Abstract
This study investigated the abundance, distribution, and composition of microbial communities at the watershed scale in a boreal peatland within the Marcell Experimental Forest (MEF), Minnesota, USA. Through a close coupling of next-generation sequencing, biogeochemistry, and advanced analytical chemistry, a biogeochemical hot spot was revealed in the mesotelm (30- to 50-cm depth) as a pronounced shift in microbial community composition in parallel with elevated peat decomposition. The relative abundance of Acidobacteria and the Syntrophobacteraceae, including known hydrocarbon-utilizing genera, was positively correlated with carbohydrate and organic acid content, showing a maximum in the mesotelm. The abundance of Archaea (primarily crenarchaeal groups 1.1c and 1.3) increased with depth, reaching up to 60% of total small-subunit (SSU) rRNA gene sequences in the deep peat below the 75-cm depth. Stable isotope geochemistry and potential rates of methane production paralleled vertical changes in methanogen community composition to indicate a predominance of acetoclastic methanogenesis mediated by the Methanosarcinales in the mesotelm, while hydrogen-utilizing methanogens predominated in the deeper catotelm. RNA-derived pyrosequence libraries corroborated DNA sequence data to indicate that the above-mentioned microbial groups are metabolically active in the mid-depth zone. Fungi showed a maximum in rRNA gene abundance above the 30-cm depth, which comprised only an average of 0.1% of total bacterial and archaeal rRNA gene abundance, indicating prokaryotic dominance. Ratios of C to P enzyme activities approached 0.5 at the acrotelm and catotelm, indicating phosphorus limitation. In contrast, P limitation pressure appeared to be relieved in the mesotelm, likely due to P solubilization by microbial production of organic acids and C-P lyases. Based on path analysis and the modeling of community spatial turnover, we hypothesize that P limitation outweighs N limitation at MEF, and microbial communities are structured by the dominant shrub, Chamaedaphne calyculata, which may act as a carbon source for major consumers in the peatland.
INTRODUCTION
Peatlands store approximately one-third of all soil carbon (C) and currently act as major sinks for atmospheric C (1, 2). The future role of peatlands in C sequestration remains uncertain and will depend on the impact of global climate change on the microbial C cycle (3). The majority of field and modeling studies predict that climate change (warming and CO2 enrichment) will result in an acceleration of organic matter decomposition in peatlands and increased flux of CO2 and CH4 to atmospheric pools (4–8). Recent studies indicate that the response of soil C to climate change will depend on the C use efficiency of specific microbial groups (9, 10), and ecological and evolutionary forces such as microbial community shifts, adaptation, or selection (11, 12) could counteract the acceleration of C loss. Therefore, particularly in critical habitats such as boreal peatland soils that represent massive C pools, lack of knowledge on the in situ dynamics of decomposer communities may represent a barrier to the reliable prediction of future global fluxes of carbon to the atmosphere or the resilience of these belowground carbon reservoirs to future perturbations.
Peatlands comprise a range of terrestrial ecosystems, including ombrotrophic bogs that rely almost solely on precipitation for water and nutrients as well as fens which are in contact with mineral soil (13). Peat can be separated with depth into two main layers: the acrotelm, which is exposed to the atmosphere and dominated by living plants, and the catotelm, which tends to be anoxic and is where the majority of organic matter is stored as peat. In addition, Clymo and Bryant identified a third layer, termed the mesotelm, which represents a transition zone between the surface acrotelm and deep catotelm, where the fluctuating water table results in redox oscillations and elevated carbon turnover (14). Carbon sequestration in peatlands results from the imbalance between plant production and the microbially mediated respiration of complex organic matter which is regulated by physicochemical parameters such as temperature, pH, O2, CO2, water potential, redox potential, and nutrient availability (15, 16).
Despite the fact that peatlands make a major contribution to the global carbon cycle that may be vulnerable to climate change, the microbiology of these complex, extreme wetland ecosystems is not well studied relative to that of other terrestrial ecosystems (17–20). To date, few next-generation-sequencing-based studies have characterized the overall diversity and environmental controls of community dynamics in northern peatlands (17, 19). Studies of microbial communities in peat soils have been conducted over relatively small scales, limited to few samples, and often with molecular approaches lacking in phylogenetic resolution or a consideration of spatial and temporal variation (6, 21–24). In particular, microbial communities in the anoxic, deep peat layers (catotelm) have been largely ignored and the mechanism of microbially mediated carbon turnover in deep peat is relatively unknown (25–27).
The objective of this study was to quantitatively link the abundance and distribution of specific microbial groups to the dynamics of specific plant-derived organic carbon compounds and environmental parameters at the ecosystem scale in a northern peatland by closely coupling analytical organic chemistry, biogeochemistry, and next-generation sequencing approaches. We hypothesized that peat decomposition as reflected in organic matter composition would directly correlate with shifts in specific heterotrophic microbial populations and that these relationships would be further impacted by the availability of nutrients and vegetation coverage. This study was conducted at the Marcell Experimental Forest (MEF) in northern Minnesota, USA, where the U.S. Department of Energy (DOE) Oak Ridge National Laboratory and the USDA Forest Service are conducting a large-scale field climate manipulation known as Spruce and Peatland Response Under Climatic and Environmental Change (SPRUCE). A secondary objective of this study was to quantify the heterotrophic microbial groups linked to carbon turnover, before the SPRUCE experiment begins.
MATERIALS AND METHODS
Study sites and sampling.
Samples were collected during four field excursions to the Marcell Experimental Forest (MEF) in August 2011, February 2012, April 2012, and July 2012. Samples were collected from the S1 bog and the Bog Lake fen for comparison (see Fig. S1 and Table S1 in the supplemental material). The S1 bog is an acidic (pH ≈3.5 to 4.0) and nutrient-deficient environment that receives water inputs primarily from precipitation and is mainly covered by Sphagnum moss, shrubs, black spruce, and eastern tamarack (28). In contrast, the Bog Lake fen (pH 4.5 to 4.8) is a poor fen, which is influenced by inputs of more nutrient-rich groundwater and is dominated by Sphagnum moss and sedges. Site hydrological and vegetation information is given in more detail in the work of Sebestyen et al. (28), and specific micrometeorological data are available from the SPRUCE website (http://mnspruce.ornl.gov). In August 2011 and April 2012, peat cores were obtained from the area of the EM2 station in the southern end of the S1 bog (see Fig. S1). In February and July 2012, peat cores were obtained from 10 sites within the S1 bog (including EM2) and the Bog Lake fen. Peat cores were sampled in hollows where the water level fluctuates seasonally from the surface to approximately 20 to 30 cm below the top of the Sphagnum layer. Pore water samples were collected using piezometers and a peristaltic pump in parallel with peat cores during the July 2012 trip as described in reference 29. Cores from each site were sectioned into increments of 0 to 10, 10 to 20, 20 to 30, 30 to 40, 40 to 50, 50 to 75, 75 to 100, 100 to 125, 125 to 150, 150 to 175, and 175 to 200 cm. Each section was homogenized in a sterile bag and subsampled for microbiological and geochemical analyses. Samples for DNA and RNA extractions were immediately frozen at −20°C and −80°C, respectively.
Physical and chemical characterization.
In situ profiling of oxygen and pH was conducted in peat pore waters during the July 2012 campaign. Oxygen concentrations were profiled in situ with an optode fiber microsensor (Microx TX3; PreSens Precision Sensing, GmbH). Pore water pH was profiled using a soil pH probe (Hanna Instruments). Methane, stable carbon isotope analysis, and fractionation factors were determined on pore waters extracted from piezometers according to a previous study (29). Determination of physical and chemical properties of the peat and organic functional groups by Fourier transform infrared spectroscopy (FT-IR) was described in reference 30. Hydraulic conductivity, a measure of the rate of water movement through soil, was calculated from dry bulk density of peat according to a regression model for peat material (31). Pore water nitrate, ammonium, and sulfate concentrations were determined by chemiluminescence detection, colorimetry, and ion chromatography, respectively (see details in the supplemental material).
Enzyme activities.
Each sample was assayed for the potential activity of seven different hydrolytic enzymes involved in C, N, and P acquisition (Table 1), according to the assay protocols in references 32 and 33. Ratios of beta-glucosidase (BG) to N-acetylglucosaminidase (NAG) and alkaline phosphatase (AP) activity were calculated to indicate microbial resource allocation to the acquisition of C, N, and P, respectively (34).
TABLE 1.
Enzymes assayed in this study, their abbreviations used in the text, nutrient cycles in which they are involved, and their target substrates
| Enzyme name | Abbreviation | Nutrient cycle | Enzyme function |
|---|---|---|---|
| Beta-glucosidase | BG | C | Hydrolysis of terminal β-d-glucosyl residues |
| Cellobiohydrolase | CB | C | Hydrolysis of β-d-glucosyl linkages |
| Xylosidase | XYL | C | Hydrolysis of β-d-xylose residues |
| N-Acetylglucosaminidase | NAG | N | Hydrolysis of chitin N-acetyl-β-d-glucosaminide |
| Alanine-aminopeptidase | AAP | N | Hydrolysis of N-terminal amino acid alanine |
| Leucine-aminopeptidase | LAP | N | Hydrolysis of N-terminal amino acid leucine |
| Acid phosphomonoesterase | PHOS | P | Hydrolysis of phosphate monoester |
DNA and RNA extraction, pyrosequencing, and qPCR.
Total genomic DNA was extracted from 0.5 g of duplicates of peat soil per sample using a MoBio PowerSoil DNA extraction kit (MoBio, Carlsbad, CA) according to the manufacturer's protocol. MoBio PowerSoil RNA extraction kits (MoBio, Carlsbad, CA) were used to extract total RNA from 2 g of peat soil per sample. For RNA samples, DNA was removed from the extracted total RNA using a Turbo DNA-free kit (Applied Biosystems, Carlsbad, CA), and an aliquot of DNA-free RNA was used for first-strand cDNA synthesis using the GoScript reverse transcription system (Promega, Madison, WI). Both genomic DNA and cDNA were sent to Research and Testing Lab (Lubbock, TX) or Molecular Research LP (Shallowater, Texas) for amplification and 454 sequencing using the standard pyrosequencing protocols (Roche 454, Branford, CT). PCR amplification of bacterial and archaeal small-subunit (SSU) rRNA genes and fungal internal transcribed spacer (ITS) regions of rRNA genes was performed using the primer pairs 515F/806R (35) and ITS1F/ITS4 (36), respectively. Quantitative real-time PCR (qPCR) was performed with 1/40-diluted genomic DNA as the templates to quantify the abundance of total bacterial, archaeal, and fungal SSU rRNA genes according to the procedures described elsewhere (23, 37).
A total of >160 samples were sequenced for 16S rRNA genes of Bacteria and Archaea. To understand seasonal and vertical variation, the sequencing was performed for samples across the whole-peat profiles at EM2 of August 2011; fen, T3M, and T3F of February 2012; EM2 of April 2012; and fen, T3M, and T3F of July 2012. Horizontal distribution patterns were analyzed by sequencing 3 representative depths (0 to 10, 30 to 40, and 75 to 100 cm) at all 10 S1 bog sites sampled in July 2012. RNA-derived amplicon sequencing was performed for July 2012 samples from the whole profiles of T3M, T3F, EM2, and fen sites. A total of 83 samples were sequenced for the fungal ITS fragment, focusing on 3 depths (0 to 10, 20 to 30, and 30 to 40 cm) near the peat surface.
Processing of pyrosequencing data.
Original sequence outputs were filtered to remove low-quality sequences and denoised with Acacia using default settings (38). The denoised fasta files were used to pick operational taxonomic units (OTUs) and representative sequences at 97% identity in QIIME (39). For bacterial SSU rRNA gene sequences, the PyNAST aligned representative sequences were used to identify chimeric sequences using ChimeraSlayer in QIIME. Chimera check of fungal ITS gene sequences was done with UCHIME de novo in a de novo mode (40). Taxonomy was assigned to each representative sequence by using BLAST with a maximum E value of 0.001 against the Greengenes core set for Bacteria and using a customized database for fungi (http://www.emerencia.org/fungalitspipeline.html). For all OTU-based analyses, the original OTU table was rarefied to depths of 1,620 bacterial and archaeal sequences and 1,000 fungal ITS sequences per sample.
Statistical analyses and modeling.
Pairwise comparisons of microbial community structure were computed as Bray-Curtis similarity and were visualized using nonmetric multidimensional scaling (NMDS) in PRIMER v6 (Primer-E Ltd., United Kingdom). A heat map of dominant microbial taxa and its clustering analysis was created with the MeV program (www.tm4.org/mev.html). This heat map included the 32 most abundant bacterial and archaeal taxa at a genus level (or level six within the Hugenholtz framework) plus the two most abundant methanotrophs (Methylocystaceae and Methylomonas) and the 10 most abundant fungal genera. In cases involving clades without a genus-level classification, sequences were summed to a defined clade of a higher taxonomic level (41).
A distance-based linear model combined with a forward model selection procedure was performed to identify key environmental variables explaining the community spatial turnover. This analysis was run in the PERMANOVA package (Primer-E). Independent variable significance (α = 0.05) was evaluated stepwise, and the order of variable evaluation was based on improvement in the model's adjusted R2. Model selection proceeded until the next independent variable was nonsignificant as determined by 1,000 permutations. Separate model selection procedures were carried out for community horizontal variation at 0 cm, 30 cm, and 75 cm and the vertical depth variation (including all 10 depths) in the system.
As a complement to the linear regression modeling, structural equation model-based path analysis (42) was employed to compare the strengths of organic matter composition, vegetation, and nutrient effects upon microbial community composition. Vegetation data were downloaded from the website of the SPRUCE project (Spruce and Peatland Response Under Climatic and Environmental Change; http://mnspruce.ornl.gov/), including total tree basal area (BA) and counts of tree species (Picea mariana and Larix laricina) and shrub species (Chamaedaphne calyculata and Ledum groenlandicum) for the nearest points to each core location. Model performance was determined via chi-square tests. Nonsignificant chi-square tests (P > 0.05) indicate that the observed covariance structure of our data matches the model, and the hypothesis cannot be rejected as a potential explanation of microbial community composition. Standardized path coefficients with values less than 0.10, around 0.30, and >0.5 generally indicate small, medium, and large effects, respectively (42). Note that the total indirect effect between two variables in a path model equals the sum of the products of each direct effect. Path analysis was done with the Lavaan package in R (43).
Seasonal differences in microbial community composition were examined using ANOSIM in PRIMER v6 (Primer-E) (see details in the supplemental material). The t tests or paired t tests were performed in PAST (http://folk.uio.no/ohammer/past/) to assess seasonal differences in enzyme activities and differences in relative abundances of specific microbial groups between samples.
RESULTS
Physical and biogeochemical characterization of peat.
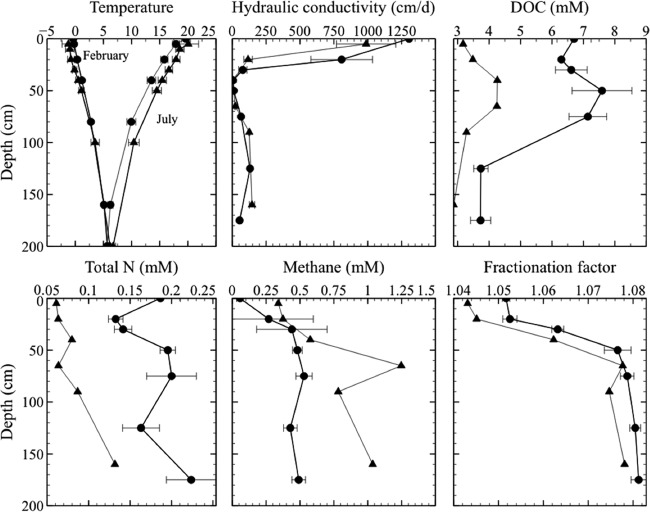
Temperature in February gradually increased from −0.8 ± 0.6°C at 0 cm to 5.9 ± 0.3°C at 200 cm, while it decreased from 19.9 ± 0.3°C at 0 cm to 6.2 ± 0.8°C at 200 cm in July (Fig. 1). Hydraulic conductivity sharply declined from >1,000 to 3 to 6 cm day−1 over the surface 30- to 40-cm depth and rebounded to 100 to 200 cm day−1 in the deeper peat (Fig. 1). Overall, higher dissolved organic carbon (DOC) and total nitrogen concentrations were observed in the bog than in the fen (Fig. 1). Maxima in DOC concentrations were observed at mid-depth near 50 cm, suggesting that the most active peat degradation and DOC release occurred in the mesotelm. Total nitrogen concentration in peat generally increased with depth at all sites, except a mid-depth peak observed in the bog (Fig. 1).
FIG 1.
Physical and chemical characterization of solid peats and pore water samples from the bog (circles) and fen (triangles) sites. Each profile was based on values averaged from multiple sites (see Table S1 in the supplemental material) and different seasons. DOC and total N are in millimolar concentrations of C and N, respectively. Temperature is in degrees Celsius.
Based on physical and chemical properties of the peat column (see Fig. S2 and S3 in the supplemental material), three distinct vertical zones, each with unique properties, were identified within the peat column (see the Discussion). All data (except temperature) indicated that changes with depth in the peat column are more pronounced than temporal or lateral spatial differences (30).
Pore water chemistry.
In situ profiles measured in July 2012 revealed that oxygen concentrations were variable in the surface peat but generally decreased to below detection within 2 to 3 cm below the water level (data not shown). Pore water pH was nearly constant in situ, ranging from 3.5 to 4.0 at the bog and from 4.5 to 4.8 at the fen sites. Nitrate and nitrite concentrations averaged below 1 μM, while sulfate concentrations were below a detection limit of 1.2 μM in the summer season. A maximum in pore water methane (CH4) concentrations was observed at mid-depth of both bog and fen sites (Fig. 1), consistent with a companion study showing the highest rates of CH4 production in this zone (30). Fractionation factors calculated from the carbon isotopic composition of methane indicated a shift in the relative importance of methane production pathways at the depth interval where the highest methane accumulation was observed (Fig. 1) (17).
Distribution and abundance of microorganisms.
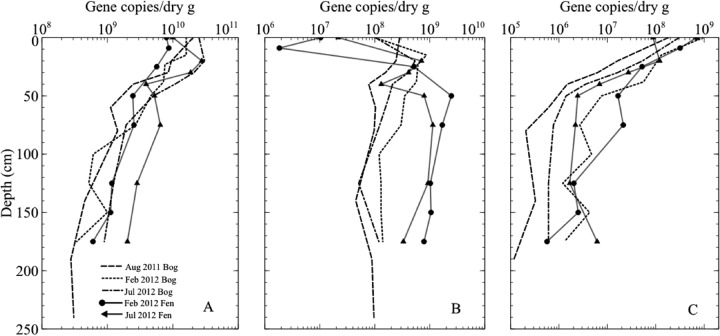
Among the 3 microbial domains, bacterial rRNA gene abundance was highest by at least an order of magnitude in the surface peat, followed by Archaea and then fungi as determined by qPCR (Fig. 2). The rRNA gene abundance of bacteria and fungi declined with depth, with a much sharper decline observed for fungi. The relative abundance of Archaea increased from <1% of total rRNA gene copies at the surface to up to 60% below the 75-cm depth. Archaeal abundance was on average 10 times higher (Tukey's pairwise test, P = 0.003) in the deep fen than in the deep bog. Fungal rRNA gene abundance above 30 cm comprised on average 0.1% of total bacterial and archaeal rRNA gene abundance, indicating archaeal and bacterial dominance. Seasonal differences in bacterial and archaeal biomass were not significant (P > 0.05). However, fungal biomass at the surface was 4.3 times higher (Tukey's pairwise test, P = 0.008) in winter than in summer at both bog and fen sites.
FIG 2.
Microbial SSU rRNA gene abundance across depths in different seasons. Each profile is based on mean values averaged from samples taken from different sites. (A) Bacteria. (B) Archaea. (C) Fungi.
Activities of enzymes that mediate the mineralization of carbon, nitrogen, and phosphorus.
Activities of all 7 enzymes consistently decreased with depth (Tables 1 and 2). Seasonal trends for the activities of beta-glucosidase (BG) and N-acetylglucosaminidase (NAG) were not apparent (P > 0.2; see Fig. S4 in the supplemental material). Ratios of enzyme activity associated with C, N, and P acquisition are used to indicate nutrient availability. For all samples, both C/N and N/P enzyme ratios were close to the mean of global soil ecosystems (34). However, the C/P ratio was 0.52, which is similar to those published for lentic ecosystems (34), indicating P limitation as more energy is allocated to PHOS (acid phosphomonoesterase) activity relative to BG activity compared to global ecosystem averages. Calculation of C/P ratios in each layer showed that values were 0.56, 0.78, and 0.42 in the surface, mid-, and deep layers, respectively.
TABLE 2.
Enzyme activity and ratios averaged from each and all layers across the peat column in July 2012
| Enzyme activity or ratio | Avg (SD) from layer: |
|||
|---|---|---|---|---|
| 0–30 cm | 30–75 cm | 75–200 cm | All | |
| Activity (nmol g−1 dry soil h−1) of enzyme | ||||
| BG | 24,488 (16,931) | 4,832 (5,993) | 2,952 (3,017) | 12,118 (3,017) |
| CB | 5,353 (3,665) | 1,248 (1,460) | 3,109 (2,107) | 3,460 (2,107) |
| XYL | 3,742 (2,616) | 2,393 (3,753) | 3,227 (4,524) | 3,188 (4,524) |
| NAG | 14,983 (12,849) | 4,057 (4,548) | 2,937 (1,755) | 8,084 (1,755) |
| AAP | 668 (223) | 236 (203) | 239 (95) | 410 (95) |
| LAP | 428 (139) | 168 (123) | 192 (74) | 280 (74) |
| PHOS | 59,150 (60,630) | 7,442 (8,612) | 7,894 (9,650) | 28,264 (9,650) |
| Ratio | ||||
| C/N | 2.01 (1.17) | 1.13 (0.36) | 0.86 (0.34) | 1.40 (0.34) |
| C/P | 0.56 (0.28) | 0.78 (0.34) | 0.42 (0.17) | 0.52 (0.17) |
| N/P | 0.36 (0.24) | 0.73 (0.35) | 0.55 (0.24) | 0.53 (0.24) |
Microbial community composition: bog versus fen, total versus active, and attached versus planktonic.
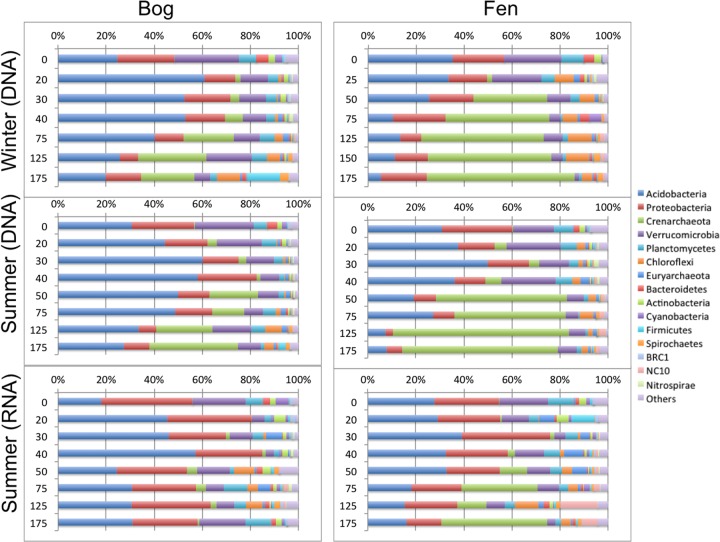
A total of 65 bacterial and archaeal phyla were detected in microbial communities. Acidobacteria (35%), Proteobacteria (18%), Verrucomicrobia (14%), and Crenarchaeota (14%) dominated the DNA-derived microbial communities of both bog and fen when relative abundance was averaged across all samples (Fig. 3). In agreement with qPCR results, bacterial phyla showed the highest relative abundance of amplicon sequences in the acrotelm and mesotelm, whereas the Crenarchaeota showed a higher relative abundance in the catotelm. The relative abundance of Acidobacteria was significantly (paired t test, P = 0.002) higher in bog than in fen, while Crenarchaeota below 30 cm were two times more abundant (paired t test, P = 0.016) in the fen than in the bog. The most striking disparity in community composition between DNA- and RNA-derived communities was that the DNA-derived microbial community relative to the RNA-derived one contained on average a >2-times-higher abundance (paired t test, P = 0.002) of Proteobacteria and approximately a 44% lower abundance (paired t test, P = 0.002) of Crenarchaeota (Fig. 3). Interestingly, the NC10 phylum, containing known members of denitrifying methanotrophs, had on average a 4-times-higher representation (paired t test, P = 0.008) in RNA-derived communities than in DNA-derived communities below 30 cm (Fig. 3).
FIG 3.
DNA- and RNA-derived bacterial and archaeal community composition across depths in bog and fen sites, compared between winter (February) and summer (July) samples. Data are averaged from specific depths across sites. The x axis is the sequence percentage of each phylum relative to the total sequences. The y axis is the depth in centimeters.
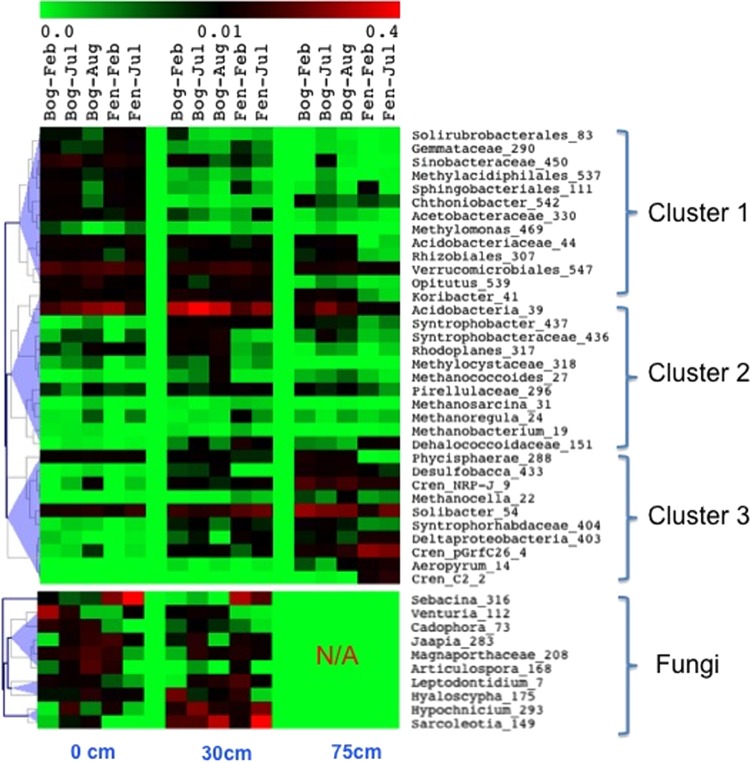
In parallel with stratified physical and chemical properties in peat columns, dominant microbial taxa also showed distinct clustering with depth (Fig. 4). Cluster 1 contains diverse heterotrophs (e.g., Sinobacteraceae OTU450 and Chthoniobacter) and methanotrophs prevalent in the surface peat. A large diversity of methanotrophs belonging to Alpha-, Beta-, and Gammaproteobacteria was detected. Methylocystaceae and Methylomonas codominated both DNA- and RNA-derived methanotrophs across seasons in the bog and fen sites (see Fig. S5 in the supplemental material). OTUs assigned to Methylacidiphilales (family LD19 of Verrucomicrobia) were as abundant (1.5% of total community) as were total proteobacterial methanotrophs (on average 1.1%), but these phylotypes are distantly (sequence identity, <0.91) related to sequences of the four cultured verrucomicrobial methanotrophs (see Fig. S6). Cluster 2 mainly harbors acidobacterial OTUs, Syntrophobacter, and putative acetoclastic methanogen Methanococcoides, peaking at 30 cm. Microorganisms in cluster 3 showed increased abundance with depth, including OTUs belonging to Crenarchaeota, hydrogenotrophic Methanocella, and Solibacter of the Acidobacteria (Fig. 4). Interestingly, the uncultivated Crenarchaeota lineages in the bog and fen mainly fell into crenarchaeotal groups 1.1c and 1.3, which showed a maximum abundance at 50 to 100 cm and below 100 cm, respectively (see Fig. S7 and S8).
FIG 4.
Distribution of dominant microbial taxa (genus level or higher) at three peat layers over seasons. The color bar indicates the fraction of each taxon in the total community. Numbers after the taxonomic names indicate the OTU numbers.
Fungal community composition was dominated by members of the Ascomycota and Basidiomycota, which on average accounted for 63% and 33% of the total fungal community, respectively (see Fig. S9 in the supplemental material). Saccharomyces and Agaricomycotina were two dominant subphyla among the detected Ascomycota and Basidiomycota, respectively. Saccharomyces (mainly yeasts) increased with depth, while Agaricomycotina decreased with depth in relative abundance. No seasonal trends were detected at subphylum level (data not shown). Interestingly, on average a 37-times-higher (t test, P = 0.001) proportion of Sebacina (mycorrhizal fungi) sequences was observed in the fen than in the bog during both seasons (Fig. 4).
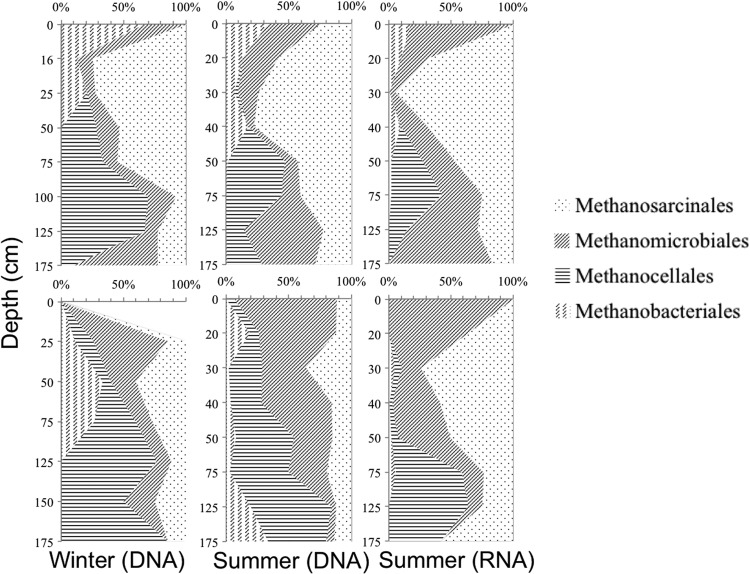
Methanogen community composition also showed a vertically stratified distribution (Fig. 5). Putative acetoclastic Methanosarcinales showed the highest relative abundance near 30 cm, while hydrogenotrophic methanogens (Methanomicrobiales and Methanocellales) comprised the majority of methanogen sequences below 50 cm. For example, below 50 cm, Methanocellales were 3 times (paired t test, P < 0.001) more abundant than Methanosarcinales. The relative abundance of Methanosarcinales was 1.5 times (paired t test, P = 0.028) higher in RNA-derived than in DNA-derived communities, suggesting a potentially active role of the Methanosarcinales near 30 cm in both bog and fen. However, the vertical profile of methane concentration was mainly correlated (R2 = 0.39) with the distribution of Methanocellales abundance. In addition, increasing fractionation from CO2 to CH4 with depth suggests the greater relative importance of the acetoclastic pathway in the mesotelm and hydrogenotrophic methanogenesis in the deeper catotelm.
FIG 5.
Temporal and vertical distribution patterns of methanogen community in bog (upper panels) and fen (lower panels) sites. Data are averaged from specific depths across sites. The x axis is the sequence percentage of each order relative to the total methanogen sequences.
Beta diversity and environmental controls over microbial community composition.
NMDS analysis of bacterial and archaeal communities identified 3 continuous clusters along depth gradients (Fig. 6A). Deep fen samples are significantly (ANOSIM, R = 0.624, P = 0.001) different from deep bog samples (Fig. 6A), mainly due to a much higher abundance of Crenarchaeota in the deep fen. ANOSIM analysis suggested no significant difference in microbial community composition between winter and summer in the surface (R = 0.11, P = 0.30). Fungal community composition also showed clear separation of surface samples from subsurface peats (Fig. 6B), with high dispersal of the surface community along the horizontal axis of the ordination space.
FIG 6.
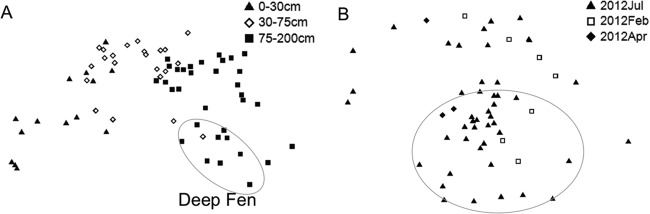
Nonmetric multidimensional scaling (NMDS) comparing similarities of bacterial and archaeal (A) (stress = 0.12) and fungal (B) (stress = 0.20) community compositions across depths in different seasons. Symbols inside the circle in panel A indicate samples from the deep fen. Symbols inside the circle in panel B indicate samples from below a 20-cm depth, and those outside the circle are surface samples.
Complex interrelationships between environmental variables and microbial community composition were validated by path analysis (Fig. 7). For vegetation cover, tree counts can predict tree basal area, whose shading blocks sunlight and negatively affects the distribution of shrub species such as Chamaedaphne calyculata and Ledum groenlandicum. Our data had significant fit with the hypothesized structure model (P = 0.13, chi-square = 25.89). The strongest factor directly affecting changes in microbial community composition was the C/N ratio (path coefficients, 0.52 versus 0.08 by Chamaedaphne), consistent with the above results from the linear model. In addition, carbohydrate content was the most influential among those of 4 organic compounds to predict peat decomposition, followed by aromatic compounds. Although weak, an indirect effect (0.09) of nutrient availability (indicated by C/P enzyme ratio) on community composition was similar to the direct effect (0.08) from Chamaedaphne.
FIG 7.
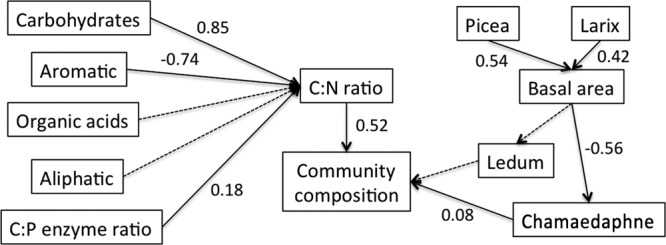
Path analysis to analyze direct and indirect effects of organic matter composition, nutrient availability, and vegetation cover on microbial community structure. Chi-square test, 25.89; P = 0.13. Solid and dashed arrows indicate significant (P < 0.05) and nonsignificant (P > 0.05) causal relations, respectively. Numbers adjacent to each arrow indicate standardized path coefficients. Values in parentheses are fractions of the variance explained by the respective variable in the model.
Distance-based linear modeling was performed for each of the three key zones and revealed that the distribution of Chamaedaphne (see Fig. S10 in the supplemental material), an Ericaceae shrub species, significantly affected bacterial and archaeal community composition at the 0- and 30-cm-depth horizons (Table 3; see also Table S2 in the supplemental material). At the 75-cm horizon, archaeal biomass (A16S) is the most important factor explaining community spatial turnover, suggesting that undetermined factors controlling archaeal distribution are important in structuring microbial community composition in deep peat. When considering all peat layers, both C/N ratio and archaeal biomass explained the most variation for microbial spatial turnover.
TABLE 3.
Environmental variables significantly explaining the bacterial and archaeal community spatial turnover at 0 cm, 30 cm, and 75 cm and all layers, identified by distance-based linear modela
| Layer | Variable | Adj R2 | P | Prop. |
|---|---|---|---|---|
| 0–10 cm | Chamaedaphne | 0.12 | 0.03 | 0.26 |
| 30–40 cm | Chamaedaphne | 0.09 | 0.02 | 0.19 |
| 75–100 cm | Archaea 16S | 0.10 | 0.03 | 0.23 |
| All layers | C/N ratio | 0.20 | 0.00 | 0.23 |
| Archaea 16S | 0.26 | 0.00 | 0.09 |
Prop., proportion of variation explained; Adj, adjusted. A complete list of statistics for each variable is provided in Table S2 in the supplemental material.
In contrast to the model containing prokaryotic communities, no environmental variables can be significantly loaded into the model to explain horizontal variation in fungal community composition at the 0-cm and 20-cm horizons (Table 4; see also Table S3 in the supplemental material). Aliphatic compounds significantly explained 22% of horizontal variation at 30 cm. In all layers, C/N ratio and Chamaedaphne, phosphatase activity, and C/P enzyme ratio are significant factors to explain fungal spatial turnover, although each of them explained less than 10% of variation.
TABLE 4.
Environmental variables significantly explaining the fungal community spatial turnover at 30-cm depth and all layers, identified by distance-based linear modela
| Layer | Variable | Adj R2 | P | Prop. |
|---|---|---|---|---|
| 30–40 cm | Aliphat2920 | 0.11 | 0.03 | 0.22 |
| All layers | C/N ratio | 0.02 | 0.05 | 0.06 |
| Chamaedaphne | 0.05 | 0.05 | 0.06 | |
| Phosphatase | 0.09 | 0.01 | 0.07 | |
| C/P enzyme activity | 0.13 | 0.01 | 0.06 |
No variables are significantly retained in the forward model selection for the 0- cm and 20-cm depths; thus, they are not shown in the table. Prop., proportion of variation explained; Adj, adjusted. A complete list of statistics for each variable is provided in Table S3 in the supplemental material.
DISCUSSION
Using a combination of advanced analytical chemistry, biogeochemistry, and microbiology approaches, organic matter decomposition and microbial community dynamics were shown to be closely coupled in an extensive field data set compiled across two seasons and 3 transects in peatlands of the Marcell Experimental Forest, where the U.S. DOE and USDA Forest Service plan to initiate a large-scale climate manipulation in late 2014. Multiple independent lines of evidence point to three distinct, vertical zones of organic matter transformation within the peat column: (i) the acrotelm, consisting of living mosses, root material, and newly formed litter (0 to 30 cm); (ii) the mesotelm, a mid-depth transition zone (30 to 75 cm) characterized by labile organic C compounds and intense decomposition; and (iii) the underlying catotelm (below 75 cm), characterized by refractory organic compounds as well as relatively low decomposition rates (30). These zones are at least in part defined by physical changes in hydraulic conductivity and water table depth. FT-IR analysis calibrated by 13C nuclear magnetic resonance (NMR) indicates that plant-derived compounds, primarily complex carbohydrates, are degraded from the acrotelm to the catotelm, resulting in the increased accumulation of lignin-like aromatic residues with depth. Similar vertical patterns in organic matter decomposition (44, 45) and microbial community stratification (19) were observed in other peatlands and are supported by concomitant changes in hydraulic conductivity, C/N ratio, and humification index.
In contrast to vertical stratification, results point to relatively minor lateral or seasonal variation in microbial community dynamics and peat organic matter composition in the MEF peatland (30), with the exception that modeling results showed that the spatial distribution pattern of the shrub Chamaedaphne calyculata plays a key role in structuring microbial community composition in the acrotelm and mesotelm, where the most intense decomposition occurs. Shrub species affect local community composition by releasing dissolved organic matter and increasing soil water retention under the shrub canopy (46). In the MEF peatland, shrub species Chamaedaphne is more abundant than other shrubs and produces more roots, and the roots penetrate deeper than those of tree species (C. M. Iversen, unpublished data), to some extent explaining the role of shrubs (relative to trees) in the structuring of microbial communities. Direct and indirect roles of vegetation cover (tree and shrub) in microbial community composition have been further confirmed through path analysis. Our results revealed that seasonal dynamics could be masked by lateral variation (caused by vegetation distribution) at the ecosystem level. Similar findings have been revealed in other temperate forests (47). On the other hand, variations in biogeochemical parameters were relatively higher in the top 50 cm of the peat column and diminished with depth, which suggests that diffusion and substrate transportation to deep peat are limited due to the existence of the midlayer, with the lowest (3 to 6 cm day−1) hydraulic conductivity. Apparently, this poor permeability combined with other factors discussed below determines the slow and stable C decomposition and enzyme activity in the catotelm.
Acrotelm.
The surface layer or acrotelm is largely oxic and receives a large amount of carbon input from plant production. At MEF, the highest microbial diversity, enzyme activity, and CO2 production rates (30) correspond to an abundance of carbohydrate compounds and low aromatic content in the acrotelm. Our results show evidence for prokaryotic dominance over microbial biomass, suggesting that peatland bacteria may outcompete fungi in utilizing plant-derived substrates, including both simple and recalcitrant organic matter, as noted in our accompanying article (48; see also reference 49). The dominant microbial taxa detected in the surface are members of the Acidobacteria, Gammaproteobacteria (Sinobacteraceae), Alphaproteobacteria (Acetobacteraceae), and Verrucomicrobia (Chthoniobacter) (Fig. 4). A substantial number of carbohydrate-utilizing Acidobacteria and Alphaproteobacteria have been cultivated under aerobic conditions from acidic peatlands (17), and many of these taxa are well represented in our sequence data set from peat depths where an abundance of carbohydrates was observed.
The Acidobacteria and Alphaproteobacteria likely play an important role in the degradation of carbohydrates in the acrotelm at MEF, as these microbial groups are abundant in RNA-derived sequence libraries and culture studies show that they are well adapted to acidic, nutrient-poor conditions (17, 50, 51). Specifically, a potential role for the Acidobacteria in degrading cellulose has been suggested based on the analysis of genome sequences (52) and laboratory tests with novel acidobacterial isolates (53, 54). Acidobacterial isolates from acidic bogs were shown to utilize a range of carbohydrate compounds, including most sugars, some heteropolysaccharides, glucuronic acid, and gluconic acid (17). In addition, community composition at the surface was highly correlated with the distribution of the dominant shrub, Chamaedaphne (see Table S2 in the supplemental material), suggesting that they may be major consumers for shrub exudates and other plant-derived carbon sources (such as dead root material and secondary metabolites like terpenoids in litter). Less information is available on the physiological ecology of Alphaproteobacteria from peatlands, but this group has been shown to utilize most sugars and a range of other low-molecular-weight carbon substrates under acidic conditions (17).
As expected, bacteria known to carry out aerobic methanotrophy were detected in the highest relative abundance in the acrotelm. However, although the relative abundance of type I and type II methanotrophs detected in our study was similar to that of other peatlands (55), a broader sequence diversity (7 genera) of putative methanotrophs belonging to the Alpha-, Beta-, and Gammaproteobacteria was observed at MEF. The role of acidophilic methanotrophs of the Verrucomicrobia remains in question in peatlands. Interestingly, we retrieved an abundance of OTUs affiliated with the Methylacidiphilales similar to that of OTUs affiliated with proteobacterial methanotrophs. However, phylogenetic analysis of these OTUs could not confirm a close relationship with cultured methanotrophic Verrucomicrobia (56), similar to the results from other acidic peatlands (24). Methylacidiphilales-like pmoA genes were also undetectable in our metagenomic libraries (48). We conclude that either the sequence diversity of methanotrophic Verrucomicrobia is broader than previously perceived or the Methylacidiphilales-like bacteria detected in acidic peatlands may not be methanotrophic.
Peatlands represent extremely nutrient-poor environments, and the microbially mediated carbon cycle has been shown to be limited by a paucity of major nutrients (3). At MEF, rates of enzyme activity indicate that microbial communities are allocating more resources to P acquisition than to C and N acquisition in the surface, thereby indicating that microbial growth and organic matter decomposition are limited by P availability. Evidence from enzyme activity is corroborated by our path analysis, which shows that P limitation and vegetation explained similar amounts of variation in microbial community composition at the ecosystem level. Moreover, metagenomic analysis provided further support for P limitation, as a much higher abundance of genes associated with P acquisition and starvation regulons was detected in the surface in comparison to other ecosystems (48). Thus, we hypothesize that P limitation outweighs N limitation at the ecosystem level at MEF. Nutrient acquisition requires further study in peatland microbial communities. For example, the biogeochemical cycles of P and N could be closely coupled in peatland soils as P limits the activity of nitrogen-fixing bacteria, which experience greater demands for P than do nonfixers, and all peatland microorganisms are likely constrained by N supply as well (57).
In general, fewer studies of fungal community composition have been conducted in peatlands using cultivation-independent molecular techniques (19, 23). In this study, fungal communities were characterized by high horizontal variation, an increasing proportion of yeast (Saccharomyces), and a reduction of the white-rot fungi (Agaricomycotina) with depth. The high horizontal variation suggests that fungi are very sensitive to vegetation cover, chemical content, and structure of litter (58), as reflected in the modeling results showing multiple drivers of fungal spatial turnover. The distribution contrast between yeast and white-rot fungi emphasizes the declining potential of phenolic compound degradation with depth and a potential role for yeast in anaerobic fermentation and decomposition of low-molecular-weight aromatic compounds (59).
Mesotelm.
The mesotelm represents a transition zone between the largely oxic acrotelm and the anoxic catotelm (14, 28). The mesotelm in the MEF peatland is associated with a fluctuating water table at the 30- to 50-cm depth (28) and is characterized by intense decomposition as determined by the consumption of carbohydrates along with the accumulation of lignin-like aromatic compounds (30). As has been shown in marine sediments, organic matter degradation in peats may be facilitated by redox oscillations (60) associated with cycles of wetting and drying at the surface of the water table. Our results indicate that the microbial C cycle may also be stimulated by the availability of P in this layer. In contrast to the surrounding vertical zones, enzyme activity data point to a lack of P limitation in the MEF mesotelm in comparison to global ecosystem averages (34). One explanation could be that relatively abundant organic acids generated in this layer (30) solubilize P from solid-phase peat for utilization by microorganisms, as microbial production of organic acids is believed to be a principal mechanism for phosphate solubilization (61). In addition, abundant C-P lyase genes were detected in mesotelm metagenomes, indicating that microbes here can effectively recycle alkylphosphonate and other forms of P bound to recalcitrant organic matter such as alkyl and aromatic compounds (48).
The physical and chemical properties of the mesotelm create a unique ecological niche for microorganisms residing there. Relative to the acrotelm, we observed a dramatic reduction in fungal biomass and a shift in microbial community toward codominance of acidobacterial OTUs, methanogenic archaea, and Syntrophobacteraceae in the mesotelm. Similar to the acrotelm, the Acidobacteria likely play an important role in the degradation of carbohydrates under aerobic conditions in the mesotelm; however, the metabolism of this group under anaerobic conditions is virtually unknown (17). As has been observed in other peatlands (18), the Syntrophobacteraceae likely catalyze secondary fermentation that supports methanogenesis under anaerobic conditions.
The majority of past studies indicate that hydrogenotrophic methanogens dominate in acidic bogs, whereas acetoclastic methanogens show increased abundance in fens (62). In corroboration of our previous work in another northern Minnesota peatland (23), sequences from members of the Methanosarcinales, known primarily as acetoclastic methanogens, predominated in the bog and fen at MEF. The relative abundance of Methanosarcinales was 1.5 times higher in RNA-derived than in DNA-derived sequence libraries, and the highest potential rates of methanogenesis were also observed in the mesotelm, further suggesting an active role for this group in methane production. The Methanosarcinales are the most metabolically versatile group of methanogens, capable of conserving energy from the consumption of acetate, methanol, methylamines, and even H2 (63, 64). However, no evidence to date indicates that the Methanosarcinales play a quantitatively important role in hydrogenotrophic methanogenesis in wetlands (62). In this study, an isotope mass balance derived from microbial respiration products, δ-13C-CH4 and δ-13C-CO2, provides evidence for a system in which methanogenesis is shifted strongly toward acetate utilization in the mesotelm (Fig. 1). Thus, we hypothesize that members of the Methanosarcinales mediate the majority of methane production through acetate utilization at MEF. This hypothesis should be further tested through the examination of expressed genes for methanogenesis and the generation of metatranscriptomes from peat.
Sulfate reduction is thermodynamically more favorable than methanogenesis, and genes associated with sulfate reduction and sulfur oxidation were observed in abundance in this layer based on our metagenomic analysis (48). In corroboration of the SSU rRNA sequence data, a high proportion of sulfite reductase genes in our metagenomes are affiliated with the Syntrophobacteraceae, suggesting a potential role in both syntrophic fermentation and sulfate respiration. Although several studies suggested the global significance of sulfate reduction and the coupling of humic reduction to organic matter mineralization in peatlands (65, 66), the identity and ecology of microorganisms responsible for these anaerobic processes are largely unknown.
Catotelm.
In the deep anoxic peat layer, or catotelm, of the MEF peatland, we observed a more homogenous system with regard to organic carbon functionality and microbial community composition. Potential rates of microbial respiration were approximately 2 orders of magnitude lower in the catotelm than in the mesotelm, and several lines of evidence indicated a high degree of humification and organic matter recalcitrance (30, 48). Similarly to the acrotelm, low ratios of C to P enzyme activity pointed to P limitation, which is likely associated with low rates of organic matter mineralization and a lack of organic acids to release P immobilized in the solid peat. Nutrient limitation and a lack of electron acceptors comprise two major factors controlling microbial activity and decomposition in deep peats (57, 66). This is supported by our metagenomic findings, which showed that the catotelm harbors a high abundance of Lon ATP-dependent protease genes for dealing with environmental stress, including starvation and acidity (48).
The catotelm has been termed the “carbon bank” of peatlands, since the bulk of carbon is stored in these deeper layers (67). The role of carbon stored deep in terrestrial soils (68) and the metabolic pathways by which this carbon may be released to the atmosphere by microbial processes have just begun to be incorporated into models of the global C cycle (69). The limited availability of labile organic matter in the deeper bog and fen samples explains why H2- and CO2-dependent methanogenesis is favored in the catotelm at MEF (62, 70). However, as has been observed in other peatlands (27, 71–73), radiocarbon signatures of microbial respiration products in deeper pore waters at MEF resembled the signatures of more modern dissolved organic carbon rather than solid-phase peat, indicating that organic matter derived from recent photosynthesis fueled the bulk of microbial respiration in the catotelm. The high ratio of CO2 to CH4 in pore waters and microcosm incubations further indicates that pathways other than methanogenesis are driving the bulk of carbon oxidation in this zone. Thus, the source of mineralized carbon is unresolved in the MEF peatland.
As indicated by qPCR and amplicon sequencing data, members of the Archaea appear to dominate in the catotelm, comprising up to 60% of the microbial community. Sequences of the Crenarchaeota, which are not known to produce methane, were particularly abundant, suggesting that this group plays an important role in the carbon cycle of deep peat layers. A predominance of Crenarchaeota sequences in deep soils and sediments has also been reported in other terrestrial ecosystems (74) and in the deep marine biosphere (45). Crenarchaeota lineages in our samples mainly fall into crenarchaeal groups 1.1c and 1.3, which peaked at 50 to 100 cm and below 100 cm, respectively, suggesting their niche stratification. The carbon metabolism and energy sources for these uncultivated Crenarchaea are largely unknown. Studies thus far suggest a role for this microbial group in the mineralization of detrital proteins (75), in degradation of fossil organic matter (76), and in anaerobic methane oxidation (76, 77). A small but growing database indicates that humic substances could serve as an electron acceptor or electron shuttle to facilitate the carbon cycle in wetlands (62). Therefore, based on our study of the carbon cycle at MEF, we propose that the crenarchaeal groups mediate anaerobic fermentation or the coupling of carbon oxidation to the respiration of some unidentified organic electron acceptor. Given the lack of cultivated representatives of this group, the importance of Crenarchaea in the peatland C cycle and their sensitivity to environmental change (78, 79) should be further investigated using single-cell genomics and other techniques that do not require cultivation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Randall K. Kolka, USDA Forest Service; Paul J. Hanson; Colleen Iversen; and other members of the SPRUCE project team for facilitating access to the Marcell Experimental Forest and the SPRUCE experimental facilities and for helping with sample handling and laboratory space support.
This work was supported by the Office of Biological and Environmental Research, Terrestrial Ecosystem Science Program, under U.S. DOE contract number DE-SC0007144, and the U.S. National Science Foundation (NSF-EAR-0628349).
The authors declare no conflict of interest.
Footnotes
Published ahead of print 28 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00205-14.
REFERENCES
- 1.Gorham E. 1991. Northern peatlands—role in the carbon-cycle and probable responses to climatic warming. Ecol. Appl. 1:182–195. 10.2307/1941811 [DOI] [PubMed] [Google Scholar]
- 2.Bridgham SD, Megonigal JP, Keller JK, Bliss NB, Trettin C. 2006. The carbon balance of North American wetlands. Wetlands 26:889–916. 10.1672/0277-5212(2006)26[889:TCBONA]2.0.CO;2 [DOI] [Google Scholar]
- 3.Limpens J, Berendse F, Blodau C, Canadell JG, Freeman C, Holden J, Roulet N, Rydin H, Schaepman-Strub G. 2008. Peatlands and the carbon cycle: from local processes to global implications—a synthesis. Biogeosciences 5:1475–1491. 10.5194/bg-5-1475-2008 [DOI] [Google Scholar]
- 4.Davidson EA, Janssens IA. 2006. Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440:165–173. 10.1038/nature04514 [DOI] [PubMed] [Google Scholar]
- 5.Dorrepaal E, Toet S, van Logtestijn RSP, Swart E, van de Weg MJ, Callaghan TV, Aerts R. 2009. Carbon respiration from subsurface peat accelerated by climate warming in the subarctic. Nature 460:616–U679. 10.1038/nature08216 [DOI] [Google Scholar]
- 6.Kim SY, Freeman C, Fenner N, Kang H. 2012. Functional and structural responses of bacterial and methanogen communities to 3-year warming incubation in different depths of peat mire. Appl. Soil Ecol. 57:23–30. 10.1016/j.apsoil.2012.02.015 [DOI] [Google Scholar]
- 7.Schuur EAG, Vogel JG, Crummer KG, Lee H, Sickman JO, Osterkamp TE. 2009. The effect of permafrost thaw on old carbon release and net carbon exchange from tundra. Nature 459:556–559. 10.1038/nature08031 [DOI] [PubMed] [Google Scholar]
- 8.Pearson RG, Phillips SJ, Loranty MM, Beck PSA, Damoulas T, Knight SJ, Goetz SJ. 2013. Shifts in Arctic vegetation and associated feedbacks under climate change. Nat. Climate Change 3:673–677. 10.1038/nclimate1858 [DOI] [Google Scholar]
- 9.Manzoni S, Taylor P, Richter A, Porporato A, Agren GI. 2012. Environmental and stoichiometric controls on microbial carbon-use efficiency in soils. New Phytol. 196:79–91. 10.1111/j.1469-8137.2012.04225.x [DOI] [PubMed] [Google Scholar]
- 10.Allison SD, Wallenstein MD, Bradford MA. 2010. Soil-carbon response to warming dependent on microbial physiology. Nat. Geosci. 3:336–340. 10.1038/ngeo846 [DOI] [Google Scholar]
- 11.Wallenstein MD, Hall EK. 2012. A trait-based framework for predicting when and where microbial adaptation to climate change will affect ecosystem functioning. Biogeochemistry 109:35–47. 10.1007/s10533-011-9641-8 [DOI] [Google Scholar]
- 12.Bardgett RD, Freeman C, Ostle NJ. 2008. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2:805–814. 10.1038/ismej.2008.58 [DOI] [PubMed] [Google Scholar]
- 13.Frolking S, Talbot J, Jones MC, Treat CC, Kauffman JB, Tuittila E-S, Roulet N. 2011. Peatlands in the Earth's 21st century climate system. Environ. Rev. 19:371–396. 10.1139/a11-014 [DOI] [Google Scholar]
- 14.Clymo RS, Bryant CL. 2008. Diffusion and mass flow of dissolved carbon dioxide, methane, and dissolved organic carbon in a 7-m deep raised peat bog. Geochim. Cosmochim. Acta 72:2048–2066. 10.1016/j.gca.2008.01.032 [DOI] [Google Scholar]
- 15.Freeman C, Ostle N, Kang H. 2001. An enzymic ‘latch' on a global carbon store—a shortage of oxygen locks up carbon in peatlands by restraining a single enzyme. Nature 409:149. 10.1038/35051650 [DOI] [PubMed] [Google Scholar]
- 16.Moore TR, Basiliko N. 2006. Decomposition in boreal peatlands, p 125–143 In Wieder RK, Vitt DH. (ed), Ecological studies, vol 188 Boreal peatland ecosystems Springer-Verlag, Berlin, Germany [Google Scholar]
- 17.Dedysh SN. 2011. Cultivating uncultured bacteria from northern wetlands: knowledge gained and remaining gaps. Front. Microbiol. 2:184. 10.3389/fmicb.2011.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drake HL, Horn MA, Wust PK. 2009. Intermediary ecosystem metabolism as a main driver of methanogenesis in acidic wetland soil. Environ. Microbiol. Rep. 1:307–318. 10.1111/j.1758-2229.2009.00050.x [DOI] [PubMed] [Google Scholar]
- 19.Andersen R, Chapman SJ, Artz RRE. 2013. Microbial communities in natural and disturbed peatlands: a review. Soil Biol. Biochem. 57:979–994. 10.1016/j.soilbio.2012.10.003 [DOI] [Google Scholar]
- 20.Artz RRE. 2009. Microbial community structure and carbon substrate use in Northern peatlands, p 111–129 In Baird A, Belyea L, Comas X, Reeve A, Slater L. (ed), Carbon cycling in northern peatlands. American Geophysical Union, Washington, DC [Google Scholar]
- 21.Dedysh SN, Pankratov TA, Belova SE, Kulichevskaya IS, Liesack W. 2006. Phylogenetic analysis and in situ identification of bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 72:2110–2117. 10.1128/AEM.72.3.2110-2117.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preston MD, Smemo KA, McLaughlin JW, Basiliko N. 2012. Peatland microbial communities and decomposition processes in the James Bay Lowlands, Canada. Front. Microbiol. 3:70. 10.3389/fmicb.2012.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin X, Green S, Tfaily MM, Prakash O, Konstantinidis KT, Corbett JE, Chanton JP, Cooper WT, Kostka JE. 2012. Microbial community structure and activity linked to contrasting biogeochemical gradients in bog and fen environments of the Glacial Lake Agassiz Peatland. Appl. Environ. Microbiol. 78:7023–7031. 10.1128/AEM.01750-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serkebaeva YM, Kim Y, Liesack W, Dedysh SN. 2013. Pyrosequencing-based assessment of the bacteria diversity in surface and subsurface peat layers of a northern wetland, with focus on poorly studied phyla and candidate divisions. PLoS One 8:e63994. 10.1371/journal.pone.0063994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye RZ, Jin QS, Bohannan B, Keller JK, McAllister SA, Bridgham SD. 2012. pH controls over anaerobic carbon mineralization, the efficiency of methane production, and methanogenic pathways in peatlands across an ombrotrophic-minerotrophic gradient. Soil Biol. Biochem. 54:36–47. 10.1016/j.soilbio.2012.05.015 [DOI] [Google Scholar]
- 26.Rumpel C, Kogel-Knabner I. 2011. Deep soil organic matter—a key but poorly understood component of terrestrial C cycle. Plant Soil 338:143–158. 10.1007/s11104-010-0391-5 [DOI] [Google Scholar]
- 27.Fontaine S, Barot S, Barre P, Bdioui N, Mary B, Rumpel C. 2007. Stability of organic carbon in deep soil layers controlled by fresh carbon supply. Nature 450:277–U210. 10.1038/nature06275 [DOI] [PubMed] [Google Scholar]
- 28.Sebestyen SD, Dorrance C, Olson D, Verry ES, Kolka RK, Elling AE, Kyllander R. 2011. Long-term monitoring sites and trends at the Marcell Experimental Forest, p 15–71 In Kolka RK, Sebestyen SD, Verry ES, Brooks KN. (ed), Peatland biogeochemistry and watershed hydrology at the Marcell Experimental Forest. CRC Press, Boca Raton, FL [Google Scholar]
- 29.Chasar LS, Chanton JP, Glaser PH, Siegel DI. 2000. Methane concentration and stable isotope distribution as evidence of rhizospheric processes: comparison of a fen and bog in the Glacial Lake Agassiz Peatland complex. Ann. Bot. 86:655–663. 10.1006/anbo.2000.1172 [DOI] [Google Scholar]
- 30.Tfaily MM, Cooper WT, Kostka J, Chanton PR, Schadt CW, Hanson PJ, Iversen CM, Chanton JP. 28 March 2014. Organic matter transformation in the peat column at Marcell Experimental Forest: humification and vertical stratification. J. Geophys. Res. Biogeosci. 10.1002/2013JG002492 [DOI] [Google Scholar]
- 31.Boelter DH. 1968. Important physical properties of peat materials. Proc. Third Int. Peat Congr. 1:150–154 [Google Scholar]
- 32.Saiya-Cork KR, Sinsabaugh RL, Zak DR. 2002. The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol. Biochem. 34:1309–1315. 10.1016/S0038-0717(02)00074-3 [DOI] [Google Scholar]
- 33.Steinweg JM, Jagadamma S, Frerichs J, Mayes MA. 2013. Activation energy of extracellular enzymes in soils from different biomes. PLoS One 8:e59943. 10.1371/journal.pone.0059943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinsabaugh RL, Hill BH, Shah JJF. 2009. Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature 462:795–798. 10.1038/nature08632 [DOI] [PubMed] [Google Scholar]
- 35.Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. 2011. Examining the global distribution of dominant archaeal populations in soil. ISME J. 5:908–917. 10.1038/ismej.2010.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- 37.Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, Delgardio J, Norton N, Hazen TC, Huettel M. 2011. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Appl. Environ. Microbiol. 77:7962–7974. 10.1128/AEM.05402-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. 2012. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat. Methods 9:425–426. 10.1038/nmeth.1990 [DOI] [PubMed] [Google Scholar]
- 39.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin X, McKinley J, Resch CT, Kaluzny R, Lauber CL, Fredrickson J, Knight R, Konopka A. 2012. Spatial and temporal dynamics of the microbial community in the Hanford unconfined aquifer. ISME J. 6:1665–1676. 10.1038/ismej.2012.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grace JB. 2006. Structural equation modeling and natural systems. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 43.Rosseel Y. 2012. Lavaan: an R package for structural equation modeling. J. Stat. Softw. 48:1–36 [Google Scholar]
- 44.Franzen LG. 2006. Increased decomposition of subsurface peat in Swedish raised bogs: are temperate peatlands still net sinks of carbon? Mires Peat 1:article 3 http://mires-and-peat.net/pages/volumes/map01/map0103.php [Google Scholar]
- 45.Cocozza C, D'Orazio V, Miano TM, Shotyk W. 2003. Characterization of solid and aqueous phases of a peat bog profile using molecular fluorescence spectroscopy, ESR and FT-IR, and comparison with physical properties. Org. Geochem. 34:49–60. 10.1016/S0146-6380(02)00208-5 [DOI] [Google Scholar]
- 46.Norton U, Saetre P, Hooker TD, Stark JM. 2012. Vegetation and moisture controls on soil carbon mineralization in semiarid environments. Soil Sci. Soc. Am. J. 76:1038–1047. 10.2136/sssaj2011.0270 [DOI] [Google Scholar]
- 47.Kuffner M, Hai B, Rattei T, Melodelima C, Schloter M, Zechmeister-Boltenstern S, Jandl R, Schindlbacher A, Sessitsch A. 2012. Effects of season and experimental warming on the bacterial community in a temperate mountain forest soil assessed by 16S rRNA gene pyrosequencing. FEMS Microbiol. Ecol. 82:551–562. 10.1111/j.1574-6941.2012.01420.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin X, Tfaily MM, Green SJ, Steinweg JM, Chanton P, Imvittaya A, Chanton JP, Cooper W, Schadt C, Kostka JE. 2014. Microbial metabolic potential for carbon degradation and nutrient (nitrogen and phosphorus) acquisition in an ombrotrophic peatland. Appl Environ. Microbiol. 80:3531–3540. 10.1128/AEM.00206-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winsborough C, Basiliko N. 2010. Fungal and bacterial activity in northern peatlands. Geomicrobiol. J. 27:315–320. 10.1080/01490450903424432 [DOI] [Google Scholar]
- 50.Sait M, Davis KE, Janssen PH. 2006. Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl. Environ. Microbiol. 72:1852–1857. 10.1128/AEM.72.3.1852-1857.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eichorst SA, Breznak JA, Schmidt TM. 2007. Isolation and characterization of soil bacteria that define Teniglobus gen. nov., in the phylum Acidobacteria. Appl. Environ. Microbiol. 73:2708–2717. 10.1128/AEM.02140-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M, Xie G, Haft DH, Sait M, Badger J, Barabote RD, Bradley B, Brettin TS, Brinkac LM, Bruce D, Creasy T, Daugherty SC, Davidsen TM, Deboy RT, Detter JC, Dodson RJ, Durkin AS, Ganapathy A, Gwinn-Giglio M, Han CS, Khouri H, Kiss H, Kothari SP, Madupu R, Nelson KE, Nelson WC, Paulsen I, Penn K, Ren QH, Rosovitz MJ, Selengut JD, Shrivastava S, Sullivan SA, Tapia R, Thompson LS, Watkins KL, Yang Q, Yu CH, Zafar N, Zhou LW, Kuske CR. 2009. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75:2046–2056. 10.1128/AEM.02294-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pankratov TA, Ivanova AO, Dedysh SN, Liesack W. 2011. Bacterial populations and environmental factors controlling cellulose degradation in an acidic Sphagnum peat. Environ. Microbiol. 13:1800–1814. 10.1111/j.1462-2920.2011.02491.x [DOI] [PubMed] [Google Scholar]
- 54.Pankratov TA, Kirsanova LA, Kaparullina EN, Kevbrin VV, Dedysh SN. 2011. Telmatobacter bradus gen. nov., sp. nov., a cellulolytic facultative anaerobe from subdivision 1 of the Acidobacteria and emended description of Acidobacterium capsulatum Kishimoto et al. 1991. Int. J. Syst. Evol. Microbiol. 62:430–437. 10.1099/ijs.0.029629-0 [DOI] [PubMed] [Google Scholar]
- 55.Kip N, van Winden JF, Pan Y, Bodrossy L, Reichart GJ, Smolders AJP, Jetten MSM, Damste JSS, Op den Camp HJM. 2010. Global prevalence of methane oxidation by symbiotic bacteria in peat-moss ecosystems. Nat. Geosci. 3:617–621. 10.1038/ngeo939 [DOI] [Google Scholar]
- 56.Op den Camp HJM, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland NK, Pol A, Dunfield PF. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1:293–306. 10.1111/j.1758-2229.2009.00022.x [DOI] [PubMed] [Google Scholar]
- 57.Vitousek PM, Menge DN, Reed SC, Cleveland CC. 2013. Biological nitrogen fixation: rates, patterns and ecological controls in terrestrial ecosystems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20130119. 10.1098/rstb.2013.0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osono T. 2007. Ecology of ligninolytic fungi associated with leaf litter decomposition. Ecol. Res. 22:955–974. 10.1007/s11284-007-0390-z [DOI] [Google Scholar]
- 59.Botha A. 2011. The importance and ecology of yeasts in soil. Soil Biol. Biochem. 43:1–8. 10.1016/j.soilbio.2010.10.001 [DOI] [Google Scholar]
- 60.Burdige DJ. 2007. Preservation of organic matter in marine sediments: controls, mechanisms, and an imbalance in sediment organic carbon budgets? Chem. Rev. 107:467–485. 10.1021/cr050347q [DOI] [PubMed] [Google Scholar]
- 61.Richardson AE, Simpson RJ. 2011. Soil microorganisms mediating phosphorus availability. Plant Physiol. 156:989–996. 10.1104/pp.111.175448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bridgham SD, Cadillo-Quiroz H, Keller JK, Zhuang Q. 2013. Methane emissions from wetlands: biogeochemical, microbial, and modeling perspectives from local to global scales. Global Change Biol. 19:1325–1346. 10.1111/gcb.12131 [DOI] [PubMed] [Google Scholar]
- 63.Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, Brown A, Allen N, Naylor J, Stange-Thomann N, DeArellano K, Johnson R, Linton L, McEwan P, McKernan K, Talamas J, Tirrell A, Ye WJ, Zimmer A, Barber RD, Cann I, Graham DE, Grahame DA, Guss AM, Hedderich R, Ingram-Smith C, Kuettner HC, Krzycki JA, Leigh JA, Li WX, Liu JF, Mukhopadhyay B, Reeve JN, Smith K, Springer TA, Umayam LA, White O, White RH, de Macario EC, Ferry JG, Jarrell KF, Jing H, Macario AJL, Paulsen I, Pritchett M, Sowers KR, Swanson RV, Zinder SH, Lander E, Metcalf WW, Birren B. 2002. The genome of M-acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532–542. 10.1101/gr.223902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu YC, Whitman WB. 2008. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 1125:171–189. 10.1196/annals.1419.019 [DOI] [PubMed] [Google Scholar]
- 65.Keller JK, Takagi KK. 2013. Solid-phase organic matter reduction regulates anaerobic decomposition in bog soil. Ecosphere 4:54. 10.1890/ES12-00382.1 [DOI] [Google Scholar]
- 66.Pester M, Knorr KH, Friedrich MW, Wagner M, Loy A. 2012. Sulfate-reducing microorganisms in wetlands—fameless actors in carbon cycling and climate change. Front. Microbiol. 3:72. 10.3389/fmicb.2012.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clymo RS. 1984. The limits to peat bog growth. Philos. Trans. R. Soc. B 303:605–654. 10.1098/rstb.1984.0002 [DOI] [Google Scholar]
- 68.Schmidt MWI, Torn MS, Abiven S, Dittmar T, Guggenberger G, Janssens IA, Kleber M, Kogel-Knabner I, Lehmann J, Manning DAC, Nannipieri P, Rasse DP, Weiner S, Trumbore SE. 2011. Persistence of soil organic matter as an ecosystem property. Nature 478:49–56. 10.1038/nature10386 [DOI] [PubMed] [Google Scholar]
- 69.Koven CD. 2013. Boreal carbon loss due to poleward shift in low-carbon ecosystems. Nat. Geosci. 6:452–456. 10.1038/ngeo1801 [DOI] [Google Scholar]
- 70.Hornibrook ERC, Longstaffe FJ, Fyfe WS. 1997. Spatial distribution of microbial methane production pathways in temperate zone wetland soils: stable carbon and hydrogen isotope evidence. Geochim. Cosmochim. Acta 61:745–753. 10.1016/S0016-7037(96)00368-7 [DOI] [Google Scholar]
- 71.Chanton JP, Bauer JE, Glaser PA, Siegel DI, Kelley CA, Tyler SC, Romanowicz EH, Lazrus A. 1995. Radiocarbon evidence for the substrates supporting methane formation within northern Minnesota peatlands. Geochim. Cosmochim. Acta 59:3663–3668. 10.1016/0016-7037(95)00240-Z [DOI] [Google Scholar]
- 72.Chanton JP, Glaser PH, Chasar LS, Burdige DJ, Hines ME, Siegel DI, Tremblay LB, Cooper WT. 2008. Radiocarbon evidence for the importance of surface vegetation on fermentation and methanogenesis in contrasting types of boreal peatlands. Global Biogeochem. Cycles 22:GB4022. 10.1029/2008GB003274 [DOI] [Google Scholar]
- 73.Corbett JE, Tfaily MM, Burdige DJ, Cooper WT, Glaser PH, Chanton JP. 2013. Partitioning pathways of CO2 production in peatlands with stable carbon isotopes. Biogeochemistry 114:327–340. 10.1007/s10533-012-9813-1 [DOI] [Google Scholar]
- 74.Borrel G, Lehours AC, Crouzet O, Jezequel D, Rockne K, Kulczak A, Duffaud E, Joblin K, Fonty G. 2012. Stratification of archaea in the deep sediments of a freshwater meromictic lake: vertical shift from methanogenic to uncultured archaeal lineages. PLoS One 7:e43346. 10.1371/journal.pone.0043346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, Stepanauskas R, Richter M, Kleindienst S, Lenk S, Schramm A, Jorgensen BB. 2013. Predominant archaea in marine sediments degrade detrital proteins. Nature 496:215–218. 10.1038/nature12033 [DOI] [PubMed] [Google Scholar]
- 76.Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sorensen KB, Anderson R, Fredricks HF, Elvert M, Kelly TJ, Schrag DP, Sogin ML, Brenchley JE, Teske A, House CH, Hinrichs KU. 2006. Heterotrophic archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U. S. A. 103:3846–3851. 10.1073/pnas.0600035103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bomberg M, Montonen L, Timonen S. 2010. Anaerobic Eury- and Crenarchaeota inhabit ectomycorrhizas of boreal forest Scots pine. Eur. J. Soil Biol. 46:356–364. 10.1016/j.ejsobi.2010.09.002 [DOI] [Google Scholar]
- 78.Hoj L, Olsen RA, Torsvik VL. 2008. Effects of temperature on the diversity and community structure of known methanogenic groups and other archaea in high Arctic peat. ISME J. 2:37–48. 10.1038/ismej.2007.84 [DOI] [PubMed] [Google Scholar]
- 79.Lehtovirta LE, Prosser JI, Nicol GW. 2009. Soil pH regulates the abundance and diversity of group 1.1c Crenarchaeota. FEMS Microbiol. Ecol. 70:367–376. 10.1111/j.1574-6941.2009.00748.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.