Abstract
Adaptation is the ability of a system to respond and reset itself even in the continuing presence of a stimulus. On one hand, adaptation is a physiological necessity that enables proper neuronal signaling and cell movement. On the other hand, adaptation can be a source of annoyance, as it can make biological systems resistant to experimental perturbations. Here we speculate where adaptation may live in eukaryotic chemotaxis and how it can be encoded in the signaling network. We then discuss tools and strategies that can be used to both understand and outwit adaptation in a wide range of cellular contexts.
Introduction
They say change is inevitable, but what matters is how you deal with it. And, within limits, nature is very good at coping with change. Many biological systems can maintain core functions at a steady level, even when faced with a change in conditions around them. Here we focus on adaptation, the ability of a system to respond and, over time, return to its baseline activity even when the influence that caused the response, persists. We regard this behavior as a subset of all homeostatic mechanisms that deal with fluctuating environments, and the conceptual and experimental tools we outline here will be relevant to the study of both.
Adaptation is a remarkable behavior that is easy to demonstrate with the following example: put your hand on a table, and you will immediately feel the table’s surface on your skin. Within a few seconds, however, you will cease to feel the table’s surface. Your sensory neurons responded to the stimulus momentarily, but over time less and less, until they may not respond at all (Figure 1A, left). Now, only pushing down harder - a stronger stimulus - will trigger another response. Hence adaptive systems can sense changes in conditions, a property that extends the dynamic range of a system to interpret the strengths of stimuli. For cellular responses this ability is encoded in protein networks. Adaptation is a widespread phenomenon and occurs in the context of many physiological functions. Nevertheless, there are only a few cellular contexts for which the relevant proteins have been identified and where it is understood how they interact with one another to generate adaptation (Alon et al., 1999; Burns and Baylor, 2001; Krupnick and Benovic, 1998; Lohse et al., 1992; Yi et al., 2000)
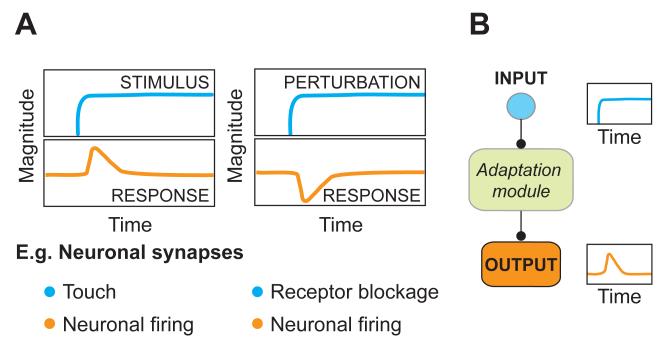
Figure 1. Some systems can sense changes yet maintain constant function.
A.) A stimulus as well as a perturbation can trigger an adapting signaling response. An adapting response returns to its pre-stimulus value despite the continuing presence of a trigger. For example, neuronal firing transiently increases and adapts to the sensation of touch. Similarly, the ability of neurons to fire transiently decreases when receptors are blocked but over time adapts back to the pre-stimulus baseline.
B.) In both cases a signal is interpreted by a signal transduction network - the ‘adaptation module’ – which directs the dynamics of its output .
Adaptation can also simply be a nuisance: consider a scientist interested in understanding how a neuron communicates via its neuronal synapse. Such scientist may add an inhibitor to attenuate the activity of receptors on a connected postsynaptic neuron. Surprisingly, this inhibitory effect turns out to be only transient. Over time, the postsynaptic neuron is able to adapt to this perturbation, a ‘stimulus’ of different sorts, and return to its original ability to fire (Frank et al., 2006) (Figure 1A, right). If not visualized at the right time, it’s easy to see how this transient behavior could have been missed and the perturbation classified as having no effect at all. Later we show how less acute perturbations may be even more susceptible to this problem (Murthy et al., 2001; Thiagarajan et al., 2002; Turrigiano et al., 1994). Annoying.
In both of these examples, and for adaptation in general, a step input is interpreted by a signal transduction circuit - the ‘adaptation module’ – which mounts a transient response that returns to its pre-stimulus level even if the stimulus persists (Figure 1B). In this Perspective we present recent advances on how adaptation is achieved. We begin with a study of bacterial chemotaxis where adaptation is understood best. Here we see how multiple, discrete adaptation modules, with distinct functions, can be found in a single signaling network. We discuss how the architecture of a protein network can form such an adaptation module. Inspired by these findings, we apply these lessons to eukaryotic chemotaxis, where adaptation is essential for proper cell behavior, yet our understanding of how it arises is limited. We end with examples of tools and strategies that will be instrumental to outwit adaptation in any cellular context - no matter whether your goal is to understand or avoid adaptation.
One system with two adaptation modules as an inspirational case study
Adaptation is a ubiquitous feature of signaling cascades throughout the natural world. Even ‘simple’ organisms, like bacteria, make full use of it to steer their tiny bodies up gradients of attractive chemicals such as nutrients and away from harmful compounds. Adaptation enables them to follow an effective strategy. If life is getting better, keep going. If life is getting worse, try a new direction (Berg and Brown, 1972; Macnab and Koshland, 1972).
E.coli alternate between ‘runs’, periods of smooth and straight swimming when they are propelled by counterclockwise (CCW) rotation of their flagella, and periods of ‘tumbling’, where clockwise (CW) rotation of their flagella reorients the cell in a random new direction. Information about changes in the environment is sensed by chemoreceptors, which regulate the directional bias of the beating flagella (Figure 2A). The key intermediate that controls this decision is phosphorylation of the protein Che-Y (CheY-P), which directly modifies tumbling frequency by binding to the motor protein FliM. A decrease in attractant concentration leads to a more active receptor, higher levels of CheY-P, and more tumbling. The opposite is true for an increase of attractant concentration, which decreases concentration of CheY-P and results in less tumbling and more runs (Porter et al., 2011) (Figure 2A).
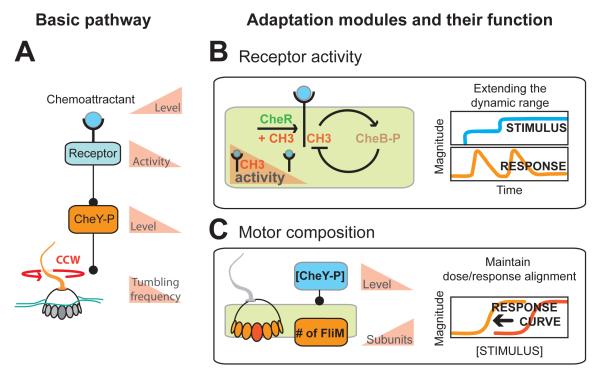
Figure 2. Adaptation modules in bacterial chemotaxis.
Bacterial chemotaxis is an example where multiple, separate adaptation modules are at work in one signaling cascade.
A.) In the basic chemosensory pathway, increases in ligand decrease the concentration of intracellular CheY-P, which regulates the direction the flagellar motor rotates. On top of this, two adaptation modules have been identified.
B.) The fast acting adaptation module 1 regulates methylation levels of the receptor and ultimately sets pathway activity. High methylation levels engage CheB in a negative feedback loop and reset receptor activity. This adaptation module serves to extend the dynamic range towards chemoattractant. Once adapted, only a stronger stimulus can trigger another response.
C.) A second, slow acting adaptation module regulates the number of FliM subunits the bacterial motor is comprised of. This module keeps the sensitivity of the motor aligned with the steady-state level of phosphorylated CheY, which can vary between cells. This module adds robustness to the signaling pathway.
A sensory adaptation module regulates the top of this cascade and is particularly well understood. Active receptors use negative feedback to reset their activity in the continuing presence of chemoattractant (Figure 2B, left). This is achieved by the methylase CheR and the demethylase CheB, which control methylation levels of the receptor. According to the prominent Barkai-Leibler model (Barkai and Leibler, 1997), CheR acts with a constant rate on all receptors while CheB is regulated by receptor activity. In this way an increase in receptor activity enhances the CheB mediated negative feedback loop, brings receptor activity back down, and prepares the system for another round of comparing concentrations (Figure 2B, left). Adaptation acts rapidly (on the order of seconds) to turn off receptor signaling, and now only an increase in stimulus concentration will trigger another response (Figure 2B, right).
Recently, a second adapting module, deeply buried in the chemosensing system, was uncovered. This module plays an essential role for the signaling cascade to function robustly. A conundrum demanded further investigation. The bacterial motor has a very narrow operating range with regards to its regulator CheY-P. A two-fold change in the concentration of CheY-P pushes the motor to extremes and results in essentially tumbles or runs only (Cluzel et al., 2000). However basal levels of CheY-P can vary widely between individual cells (Kollmann et al., 2005), and hence a significant portion of cells should be stuck running or tumbling. But this is not the case. Robust switching responses are observed for the whole population (Alon et al., 1999). How do bacteria match the sensitivity of the motor to their CheY-P levels? Adaptation comes to the rescue - this time adaptation to an ambient internal protein concentration. A recent paper from the Berg lab shows that the steady state concentration of CheY-P itself can be sensed and serve as an input signal for a slow adapting module (minute timescale) that regulates how many subunits of FliM the motor consists of (Yuan and Berg, 2013; Yuan et al., 2012) (Figure 2B, left). In this way the motor itself adapts to ambient CheY-P concentrations. As a result, responses triggered by the receptor are always well aligned with the sensitivity of the motor (Figure 2B, right).
This section on bacterial chemotaxis demonstrates that adaptation isn’t picky and can happen at several levels. One upstream adaptation module is centered on the receptor, and another downstream module regulates the composition of the motor. In both cases these modules confer the ability to sense relative changes. One module enables the receptor to remain sensitive to increases of an extracellular signal, the chemoattractant, while similarly, the second module enables the motor to remain sensitive to changes in concentrations of the intracellular signal, CheY-P.
Moving forward, we appreciate that despite their similarities, these modules are another example for the two faces of adaptation. The adaptation module at the receptor has been at the focus point of experiments aimed at understanding chemotactic sensing. In contrast, the adaptation module conferring robustness to variable CheY-P levels could easily have been missed and classified as an annoying case where a reduction in protein levels doesn’t give a ‘phenotype’.
Inspired by bacterial chemotaxis we take a look at the less understood signaling network of eukaryotic chemotaxis and suggest that adaptation, as in bacteria, may happen at several places in the signaling cascade. We speculate where such adaptation modules may lie and present the difficulties in defining them and teasing them apart.
Adaptation in eukaryotic chemotaxis
Most of what we know about single cell chemotaxis in eukaryotes is based on Dictyostelium amoeba and neutrophils. While bacteria are thought to rely exclusively on temporal comparisons to guide their movements, the bigger size of eukaryotic cells allows them to also sense spatial differences of ligand over their surface (Berg, 1988). These cells are able to make accurate spatial comparisons over a wide range of external agonist concentrations and employ adaptation to control appropriate responses in gradients (Zigmond, 1977; Zigmond and Sullivan, 1979). However, spatial gradients are not required to study adaptive behavior in these systems. Uniform addition of agonist suffices.
Following a uniform increase in external agonist there is no shortage of adaptive behaviors. Cells stop in their track when hit by uniform chemoattractant, but after a while adapt and continue along their path (Zigmond and Sullivan, 1979). This is recapitulated by transient activation of signaling pathways and transient accumulation or depletion of their outputs such as the signaling lipid PIP3 (Janetopoulos et al., 2004; Meili et al., 1999; Parent et al., 1998; Servant et al., 2000; Stephens et al., 1991), cGMP (Van Haastert and Van der Heijden, 1983), cAMP (Devreotes and Steck, 1979; Dinauer et al., 1980), actin polymerization (Hall et al., 1988) and activation of small GTPases Ras (Kae et al., 2004), Rac (Benard et al., 1999; Li et al., 2002; Park et al., 2004), Cdc42 (Benard et al., 1999), Rho (Wong et al., 2006), and Rap (Jeon et al., 2007), most of which can be followed with reporters in living cells.
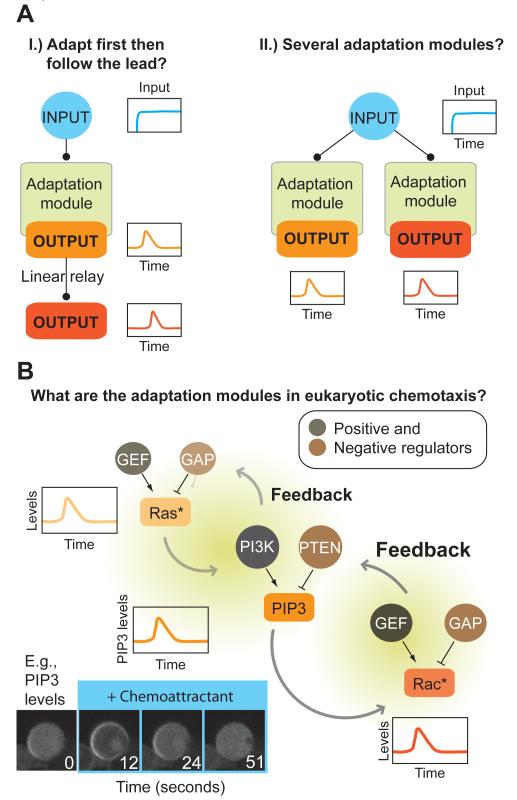
How are these transient responses related to one another? Is adaptation generated in one place and successive downstream outputs follow (Figure 3Ai), or are there several independent adaptation modules (Figure 3Aii)?
Figure 3. Adaptation modules in eukaryotic chemotaxis.
A.) Similar to bacterial chemotaxis, an adaptation module may be present early in the signaling pathway. Its output may then become transmitted to downstream nodes, relaying activity at each moment in time.
B.) Several adaptation modules may exist. Shown is a case where adaptation modules operate in parallel.
C.) Adapting nodes are highly interconnected, making it challenging to tease apart the underlying adaptation module(s). The minimum circuits necessary for each individual adapting node are currently unknown. Shown is one example of adapting nodes, along with some of their regulators and linkages. Positive and negative regulators, GTPase Exchange Factors (GEFs) and GTPase activating factors (GAPs) determine the GTP binding state and therefore activities of Ras and Rac. Similarly, PI3kinases and lipid phosphatases like PTEN regulate levels of PIP3 in the membrane. Each of these outputs shows adaptive behavior and can feed both forward and backward in the signaling cascade. An example is shown for the PIP3 response in chemoattractant-stimulated Dictyostelium.
Based on studies in bacteria and other sensory systems, the simplest assumption is that sensory adaptation occurs at the level of the receptor. In this case, the downstream signaling cascade might only need to transmit the transient signal emanating from the receptor (Figure 3Ai). Unfortunately, unlike bacteria, known post–translational modifications of GPCRs are not required for adaptation of many downstream signals involved in chemotaxis (Arai et al., 1997; Brzostowski et al., 2013; Hsu et al., 1997; Kim et al., 1997). This is surprising, because stimulus-dependent phosphorylation of a number of GPCRs (rhodopsin, B2-adrenergic receptor) leads to downregulation of these receptors, often accompanied by their internalization (Goodman et al., 1996). The nature of the first and most upstream component displaying adaptive behavior in eukaryotic chemotaxis is still uncertain. FRET measurements in Dictyostelium indicate that the alpha and beta/gamma subunits of heterotrimeric G-proteins become dissociated by receptor stimulation and remain in this state for as long as chemoattractant is present, even while downstream signals adapt (Janetopolous et al., 2001; Xu et al., 2005). The first downstream signal known to adapt is Ras activity. A number of Ras isoforms are activated (GTP loaded) by chemoattractant in Dictyostelium (Kae et al., 2004; Zhang et al., 2008) and neutrophils (Zheng et al., 1997), two of which, RasC and RasG, have been most carefully studied in Dictyostelium with pulldowns and live-cell reporters (Kae et al., 2004; Sasaki et al., 2004). Supplemented by genetic data (Bolourani et al., 2006; Sasaki et al., 2004), they appear to sit at a branching point controlling several chemotaxis-relevant downstream responses, including the activation of the TOR complex 2 (Cai et al., 2010; Charest et al., 2010) and PI3kinase signaling (Funamoto et al., 2002). RasC and RasG have distinct positive (GTP exchange factors (GEFs)) (Kae et al., 2007) and negative (GTPase activating proteins (GAPs)) regulators, only some of which have been identified. Whether any Ras’ immediate upstream regulators shows adaptive behavior has not yet been investigated. These studies suggests that, if signaling is linear and only one adaptation module exists in the cascade, it lies downstream or parallel to the activation of heterotrimeric G-proteins, and upstream of or at the level of Ras activation.
Wherever it originates, some signaling nodes appear to efficiently transmit the kinetics of upstream activation. Studies in Dictyostelium show that indeed a change in adaptation dynamics can be relayed through several nodes down the signaling network (Figure 3Ai). Dominant active RasC extends the activation timecourse of both immediate downstream effectors PKB/PKBR1 and their respective downstream substrates (Cai et al., 2010). Similarly, genetic lack of the RasG GAP DdNF1, not only extends the adaptation timecourse of RasG activity but also that of the downstream PIP3 response (Zhang et al., 2008).
Is there more than one adaptation module in eukaryotic chemotaxis? Even bacteria, with their relatively simple signaling cascade have more than one adaptation module. Chemotaxing eukaryotes, with their significantly more numerous signaling components (Ridley et al., 2003), molecular redundancies (Hoeller and Kay, 2007; Hoeller et al., 2013; Ku et al., 2012; Sun et al., 2004; Vlahou and Rivero, 2006), parallel pathways (Chen et al., 2007; van Haastert et al., 2007; Veltman et al., 2008), and feedback loops (Charest et al., 2010; Sasaki et al., 2007; Weiner et al., 2002, 2006) are similarly likely to encode more than one adaptation module (Figure 3Aii). One line of evidence supporting this hypothesis is that different components adapt at different timescales. In Dictyostelium, adapting responses can be roughly grouped into early (timescale: ~30sec - Ras activation, PIP3 production, PKB activation, cGMP production) and late (timescale: minutes - PLC activation, Ca2+ influx, cAMP production, myosin II light chain phosphorylation, PakA activation) (Franca-Koh et al., 2006). A second and stronger piece of evidence supporting multiple adaptation modules is that perturbations can affect the ability of some responses to adapt, while leaving other responses intact. For example, stimulating cells that express a mutant, non-phosphorylatable receptor, results in a failure to generate adaptation for cAMP production, yet the adapting PIP3 response is left intact (Brzostowski et al., 2013).
How can we disentangle the relation between adapting outputs to uncover and define the circuits that form adaptation modules? Let’s consider a specific example to appreciate where the difficulty lies (Figure 3B).
In Dictyostelium, PIP3 levels, balanced by PI3kinase (PI3K) and lipid phosphatases like PTEN (Funamoto et al., 2002; Iijima and Devreotes, 2002), rise and adapt to a step input of chemoattractant. PI3K produces PIP3, and this enzyme is activated by binding to GTP-loaded Ras. Importantly, the Ras-mediated effect on PIP3 production will not be constant (Huang et al., 2003), because Ras activity itself temporarily peaks after uniform stimulation with chemoattractant. Hence, do PIP3 levels just track the earlier Ras activity? Are Ras dynamics required for PIP3 transients?
Feedback regulation further complicates the identification of adaptation modules. How can we know what constitutes the relevant subcircuit when ‘downstream’ feeds back to ‘upstream’? In our example, ‘upstream’ Ras activity has been shown to be sensitive to changes in PIP3 levels ‘downstream’ (Huang et al., 2013; Sasaki et al., 2004) (Figure 3B). A similar exuberance of connectivity has been shown for PIP3 and Rac in neutrophils (Brachmann et al., 2005; Inoue and Meyer, 2008; Weiner et al., 2006; Welch et al., 2002; Yang et al., 2012) and is found in many signaling systems.
Activation of the chemoattractant receptor, and by consequence its many downstream effectors, has revealed a multitude of adapting responses. However, to identify the minimal circuits that are capable of carrying out adaptation, we need tools that can drive activation of specific subcircuits downstream of the receptor. Recent years have seen an exciting expansion of the toolkit available to perturb cellular systems with high specificity and temporal resolution. In the next section we describe some of these tools and show how they allow us to trigger signaling at user-defined nodes, watch adaptation as it happens, and avoid the complicating effects of compensation.
Methods for outwitting adaptation
No matter whether you regard adaptation as an interesting feature or an annoyance, weak, slow acting perturbations are your enemies, as adaptation can make their effect invisible. In contrast, an acute and strong perturbation is most likely to elicit a clear, interpretable response (Figure 4A).
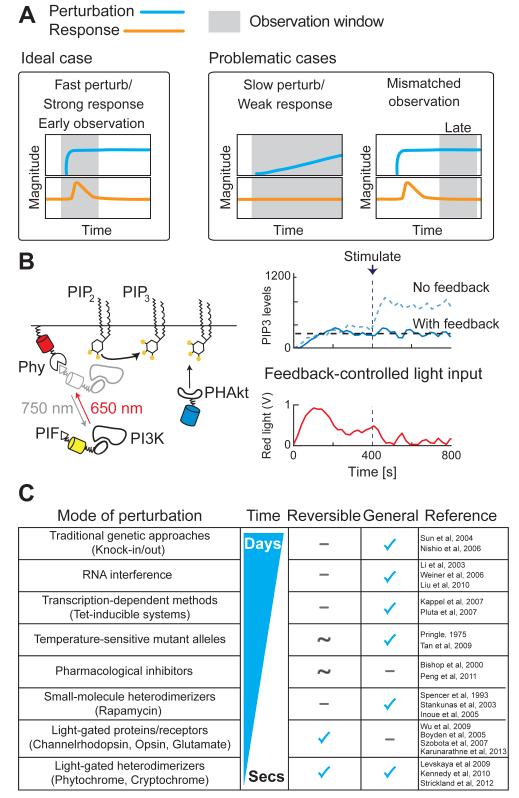
Figure 4. Tools for rapid perturbation of molecular networks.
A.) An ideal perturbation acts quickly, and its effect can be observed immediately (left). Weak phenotypes may be observed if perturbations are slow to take effect or if the observation is made at the wrong time (right).
B.) The Phy-PIF optogenetic dimerization system. Red-light induces a conformational change in the Phy protein to allow the PIF protein to bind, while infra-red light reverses this interaction, providing a means for light-gated control of dimerization (left). When Phy is localized to the site of activity for a signaling protein that is fused to PIF, this leads to optogenetic control of protein activity. For example, light-gated recruitment of PI3K to the plasma membrane leads to the production of PIP3, the lipid product of PI3K (left). Paired with a live-cell readout of PIP3 production, a computational feedback controller can measure PIP3 production and deliver the appropriate amount of light input to drive the proper amount of PI3K to the membrane to hold PIP3 levels steady, even in the presence of a low dose of stimulus (right).
C.) Table of commonly-used perturbation tools. Some examples of their uses are mentioned in the text, and references are indicated in the rightmost column.
If the perturbation is weak or slow to take effect, adaptation can catch up with it, and the system may not show any response or phenotype (Figure 4A, middle). Genetic perturbations such as knockout and RNAi have identified a number of molecules that mediate chemotaxis (Sun et al., 2004; Gu et al., 2003; Weiner et al., 2006; Liu et al., 2010; Yoo et al., 2010; Nishio et al., 2007; Li et al., 2003; Artemenko et al., 2012, 2011; Chen et al., 2007; Iijima and Devreotes, 2002; Zhao et al., 2002). While RNAi allows researchers to survey large numbers of candidate molecules in a reasonable time frame, phenotypes elicited by this approach are often weak or absent due to insufficient reduction of target proteins owing to inefficient silencing (Pankov et al., 2005) and or long protein half-lives (D’Angelo et al., 2009). These limited and chronic perturbations may allow cells to compensate through the modulation of intracellular signaling, an outcome that may mask the effect of the target molecule’s inhibition (Figure 4A, middle).
A second important consideration is the choice of an appropriate observation window: look too late and the response is missed, leaving only the compensated state to be observed (Figure 4A, right). This applies particularly to non-conditional genetic knockouts where no observation is possible immediately after the perturbation. As a result of these technical and biological constraints, tools with greater temporal acuity are required to probe adaptive signaling processes. Only with such tools can one truly see how signaling molecules form adaptive circuits and how these circuits relate to each other within the signaling network. Of particular value are methods that allow for fast switching between off- and on-states, because these enable the researcher to make observations before the system has the chance to adapt (Figure 4A, left).
Pharmacological manipulation provides an excellent means of acute and potentially reversible perturbation of endogenous molecules and can act on the order of seconds. While extant and well-characterized small molecules are very useful tools, many important signaling molecules are not currently known to be small-molecule targets. Furthermore, even where drug like molecules are available, isoform specificity and off-target effects are significant concerns. This is especially true for small molecules that target conserved binding domains, regions of conservation within a protein family, or share structural similarity with other molecules. For example, drugs that target protein kinases and function as ATP competitors run the risk of targeting other kinases with similarly structured ATP binding pockets. To get around this problem of specificity, Shokat and colleagues took a chemical genetic approach in which the ATP-binding site of a Src-family tyrosine kinase was engineered to have a unique ATP-binding pocket not found in any wild type kinases (Bishop et al., 2000). By screening through rationally designed ATP analogs, Shokat and colleagues identified a potent and specific inhibitor for this modified kinase. Furthermore, the generality of this approach enables the generation of conditional alleles for many kinases, allowing the possibility of understanding the specific signaling roles of individual kinases in signaling systems rich with crosstalk (Bishop et al., 2000). While such an approach is broadly applicable to kinases, analogous strategies are not readily apparent for many other important protein families for which we need more general perturbative tools.
To overcome these shortcomings, many groups have recently begun to utilize several different flavors of genetically encoded systems that allow reversible control of protein-protein interactions. These ‘Induced dimerization systems’ have been a big hit, owing to their generalizability, stability, fast switching times (~ secs) and orthogonality to cellular processes (Bayle et al., 2006; Kennedy et al., 2010; Levskaya et al., 2009; Stankunas et al., 2007; Strickland et al., 2012). A founding member of this family is the rapamycininducible protein-protein interaction system, in which the small molecule rapamycin forms a ternary complex with two protein domains: FKBP, a domain from the FK506 binding protein, and FRB, the FKBP12 rapamycin binding domain of mTOR. The rapamycin system has been used extensively with success in a variety of cell types, with one protein domain (usually FRB) targeted to a subcellular location and serving as the anchor point for recruitment of a signaling domain fused to the other (FKBP). Since the first application of this system to oligomerize artificial receptors to initiate signal transduction in live cells (Spencer et al., 1993), its uses have extended to other cell types such as yeast (Geda et al., 2008; Haruki et al., 2008), neutrophils, and other mammalian cells (Inoue and Meyer, 2008; Inoue et al., 2005) and mouse models (Stankunas et al., 2003). For signaling molecules regulated by localization, this method allows for the fast recruitment of proteins to sites of activity for gain-of-function phenotypes. Conversely, rapidly recruiting molecules away from their physiologically relevant location can enable the study of acute loss-of-function phenotypes (Haruki et al., 2008; Komatsu et al., 2010; Robinson et al., 2010).
More recently, optogenetic systems have been developed that make use of light-gated conformational changes in naturally occurring light-responsive proteins. Several approaches have been developed to control cell signaling with light, including receptors such as channelrhodopsin (Boyden et al., 2005; Han and Boyden, 2007), synthetic light-gated glutamate receptor (Szobota et al., 2007), and mammalian visual blue opsin (Karunarathne et al., 2013), the homodimerizer FP Dronpa (Zhou et al., 2012), and protein heterodimerizers, which include cryptochromes (Kennedy et al., 2010), light-oxygen-voltage sensing (LOV) domains (Strickland et al., 2012; Wu et al., 2009), and phytochromes (Quail, 2002) from plants. Our lab has focused on the phytochrome red/infrared light sensing system from plants. Red light induces the association of the light-responsive phytochrome (Phy) protein to its interacting factor, PIF, and this association is reversed in the presence of infrared light (Figure 4B, left) (Quail, 2002). This Phy-PIF light-gated protein heterodimerization system has now been optimized as an optogenetic tool that can deliver precise, time-varying, intracellular signaling inputs into individual cells (Figure 4B, right). Importantly, image-based live-cell readouts of intracellular activities can be used with the Phy-PIF system to titrate the amount of light input required to drive a defined timecourse of signaling activity, or to clamp intracellular signaling activities at a desired level. Light-gated dimerization of the yeast mitotic cyclin Clb2 to different subcellular locations during mitosis revealed different functions for Clb2 in coordinating cell division (Yang et al., 2013). Additionally, a computational feedback controller that measures PIP3 production and drives light-gated PI3K recruitment can hold intracellular levels of PIP3 steady even in the face of changing pathway activation in single cells (Figure 4B, right) (Toettcher et al., 2011). Coupling automated control of light inputs with the Phy-PIF system promises to be a transformative tool for studying complex signaling systems. By probing how the activities of individual signaling molecules within a circuit vary over time in response to such synthetic signaling inputs, we can watch the information flow and processing in signaling systems. Figure 4C lists the characteristics of some of the aforementioned commonly used tools for perturbation studies.
One can now begin to dissect signaling circuits by first selecting the input and output nodes for characterization. Using an acute perturbation tool such as the Phy-PIF system, a light-gated step input can be delivered to activate signaling at one node, while at another node downstream the output may be measured (e.g., live-cell readout, collecting timepoints for biochemical analysis, etc.) (Toettcher et al., 2013) (Figure 5A). By mapping input-output relationships throughout small portions of the network, functional signaling units capable of adaptation can be identified. Used in combination with other pharmacologic or genetic tools, the circuit in question can further be isolated from complicating feedback mechanisms.
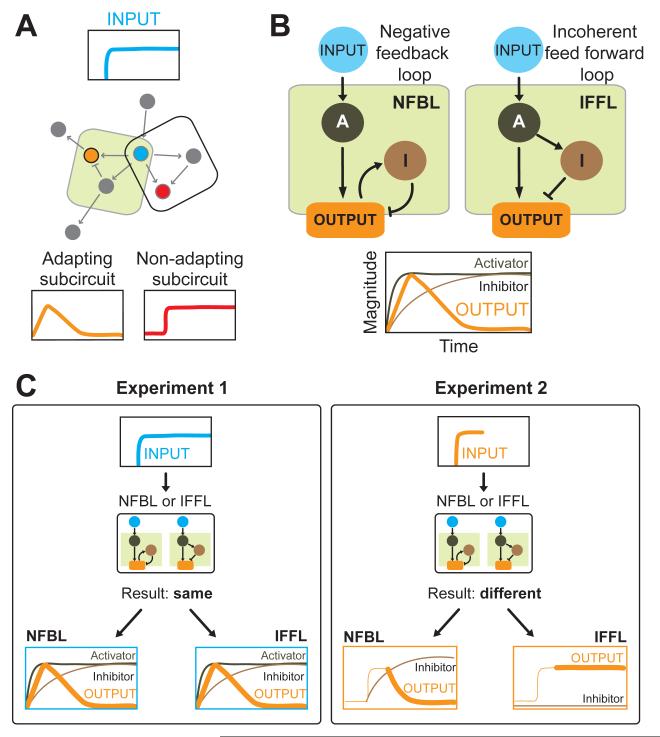
Figure 5. Strategies to identify and dissect adapting circuits.
A.) Using an acute perturbation tool such as the Phy-PIF system and a near-immediate observation window, one can deliver a step input to one node and measure the signaling response of another node. In this manner, systematic mapping of input-output relationships can help identify subcircuits capable of adaptation.
B.) Computational experiments show that only two three-node enzymatic networks can achieve robust adaptation. In both cases the stimulation of an activator causes an output to initially rise. The signal turnoff is either regulated by a negative feedback loop triggered by the output itself (left), or an incoherent feed forward loop where the activator also triggers an inhibitor proportional to its activity but delayed in action (right).
C.) One way to distinguish between the two network architectures capable of adaptation is to increase the activity of the output. In the negative feedback loop model (NFBL), the output triggers the inhibitor. A sudden bolus of activity administered to the output node engages the negative feedback loop and decreases the activity of the output node over time. For the incoherent feedforward loop model (IFFL), since the inhibitor is independent of the output, output activity remains high.
Whether a researcher is interested in studying adaptation or avoiding it altogether, combining a fast perturbation with the appropriate time window of observation will increase the chances of visualizing the strongest phenotype prior to adaptation kicking back in.
For a researcher interested in adaptation, once an adaptation module has been identified, the next question will be how the components are linked together to achieve adaptation. In the next section we discuss the network topologies capable of carrying out adaptive responses.
The wiring of an adaptation module
It appears that only a limited number of simple network topologies can support adaptation. A recent computational study employed a reverse engineering approach to comprehensively elucidate the number of ways a simple network can be wired to achieve robust and precise adaptation. This analysis revealed that at least three nodes are required, and that even for a three-node network only a limited set of architectures give adaptive behavior to a stimulus. With one node receiving input, one node transmitting output and one regulatory node, only two overall architectures emerged (Figure 5B) (Ma et al., 2009).
The first architecture is a negative feedback loop (NFBL), where an inhibitor acts as a ‘buffer’ to integrate the difference between steady-state and response and then feeds this back into the output node. In this architecture, the stimulus quickly turns on an output, but negative feedback triggered by the output acts over time to return the output to baseline (Figure 2B left panel and graph). This topology is used to regulate receptor activity in many systems including sensory receptors in bacterial chemotaxis, the G-protein coupled receptor (GPCR) rhodopsin in visual transduction (Arshavsky, 2002), as well as other eukayotic systems ranging from osmoregulation in yeast (Muzzey et al., 2009) to Ca2+ homeostasis (El-Samad et al., 2002).
The second architecture is given by an incoherent feed forward loop (IFFL), where adaptation is achieved without directly monitoring the level of output. Here a stimulus turns on an output quickly, but also directly and, proportional to the strength of stimulus, turns on an inhibitor. Two conditions need to be met for this circuit to produce adaptation. First, the inhibitor needs to act on a slower timescale to give the output time for the initial rise. Second, the inhibitor and activator must balance perfectly in strength at later timepoints (Figure 5B). This architecture is the leading candidate for an overall adaptation architecture in eukaryotic chemotaxis (Iglesias and Devreotes, 2008, 2012; Takeda et al., 2012; Wang et al., 2012; Xiong et al., 2010), but continues to be scrutinized because definitive molecular evidence is lacking.
Both the NFBL and the IFFL topologies yield similar adaptive behaviors to step inputs of external agonist, but selective activation of nodes at different levels in the pathway and more complex time-varying inputs can help to distinguish the underlying network architecture.
Acute perturbation tools enable elucidation of network circuitry
Acute perturbation tools enable us to tease apart the two network architectures capable of perfect adaptation (Figure 5B) (Ma et al., 2009). For eukaryotic chemotaxis, it has been suggested that both negative feedback as well as incoherent feed forward can regulate portions of the signaling cascades (Charest et al., 2010; Takeda et al., 2012). Both topologies produce a transient output from a step input at the top node (Figure 5C, plots outlined in blue). What sort of input might be used to distinguish between these two models? One possibility is to apply a step input at the level of the output. In the IFFL model, inhibitor activation is independent of the output, whereas the NFBL model relies on the output to drive the inhibitor. By applying a bolus of output activity and determining which trajectory the activity will take, the two models can be distinguished (Figure 5C, plots outlined in orange). For NFBL, a step input of output activity will trigger the activation of the inhibitor. Since the output and the inhibitor are linked by negative feedback, output levels will decline back to baseline. The IFFL is different - since the inhibitor is independent of the output, the output itself remains in its sustained, high activity level.
This is only the beginning, as more sophisticated time-varying inputs such as linear ramps (Chang and Levchenko, 2013), noise fluctuation (Becskei and Serrano, 2000; Weinberger et al., 2008), frequency responses (Purvis et al., 2012; Toettcher et al., 2013) and multiple inputs are ported from engineering into biology to interrogate signaling circuits. For example, Chang and Levchenko propose that inputs that increase with constant rates (‘ramps’) can be used to distinguish between IFFL and NFBL networks (Chang and Levchenko, 2013). Firtel and colleagues found that varying the stimulus step size in Dictyostelium led to RasG adaptation kinetics that is predicted by an incoherent feedforward mechanism (Takeda et al., 2012). Weinberger et al have successfully used the autocorrelation of noise in gene expression to identify positive feedback as a means of regulating gene expression lifetime (Weinberger et al., 2008).
As cell biology moves into an era of identifying higher order network behavior, exciting times are ahead. Old questions, like how moving cells deal with chemical stimuli, may be solved, and undoubtedly the new wave of tools will open new questions. And if this nagging doubt about a mutant you always thought should have a phenotype creeps back in your mind, maybe it is time to test some of these tools for yourself?
Acknowledgements
We would like to thank the members of the Weiner lab for helpful discussion. This work was supported by an EMBO postdoctoral fellowship (O.H.), an NIH T32 Cardiovascular Research Institute postdoctoral fellowship (D.G.), and NIH grants GM084040 and GM096164 (O.D.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- Arai H, Monteclaro FS, Tsou CL, Franci C, Charo IF. Dissociation of chemotaxis from agonist-induced receptor internalization in a lymphocyte cell line transfected with CCR2B. Evidence that directed migration does not require rapid modulation of signaling at the receptor level. J. Biol. Chem. 1997;272:25037–25042. doi: 10.1074/jbc.272.40.25037. [DOI] [PubMed] [Google Scholar]
- Arshavsky VY. Rhodopsin phosphorylation: from terminating single photon responses to photoreceptor dark adaptation. Trends Neurosci. 2002;25:124–126. doi: 10.1016/s0166-2236(00)02094-4. [DOI] [PubMed] [Google Scholar]
- Artemenko Y, Swaney KF, Devreotes PN. Assessment of development and chemotaxis in Dictyostelium discoideum mutants. Methods Mol. Biol. Clifton NJ. 2011;769:287–309. doi: 10.1007/978-1-61779-207-6_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artemenko Y, Batsios P, Borleis J, Gagnon Z, Lee J, Rohlfs M, Sanséau D, Willard SS, Schleicher M, Devreotes PN. Tumor suppressor Hippo/MST1 kinase mediates chemotaxis by regulating spreading and adhesion. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13632–13637. doi: 10.1073/pnas.1211304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- Bayle JH, Grimley JS, Stankunas K, Gestwicki JE, Wandless TJ, Crabtree GR. Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem. Biol. 2006;13:99–107. doi: 10.1016/j.chembiol.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Benard V, Bohl BP, Bokoch GM. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J. Biol. Chem. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- Berg HC. A physicist looks at bacterial chemotaxis. Cold Spring Harb. Symp. Quant. Biol. 1988;53(Pt 1):1–9. doi: 10.1101/sqb.1988.053.01.003. [DOI] [PubMed] [Google Scholar]
- Berg HC, Brown DA. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Bolourani P, Spiegelman GB, Weeks G. Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol. Biol. Cell. 2006;17:4543–4550. doi: 10.1091/mbc.E05-11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Brachmann SM, Yballe CM, Innocenti M, Deane JA, Fruman DA, Thomas SM, Cantley LC. Role of phosphoinositide 3-kinase regulatory isoforms in development and actin rearrangement. Mol. Cell. Biol. 2005;25:2593–2606. doi: 10.1128/MCB.25.7.2593-2606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzostowski JA, Sawai S, Rozov O, Liao X-H, Imoto D, Parent CA, Kimmel AR. Phosphorylation of chemoattractant receptors regulates chemotaxis, actin reorganization and signal relay. J. Cell Sci. 2013;126:4614–4626. doi: 10.1242/jcs.122952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns ME, Baylor DA. Activation, deactivation, and adaptation in vertebrate photoreceptor cells. Annu. Rev. Neurosci. 2001;24:779–805. doi: 10.1146/annurev.neuro.24.1.779. [DOI] [PubMed] [Google Scholar]
- Cai H, Das S, Kamimura Y, Long Y, Parent CA, Devreotes PN. Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J. Cell Biol. 2010;190:233–245. doi: 10.1083/jcb.201001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Levchenko A. Adaptive molecular networks controlling chemotactic migration: dynamic inputs and selection of the network architecture. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013;368:20130117. doi: 10.1098/rstb.2013.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest PG, Shen Z, Lakoduk A, Sasaki AT, Briggs SP, Firtel RA. A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev. Cell. 2010;18:737–749. doi: 10.1016/j.devcel.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Iijima M, Tang M, Landree MA, Huang YE, Xiong Y, Iglesias PA, Devreotes PN. PLA2 and PI3K/PTEN pathways act in parallel to mediate chemotaxis. Dev. Cell. 2007;12:603–614. doi: 10.1016/j.devcel.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes PN, Steck TL. Cyclic 3′,5′ AMP relay in Dictyostelium discoideum. II. Requirements for the initiation and termination of the response. J. Cell Biol. 1979;80:300–309. doi: 10.1083/jcb.80.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinauer MC, Steck TL, Devreotes PN. Cyclic 3′,5′-AMP relay in Dictyostelium discoideum V. Adaptation of the cAMP signaling response during cAMP stimulation. J. Cell Biol. 1980;86:554–561. doi: 10.1083/jcb.86.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Samad H, Goff JP, Khammash M. Calcium homeostasis and parturient hypocalcemia: an integral feedback perspective. J. Theor. Biol. 2002;214:17–29. doi: 10.1006/jtbi.2001.2422. [DOI] [PubMed] [Google Scholar]
- Franca-Koh J, Kamimura Y, Devreotes P. Navigating signaling networks: chemotaxis in Dictyostelium discoideum. Curr. Opin. Genet. Dev. 2006;16:333–338. doi: 10.1016/j.gde.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Frank CA, Kennedy MJ, Goold CP, Marek KW, Davis GW. Mechanisms underlying the rapid induction and sustained expression of synaptic homeostasis. Neuron. 2006;52:663–677. doi: 10.1016/j.neuron.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- Geda P, Patury S, Ma J, Bharucha N, Dobry CJ, Lawson SK, Gestwicki JE, Kumar A. A small molecule-directed approach to control protein localization and function. Yeast Chichester Engl. 2008;25:577–594. doi: 10.1002/yea.1610. [DOI] [PubMed] [Google Scholar]
- Goodman OB, Jr, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- Gu Y, Filippi M-D, Cancelas JA, Siefring JE, Williams EP, Jasti AC, Harris CE, Lee AW, Prabhakar R, Atkinson SJ, et al. Hematopoietic cell regulation by Rac1 and Rac2 guanosine triphosphatases. Science. 2003;302:445–449. doi: 10.1126/science.1088485. [DOI] [PubMed] [Google Scholar]
- Van Haastert PJM, Keizer-Gunnink I, Kortholt A. Essential role of PI3-kinase and phospholipase A2 in Dictyostelium discoideum chemotaxis. J. Cell Biol. 2007;177:809–816. doi: 10.1083/jcb.200701134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert PJ, Van der Heijden PR. Excitation, adaptation, and deadaptation of the cAMP-mediated cGMP response in Dictyostelium discoideum. J. Cell Biol. 1983;96:347–353. doi: 10.1083/jcb.96.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AL, Schlein A, Condeelis J. Relationship of pseudopod extension to chemotactic hormone-induced actin polymerization in amoeboid cells. J. Cell. Biochem. 1988;37:285–299. doi: 10.1002/jcb.240370304. [DOI] [PubMed] [Google Scholar]
- Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PloS One. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr. Biol. CB. 2007;17:813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Hoeller O, Bolourani P, Clark J, Stephens LR, Hawkins PT, Weiner OD, Weeks G, Kay RR. Two distinct functions for PI3-kinases in macropinocytosis. J. Cell Sci. 2013;126:4296–4307. doi: 10.1242/jcs.134015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MH, Chiang SC, Ye RD, Prossnitz ER. Phosphorylation of the N-formyl peptide receptor is required for receptor internalization but not chemotaxis. J. Biol. Chem. 1997;272:29426–29429. doi: 10.1074/jbc.272.47.29426. [DOI] [PubMed] [Google Scholar]
- Huang C-H, Tang M, Shi C, Iglesias PA, Devreotes PN. An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration. Nat. Cell Biol. 2013;15:1307–1316. doi: 10.1038/ncb2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YE, Iijima M, Parent CA, Funamoto S, Firtel RA, Devreotes P. Receptor-mediated regulation of PI3Ks confines PI(3,4,5)P3 to the leading edge of chemotaxing cells. Mol. Biol. Cell. 2003;14:1913–1922. doi: 10.1091/mbc.E02-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias PA, Devreotes PN. Navigating through models of chemotaxis. Curr. Opin. Cell Biol. 2008;20:35–40. doi: 10.1016/j.ceb.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Iglesias PA, Devreotes PN. Biased excitable networks: how cells direct motion in response to gradients. Curr. Opin. Cell Biol. 2012;24:245–253. doi: 10.1016/j.ceb.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- Inoue T, Meyer T. Synthetic activation of endogenous PI3K and Rac identifies an AND-gate switch for cell polarization and migration. PloS One. 2008;3:e3068. doi: 10.1371/journal.pone.0003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetopoulos C, Jin T, Devreotes P. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science. 2001;291:2408–11. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- Janetopoulos C, Ma L, Devreotes PN, Iglesias PA. Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8951–8956. doi: 10.1073/pnas.0402152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon TJ, Lee D-J, Merlot S, Weeks G, Firtel RA. Rap1 controls cell adhesion and cell motility through the regulation of myosin II. J. Cell Biol. 2007;176:1021–1033. doi: 10.1083/jcb.200607072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kae H, Lim CJ, Spiegelman GB, Weeks G. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 2004;5:602–606. doi: 10.1038/sj.embor.7400151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kae H, Kortholt A, Rehmann H, Insall RH, Van Haastert PJM, Spiegelman GB, Weeks G. Cyclic AMP signalling in Dictyostelium: G-proteins activate separate Ras pathways using specific RasGEFs. EMBO Rep. 2007;8:477–482. doi: 10.1038/sj.embor.7400936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel S, Matthess Y, Kaufmann M, Strebhardt K. Silencing of mammalian genes by tetracycline-inducible shRNA expression. Nat. Protoc. 2007;2:3257–3269. doi: 10.1038/nprot.2007.458. [DOI] [PubMed] [Google Scholar]
- Karunarathne WKA, Giri L, Patel AK, Venkatesh KV, Gautam N. Optical control demonstrates switch-like PIP3 dynamics underlying the initiation of immune cell migration. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1575–1583. doi: 10.1073/pnas.1220755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Soede RD, Schaap P, Valkema R, Borleis JA, Van Haastert PJ, Devreotes PN, Hereld D. Phosphorylation of chemoattractant receptors is not essential for chemotaxis or termination of G-protein-mediated responses. J. Biol. Chem. 1997;272:27313–27318. doi: 10.1074/jbc.272.43.27313. [DOI] [PubMed] [Google Scholar]
- Kollmann M, Løvdok L, Bartholomé K, Timmer J, Sourjik V. Design principles of a bacterial signalling network. Nature. 2005;438:504–507. doi: 10.1038/nature04228. [DOI] [PubMed] [Google Scholar]
- Komatsu T, Kukelyansky I, McCaffery JM, Ueno T, Varela LC, Inoue T. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat. Methods. 2010;7:206–208. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupnick JG, Benovic JL. The role of receptor kinases and arrestins in G protein-coupled receptor regulation. Annu. Rev. Pharmacol. Toxicol. 1998;38:289–319. doi: 10.1146/annurev.pharmtox.38.1.289. [DOI] [PubMed] [Google Scholar]
- Ku C-J, Wang Y, Weiner OD, Altschuler SJ, Wu LF. Network crosstalk dynamically changes during neutrophil polarization. Cell. 2012;149:1073–1083. doi: 10.1016/j.cell.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Yamauchi A, Marchal CC, Molitoris JK, Quilliam LA, Dinauer MC. Chemoattractant-stimulated Rac activation in wild-type and Rac2-deficient murine neutrophils: preferential activation of Rac2 and Rac2 gene dosage effect on neutrophil functions. J. Immunol. Baltim. Md. 2002;169:5043–5051. doi: 10.4049/jimmunol.169.9.5043. 1950. [DOI] [PubMed] [Google Scholar]
- Li Z, Hannigan M, Mo Z, Liu B, Lu W, Wu Y, Smrcka AV, Wu G, Li L, Liu M, et al. Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of Cdc42. Cell. 2003;114:215–227. doi: 10.1016/s0092-8674(03)00559-2. [DOI] [PubMed] [Google Scholar]
- Liu L, Das S, Losert W, Parent CA. mTORC2 regulates neutrophil chemotaxis in a cAMP- and RhoA-dependent fashion. Dev. Cell. 2010;19:845–857. doi: 10.1016/j.devcel.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MJ, Andexinger S, Pitcher J, Trukawinski S, Codina J, Faure JP, Caron MG, Lefkowitz RJ. Receptor-specific desensitization with purified proteins. Kinase dependence and receptor specificity of beta-arrestin and arrestin in the beta 2-adrenergic receptor and rhodopsin systems. J. Biol. Chem. 1992;267:8558–8564. [PubMed] [Google Scholar]
- Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138:760–773. doi: 10.1016/j.cell.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM, Koshland DE., Jr The gradient-sensing mechanism in bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 1972;69:2509–2512. doi: 10.1073/pnas.69.9.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili R, Ellsworth C, Lee S, Reddy TB, Ma H, Firtel RA. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy VN, Schikorski T, Stevens CF, Zhu Y. Inactivity produces increases in neurotransmitter release and synapse size. Neuron. 2001;32:673–682. doi: 10.1016/s0896-6273(01)00500-1. [DOI] [PubMed] [Google Scholar]
- Muzzey D, Gómez-Uribe CA, Mettetal JT, van Oudenaarden A. A systems-level analysis of perfect adaptation in yeast osmoregulation. Cell. 2009;138:160–171. doi: 10.1016/j.cell.2009.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio M, Watanabe K, Sasaki J, Taya C, Takasuga S, Iizuka R, Balla T, Yamazaki M, Watanabe H, Itoh R, et al. Control of cell polarity and motility by the PtdIns(3,4,5)P3 phosphatase SHIP1. Nat. Cell Biol. 2007;9:36–44. doi: 10.1038/ncb1515. [DOI] [PubMed] [Google Scholar]
- Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E, Matsumoto K, Yamada KM. A Rac switch regulates random versus directionally persistent cell migration. J. Cell Biol. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- Park KC, Rivero F, Meili R, Lee S, Apone F, Firtel RA. Rac regulation of chemotaxis and morphogenesis in Dictyostelium. EMBO J. 2004;23:4177–4189. doi: 10.1038/sj.emboj.7600368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng GE, Wilson SR, Weiner OD. A pharmacological cocktail for arresting actin dynamics in living cells. Mol. Biol. Cell. 2011;22:3986–3994. doi: 10.1091/mbc.E11-04-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta K, Diehl W, Zhang X-Y, Kutner R, Bialkowska A, Reiser J. Lentiviral vectors encoding tetracycline-dependent repressors and transactivators for reversible knockdown of gene expression: a comparative study. BMC Biotechnol. 2007;7:41. doi: 10.1186/1472-6750-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SL, Wadhams GH, Armitage JP. Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 2011;9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- Pringle JR. Induction, selection, and experimental uses of temperature-sensitive and other conditional mutants of yeast. Methods Cell Biol. 1975;12:233–272. doi: 10.1016/s0091-679x(08)60959-0. [DOI] [PubMed] [Google Scholar]
- Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Robinson MS, Sahlender DA, Foster SD. Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev. Cell. 2010;18:324–331. doi: 10.1016/j.devcel.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 2004;167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki AT, Janetopoulos C, Lee S, Charest PG, Takeda K, Sundheimer LW, Meili R, Devreotes PN, Firtel RA. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J. Cell Biol. 2007;178:185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, Bourne HR. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- Stankunas K, Bayle JH, Gestwicki JE, Lin Y-M, Wandless TJ, Crabtree GR. Conditional protein alleles using knockin mice and a chemical inducer of dimerization. Mol. Cell. 2003;12:1615–1624. doi: 10.1016/s1097-2765(03)00491-x. [DOI] [PubMed] [Google Scholar]
- Stankunas K, Bayle JH, Havranek JJ, Wandless TJ, Baker D, Crabtree GR, Gestwicki JE. Rescue of degradation-prone mutants of the FK506-rapamycin binding (FRB) protein with chemical ligands. Chembiochem Eur. J. Chem. Biol. 2007;8:1162–1169. doi: 10.1002/cbic.200700087. [DOI] [PubMed] [Google Scholar]
- Stephens LR, Hughes KT, Irvine RF. Pathway of phosphatidylinositol(3,4,5)-trisphosphate synthesis in activated neutrophils. Nature. 1991;351:33–39. doi: 10.1038/351033a0. [DOI] [PubMed] [Google Scholar]
- Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CX, Downey GP, Zhu F, Koh ALY, Thang H, Glogauer M. Rac1 is the small GTPase responsible for regulating the neutrophil chemotaxis compass. Blood. 2004;104:3758–3765. doi: 10.1182/blood-2004-03-0781. [DOI] [PubMed] [Google Scholar]
- Szobota S, Gorostiza P, Del Bene F, Wyart C, Fortin DL, Kolstad KD, Tulyathan O, Volgraf M, Numano R, Aaron HL, et al. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54:535–545. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Takeda K, Shao D, Adler M, Charest PG, Loomis WF, Levine H, Groisman A, Rappel W-J, Firtel RA. Incoherent feedforward control governs adaptation of activated ras in a eukaryotic chemotaxis pathway. Sci. Signal. 2012;5:ra2. doi: 10.1126/scisignal.2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G, Chen M, Foote C, Tan C. Temperature-sensitive mutations made easy: generating conditional mutations by using temperature-sensitive inteins that function within different temperature ranges. Genetics. 2009;183:13–22. doi: 10.1534/genetics.109.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiagarajan TC, Piedras-Renteria ES, Tsien RW. alpha- and betaCaMKII. Inverse regulation by neuronal activity and opposing effects on synaptic strength. Neuron. 2002;36:1103–1114. doi: 10.1016/s0896-6273(02)01049-8. [DOI] [PubMed] [Google Scholar]
- Toettcher JE, Gong D, Lim WA, Weiner OD. Light-based feedback for controlling intracellular signaling dynamics. Nat. Methods. 2011;8:837–839. doi: 10.1038/nmeth.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Weiner OD, Lim WA. Using Optogenetics to Interrogate the Dynamic Control of Signal Transmission by the Ras/Erk Module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano G, Abbott LF, Marder E. Activity-dependent changes in the intrinsic properties of cultured neurons. Science. 1994;264:974–977. doi: 10.1126/science.8178157. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Keizer-Gunnik I, Van Haastert PJM. Four key signaling pathways mediating chemotaxis in Dictyostelium discoideum. J. Cell Biol. 2008;180:747–753. doi: 10.1083/jcb.200709180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahou G, Rivero F. Rho GTPase signaling in Dictyostelium discoideum: insights from the genome. Eur. J. Cell Biol. 2006;85:947–959. doi: 10.1016/j.ejcb.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Wang CJ, Bergmann A, Lin B, Kim K, Levchenko A. Diverse sensitivity thresholds in dynamic signaling responses by social amoebae. Sci. Signal. 2012;5:ra17. doi: 10.1126/scisignal.2002449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger LS, Dar RD, Simpson ML. Transient-mediated fate determination in a transcriptional circuit of HIV. Nat. Genet. 2008;40:466–470. doi: 10.1038/ng.116. [DOI] [PubMed] [Google Scholar]
- Weiner OD, Neilsen PO, Prestwich GD, Kirschner MW, Cantley LC, Bourne HR. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat. Cell Biol. 2002;4:509–513. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner OD, Rentel MC, Ott A, Brown GE, Jedrychowski M, Yaffe MB, Gygi SP, Cantley LC, Bourne HR, Kirschner MW. Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 2006;4:e38. doi: 10.1371/journal.pbio.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch HCE, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, Tempst P, Hawkins PT, Stephens LR. P-Rex1, a PtdIns(3,4,5)P3- and Gbetagamma-regulated guanine-nucleotide exchange factor for Rac. Cell. 2002;108:809–821. doi: 10.1016/s0092-8674(02)00663-3. [DOI] [PubMed] [Google Scholar]
- Wong K, Pertz O, Hahn K, Bourne H. Neutrophil polarization: spatiotemporal dynamics of RhoA activity support a self-organizing mechanism. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3639–3644. doi: 10.1073/pnas.0600092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Huang C-H, Iglesias PA, Devreotes PN. Cells navigate with a local-excitation, global-inhibition-biased excitable network. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17079–17086. doi: 10.1073/pnas.1011271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Meier-Schellersheim M, Jiao X, Nelson LE, Jin T. Quantitative imaging of single live cells reveals spatiotemporal dynamics of multistep signaling events of chemoattractant gradient sensing in Dictyostelium. Mol. Biol. Cell. 2005;16:676–688. doi: 10.1091/mbc.E04-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, Shin M-G, Lee S, Kim J-R, Park WS, Cho K-H, Meyer T, Heo WD. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol. Cell. 2012;47:281–290. doi: 10.1016/j.molcel.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Jost AP-T, Weiner OD, Tang C. A light-inducible organelle-targeting system for dynamically activating and inactivating signaling in budding yeast. Mol. Biol. Cell. 2013;24:2419–2430. doi: 10.1091/mbc.E13-03-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi TM, Huang Y, Simon MI, Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, Huttenlocher A. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev. Cell. 2010;18:226–236. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Berg HC. Ultrasensitivity of an adaptive bacterial motor. J. Mol. Biol. 2013;425:1760–1764. doi: 10.1016/j.jmb.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Branch RW, Hosu BG, Berg HC. Adaptation at the output of the chemotaxis signalling pathway. Nature. 2012;484:233–236. doi: 10.1038/nature10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Charest PG, Firtel RA. Spatiotemporal regulation of Ras activity provides directional sensing. Curr. Biol. CB. 2008;18:1587–1593. doi: 10.1016/j.cub.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Jin T, McCaig CD, Forrester JV, Devreotes PN. Genetic analysis of the role of G protein-coupled receptor signaling in electrotaxis. J. Cell Biol. 2002;157:921–927. doi: 10.1083/jcb.200112070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Eckerdal J, Dimitrijevic I, Andersson T. Chemotactic peptide-induced activation of Ras in human neutrophils is associated with inhibition of p120-GAP activity. J. Biol. Chem. 1997;272:23448–23454. doi: 10.1074/jbc.272.37.23448. [DOI] [PubMed] [Google Scholar]
- Zhou XX, Chung HK, Lam AJ, Lin MZ. Optical control of protein activity by fluorescent protein domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J. Cell Biol. 1977;75:606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH, Sullivan SJ. Sensory adaptation of leukocytes to chemotactic peptides. J. Cell Biol. 1979;82:517–527. doi: 10.1083/jcb.82.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]