ABSTRACT
STING (stimulator of interferon genes) is known to control the induction of innate immune genes in response to the recognition of cytosolic DNA species, including the genomes of viruses such as herpes simplex virus 1 (HSV-1). However, while STING is essential for protection of the host against numerous DNA pathogens, sustained STING activity can lead to lethal inflammatory disease. It is known that STING utilizes interferon regulatory factor 3 (IRF3) and nuclear factor κB (NF-κB) pathways to exert its effects, although the signal transduction mechanisms remain to be clarified fully. Here we demonstrate that in addition to the activation of these pathways, potent induction of the Jun N-terminal protein kinase/stress-activated protein kinase (JNK/SAPK) pathway was similarly observed in response to STING activation by double-stranded DNA (dsDNA). Furthermore, TANK-binding kinase 1 (TBK1) associated with STING was found to facilitate dsDNA-mediated canonical activation of NF-κB as well as IRF3 to promote proinflammatory gene transcription. The triggering of NF-κB function was noted to require TRAF6 activation. Our findings detail a novel dsDNA-mediated NF-κB activation pathway facilitated through a STING-TRAF6-TBK1 axis and suggest a target for therapeutic intervention to plausibly stimulate antiviral activity or, alternatively, avert dsDNA-mediated inflammatory disease.
IMPORTANCE The IKK complex, which is composed of two catalytic subunits, IKKα and IKKβ, has been suggested to be essential for the activation of canonical NF-κB signaling in response to various stimuli, including cytokines (e.g., interleukin-1α [IL-1α] and tumor necrosis factor alpha [TNF-α]), Toll-like receptor (TLR) ligands (e.g., lipopolysaccharide [LPS]), and dsRNAs derived from viruses, or a synthetic analog. STING has been identified as a critical signaling molecule required for the detection of cytosolic dsDNAs derived from pathogens and viruses. However, little is known about how cytosolic dsDNA triggers NF-κB signaling. In the present study, we demonstrate that TBK1, identified as an IKK-related kinase, may predominantly control the activation of NF-κB in response to dsDNA signaling via STING through the IKKαβ activation loop. Thus, our results establish TBK1 as a downstream kinase controlling dsDNA-mediated IRF3 and NF-κB signaling dependent on STING.
INTRODUCTION
The detection of cytosolic nucleic acids derived from pathogens such as herpes simplex virus 1 (HSV-1) is a central cellular host defense strategy which triggers essential innate immune responses required for protection of the host. However, inappropriate recognition of self nucleic acids, predominantly involving double-stranded DNA (dsDNA) species, can result in debilitating inflammatory diseases, such as systemic lupus erythematosus (SLE) (1–3). How the cell triggers the induction of innate immune genes in response to nucleic acids derived from microbes, such as DNA viruses, intracellular bacteria, and parasites, or in response to self DNA, has not been elucidated fully. However, we previously reported that an endoplasmic reticulum (ER)-associated multiple-transmembrane protein, STING (stimulator of interferon genes; also referred to as TMEM173, MPYS, ERIS, and MITA), functions as an essential innate immune signaling molecule required for triggering dsDNA-mediated gene induction (4–8). STING may directly associate with stimulatory ligands, which include dsDNA, as well as with cyclic dinucleotides (CDNs) (9), produced directly by pathogens or metabolized from dsDNA by cyclic-GMP-AMP (cGAMP) synthase (cGAS) (10, 11). The repertoire of cytosolic dsDNA-dependent induced genes, including those for type I interferon (IFN) and proinflammatory genes such as those for interleukin-6 (IL-6), IL-1β, and tumor necrosis factor alpha (TNF-α), is known to require the IRF3 and NF-κB pathways (12). In resting cells, NF-κB remains in the cytoplasm through its association with inhibitory IκB. A central step in the activation of NF-κB is phosphorylation of inhibitory IκB by the IκB kinases (IKKs) α and β. Phosphorylation of IκB results in its degradation by the proteasome, followed by the release and nuclear translocation of NF-κB transcription factor subunits, such as p50 and NF-κBp65/RelA (13). However, recent evidence suggests that NF-κB activity can also be controlled by alternate kinases. For example, cells lacking glycogen synthase kinase 3β (GSK3β), the IKK-related kinase TANK-binding kinase 1 (TBK1; also referred to as NAK or T2K) or inducible IκB kinase (IKKi; also referred to as IKKε), or protein kinase ζ (PKC ζ) while exhibiting normal IκB degradation in response to select stimuli are evidently impaired in the activation of NF-κB-dependent gene transcription (14–16). AKT/phosphatidylinositol 3-kinase (PI3K) has also been reported to be involved in NF-κBp65/RelA activation through IKKβ activation (17). TBK1 was originally identified in the context of its ability to regulate NF-κB in vitro (14, 18, 19). However, subsequent studies using mouse embryonic fibroblast cells (MEFs) lacking TBK1 or IKKi did not strongly support this evidence (20, 21). Rather, cells deficient in TBK1 indicated that TBK1 is an essential factor for the production of type I IFN through control of the IRF3 pathway. The complexity of this regulation is exemplified by recent progress that has identified four serine residues in the NF-κBp65/RelA subunit, namely, Ser276, Ser311, Ser529, and Ser536, that are inducibly phosphorylated through mechanisms regulated by cytokines such as TNF-α or IL-1 (13).
In this study, we demonstrate that TBK1 may predominantly control the activation of NF-κBp65/RelA following cellular detection of cytosolic dsDNA. The activation of NF-κBp65, including the phosphorylation of Ser536, nuclear translocation, and the expression of NF-κB target genes by stimulation with dsDNA, was significantly reduced in TBK1-deficient or RNA interference (RNAi)-silenced MEFs. We also demonstrate that TRAF6 plays a key role in facilitating STING-mediated NF-κB activation. Thus, our results establish TBK1 as a downstream kinase controlling dsDNA-mediated IRF3 and NF-κB signaling pathways dependent upon STING.
MATERIALS AND METHODS
Cells.
STING-deficient MEFs were described previously (4). Immortalized MEFs derived from IKKα-, IKKβ-, and TBK1-deficient mice were described previously (22). MEFs reconstituted with Myc-tagged human TBK1 (hTBK1) were generated by the transduction of retroviral vector systems. 293T cells were obtained from the ATCC. All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Gemini Bio-Products) and were cultured at 37°C in a humidified atmosphere with 5% CO2.
Antibodies, reagents, and plasmids.
A rabbit polyclonal antibody against STING was described previously (4). Other antibodies were purchased from the indicated sources. Antibodies specific to phosphorylated IRF3 (Ser396) (4D4G), phosphorylated NF-κBp65 (Ser536) (93H1), NF-κBp65 (D14E12), extracellular signal-regulated kinase 1/2 (ERK1/2) (137F5), phosphorylated ERK1/2 (Thr202/Tyr204) (D13.14.4E), p38 (D13E1), phosphorylated p38 (Thr180/Tyr182) (D3F9), stress-activated protein kinase/Jun N-terminal protein kinase (SAPK/JNK) (56G8), phosphorylated SAPK/JNK (Thr183/Tyr185) (81E11), phosphorylated c-Jun (Ser63) (54B3), IκBα (L35A5), IKKα and IKKβ (2C8), and phosphorylated TBK1/NAK (Ser172) (D52C2) were purchased from Cell Signaling. TBK1/NAK (EP611Y), HSV-1 ICP4 (ab6514), and HSV-1 glycoprotein D (gD) (ab6507) antibodies were purchased from Abcam. TRAF3 (M-20) and TRAF6 antibodies were purchased from Santa Cruz Biotechnology and Stratagene, respectively. Poly(I·C), 5,6-dimethylxanthenone-4-acetic acid (DMXAA), canonical 3′-5′ cyclic-GMP-AMP (cGAMP), and poly(dA-dT) were purchased from Invivogen. dsDNA90 was prepared as follows: a single-stranded DNA (ssDNA) sense strand (5′-TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACATACATACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACA-3′) was annealed to an ssDNA90 antisense sequence. The Quantikine ELISA (enzyme-linked immunosorbent assay) murine IL-6 immunoassay and VeriKine mouse IFN-β ELISA kits were purchased from R&D Systems and PBL Biomedical Laboratories, respectively. The phosphorylation status of NF-κBp65 was determined by use of a PathScan phospho-NF-κB p65 (Ser536) sandwich ELISA kit (Cell Signaling). DNA fragments encoding murine TRAF3 and TRAF6 were amplified by reverse transcription-PCR (RT-PCR) from total RNA of primary MEFs and cloned into pcDNA3.1-A-Myc-His (Invitrogen). The C-terminal deletion mutant of TRAF3 (TRAF3 mut) was amplified by a PCR using AccuPrimePfx SuperMix (Invitrogen) and subcloned into pcDNA3.1-A-Myc-His. The construction of hemagglutinin (HA)-tagged murine STING (STING-HA) and FLAG-tagged human TBK1 (TBK1-FL) was performed as described previously (4).
RNA interference.
MEFs were transfected with corresponding small interfering RNAs (siRNAs) (ON-Target Plus siRNA-SMARTpool; Dharmacon) by use of Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Seventy-two hours after transfection, cells were used for further experiments. Experiments were done in triplicate, using more than one siRNA for each gene.
Reporter analysis.
293T cells were transfected with a plasmid for STING-HA in combination with empty vector (EV), pcDNA3.1-murine TRAF3-Myc-His (TRAF3-His), or pcDNA3.1-murine TRAF6-Myc-His (TRAF6-His), together with a reporter plasmid carrying the luciferase gene under the control of the IFN-β, NF-κB, pRDIII, or ISRE promoter. Luciferase activity was determined at 24 h posttransfection (Promega).
Immunoblotting and cell fractionation.
Cells were seeded onto a 24-well tissue culture plate for 24 h and stimulated with dsDNA90 (10 μg/ml), poly(dA-dT) (5 μg/ml), poly(I·C) (5 μg/ml), or DMXAA (200 μM) for the indicated time. Cell lysis was performed with RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% deoxycholate [DOC], and protease inhibitors) at 4°C for 60 min. The samples were boiled in 20 μl of SDS sample buffer and then subjected to SDS-10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Thermo Scientific). These membranes were blocked with Tris-buffered saline with Tween 20 (TBST) containing 5% skim milk at room temperature for 1 h and then incubated with corresponding antibodies. Membranes were visualized with Supersignal West Femto maximum-sensitivity substrate (Thermo Scientific). Cell fractionation assays were performed by using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific) according to the manufacturer's instructions. IκBα (Cell Signaling) and RFC1 (Abcam) or histone H3 (Abcam) were used as cytoplasmic and nuclear markers, respectively.
Real-time PCR.
Total RNA was prepared from each cell line by using an RNeasy minikit (Qiagen). First-strand cDNA was synthesized by using a QuantiTech reverse transcription kit (Qiagen). Real-time PCR was performed by using Sybr green (Finnzyme; Thermo Scientific) for the murine TRAF3 (5′-AAGCATCATCAAAGACAAGG-3′ and 5′-ATTCCGACAGTAGACCTGAA-3′), murine TRAF6 (5′-TGTTCTTAGCTGCTGGGGTGT-3′ and 5′-GAAGGAGCTGGAGAGAGGTTCC-3′), and murine IKKβ (5′-GGAGTACTGCCAAGGAGGAGAT-3′ and 5′-ACAGGCTGCCAGTTAGGGAGGAAG-3′) genes and the TaqMan gene expression assay (Applied Biosystems) for innate immune genes and inflammatory cytokine genes (for IFN-β, primer Mm010439546; for CXCL10, primer Mm00445235; for Ccl5, primer Mm01302427; for Ccl2, primer Mm99999056; and for IL-6, primer Mm00446190).
Confocal microscopy.
Silencing of TRAF3 or TRAF6 in STING-deficient MEFs reconstituted with HA-tagged murine STING was performed with dsDNA90 for 3 h. MEFs were fixed in 4% paraformaldehyde for 10 min at room temperature and permeabilized with 0.5% Triton X-100. After blocking with 2% bovine serum albumin (BSA) in phosphate-buffered saline containing 0.1% Tween 20 (PBST) for 2 h at room temperature, the cells were incubated with anti-HA mouse monoclonal antibody (HA-7) (Sigma) and anti-NF-κBp65 rabbit monoclonal antibody (D14E12) (Cell Signaling) for 1 h at room temperature, followed by incubation with anti-mouse IgG conjugated with Alexa Fluor 488 and anti-rabbit IgG conjugated with Alexa Fluor 555, respectively. The stained cells were analyzed by use of an SP5 confocal microscope (Leica) at the University of Miami Imaging Core Facility.
Statistics.
Results are expressed as means ± standard deviations (SD). Student's t test was used to analyze data. P values of ≤0.05 were considered to denote significance.
RESULTS
MAPKs and NF-κB are robustly activated in MEFs following exposure to STING ligands.
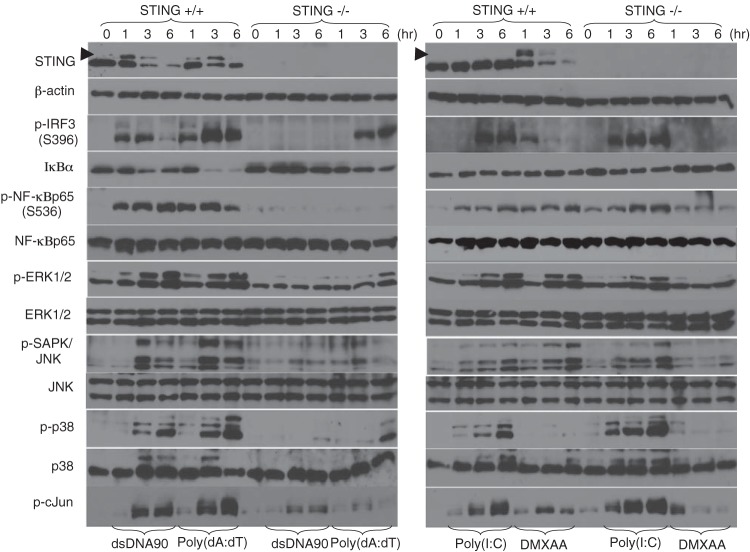
Previous reports have shown that the activation of NF-κB is reduced in MEFs or macrophages in response to STING-dependent dsDNA signaling (3, 5, 23, 24). To further evaluate the mechanisms of STING function, we analyzed select signaling profiles in STING−/− and wild-type (STING+/+) MEFs in response to cytosolic dsDNA. We observed that the transduction of dsDNA90 into wild-type MEFs induced the phosphorylation and subsequent degradation of STING after approximately 3 to 6 h (Fig. 1, top left panel). We also observed robust phosphorylation of IRF3 (on Ser396) and NF-κBp65/RelA (on Ser536) (Fig. 1, left panels). Similarly, the activation of mitogen-activated protein kinases (MAPKs), including ERK1/2, JNK, c-Jun, and p38, was also noted following the transduction of dsDNA90 into wild-type MEFs (Fig. 1, left panels). These events were greatly reduced or eliminated in STING−/− MEFs (Fig. 1, left panels). Treatment of cells with DMXAA, which was recently classified as a novel STING activator (25, 26), also exhibited an effect similar to that of dsDNA, except for p38 MAPK activation (Fig. 1, right panels). The transduction of canonical 3′-5′ cGAMP, which is known as a new STING ligand produced by cGAS (10, 11), also activated STING-dependent signaling in MEFs (see Fig. S1A in the supplemental material). Poly(dA-dT)-mediated phosphorylation of IRF3 or ERK1/2 was partially retained in STING-deficient MEFs at 3 to 6 h postransduction (Fig. 1, left panels), likely due to the involvement of RIG-I-dependent sensing through the cytosolic DNA-dependent RNA polymerase III response (27). In contrast to known STING ligands, the use of poly(I·C), which does not trigger STING activity, showed robust activation of IRF3, NF-κBp65, and MAPKs, in a STING-independent manner (Fig. 1, right panels). Cell fractionation analysis revealed that dsDNA-mediated IRF3 and NF-κBp65 nuclear translocation was substantially reduced in STING-deficient MEFs (see Fig. S1B). Similarly, quantitative RT-PCR analysis established that the STING pathway controlled dsDNA- and DMXAA-mediated IFN-β, CXCL10, Ccl5, and IL-6 induction, which was not stimulated by poly(I·C) (see Fig. S1C). However, poly(dA-dT) was able to trigger RIG-I-dependent gene expression (see Fig. S1C).
FIG 1.
STING ligand-mediated signaling response in MEFs. Primary MEFs (1 × 105 cells/well) derived from wild-type (STING+/+) or STING-deficient (STING−/−) mice were stimulated with 10 μg/ml of dsDNA90, 5 μg/ml of poly(dA-dT), 5 μg/ml of poly(I·C), or 200 μM DMXAA for the indicated times. The expression levels of IκBα, STING, IRF3 phosphorylated at Ser396 (p-IRF3), NF-κBp65 phosphorylated at Ser536 (p-NF-κBp65), NF-κBp65, ERK1/2 phosphorylated at Thr202/Tyr204 (p-ERK1/2), ERK1/2, SAPK/JNK phosphorylated at Thr183/Tyr185 (p-SAPK/JNK), JNK1/2, p38 phosphorylated at Thr180/Tyr182 (p-p38), p38, c-Jun phosphorylated at Ser63 (p-cJun), and β-actin were determined by immunoblotting. The phosphorylation state of STING is indicated by an arrowhead.
Previously, STING was shown to be essential for host defense against HSV-1 infection (5). HSV-1 infection has also been shown to activate NF-κBp65 through multiple pathways, including via IκB kinases such as IKKα and IKKβ, through PKR, and through the Toll-like receptor 2 (TLR2)–MyD88 axis (28–30). To further elucidate the role of STING in HSV-1-mediated NF-κBp65 activation, MEFs derived from wild-type (STING+/+) or STING-deficient (STING−/−) MEFs were stimulated with purified HSV-1 virions or dsDNA representing the genome of HSV-1 at the indicated times, and cell lysates were analyzed by immunoblotting. Purified HSV-1 virions or dsDNA derived from HSV-1 induced phosphorylation of IRF3 and TBK1 that was totally dependent upon STING (see Fig. S2 in the supplemental material). Our data indicate that STING is required for the efficient induction of SAPK/JNK signaling, as well as IRF3 and NF-κB signaling, in response to HSV-1 or cytosolic DNA.
IKKαβ is involved in dsDNA-mediated NF-κB activation in MEFs.
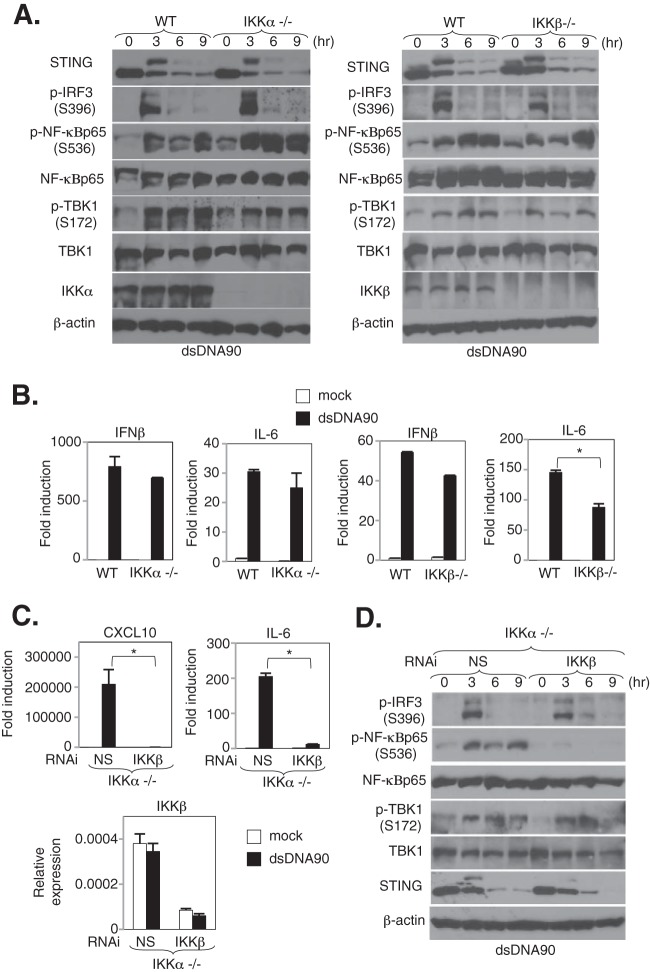
The IKK complex, which is composed of two catalytic subunits, IKKα and IKKβ, and a regulatory subunit, IKKγ/NEMO, has been suggested to be essential for the activation of canonical NF-κB signaling in response to various stimuli, including cytokines (e.g., IL-1α and TNF-α), TLR ligands (e.g., lipopolysaccharide [LPS] and CpG oligonucleotides), and RIG-I-like receptor (RLR) ligands (e.g., RNA viruses or a synthetic analog of dsRNA) (13, 31). However, little is known about how cytosolic dsDNA triggers NF-κB signaling. Thus, we further evaluated the mechanisms of dsDNA-mediated NF-κB activation in MEFs lacking several NF-κB-related genes. In IKKα-deficient MEFs treated with dsDNA90, we observed that the activation of NF-κBp65, as determined by analyzing phosphorylation levels, nuclear translocation, and the transcription of cytokines such as IFN-β and IL-6, remained intact (Fig. 2A and B). On the other hand, we observed a partial reduction of IL-6 (*, P < 0.01) but not IFN-β expression and a slight reduction of NF-κBp65 phosphorylation 3 to 6 h following stimulation in IKKβ-deficient MEFs (Fig. 2A and B). Moreover, to elucidate the effects of both IKKα and IKKβ on dsDNA-mediated signaling responses, IKKα-deficient MEFs were transduced with nonspecific (NS) or IKKβ siRNA for 72 h, followed by dsDNA90 stimulation at the indicated times. In contrast to the evaluation of single-knockout MEFs derived from IKKα- or IKKβ-deficient mice, the depletion of IKKβ in IKKα-deficient MEFs showed substantial reductions of both NF-κBp65 phosphorylation and NF-κBp65-mediated gene expression, such as that of IL-6 and CXCL10, after stimulation with dsDNA90 (Fig. 2C and D). These results may suggest that classical IκB kinases, such as IKKα and IKKβ, are involved in STING-dependent, dsDNA-mediated NF-κBp65 activation in MEFs.
FIG 2.
IKKαβ is involved in dsDNA-mediated NF-κBp65 activation in MEFs. (A) Immortalized MEFs derived from wild-type (WT) and IKKα- or IKKβ-deficient mice were stimulated with 10 μg/ml of dsDNA90 for the indicated times. The expression levels of STING, TBK1, TBK1 phosphorylated at Ser172 (p-TBK1), IRF3 phosphorylated at Ser396 (p-IRF3), NF-κBp65 phosphorylated at Ser536 (p-NF-κBp65), NF-κBp65, IKKα, IKKβ, and β-actin were determined by immunoblotting. (B) WT and IKKα- and IKKβ-deficient MEFs were stimulated with 10 μg/ml of dsDNA90 for 6 h, the total RNAs were extracted from these cells, and the expression levels of mRNAs for IL-6 and IFN-β were determined by real-time PCR. Real-time PCR data were normalized to the amount of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. *, P < 0.05 (Student's t test). Error bars indicate standard deviations (SD). (C) IKKα-deficient MEFs subjected to RNAi by use of nonspecific (NS) and IKKβ siRNAs were stimulated with 5 μg/ml of dsDNA90 for 6 h, the total RNAs were extracted from these cells, and the expression levels of mRNAs for IL-6, CXCL10, and IKKβ were determined by real-time PCR. Real-time PCR data were normalized to the amount of GAPDH mRNA. *, P < 0.05 (Student's t test). Error bars indicate SD. (D) IKKα-deficient MEFs subjected to RNAi by use of NS and IKKβ siRNAs were stimulated with 5 μg/ml of dsDNA90 for the indicated times, and then the signaling response was determined by immunoblotting using specific antibodies for STING, IRF3 phosphorylated at Ser396 (p-IRF3), TBK1 phosphorylated at Ser172 (p-TBK1), NF-κBp65 phosphorylated at Ser536 (p-NF-κBp65), NF-κBp65, and TBK1, with β-actin serving as a loading control.
TBK1 is involved in dsDNA-mediated NF-κB activation in MEFs.
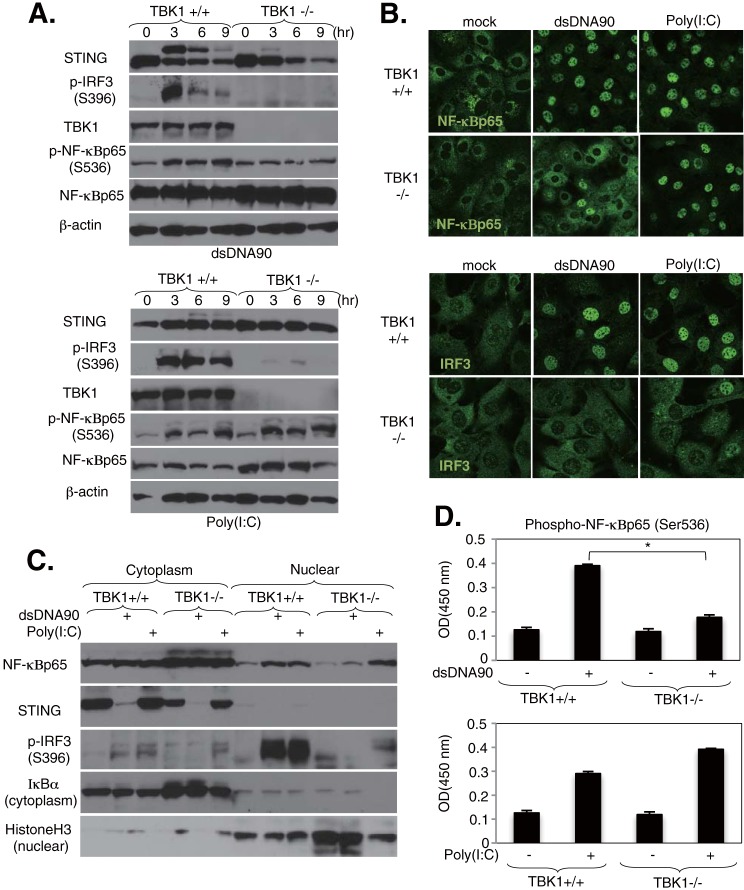
Next, we examined whether another IKK-related kinase, such as TBK1, may be involved in dsDNA-mediated NF-κBp65 activation in MEFs. This was considered a possibility because TBK1 is known to activate NF-κB in vitro and in vivo (14, 19, 32, 33). Immortalized MEFs derived from wild-type (TBK1+/+) or TBK1-deficient (TBK1−/−) mice were stimulated with dsDNA90 or poly(I·C) at the indicated times, and cell lysates were analyzed by immunoblotting. Surprisingly, the phosphorylation of NF-κBp65 in response to dsDNA90 did not significantly increase in TBK1-deficient MEFs compared with wild-type MEFs (Fig. 3A). In contrast, the same cells exposed to poly(I·C) stimulation exhibited comparable levels of NF-κBp65 activity. dsDNA90- and poly(I·C)-induced IRF3 phosphorylation was totally dependent on TBK1, as previously reported (Fig. 3A). The activation of MAPKs in TBK1-deficient MEFs following stimulation with dsDNA90 or poly(I·C) was comparable to that in wild-type MEFs (see Fig. S3A in the supplemental material). We also confirmed that ectopic expression of TBK1 in 293T cells activated the induction of both the IRF3 and NF-κB pathways, but not MAPK signaling responses (see Fig. S3B), suggesting that TBK1 is not involved in dsDNA-mediated MAPK activation. Consistent with our immunoblot analysis, dsDNA90- but not poly(I·C)-induced nuclear translocation of NF-κBp65 was substantially reduced in TBK1-deficient MEFs (Fig. 3B). Alternatively, dsDNA90- and poly(I·C)-induced nuclear translocation of IRF3 was totally dependent upon TBK1 (Fig. 3B). Further support showing that TBK1 may be involved in dsDNA-mediated NF-κB activation in MEFs was obtained by cell fractionation analysis and ELISA. The presence of NF-κBp65 in the nuclear fraction of MEFs following stimulation with dsDNA90 was substantially reduced in TBK1-deficient cells, while that after poly(I·C) stimulation remained comparable (Fig. 3C). In addition, quantitative ELISA also showed a significant reduction of NF-κBp65 phosphorylation in TBK1-deficient MEFs after stimulation with dsDNA90, while poly(I·C) stimulation did not significantly alter the level (Fig. 3D). We also examined the effects of IKKi/IKKε, another IKK-related kinase that is involved in IRF3 and NF-κB signaling, on STING-mediated signaling responses in MEFs. In IKKi/IKKε knockdown MEFs treated with dsDNA90, we observed that the activation of NF-κBp65, as determined by analyzing phosphorylation levels and the transcription of cytokines such as IL-6 and CXCL10, remained intact (see Fig. S4).
FIG 3.
TBK1 regulates dsDNA-mediated NF-κB activation in MEFs. (A) Immortalized MEFs derived from wild-type (TBK1+/+) or TBK1-deficient (TBK1−/−) mice were stimulated with 10 μg/ml of dsDNA90 or 5 μg/ml of poly(I·C) for the indicated times. The expression levels of TBK1, STING, IRF3 phosphorylated at Ser396 (p-IRF3), NF-κBp65 phosphorylated at Ser536 (p-NF-κBp65), NF-κBp65, and β-actin were determined by immunoblotting. (B) TBK1+/+ and TBK1−/− MEFs were stimulated with 10 μg/ml of dsDNA90 or 5 μg/ml of poly(I·C) for 3 h, and then the cells were stained with antibodies against NF-κBp65 or IRF3. (C) Detection of nuclear translocation of NF-κBp65 by fractionation assay. TBK1+/+ and TBK1−/− MEFs were stimulated with 10 μg/ml of dsDNA90 or 5 μg/ml of poly(I·C) for 3 h, and then cell lysates were separated into cytosolic and nuclear fractions. Each fraction was concentrated and subjected to immunoblotting with the indicated antibodies. (D) Quantitative analysis of NF-κBp65 phosphorylated at Ser536 by ELISA. TBK1+/+ and TBK1−/− MEFs were stimulated with 10 μg/ml of dsDNA90 or 5 μg/ml of poly(I·C) for 3 h, and then endogenous levels of NF-κBp65 phosphorylated at Ser536 were determined by ELISA. *, P < 0.05 (Student's t test). Error bars indicate SD. OD, optical density.
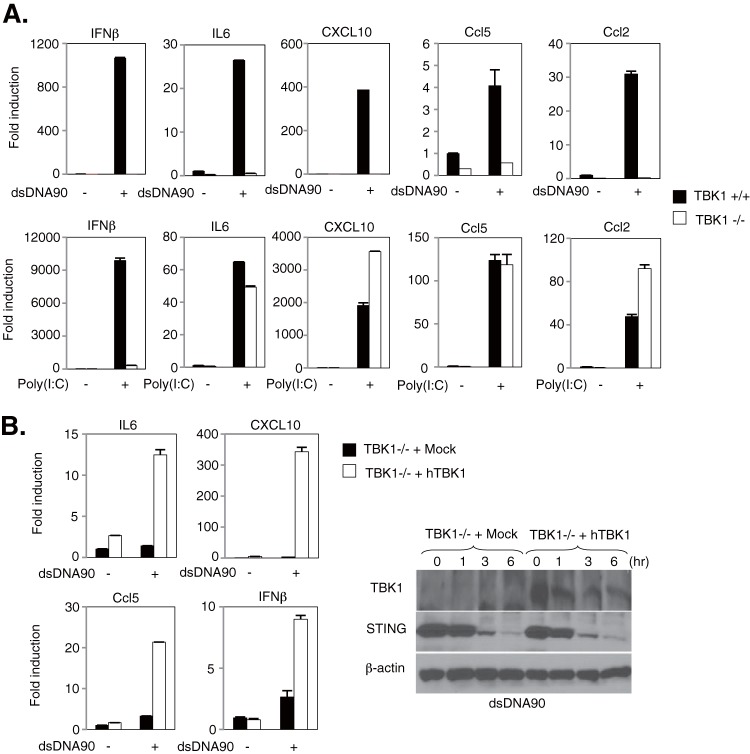
To further evaluate the role of NF-κBp65 activation in dsDNA-mediated signaling, wild-type and TBK1-deficient MEFs were stimulated with dsDNA90 or poly(I·C), and the expression of NF-κB-related cytokines was measured by quantitative RT-PCR. Induction of IL-6, CXCL10, Ccl5, and Ccl2 following stimulation with dsDNA90, but not poly(I·C), was completely abolished in TBK1-deficient MEFs, whereas IFN-β induction was abolished regardless of the type of nucleic acid stimulation (Fig. 4A). These results may imply that the expression of poly(I·C)-mediated, NF-κB-related cytokines may be regulated by the classical IκB kinases rather than through TBK1. Finally, we carried out transcomplementation experiments in TBK1−/− MEFs transduced with retroviral vectors encoding Myc-tagged human TBK1 (hTBK1) and then evaluated the expression of NF-κB-related cytokines by quantitative RT-PCR. We observed that TBK1-deficient MEFs transduced with hTBK1 showed rescued expression of some NF-κB-related cytokines, in addition to IFN-β, following stimulation with dsDNA90, while the expression in mock-transduced cells was not restored (Fig. 4B). Collectively, these results suggest that TBK1 dominantly regulates dsDNA-mediated NF-κBp65 activation through the IKKαβ activation loop in MEFs.
FIG 4.
Regulation of dsDNA-mediated gene expression by TBK1 in MEFs. (A) TBK1+/+ and TBK1−/− MEFs were stimulated with 10 μg/ml of dsDNA90 or 5 μg/ml of poly(I·C) for 6 h, the total RNAs were extracted from these cells, and the expression levels of mRNAs for IFN-β, IL-6, CXCL10, Ccl5, and Ccl2 were determined by qRT-PCR. (B) TBK1−/− MEFs reconstituted by transduction with retroviral vectors encoding Myc-tagged human TBK1 (hTBK1) or mock-transduced cells (Mock) were stimulated with 10 μg/ml of dsDNA90 for 6 h, the total RNAs were extracted from these cells, and the expression levels of mRNAs for IFN-β, IL-6, CXCL10, and Ccl5 were determined by qRT-PCR. Data were normalized to the amount of GAPDH mRNA. Error bars indicate SD. The expression levels of TBK1 and STING were determined by immunoblotting using specific antibodies, with β-actin serving as a loading control.
DMXAA similarly activates NF-κB through TBK1.
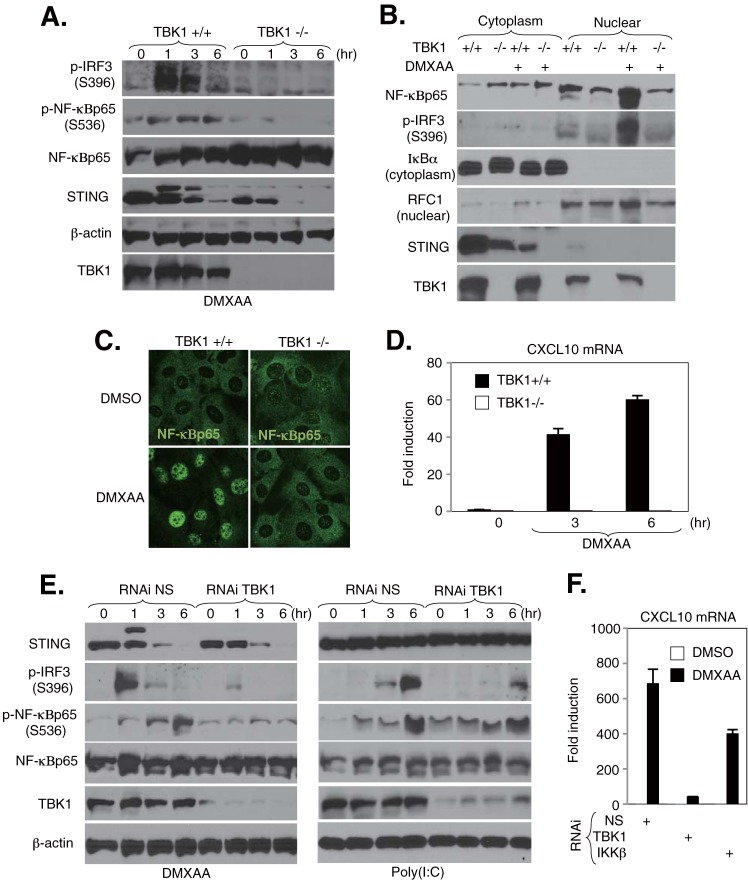
Previous studies have shown that DMXAA acts as a potent IFN inducer through the TBK1-IRF3 axis in MEFs and murine macrophages (34). Recently, it was clarified that DMXAA achieves this through functioning as a STING ligand (25, 26). To evaluate whether DMXAA similarly activates NF-κB through TBK1, we examined the effects of DMXAA-mediated NF-κB activation in MEFs lacking TBK1 as described for Fig. 3 and 4. Similar to our previous observations, we noted that TBK1-deficient MEFs exhibited substantially reduced DMXAA-mediated activation of both IRF3 and NF-κBp65 phosphorylation (Fig. 5A). Moreover, consistent with our immunoblotting analysis, we observed a reduction in the nuclear translocation of NF-κBp65 in TBK1-deficient MEFs in response to DMXAA treatment by cell fractionation and confocal imaging (Fig. 5B and C). In addition, abolishment of CXCL10 mRNA expression was observed in response to DMXAA in MEFs lacking TBK1 (Fig. 5D). Next, to confirm the effect of TBK1 on DMXAA-mediated NF-κB activation, primary MEFs were transduced with nonspecific (NS) or TBK1 siRNA for 72 h, followed by DMXAA or poly(I·C) stimulation at the indicated times. Cell lysates were analyzed by immunoblotting. Knockdown of TBK1 resulted in a substantial reduction in DMXAA-mediated NF-κBp65 phosphorylation, while levels of NF-κBp65 phosphorylation in cells stimulated with poly(I·C) remained intact (Fig. 5E). DMXAA- and poly(I·C)-induced IRF3 phosphorylation was totally dependent upon TBK1 (Fig. 5E). A significant reduction in the level of CXCL10 mRNA in response to DMXAA was also observed in MEFs treated with siRNA specific to TBK1 (Fig. 5F). These results confirm that DMXAA activates the NF-κBp65 signaling response via TBK1 in MEFs, similar to dsDNA.
FIG 5.
TBK1 regulates DMXAA-mediated NF-κB activation in MEFs. (A) TBK1+/+ and TBK1−/− MEFs were stimulated with 200 μM DMXAA for the indicated times. The expression levels of TBK1, STING, IRF3 phosphorylated at Ser396 (p-IRF3), NF-κBp65 phosphorylated at Ser536 (p-NF-κBp65), NF-κBp65, and β-actin were determined by immunoblotting. (B) Detection of nuclear translocation of NF-κBp65 by fractionation assay. TBK1+/+ and TBK1−/− MEFs were stimulated with 200 μM DMXAA for 3 h, and then cell lysates were separated into cytosolic and nuclear fractions. Each fraction was concentrated and subjected to immunoblotting with the indicated antibodies. (C) TBK1+/+ and TBK1−/− MEFs were stimulated with 200 μM DMXAA for 3 h, and then cells were stained with antibodies against NF-κBp65. (D) TBK1+/+ and TBK1−/− MEFs were stimulated with 200 μM DMXAA for 6 h, the total RNAs were extracted from these cells, and the expression levels of mRNA for CXCL10 were determined by qRT-PCR. Data were normalized to the levels of GAPDH mRNA. (E) MEFs subjected to RNAi by use of nonspecific (NS) and TBK1 siRNAs were stimulated with 200 μM DMXAA or 5 μg/ml of poly(I·C) for the indicated times. The cell lysates were subjected to immunoblotting with the indicated antibodies. (F) MEFs subjected to RNAi by use of NS, TBK1, and IKKβ siRNAs were stimulated with 200 μM DMXAA for 6 h, the total RNAs were extracted from these cells, and the expression levels of mRNA for CXCL10 were determined by qRT-PCR. Data were normalized to the levels of GAPDH mRNA.
dsDNA-mediated NF-κBp65 activation is essential for antiviral activity.
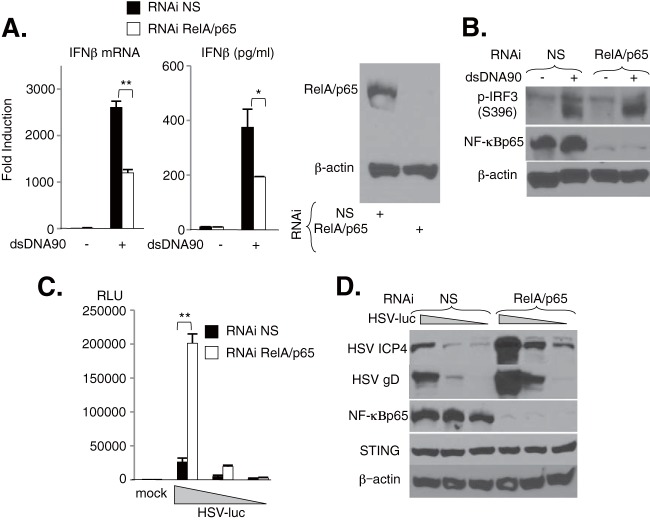
To elucidate the antiviral importance of STING-dependent activation of NF-κB, primary MEFs were treated with nonspecific (NS) or NF-κBp65/RelA siRNA for 72 h. Cells were then transfected with dsDNA90 or infected with HSV-1 expressing luciferase (HSV-luc), and IFN-β production or viral propagation was measured. Silencing of NF-κBp65/RelA in MEFs reduced the production of IFN-β mRNA and protein in response to dsDNA90 transfection by 50% (Fig. 6A), but levels of IRF3 phosphorylation remained approximately the same (Fig. 6B). These results may suggest a contribution of NF-κBp65/RelA to STING-dependent, dsDNA-mediated IFN production. Moreover, suppression of NF-κBp65/RelA also facilitated the replication of HSV-luc in MEFs (Fig. 6C). Immunoblot analysis further confirmed that the expression of select viral proteins, such as ICP4 and glycoprotein D (gD), was increased in NF-κBp65/RelA-lacking cells compared to MEFs treated with NS siRNA (Fig. 6D). Thus, activation of NF-κBp65/RelA by STING induces an anti-DNA viral host defense.
FIG 6.
Role of NF-κBp65 in dsDNA-mediated IFN production in MEFs. (A) MEFs subjected to RNAi by use of nonspecific (NS) and NF-κBp65 (RelA/p65) siRNAs were stimulated with 5 μg/ml of dsDNA90, and the expression levels of IFN-β mRNA and production of IFN-β in the supernatant were determined by real-time PCR and ELISA, respectively. Real-time PCR data were normalized to the amount of GAPDH mRNA. Silencing of RelA/p65 expression was demonstrated by immunoblotting, with β-actin serving as a loading control. (B) MEFs subjected to RNAi by use of NS and RelA/p65 siRNAs were stimulated with 5 μg/ml of dsDNA90 for 3 h, and the expression levels of IRF3 phosphorylated at Ser396 (p-IRF3), NF-κBp65, and β-actin were determined by immunoblotting. (C) MEFs subjected to RNAi by use of NS and RelA/p65 siRNAs were inoculated with HSV-luc at a multiplicity of infection (MOI) of 1, 0.2, or 0.04 PFU/ml for 24 h, and then cell lysates were analyzed by luciferase assay. RLU, relative light units. (D) Following silencing of RelA/p65, the expression levels of viral proteins, such as ICP4 and gD, or of endogenous STING were demonstrated by immunoblotting, with β-actin serving as a loading control. **, P < 0.01; *, P < 0.05 (Student's t test). Error bars indicate SD.
TRAF3 and TRAF6 predominantly facilitate STING-mediated innate immune signaling.
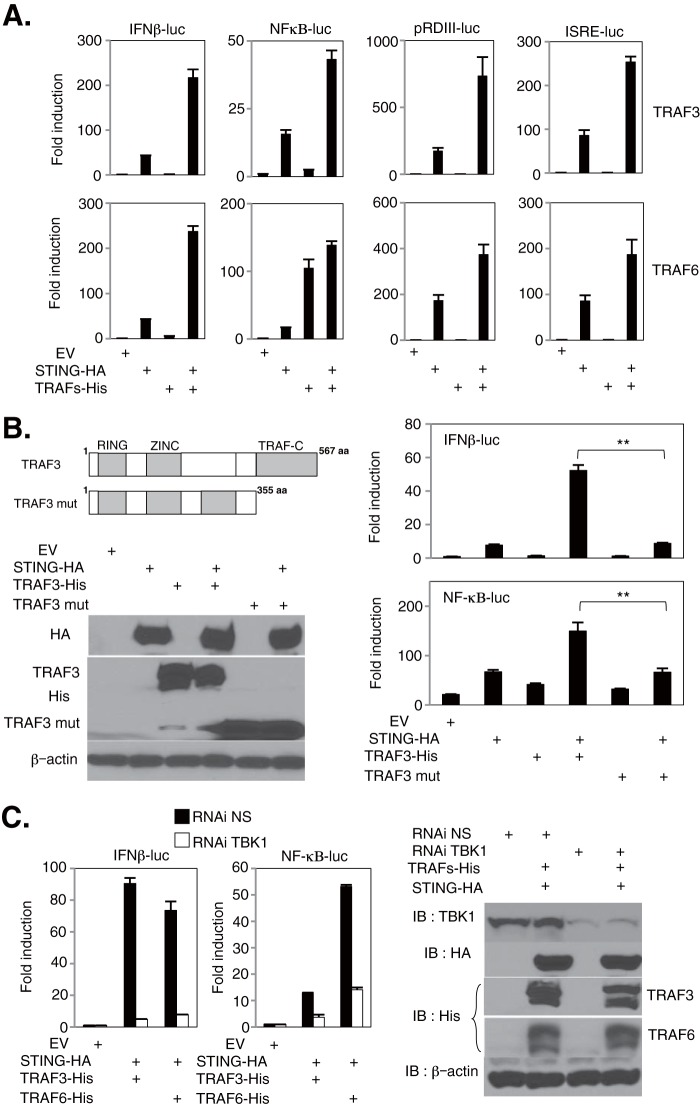
Members of the TNF receptor-associated factor (TRAF) family are known to facilitate NF-κB and IRF activity (35). For example, it has been reported that TRAF3 and TRAF6 are involved in dsRNA- or dsDNA-mediated signaling in a variety of cell types (36–39). However, it is unclear whether these TRAF members participate in STING-dependent signaling (7, 40). Thus, to determine the involvement of TRAF molecules in STING function, 293T cells were cotransfected with HA-tagged murine STING (STING-HA), members of the TRAF family (TRAFs-His), and reporter plasmids encoding luciferase genes under the control of the IFN-β, ISRE, pRDIII, or NF-κB promoter. This analysis revealed that the expression of TRAF3 and TRAF6 enhanced promoter activity when the proteins were coexpressed with STING, except for that of the NF-κB promoter in cells coexpressing STING and TRAF6 alone (Fig. 7A). We hypothesized that the observed diminutive additive effect of the NF-κB promoter coexpressed with STING and TRAF6 was due to TRAF6 already activating the NF-κB pathway (35). We did not observe the same induction with TRAF1 or -2 (data not shown). Recently, it was also reported that TRAF4 plays a role as a substrate for IKKα in the NOD-dependent signaling pathway (41). However, single IKKα knockout MEFs did not appear to be involved in dsDNA-mediated IRF3 or NF-κB activation (Fig. 2A and B). We also noticed that the enhancement of IFN-β and NF-κB promoter activation in cells coexpressing TRAF3 was reduced using defective TRAF variants lacking the C-terminal TRAF domain important for TRAF oligomerization and interactions with upstream regulators (Fig. 7B). Interestingly, we observed an enhancement of IFN-β and NF-κB promoter activation in cells coexpressing STING in combination with TRAF3 or TRAF6, which was significantly reduced in cells with TBK1 knocked down (Fig. 7C). This suggests that these TRAFs contribute to STING-mediated signaling responses upstream of TBK1. Thus, TRAF3 and TRAF6 may predominantly facilitate STING-dependent activity.
FIG 7.
TRAF3 and TRAF6 may contribute to the STING pathway upstream of TBK1. (A) 293T cells were transfected with a plasmid for HA-tagged STING (STING-HA) in combination with empty vector (EV) or a plasmid encoding His-tagged TRAF3 (TRAF3-His) or TRAF6 (TRAF6-His), together with a reporter plasmid carrying the luciferase gene under the control of the IFN-β, NF-κB, pRDIII, or ISRE promoter. Luciferase activity was determined at 24 h posttransfection. (B) Structure of murine TRAF3 and a deletion mutant of the TRAF domain (TRAF3 mut). The E3 ring finger ubiquitin ligase domain, zinc finger domain, and C-terminal TRAF-C domain (also known as a meprin and TRAF homology domain) are indicated. 293T cells were transfected with a STING-HA vector in combination with EV or a TRAF3-His or TRAF3 mut vector, together with a reporter plasmid carrying the luciferase gene under the control of the IFN-β or NF-κB promoter, and luciferase activity was determined at 24 h posttransfection. TRAF3, TRAF3 mut, or STING expression was demonstrated by immunoblotting, with β-actin serving as a loading control. (C) 293T cells subjected to RNAi by use of nonspecific (NS) or TBK1 siRNA were cotransfected with the STING-HA vector in combination with EV or a TRAF3-His or TRAF6-His vector, together with a reporter plasmid carrying the luciferase gene under the control of the IFN-β or NF-κB promoter, and luciferase activity was determined at 24 h posttransfection. **, P < 0.01 (Student's t test). Error bars indicate SD. IB, immunoblotting.
Distinct roles of TRAF3 and TRAF6 in STING-mediated signaling.
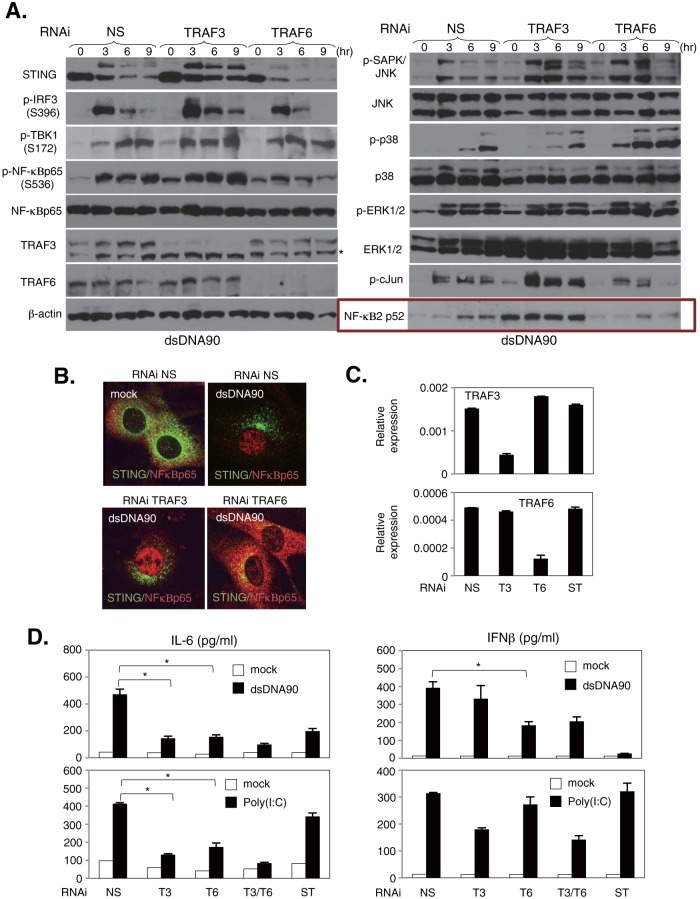
To further determine the involvement of TRAF3 and TRAF6 in dsDNA-mediated signaling responses, primary MEFs were transduced with siRNA targeting TRAF3 or TRAF6 for 72 h and then treated with dsDNA90. Phosphorylation of IRF3, NF-κBp65, TBK1, and ERK1/2 in MEFs with silenced TRAF3 was comparable to that in MEFs treated with nonspecific siRNA (NS) (Fig. 8A). Consistent with previous observations, the constitutive activation of NF-κBp52, which is facilitated by the activation of the noncanonical NF-κB pathway (42), was observed in TRAF3-silenced MEFs (Fig. 8A, boxed panel). Conversely, cells with reduced TRAF6 expression exhibited less NF-κBp65 activation, but not IRF3 and TBK1 phosphorylation, following stimulation with dsDNA90 (Fig. 8A). This further indicates that TRAF6 may play a role in STING-mediated NF-κB activation. Next, we confirmed the nuclear translocation of NF-κBp65 after stimulation with dsDNA90 in RNAi-treated cells. TRAF6- but not TRAF3-silenced MEFs lacked NF-κBp65 nuclear translocation but did not shown an influence on STING trafficking (Fig. 8B). To further address the influence of TRAF3 and TRAF6 on STING function, IFN-β and IL-6 levels were measured by ELISA in MEFs knocked down for TRAF3 or TRAF6 (Fig. 8C). Cells with knocked-down TRAF3 expression exhibited a significant reduction in IL-6 but not IFN-β production (Fig. 8D), even though the phosphorylation and nuclear translocation of NF-κBp65 remained intact (Fig. 8A). Alternatively, a reduction in IL-6 and a partial reduction in IFN-β production were observed in TRAF6-silenced MEFs (Fig. 8D), also suggesting that NF-κBp65 contributes to IRF3-mediated IFN-β production, consistent with our results in Fig. 6. MEFs treated with siRNA specific to both TRAF3 and TRAF6 similarly showed a significant reduction in poly(I·C)-mediated IL-6 but not IFN-β production (Fig. 8D). These results indicate that TRAF6 may be involved predominantly in dsDNA-mediated NF-κB activation rather than IRF3-mediated IFN-β production in MEFs.
FIG 8.
Distinct roles of TRAF3 and TRAF6 in the dsDNA-mediated signaling response in MEFs. (A) MEFs subjected to RNAi by use of nonspecific (NS), TRAF3, or TRAF6 siRNA were stimulated with 5 μg/ml of dsDNA90 for the indicated times. The expression levels of STING, IRF3 phosphorylated at Ser396 (p-IRF3), TBK1 phosphorylated at Ser172 (p-TBK1), NF-κBp65 phosphorylated at Ser536 (p-NF-κBp65), NF-κBp65, NF-κB2p52, TRAF3, TRAF6, ERK1/2 phosphorylated at Thr202/Tyr204 (p-ERK1/2), ERK1/2, SAPK/JNK phosphorylated at Thr183/Tyr185 (p-SAPK/JNK), JNK1/2, p38 phosphorylated at Thr180/Tyr182 (p-p38), p38, c-Jun phosphorylated at Ser63 (p-cJun), and β-actin were determined by immunoblotting. (B) STING-deficient MEFs reconstituted with HA-tagged murine STING and knocked down for TRAF3 or TRAF6 by RNAi were stimulated with 5 μg/ml of dsDNA90 for 3 h, and then cells were costained with antibodies against NF-κBp65 and HA. (C) Levels of TRAF3 and TRAF6 in MEFs treated with NS, TRAF3 (T3), TRAF6 (T6), or STING (ST) siRNA were determined by qRT-PCR as shown in panel D. qRT-PCR data were normalized to the amount of GAPDH mRNA. (D) MEFs treated with NS, TRAF3 (T3), TRAF6 (T6), TRAF3 and TRAF6 (T3/T6), or STING (ST) siRNA were stimulated with 5 μg/ml of dsDNA90 or 5 μg/ml of poly(I·C) for 24 h. The production of IL-6 (left) and IFN-β (right) in the supernatants was determined by ELISA. *, P < 0.05 (Student's t test). Error bars indicate SD.
STING ligands activate the noncanonical NF-κB signaling pathway.
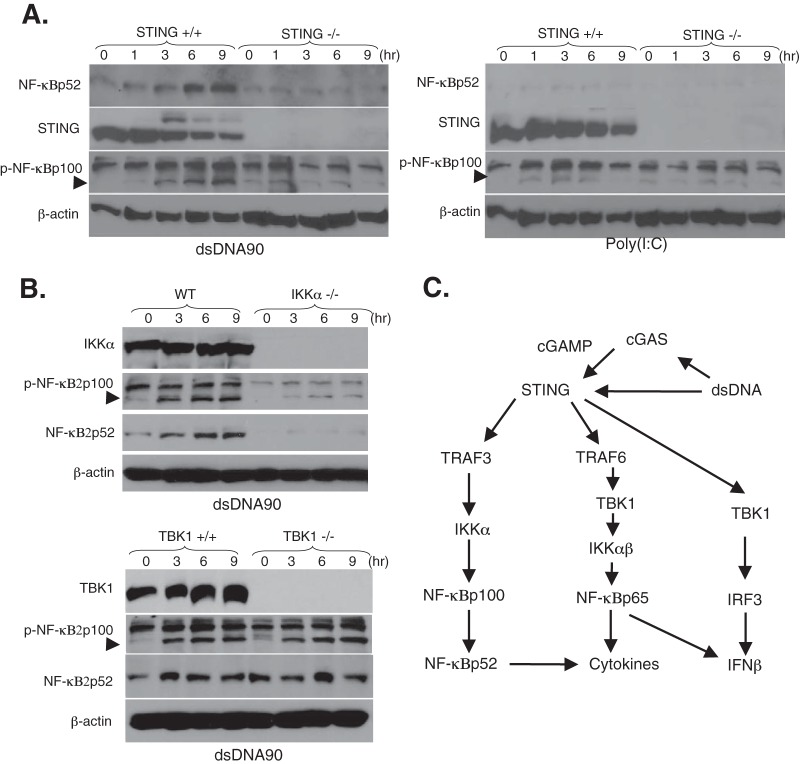
Previous reports have suggested that TRAF3 deficiency causes the constitutive activation of noncanonical NF-κB signaling (42). To evaluate the distinct role of TRAF3 in the modulation of dsDNA-mediated NF-κB activation, we compared canonical and noncanonical NF-κB activation after STING activation. We observed that the induction of NF-κBp52 expression was regulated via the TRAF3–NF-κB inducing kinase (NIK) pathway (Fig. 8A, right panels). We also confirmed that dsDNA90, but not poly(I·C), induced NF-κBp52 processing following the phosphorylation of NF-κB2p100 in a STING-dependent manner (Fig. 9A). It has been suggested that IKKα is involved in noncanonical NF-κB signaling downstream of the TRAF3-NIK axis (32). IKKα-deficient MEFs exhibited substantially reduced NF-κBp52 processing following the phosphorylation of NF-κB2p100 during stimulation with dsDNA (Fig. 9B, top panels). These results indicate that IKKα may be involved in dsDNA-mediated noncanonical NF-κB signaling. Although we observed that TBK1 was necessary for the dsDNA-mediated canonical NF-κB signaling pathway as shown in Fig. 3, TBK1-deficient MEFs exhibited a normal response to NF-κBp52 processing following the phosphorylation of NF-κB2p100 after dsDNA90 stimulation (Fig. 9B, bottom panels). In contrast to a requirement for TRAF3 in the dsDNA-mediated noncanonical NF-κB pathway, NIK, which is an essential activator of the noncanonical NF-κB pathway, was not predominantly involved in dsDNA-mediated signaling responses in MEFs (Fig. 9C). These results may suggest that STING ligands can also activate canonical and noncanonical NF-κB signaling pathways through distinct control of TRAFs.
FIG 9.
STING ligands activate the noncanonical NF-κB signaling pathway in a STING-dependent manner in MEFs. (A) STING+/+ and STING−/− MEFs were stimulated with 10 μg/ml of dsDNA90 or 10 μg/ml of poly(I·C) for the indicated times. The expression levels of STING, NF-κBp52, NF-κBp100 phosphorylated at Ser866/870 (p-NF-κBp100), and β-actin were determined by immunoblotting. (B) Immortalized MEFs derived from wild-type (WT) and IKKα-deficient (IKKα−/−) mice (top panels) or wild-type (TBK1+/+) and TBK1-deficient (TBK1−/−) mice (bottom panels) were stimulated with 10 μg/ml of dsDNA90 for the indicated times. The expression levels of IKKα, TBK1, NF-κBp52, NF-κBp100 phosphorylated at Ser866/870 (p-NF-κBp100), and β-actin were determined by immunoblotting. The phosphorylation state of NF-κBp100 is indicated by an arrowhead. (C) Model of dsDNA-mediated canonical and noncanonical NF-κB activation triggered by STING. Upon dsDNA stimulation, STING is activated and traffics with TBK1 as a signaling complex from the ER to a perinuclear endosomal compartment to activate IRF3 (right arm) and NF-κB. STING is also activated by cGAMP produced by cGAS, which was recently identified as a candidate DNA sensor. TRAF6 may be recruited to the signaling complexes with STING and TBK1, which in turn activates the canonical NF-κBp65 signaling pathway through the IKKαβ activation loop (center arm). STING-mediated NF-κBp65 activation may also contribute to dsDNA-mediated IFN-β production. On the other hand, STING may also activate the noncanonical NF-κB signaling pathway through the TRAF3-IKKα axis, leading to modulation of the dsDNA-mediated canonical NF-κB signaling pathway (left arm). Thus, TRAF3 may be involved in the modulation of canonical and noncanonical dsDNA-mediated NF-κB activation triggered upon STING activation.
DISCUSSION
STING has been identified as a critical signaling molecule required for the detection of cytosolic nucleic acids, particularly dsDNAs derived from pathogens and viruses as well as endogenous second messengers, such as cyclic-di-GMP and -AMP (43, 44). These events result in the production of innate immune response genes through the IRF3 and NF-κB pathways. In the present study, we demonstrate that TBK1 may predominantly control NF-κBp65 in response to STING ligands through the IKKαβ activation loop. Furthermore, we show that TRAF6 may control STING-mediated NF-κB function. These findings detail novel insights into STING-dependent innate immune signaling mechanisms (Fig. 9C).
TBK1 was originally identified as a kinase that activates IRF3 in response to various stimuli, such as proinflammatory cytokines, bacterial components, and RNA or DNA virus infection (20, 21, 45, 46). After dsDNA stimulation, STING traffics with the kinase TBK1 in an autophagic signaling complex, from the ER to perinuclear endosomal compartments harboring IRF3 and NF-κB (5, 23, 47). Recent reports have implicated that STING may be phosphorylated by TBK1 (48), followed by degradation to avoid the sustained production of innate immune-related proinflammatory genes (43). However, STING was not observed to be phosphorylated by TBK1 in vitro (49). Interestingly, our data here indicate that STING degradation was maintained in MEFs lacking TBK1 (Fig. 3A and 5A and E). These results imply that alternate kinases may be involved in the phosphorylation and degradation of STING (49).
Activation of IKKs depends upon the phosphorylation of IKKα/β at Ser176/180 in the activation loop of IKKβ and results in the triggering of IκBα degradation through its phosphorylation at Ser32 and Ser36. Previous reports have suggested that TBK1 facilitates the phosphorylation of IκBα at Ser36 through the IKKβ activation loop (19). We also showed that TBK1 facilitates STING-dependent, dsDNA-mediated NF-κB activation through the IKKαβ activation loop, despite a weak response of IκBα degradation following the phosphorylation of IκBα by stimulation with STING ligands. These results may suggest the direct activation of NF-κBp65 by TBK1 in response to the recognition of STING ligands.
NF-κB signaling has been shown to require members of the TRAF family. The C terminus of STING contains putative TRAF2-binding motifs (40) and has also been suggested to interact with the E3 ubiquitin ligase TRAF3 (7). In the present study, we additionally found that TRAF6 may also contribute to dsDNA-mediated canonical NF-κB signaling upstream of TBK1 (Fig. 8A, B, and D). Recently, it was reported that targeted disruption of TRAF3 results in constitutive activation of the noncanonical NF-κB pathway (42). A previous report has also shown that TRAF3 acts as a suppressor of canonical NF-κB activity through modulation of the IKK complex (50). This could cause the triggering of a negative-feedback signaling response through activation of the canonical NF-κB signaling pathway. Previous studies used classical NF-κB inducers, such as proinflammatory cytokines (IL-1β and TNF-α), or noncanonical NF-κB signaling stimulators (αLTβR) (35). Our data indicate that STING ligands may also activate noncanonical NF-κB signaling in a STING-dependent manner. Recently, Liu and colleagues suggested that MAVS recruits TRAF molecules, such as TRAF2, -5, and -6, for activation of the transcription factors IRF3 and NF-κB, leading to the induction of antiviral immune responses (51). These results may imply the possibility that STING may also require a combination of TRAF molecules for the optimal induction of type I IFN production through IRF3 activation.
Most TRAFs, with the exception of TRAF1, contain a conserved N-terminal domain that consists of several zinc finger domains and a ring finger motif. The ring finger motif is also found in many E3 ubiquitin ligases, and ring finger-mediated protein ubiquitination has emerged as a key mechanism in TRAF-dependent signal transduction. It has been suggested that STING is also subject to the ubiquitination state by E3 ligases (52–54). The ubiquitin ligase RNF5 has been reported to negatively regulate STING-mediated signaling responses, whereas other ubiquitin ligases, such as TRIM56 and TRIM32, have been implicated as positive regulators of STING signaling. Our preliminary data suggest that TRAF6, but not TRAF3, may be involved in the ubiquitination state of STING in cells (data not shown). However, further experimentation will be required to fully clarify the mechanisms of STING ubiquitination and degradation.
Based on our data, we propose that ligands such as dsDNA, cGAMP, and DMXAA activate STING, which delivers TBK1, as an autophagosome signaling complex, from the ER to perinuclear endosomal compartments to activate IRF3 and NF-κB (43). STING is subsequently phosphorylated by kinases, such as ULK1 (49), which in turn may recruit TRAF6, leading to NF-κBp65 activation. In addition, the stimulation of STING may also induce the activation of TRAF3-mediated noncanonical NF-κB signaling, leading to the processing of NF-κB2 (p100/p52) in a STING-dependent way (Fig. 9A). Although it remains to be defined, understanding the signaling cross talk between canonical and noncanonical NF-κB signaling pathways following STING activation could be useful for the design of antiviral drugs or, alternatively, therapies to control dsDNA-mediated inflammatory responses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alan Goodman and Hiroyasu Konno for helpful discussions and advice.
This work was funded in part by grants R01AI079336 and UO1AI083015.
T.A. carried out the experiments. T.A. and G.N.B. wrote the manuscript.
Footnotes
Published ahead of print 5 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00037-14.
REFERENCES
- 1.Crow MK. 2007. Type I interferon in systemic lupus erythematosus. Curr. Top. Microbiol. Immunol. 316:359–386. 10.1007/978-3-540-71329-6_17 [DOI] [PubMed] [Google Scholar]
- 2.Nagata S, Hanayama R, Kawane K. 2010. Autoimmunity and the clearance of dead cells. Cell 140:619–630. 10.1016/j.cell.2010.02.014 [DOI] [PubMed] [Google Scholar]
- 3.Ahn J, Gutman D, Saijo S, Barber GN. 2012. STING manifests self DNA-dependent inflammatory disease. Proc. Natl. Acad. Sci. U. S. A. 109:19386–19391. 10.1073/pnas.1215006109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa H, Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674–678. 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishikawa H, Ma Z, Barber GN. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461:788–792. 10.1038/nature08476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. 2008. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 28:5014–5026. 10.1128/MCB.00640-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550. 10.1016/j.immuni.2008.09.003 [DOI] [PubMed] [Google Scholar]
- 8.Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. 2009. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U. S. A. 106:8653–8658. 10.1073/pnas.0900850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478:515–518. 10.1038/nature10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paludan SR, Bowie AG. 2013. Immune sensing of DNA. Immunity 38:870–880. 10.1016/j.immuni.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen LF, Greene WC. 2004. Shaping the nuclear action of NF-kappaB. Nat. Rev. Mol. Cell Biol. 5:392–401. 10.1038/nrm1368 [DOI] [PubMed] [Google Scholar]
- 14.Bonnard M, Mirtsos C, Suzuki S, Graham K, Huang J, Ng M, Itie A, Wakeham A, Shahinian A, Henzel WJ, Elia AJ, Shillinglaw W, Mak TW, Cao Z, Yeh WC. 2000. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 19:4976–4985. 10.1093/emboj/19.18.4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, Woodgett JR. 2000. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature 406:86–90. 10.1038/35017574 [DOI] [PubMed] [Google Scholar]
- 16.Leitges M, Sanz L, Martin P, Duran A, Braun U, Garcia JF, Camacho F, Diaz-Meco MT, Rennert PD, Moscat J. 2001. Targeted disruption of the zetaPKC gene results in the impairment of the NF-kappaB pathway. Mol. Cell 8:771–780. 10.1016/S1097-2765(01)00361-6 [DOI] [PubMed] [Google Scholar]
- 17.Madrid LV, Mayo MW, Reuther JY, Baldwin AS., Jr 2001. Akt stimulates the transactivation potential of the RelA/p65 subunit of NF-kappa B through utilization of the Ikappa B kinase and activation of the mitogen-activated protein kinase p38. J. Biol. Chem. 276:18934–18940. 10.1074/jbc.M101103200 [DOI] [PubMed] [Google Scholar]
- 18.Pomerantz JL, Baltimore D. 1999. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 18:6694–6704. 10.1093/emboj/18.23.6694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tojima Y, Fujimoto A, Delhase M, Chen Y, Hatakeyama S, Nakayama K, Kaneko Y, Nimura Y, Motoyama N, Ikeda K, Karin M, Nakanishi M. 2000. NAK is an IkappaB kinase-activating kinase. Nature 404:778–782. 10.1038/35008109 [DOI] [PubMed] [Google Scholar]
- 20.Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H, Kawai T, Hoshino K, Takeda K, Akira S. 2004. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199:1641–1650. 10.1084/jem.20040520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry AK, Chow EK, Goodnough JB, Yeh WC, Cheng G. 2004. Differential requirement for TANK-binding kinase-1 in type I interferon responses to Toll-like receptor activation and viral infection. J. Exp. Med. 199:1651–1658. 10.1084/jem.20040528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balachandran S, Thomas E, Barber GN. 2004. A FADD-dependent innate immune mechanism in mammalian cells. Nature 432:401–405. 10.1038/nature03124 [DOI] [PubMed] [Google Scholar]
- 23.Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, Barber GN. 2013. STING recognition of cytoplasmic DNA instigates cellular defense. Mol. Cell 50:5–15. 10.1016/j.molcel.2013.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stetson DB, Medzhitov R. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24:93–103. 10.1016/j.immuni.2005.12.003 [DOI] [PubMed] [Google Scholar]
- 25.Conlon J, Burdette DL, Sharma S, Bhat N, Thompson M, Jiang Z, Rathinam VA, Monks B, Jin T, Xiao TS, Vogel SN, Vance RE, Fitzgerald KA. 2013. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J. Immunol. 190:5216–5225. 10.4049/jimmunol.1300097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prantner D, Perkins DJ, Lai W, Williams MS, Sharma S, Fitzgerald KA, Vogel SN. 2012. 5,6-Dimethylxanthenone-4-acetic acid (DMXAA) activates stimulator of interferon gene (STING)-dependent innate immune pathways and is regulated by mitochondrial membrane potential. J. Biol. Chem. 287:39776–39788. 10.1074/jbc.M112.382986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu YH, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591. 10.1016/j.cell.2009.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Lint AL, Murawski MR, Goodbody RE, Severa M, Fitzgerald KA, Finberg RW, Knipe DM, Kurt-Jones EA. 2010. Herpes simplex virus immediate-early ICP0 protein inhibits Toll-like receptor 2-dependent inflammatory responses and NF-kappaB signaling. J. Virol. 84:10802–10811. 10.1128/JVI.00063-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taddeo B, Luo TR, Zhang W, Roizman B. 2003. Activation of NF-kappaB in cells productively infected with HSV-1 depends on activated protein kinase R and plays no apparent role in blocking apoptosis. Proc. Natl. Acad. Sci. U. S. A. 100:12408–12413. 10.1073/pnas.2034952100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregory D, Hargett D, Holmes D, Money E, Bachenheimer SL. 2004. Efficient replication by herpes simplex virus type 1 involves activation of the IkappaB kinase-IkappaB-p65 pathway. J. Virol. 78:13582–13590. 10.1128/JVI.78.24.13582-13590.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unterholzner L, Bowie AG. 2008. The interplay between viruses and innate immune signaling: recent insights and therapeutic opportunities. Biochem. Pharmacol. 75:589–602. 10.1016/j.bcp.2007.07.043 [DOI] [PubMed] [Google Scholar]
- 32.Fujita F, Taniguchi Y, Kato T, Narita Y, Furuya A, Ogawa T, Sakurai H, Joh T, Itoh M, Delhase M, Karin M, Nakanishi M. 2003. Identification of NAP1, a regulatory subunit of IkappaB kinase-related kinases that potentiates NF-kappaB signaling. Mol. Cell. Biol. 23:7780–7793. 10.1128/MCB.23.21.7780-7793.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buss H, Dorrie A, Schmitz ML, Hoffmann E, Resch K, Kracht M. 2004. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-{kappa}B at serine 536 is mediated by multiple protein kinases including I{kappa}B kinase (IKK)-{alpha}, IKK{beta}, IKK{epsilon}, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J. Biol. Chem. 279:55633–55643. 10.1074/jbc.M409825200 [DOI] [PubMed] [Google Scholar]
- 34.Roberts ZJ, Goutagny N, Perera PY, Kato H, Kumar H, Kawai T, Akira S, Savan R, van Echo D, Fitzgerald KA, Young HA, Ching LM, Vogel SN. 2007. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J. Exp. Med. 204:1559–1569. 10.1084/jem.20061845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hacker H, Tseng PH, Karin M. 2011. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 11:457–468. 10.1038/nri2998 [DOI] [PubMed] [Google Scholar]
- 36.Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, Shahangian A, Zarnegar B, Shiba TL, Wang Y, Cheng G. 2006. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 25:3257–3263. 10.1038/sj.emboj.7601220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paz S, Vilasco M, Werden SJ, Arguello M, Joseph-Pillai D, Zhao T, Nguyen TL, Sun Q, Meurs EF, Lin R, Hiscott J. 2011. A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell Res. 21:895–910. 10.1038/cr.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida R, Takaesu G, Yoshida H, Okamoto F, Yoshioka T, Choi Y, Akira S, Kawai T, Yoshimura A, Kobayashi T. 2008. TRAF6 and MEKK1 play a pivotal role in the RIG-I-like helicase antiviral pathway. J. Biol. Chem. 283:36211–36220. 10.1074/jbc.M806576200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konno H, Yamamoto T, Yamazaki K, Gohda J, Akiyama T, Semba K, Goto H, Kato A, Yujiri T, Imai T, Kawaguchi Y, Su B, Takeuchi O, Akira S, Tsunetsugu-Yokota Y, Inoue J. 2009. TRAF6 establishes innate immune responses by activating NF-kappaB and IRF7 upon sensing cytosolic viral RNA and DNA. PLoS One 4:e5674. 10.1371/journal.pone.0005674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin L, Xu LG, Yang IV, Davidson EJ, Schwartz DA, Wurfel MM, Cambier JC. 2011. Identification and characterization of a loss-of-function human MPYS variant. Genes Immun. 12:263–269. 10.1038/gene.2010.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marinis JM, Hutti JE, Homer CR, Cobb BA, Cantley LC, McDonald C, Abbott DW. 2012. IkappaB kinase alpha phosphorylation of TRAF4 downregulates innate immune signaling. Mol. Cell. Biol. 32:2479–2489. 10.1128/MCB.00106-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He JQ, Zarnegar B, Oganesyan G, Saha SK, Yamazaki S, Doyle SE, Dempsey PW, Cheng G. 2006. Rescue of TRAF3-null mice by p100 NF-kappa B deficiency. J. Exp. Med. 203:2413–2418. 10.1084/jem.20061166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barber GN. 2011. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr. Opin. Immunol. 23:10–20. 10.1016/j.coi.2010.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burdette DL, Vance RE. 2013. STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 14:19–26. 10.1038/ni.2491 [DOI] [PubMed] [Google Scholar]
- 45.Matsui K, Kumagai Y, Kato H, Sato S, Kawagoe T, Uematsu S, Takeuchi O, Akira S. 2006. Cutting edge: role of TANK-binding kinase 1 and inducible IkappaB kinase in IFN responses against viruses in innate immune cells. J. Immunol. 177:5785–5789 http://www.jimmunol.org/content/177/9/5785.long [DOI] [PubMed] [Google Scholar]
- 46.Miyahira AK, Shahangian A, Hwang S, Sun R, Cheng G. 2009. TANK-binding kinase-1 plays an important role during in vitro and in vivo type I IFN responses to DNA virus infections. J. Immunol. 182:2248–2257. 10.4049/jimmunol.0802466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S. 2009. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. U. S. A. 106:20842–20846. 10.1073/pnas.0911267106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka Y, Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5:ra20. 10.1126/scisignal.2002521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konno H, Konno K, Barber GN. 2013. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155:688–698. 10.1016/j.cell.2013.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zarnegar B, Yamazaki S, He JQ, Cheng G. 2008. Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proc. Natl. Acad. Sci. U. S. A. 105:3503–3508. 10.1073/pnas.0707959105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, Sun L, Chen ZJ. 2013. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife 2:e00785. 10.7554/eLife.00785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30:397–407. 10.1016/j.immuni.2009.01.008 [DOI] [PubMed] [Google Scholar]
- 53.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. 2010. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33:765–776. 10.1016/j.immuni.2010.10.013 [DOI] [PubMed] [Google Scholar]
- 54.Zhang J, Hu MM, Wang YY, Shu HB. 2012. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 287:28646–28655. 10.1074/jbc.M112.362608 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.