sci. 2004;13:3077–84. doi: 10.1110/ps.04915904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cieśla J. Metabolic enzymes that bind RNA: yet another level of cellular regulatory network? Acta Biochim Pol. 2006;53:11–32. [PubMed] [Google Scholar]
- 35.Esposito G, Imperato MR, Ieno L, Sorvillo R, Benigno V, Parenti G, Parini R, Vitagliano L, Zagari A, Salvatore F. Hereditary fructose intolerance: functional study of two novel ALDOB natural variants and characterization of a partial gene deletion. Hum Mutat. 2010;31:1294–303. doi: 10.1002/humu.21359. [DOI] [PubMed] [Google Scholar]
- 36.Jason-Moller L, Murphy M, Bruno J. Overview of Biacore systems and their applications. Curr Protoc Protein Sci 2006; Chapter 19:Unit 19 3. [DOI] [PubMed] [Google Scholar]
- 37.Pezza JA, Choi KH, Berardini TZ, Beernink PT, Allen KN, Tolan DR. Spatial clustering of isozyme-specific residues reveals unlikely determinants of isozyme specificity in fructose-1,6-bisphosphate aldolase. J Biol Chem. 2003;278:17307–13. doi: 10.1074/jbc.M209185200. [DOI] [PubMed] [Google Scholar]
- 38.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.