Abstract
Francisella spp. are facultative intracellular pathogens identified in increasingly diverse hosts, including mammals. F. noatunensis subsp. orientalis and F. noatunensis subsp. noatunensis infect fish inhabiting warm and cold waters, respectively, while F. tularensis subsp. novicida is highly infectious for mice and has been widely used as a model for the human pathogen F. tularensis. Here, we established zebrafish embryo infection models of fluorescently labeled F. noatunensis subsp. noatunensis, F. noatunensis subsp. orientalis, and F. tularensis subsp. novicida at 22, 28, and 32°C, respectively. All infections led to significant bacterial growth, as shown by reverse transcription-quantitative PCR (RT-qPCR), and to a robust proinflammatory immune response, dominated by increased transcription of tumor necrosis factor alpha (TNF-α) and interleukin-1β (IL-1β). F. noatunensis subsp. orientalis was the most virulent, F. noatunensis subsp. noatunensis caused chronic infection, and F. tularensis subsp. novicida showed moderate virulence and led to formation of relatively small granuloma-like structures. The use of transgenic zebrafish strains with enhanced green fluorescent protein (EGFP)-labeled immune cells revealed their detailed interactions with Francisella species. All three strains entered preferentially into macrophages, which eventually assembled into granuloma-like structures. Entry into neutrophils was also observed, though the efficiency of this event depended on the route of infection. The results demonstrate the usefulness of the zebrafish embryo model for studying infections caused by different Francisella species at a wide range of temperatures and highlight their interactions with immune cells.
INTRODUCTION
Members of the genus Francisella have been identified from many different environments and can infect and cause disease in diverse hosts, ranging from amoebae to humans (1, 2). Francisella species are nonmotile, pleomorphic, Gram-negative, strictly aerobic, facultative intracellular coccobacilli. The genus contains species capable of causing disease in humans (F. tularensis and F. philomiragia) as well as in fish (F. noatunensis) (2). F. tularensis, the causative agent of tularemia, is further divided into four subspecies, each with a distinct infectivity and capacity to cause illness in humans. Due to its extreme virulence, low infectious dose, ease of aerosol dissemination, and capacity to cause severe illness and death, F. tularensis subsp. tularensis has been classified as a class A bioterrorism agent by the Centers for Disease Control and Prevention (3). The closely related F. tularensis subsp. novicida has low virulence in humans but is highly infectious to mice and has been widely used as a model for F. tularensis (2). F. noatunensis subspecies are essentially avirulent to mammals (4) but cause francisellosis in fish (5), resulting in large losses for the fish farming industry worldwide (6–11). F. noatunensis consists of two subspecies, which appear to be adapted to different host temperatures: F. noatunensis subsp. orientalis causes disease in “warm-water” fish (6–9), while F. noatunensis subsp. noatunensis causes disease in fish living in colder waters (10–12).
Most Francisella spp. are highly infectious organisms for their hosts, where 10 to 30 CFU is capable of establishing infections (13–15), although the virulence of the strains differs. Infection with F. tularensis results in the development of tularemia, commonly characterized by the formation of localized or systemic suppurative (pus-forming) or granulomatous lesions (3), where the route of infection contributes significantly to the clinical symptoms. F. noatunensis subsp. noatunensis infection results in the development of chronic systemic granulomatous disease in Atlantic cod (11, 12), with cumulative mortality reaching 40% (11). F. noatunensis subsp. orientalis also causes systemic granulomatosis (7, 9, 16) with reported mortality of 50 to 60%.
An important part of Francisella sp. pathogenesis is the ability of the bacteria to replicate in the intracellular environment of professional phagocytic cells. For F. tularensis it has been shown that after phagocytosis by macrophages, the bacteria are able to lyse the phagosomal membrane and escape to the cytosol (17), where they replicate extensively (18). This eventually results in release of the bacteria to the extracellular space through host cell lysis or in the formation of Francisella-containing vacuoles through interactions with the autophagic pathway (19). It has been suggested that F. noatunensis subspecies are also able to escape from phagosomes into the cytosol (20–22).
In the early days of tularemia research, challenge experiments with human volunteers were performed. These studies provided valuable information regarding the efficacy of antibiotic treatment and vaccines (23–25), and some human studies are still being performed (26, 27). Several experimental models for studying tularemia are now available (23). The nonhuman primate model most closely resembles human tularemia with regard to strain virulence and vaccine-induced protection (23). It is, however, not suitable as a standard model due to its high cost and public concern about the use of experimental animals. The mouse model has become popular for studying tularemia because of the wide availability of genetic tools. The use of the mouse model has its disadvantages, particularly with regard to susceptibility to different strains of F. tularensis and differences in vaccine-induced protection compared to the case for humans. While the rat model has gained some favor, especially in studying vaccine efficacy, it has reduced sensitivity to even the most virulent subspecies of F. tularensis. Thus, new infection models are warranted to fully understand Francisella infection and disease mechanisms.
F. noatunensis subsp. orientalis pathogenesis can be studied in its natural host Nile tilapia Oreochromis niloticus L., as it causes mortality in a dose-dependent manner within weeks after challenge (14). There have been several reports of tilapia being used for studying F. noatunensis subsp. orientalis virulence factors and vaccine efficacy (14, 28). The mortality of Atlantic cod challenged with F. noatunensis subsp. noatunensis under experimental conditions is highly variable, ranging from 75% (12) to a few percent, even many months after challenge, when gross pathology is evident in the fish (4, 29). Due to the slow progression of the disease, the inconsistent mortality of Atlantic cod, and the lack of alternate animal models (5), there is a need for establishment of models which allow faster development of the infection, giving more consistent results and hopefully providing insight into the Francisella sp. research in general. In addition, the above-mentioned models for studying francisellosis are not able to provide sufficient information about the initial steps of disease pathogenesis in vivo.
A large number of bacteria and viruses have been shown to cause disease in zebrafish (Danio rerio) (30, 31). Zebrafish has a number of valuable advantages compared to other commonly used models for studying Francisella pathogenesis. The fish are low cost, are easy to maintain, and require minimal laboratory space, in addition to the increasing availability of transgenic lines and a fully sequenced genome. The use of transparent zebrafish embryos infected with fluorescently labeled pathogens allows real-time in vivo imaging of disease progression, starting from the first minutes postinfection. The small size of adult zebrafish allows longitudinal histological sections to be made through the entire fish (30). Moreover, transgenic zebrafish lines with fluorescently labeled cells enable the study of the infected host cell repertoire. The embryological development of the zebrafish immune system also offers advantages (32). While the innate immune system, with its effector cells dominated by macrophages and neutrophilic granulocytes, is present and active within 48 h postfertilization (hpf), the adaptive arm of the immune system is not fully developed for another 4 to 6 weeks (33). This presents an opportunity to study the function of the innate immune system in a vertebrate host without the interference of adaptive immunity. The immune system of the fully immunocompetent zebrafish is closely related to those of other vertebrates, making zebrafish a more versatile model for studying pathogenesis and host-pathogen interactions than other nonmammalian models that lack an adaptive immune system, such as Drosophila melanogaster and Caenorhabditis elegans (30, 31).
Zebrafish have been shown to rapidly clear experimentally induced bacteremia within hours after infection with Escherichia coli K-12, Bacillus subtilis, and an avirulent strain of Salmonella enterica serovar Typhimurium (34, 35), suggesting a requirement for bacterial virulence to efficiently establish disease. The susceptibility of zebrafish to infection with Francisella was recently reported for F. noatunensis subsp. orientalis (36), in a study describing dose-dependent acute mortality of adult zebrafish upon intraperitoneal injections, demonstrating that Francisella can cause disease in adult zebrafish. Infected fish responded with a proinflammatory immune response characterized by increased transcription of interleukin-1β (IL-1β), gamma interferon (IFN-γ), and tumor necrosis factor alpha (TNF-α) starting at 6 h postinfection (hpi) and lasting at least 7 days. The study was done without the use of fluorescently labeled bacteria, and therefore no real-time observation of disease progression was provided. In the current study, we established and further characterized zebrafish embryo infection models for three different Francisella spp. at different temperatures.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The bacterial strains used in this study are given in Table S1 in the supplemental material. F. noatunensis subsp. noatunensis NCIMB14265, isolated from diseased Atlantic cod Gadus morhua L. in Norway, transformed with the green fluorescent protein (GFP)-expressing plasmid pKK289Km/gfp (F. noatunensis subsp. noatunensis-GFP) has been described previously as strain HWL108 (21). We additionally constructed a red fluorescent strain by transformation with the mCherry-expressing plasmid pKK289Km/mCherry (F. noatunensis subsp. noatunensis-mCh). F. noatunensis subsp. orientalis LADL 07-285A, isolated from diseased tilapia Oreochromis sp. L. in Costa Rica (16), was transformed with the mCherry-expressing plasmid pKK289Km/mCherry (F. noatunensis subsp. orientalis-mCh). F. tularensis subsp. novicida U112 was transformed with pKK289Km/gfp (F. tularensis subsp. novicida-GFP) and pKK289Km/mCherry (F. tularensis subsp. novicida-mCh). E. coli DH5α harboring the pmCherry vector (catalog no. 632522; Clontech Laboratories Inc., Mountain View, CA, USA) was used as a control in some experiments.
Bacteria were kept at −80°C for long-term storage either in BD Bacto Eugon broth (BD Diagnostic Systems, Sparks, MD, USA) supplemented with 20% glycerol (Sigma-Aldrich Co., St. Louis, MO, USA) or in autoclaved 10% skim milk (Merck KGaA, Darmstadt, Germany). Prior to experiments, bacteria were plated on Eugon chocolate agar (ECA) plates (37) supplemented with 15 μg/ml kanamycin (Sigma-Aldrich Co., St. Louis, MO, USA) and incubated at 22°C for F. noatunensis subsp. noatunensis, 28°C for F. noatunensis subsp. orientalis, and 37°C for F. tularensis subsp. novicida. E. coli pmCherry was propagated using Luria-Bertani agar with 50 μg/ml ampicillin (LAamp50). At 1 day prior to injections, the Francisella strains were inoculated into Eugon broth supplemented with 2 mM FeCl3 (Sigma-Aldrich Co., St. Louis, MO, USA) and the appropriate concentration of kanamycin (15 μg/ml for F. noatunensis subsp. orientalis and F. noatunensis subsp. noatunensis and 30 μg/ml for F. tularensis subsp. novicida) and incubated overnight. E. coli pmCherry was grown overnight on LAamp50 plates and resuspended in phosphate-buffered saline (PBS) prior to injections.
Bacterial transformation with mCherry-expressing plasmid.
The plasmid pKK289Km/mCherry was created by amplification of the mCherry gene from the pmCherry vector followed by ligation of the PCR product into the cloning vector pCR4 (Invitrogen by Life Technologies, Carlsbad, CA, USA), creating pCR4/mCherry. The resulting plasmid was digested with NdeI and EcoRI (New England BioLabs Inc., Ipswich, MA, USA) before the mCherry gene was ligated into the plasmid pKK289Km digested with the same enzymes, thereby placing the mCherry gene by translational fusion under control of the GroEL promoter. The plasmid was transformed into all Francisella strains by electroporation essentially as described by Bakkemo et al. (21).
Preparation of bacteria for microinjections.
All the strains used for infecting zebrafish embryos expressed a fluorophore in trans, either GFP or mCherry. Overnight cultures were harvested by centrifugation at 12,800 × g for 10 min at 4°C and the pellet resuspended in PBS, pH 7.4. Optical density at 600 nm (OD600) was measured with a Genesys 20 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) and adjusted to the desired value. An aliquot of phenol red sodium salt (Sigma-Aldrich Co., St. Louis, MO, USA) stock solution was added to the prepared bacterial suspensions in PBS to a final concentration of 0.01% to ensure visualization of the injection mixtures. The number of CFU per injection was estimated by plating 10 μl of 10-fold serial dilutions of the bacterial suspensions on ECA plates, which were incubated at the temperature optimal for each bacterial strain. The number of CFU was counted after at least 7 days for F. noatunensis subsp. orientalis, at least 3 weeks for F. noatunensis subsp. noatunensis, and 1 day for F. tularensis subsp. novicida.
Zebrafish embryo maintenance.
Zebrafish (Danio rerio) wild-type (wt) strain AB embryos were obtained from the Aleström Zebrafish Lab facility at the Norwegian School for Veterinary Sciences. Transgenic zebrafish lines expressing enhanced GFP (EGFP) in macrophages [Tg(mpeg1:EGPF)gl22] (38) and neutrophilic granulocytes [Tg(mpx:EGFP)i114] (39) were kept at the Griffiths Zebrafish Lab at the University of Oslo, and embryos were produced in-house. All zebrafish adults and embryos were handled according to standard protocols (40). Zebrafish embryos were manually dechorionated at the age of 30 h, transferred into fresh embryo water, and kept at 28°C prior to injections.
Zebrafish embryo microinjections.
Injections of zebrafish embryos were performed using a Narishige Inc. EG-400 microinjector with an Eppendorf Femtojet controlling unit. Borosilicate needles (Harvard apparatus, 1-mm outer diameter by 0.78-mm inner diameter) for the injections were prepared using a micropipette-pulling device (Shutter Inc., Flaming/Brown P-97) with the following parameters: heat, 610; pull, = 40; velocity, 5; and time, 5. The injection volume was adjusted by breaking the tip of an initially sealed needle at a certain point. Once the needle was opened, trial injections were made in paraffin oil, which led to the creation of droplets suspended in the oil. The diameter of the droplets was measured with a ruler incorporated into an eyepiece of the microscope, from which the injection volume was calculated. Different volumes were used for infection of the embryos at three anatomical sites (see Fig. S1 in the supplemental material): 2 to 3 nl for intravascular (duct of Cuvier), 1 to 2 nl for muscle tissue (tail muscle), and below 1 nl for the otic vesicle (ear). Wild-type strain AB or the transgenic zebrafish embryos at 48 to 52 h postfertilization (hpf) were anesthetized with approximately 170 μg/ml Tricaine methanesulfonate MS-222 (Tricaine) (pH 7.4) (Finquel; Argent Laboratories Group, Inc., Redmond, WA, USA) (41) for 1 to 2 min and placed on 2% agar (BD, Franklin Lakes, NJ, USA) plates before injection. Infected embryos were transferred into petri dishes with fresh embryo water and kept at 28°C for F. noatunensis subsp. orientalis, 22°C for F. noatunensis subsp. noatunensis, and 28°C or 32°C for F. tularensis subsp. novicida. Survival experiments were performed using at least 15 embryos per group injected with Francisella spp. in PBS with an OD600 of 2.0. PBS without bacteria was used for injections as negative controls. The survival data were processed using the Kaplan-Meier method with GraphPad Prism version 4.0 for Windows (GraphPad Software, La Jolla, CA, USA). For gene transcription studies, the group size was approximately 30 embryos to ensure a sufficient amount of genetic material.
Imaging of zebrafish embryos.
Prior to imaging, zebrafish embryos were anesthetized with Tricaine as described above. Fluorescent imaging was performed with two light microscopes, the Leica DM IRB (Leica Microsystems CMS GmbH, Wetzlar, Germany) and Leica AF 6000 (Leica Microsystems AG, Heerbrugg, Switzerland). Confocal microscopy was performed with an Olympus Fluoview FV 1000 confocal microscope (Olympus Europa Holding GmbH, Hamburg, Germany). When using the Leica DM IRB inverted microscope, embryos were transferred to a glass slide with a reservoir filled with an anesthetic mixture of embryo water and Tricaine (see above) and covered with a coverslip. While using the Leica AF 6000, the embryos were transferred into a 47-mm petri dish filled with the mixture of embryo water and Tricaine. The embryos were anesthetized and embedded into low-melting-point agarose prior to confocal imaging. The micrographs were taken using Leica Application Suite (LAS) V3.8 and V4.0.0 and Olympus Fluoview version 3.1 software and further processed on the Fiji platform (42).
Sampling and RNA extraction from infected zebrafish embryos.
At each time point, 9 randomly chosen zebrafish embryos (AB, wt) from each group were euthanized by a prolonged immersion in an overdose of Tricaine solution (200 to 300 μg/ml) (41) and transferred into 1.5-ml Eppendorf tubes. Three embryos were pooled into one sample to allow for a sufficient amount of starting material for RNA extraction, and the total number of samples per time point per strain was three. The embryo water was replaced by RNAlater (Ambion by Life Technologies, Carlsbad, CA, USA) immediately after transfer. The samples were kept at 4°C until extraction of RNA. For the extraction of total RNA, RNAlater was replaced with 1 ml of TRIzol (Ambion by Life Technologies, Carlsbad, CA, USA), the embryos were transferred together with the TRIzol to a 2.0-ml SafeLock Eppendorf tube (Eppendorf AG, Hamburg, Germany) containing a 0.5-mm-diameter steel bead (Qiagen GmBH, Hilden, Germany), and the tissue was homogenized at 25 Hz for 5 min with TissueLyserII (Qiagen GmBH, Hilden, Germany). After 5 min of incubation at room temperature, 200 μl chloroform (Sigma-Aldrich Co., St. Louis, MO, USA) was added, and the samples were incubated for 3 min at room temperature before centrifugation at 21,500 × g for 20 min at 4°C. RNA-containing supernatant was mixed with an equal volume of 70% ethanol and loaded on RNeasy Minispin columns (Qiagen GmBH, Hilden, Germany). The samples were thereafter handled according to the manufacturers' instructions, including a 15-min on-column DNase treatment using the RNase-free DNase set (Qiagen GmBH, Hilden, Germany), and eluted in 50 μl diethyl pyrocarbonate (DEPC)-treated H2O (Invitrogen by Life Technologies, Carlsbad, CA, USA).
RNA quantity and quality were assessed as previously described (37). Due to a too-low concentration of RNA extracted from each pool of zebrafish embryos, we could not use 1 μg of RNA for reverse transcription (RT). The maximum amount of 12 μl RNA per reverse transcription reaction mixture (usually corresponding to between 300 and 600 ng total RNA), was therefore used for the reverse transcription step performed with QuantiTect the reverse transcription kit (Qiagen GmBH, Hilden, Germany) using random primers. A control experiment with reverse transcriptase omitted was performed for each extraction to investigate the potential presence of contaminating genomic DNA.
Primer design and quantitative PCR.
Primers (see Table S2 in the supplemental material) were chosen to target a diverse repertoire of the immune response from zebrafish. TNF-α, IL-1β, and IL-12a were chosen to represent proinflammatory cytokines, as they are among the main proinflammatory cytokines produced by phagocytes (43). IL-10 (44) was chosen as an anti-inflammatory cytokine. Complement component 3b (C3b) was chosen to represent the complement system in zebrafish (45). IFN-γ1-2 was chosen for investigation of a potential type II interferon (IFN) response in zebrafish (46). Mx was chosen to represent genes induced by the type I IFNs in zebrafish (47). Suppressors of cytokine signaling (SOCS) were chosen due to their importance in regulation of the balance between pro- and anti-inflammatory signals during infection (48).
QuantiTect bioinformatically validated primers were obtained from Qiagen GmBH (Hilden, Germany) for most of the genes used in the study, while for the remaining immune genes, primers previously used in gene transcription studies of zebrafish (49) were obtained from Life Technologies Inc. (Carlsbad, CA, USA). Quantitative PCR (qPCR) was performed in triplicates using a LightCycler 480 (Roche, Basel, Switzerland) qPCR machine in 20-μl reaction volumes containing LightCycler 480 SYBR green I Master Mix (Roche, Basel, Switzerland), 0.5 μM each primer, and 2 μl template under the following cycling conditions: 5 min of initial denaturation at 95°C; 45 cycles of amplification using 10 s at 95°C, 30 s at 60°C, and 8 s at 72°C; and then melting curve analysis to verify single amplification peaks for the immune response experiments. For quantification of bacterial growth, a Stratagene Mx3005p (Stratagene, La Jolla, CA) qPCR machine was used and the reactions run in triplicate in 20-μl volumes using Express SYBR GreenER qPCR Supermix Universal (Life Technologies Inc., Carlsbad, CA, USA) containing 50 nM Rox reference dye and 300 nM appropriate primers under the following cycling conditions: 2 min of incubation with uracil-DNA glycosylase (UDG) at 50°C, 2 min of initial denaturation at 95°C, 40 cycles of amplification using 15 s at 95°C and 60 s at 60°C, and melting curve analysis.
Relative transcription levels for each gene and time point were determined as previously described (37) and normalized against the reference genes 18S rRNA (alternative designation, zgc:158463), eef1a1l1, and actb1, which have been previously reported to be stably transcribed during zebrafish embryonic development (50, 51). The normalized immune response data for infected fish were standardized against the transcription levels in PBS-injected fish for each time point, as the basal transcription levels of immune genes in PBS-injected fish depend on the stage of embryonic development (data not shown). Statistical analysis of the resulting data sets was performed using JMP 8.0.2. (SAS Institute Inc., Cary, NC, USA). Differences between groups were deemed statistically significant at a P value of <0.05 using Student's t test assuming unequal variance with a two-tailed test for the immune response and a one-tailed test for verification of bacterial growth.
The gyrA and the fopA genes were chosen for relative quantification of F. noatunensis subsp. orientalis and F. noatunensis subsp. noatunensis, respectively, as they have previously been used for RT-qPCR studies and were proved to be stably transcribed under various conditions (37). The fopA gene has additionally been used for diagnostic purposes (52) and was chosen for quantifying relative transcription levels for F. tularensis subsp. novicida.
RESULTS
Estimation of challenge dose.
The amount of bacteria injected into the zebrafish embryos was controlled by OD600 measurements of the bacterial cultures in phosphate-buffered saline (PBS), which were subsequently used for infection. Plating F. noatunensis suspensions with an OD600 of 2.0 resulted in an estimated CFU count of 3.5 × 109 CFU/ml, corresponding to a challenge dose of approximately 9 × 103 CFU per embryo. Plating F. tularensis subsp. novicida suspensions with an OD600 of 2.0 in PBS gave an average count of 1.25 × 1010 CFU/ml, or approximately 3.3 ×104 CFU per embryo. The maximum amount of F. tularensis subsp. novicida used for infecting zebrafish embryos was therefore 3.6-fold higher than the corresponding doses of F. noatunensis subsp. orientalis and F. noatunensis subsp. noatunensis.
Model 1, infection of zebrafish embryos with F. noatunensis subsp. orientalis at 28°C: F. noatunensis subsp. orientalis-mCh infection causes acute disease and fast mortality in zebrafish embryos.
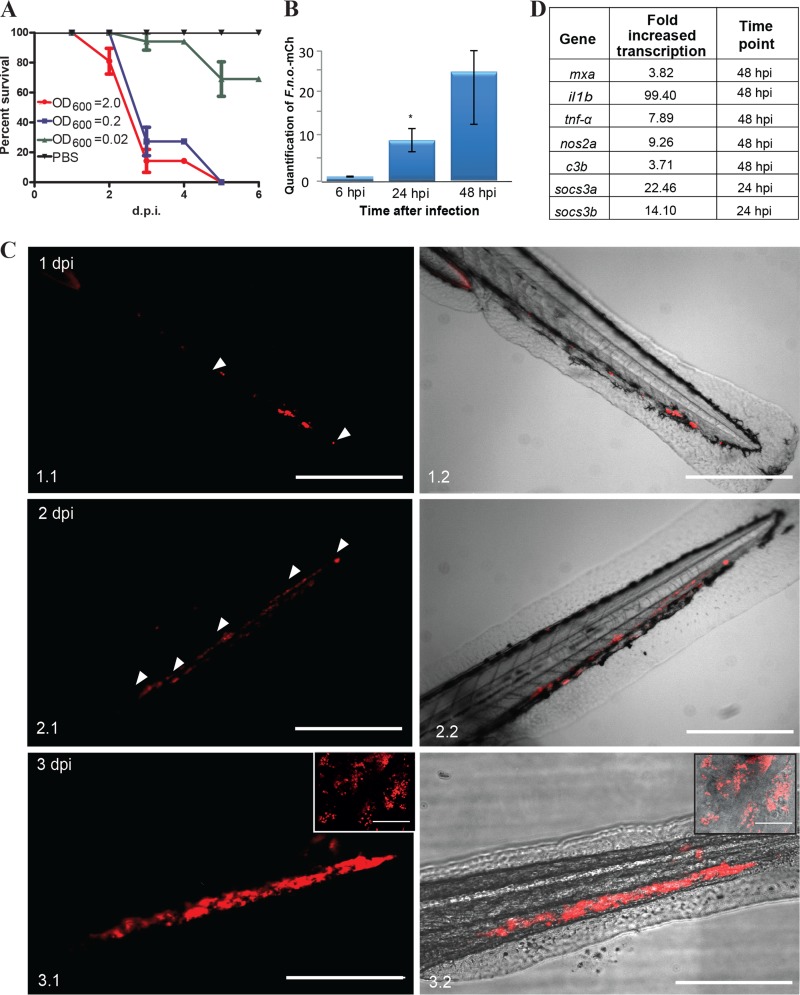
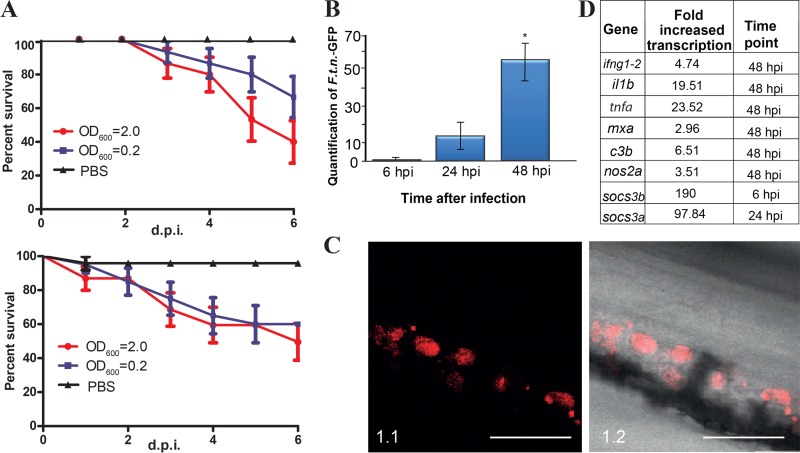
Taking into consideration that F. noatunensis subsp. orientalis-mCh has the most favorable optimal growth temperature for the fish (28°C) and that adult zebrafish have previously been shown to be susceptible to F. noatunensis subsp. orientalis infection (36) F. noatunensis subsp. orientalis-mCh was the first strain tested for zebrafish embryo infection. Zebrafish embryos (AB, wild type [wt]) at 28°C were injected with 2 to 3 nl of F. noatunensis subsp. orientalis-mCh suspension at an OD600 of 2.0, 0.2, or 0.02 into the duct of Cuvier, a wide blood vessel that spreads over the top of the yolk sac (see Fig. S1 in the supplemental material). The bacteria were able to establish infection and kill all the infected embryos within 5 days postinfection (dpi) with a challenge dose at an OD600 of 2.0 or 0.2, while an OD600 of 0.02 resulted in only around 20% mortality (Fig. 1A). The ability of the bacteria to replicate inside the embryos was next evaluated by RT-qPCR (Fig. 1B). Bacterial RNA was detected in the infected embryos at all investigated time points, and the amount of F. noatunensis subsp. orientalis-mCh increased by 8-fold by 24 h postinfection (hpi) and by 23-fold by 48 hpi. This verified strong F. noatunensis subsp. orientalis-mCh growth in the infected embryos, with an estimated doubling time of 9 h.
FIG 1.
F. noatunensis subsp. orientalis-mCh infection of zebrafish embryos. (A) Kaplan-Meier representation of cumulative survival of zebrafish embryos infected with approximately 9 × 103 (OD600 = 2), 9 × 102 (OD600 = 0.2), and 9 × 10 (OD600 = 0.02) CFU F. noatunensis subsp. orientalis-mCh at 28°C. (B) Time-dependent growth of F. noatunensis subsp. orientalis-mCh in zebrafish embryos infected with 9 × 103 CFU. The asterisk indicates a statistically significant difference (P < 0.05). (C). Development of F. noatunensis subsp. orientalis-mCh granuloma-like structures in zebrafish embryos over time: 1 dpi (panels 1.1 and 1.2), 2 dpi (panels 2.1 and 2.2), and 3 dpi (panels 3.1 and 3.2). Insets show granuloma-like structures at higher magnification. (D) The most upregulated genes of zebrafish embryos infected with F. noatunensis subsp. orientalis-mCh. Scale bars, 200 μm (insets, 20 μm).
(i) Imaging of F. noatunensis subsp. orientalis-mCh-infected zebrafish embryos reveals formation of granuloma-like structures.
The first clinical sign of infection was decreased fish motility, usually on day 1 to 2 postinfection. Within hours after infection, we observed accumulation of fluorescently labeled bacteria by fluorescence microscopy. More detailed observation revealed fluorescent bacterial aggregates that enlarged over the course of the infection and were often visible in the infected embryos at late stages of the disease as what we operationally define as granuloma-like structures (GLS) (Fig. 1C). The GLS-containing red fluorescent bacteria formed rapidly and expanded.
(ii) Zebrafish embryo immune response to F. noatunensis subsp. orientalis-mCh infection.
To investigate the immune response of zebrafish embryos to F. noatunensis subsp. orientalis-mCh infection, we used RT-qPCR to monitor the transcription of mRNA for selected immune-related genes. Single amplification peaks were obtained from all primer sets, and the absence of contaminating genomic DNA was verified using controls without reverse transcriptase. As summarized in Fig. 1D and shown in Fig. S2 in the supplemental material, the immune response was dominated by increased transcription of il1b and tnfa. Several pathogens, including Francisella, utilize the suppressors of cytokine signaling (SOCS) pathways to inhibit the host's ability to clear an infection (53). The effect of zebrafish infection with Francisella on the transcription of socs1, socs3a, and socs3b was therefore investigated. Both socs3a and socs3b showed a strong increase in transcription at all time points tested starting from the earliest one (6 hpi), when it was statistically significant only for socs3b, while transcription of socs1 remained unaffected. A similar response was previously reported for F. tularensis infection in murine macrophages (53). There was less effect on other selected gene products, although complement component 3 showed a significant increase at 48 hpi. There was no difference in transcription of ifng1-2 whatsoever. At 48 hpi there was a slight increase in nos2a, but the effect was small and variable between replicates.
(iii) F. noatunensis subsp. orientalis-mCh colocalization with zebrafish embryo phagocytes.
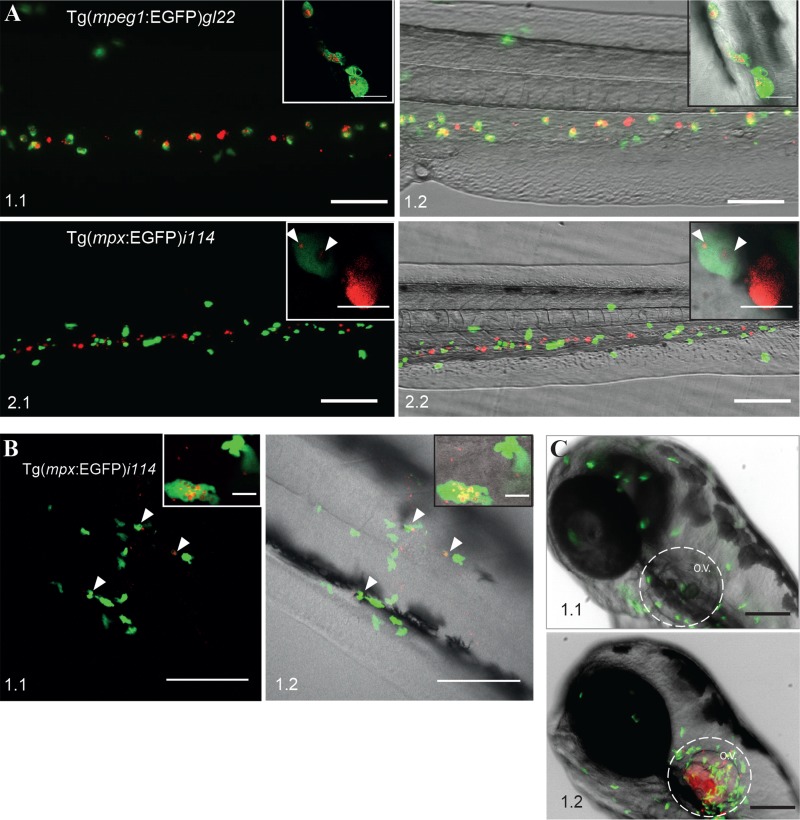
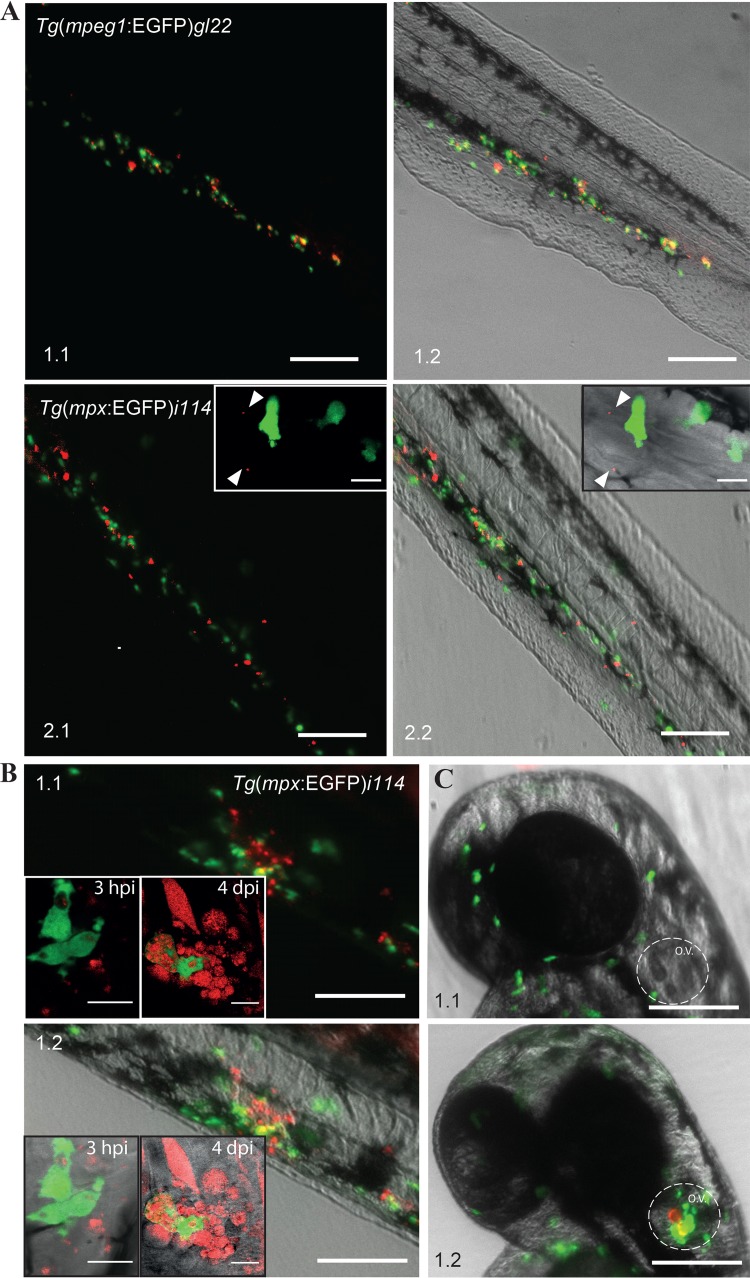
In the early embryo the two prominent phagocytic cells, macrophages and neutrophils, can potentially play a role in bacterial uptake and GLS formation (54). To investigate which cells types were involved in these processes we took advantage of the two transgenic zebrafish lines with either macrophages [Tg(mpeg1:EGFP)gl22] (38) or neutrophils [Tg(mpx:EGFP)i114] expressing EGFP (39). F. noatunensis subsp. orientalis-mCh was injected into the duct of Cuvier of transgenic zebrafish embryos with fluorescently labeled macrophages. The transgenic zebrafish embryos were less robust than the wild type, with full mortality often observed 1 day earlier (data not shown). This might be related to the different genetic background of the zebrafish lines; however, it is unlikely to influence the early colocalization of F. noatunensis subsp. orientalis-mCh with immune cells to any significant extent. It was possible to detect single red fluorescent bacteria in the blood flow, followed by phagocytosis by macrophages within minutes after infection (see Video S1 in the supplemental material). Several hours later the majority of the bacteria were phagocytized by macrophages (Fig. 2A, panels 1.1 and 1.2). Further imaging of F. noatunensis subsp. orientalis-mCh interaction with macrophages was complicated, as the fluorescent signal from the Tg(mpeg1:EGFP)gl22 cells was lost at later time points. Transcription controlled by the mpeg1 promoter has previously been shown to decrease in zebrafish when infected with S. Typhimurium (55). This could explain the lack of EGFP-expressing macrophages at 4 dpi in zebrafish infected with F. noatunensis subsp. orientalis-mCh despite the presence of bacterial aggregates in the tissue, presumably representing intracellular F. noatunensis subsp. orientalis-mCh in macrophages. In contrast to the data obtained for macrophages, when the zebrafish strain with EGFP-expressing neutrophils was injected intravascularly (duct of Cuvier) with F. noatunensis subsp. orientalis-mCh, the amount of bacteria colocalizing with neutrophils was much lower (Fig. 2A, panels 2.1 and 2.2). High-resolution confocal microscopy revealed that even though neutrophils were able to take up F. noatunensis subsp. orientalis-mCh, they were less efficient than macrophages (Fig. 2A, panels 2.1 and 2.2, insets). Nevertheless, these observations showed that both lineages of zebrafish phagocytes were able to take up the bacteria.
FIG 2.
Interaction of F. noatunensis subsp. orientalis-mCh with innate immune cells of zebrafish embryos. (A) F. noatunensis subsp. orientalis-mCh colocalizes with EGFP-expressing macrophages in Tg(mpeg1:EGFP)gl22 zebrafish embryos (panels 1.1 and 1.2; insets show single cells) at 3 hpi, whereas only a low degree of colocalization was observed with EGFP-expressing neutrophils in Tg(mpx:EGFP)i114 zebrafish embryos (panels 2.1 and 2.2; inserts show a single neutrophil with few phagocytized bacteria and a highly loaded macrophage). (B) Neutrophils are attracted to infected tail muscle of Tg(mpx:EGFP)i114 zebrafish embryos and efficiently phagocytize F. noatunensis subsp. orientalis-mCh (panels 1.1 and 1.2; insets show single cells) at 3 hpi. (C) Neutrophils are attracted to F. noatunensis subsp. orientalis-mCh infected otic vesicle of Tg(mpx:EGFP)i114 zebrafish embryos (panel 1.2), in contrast to the control zebrafish injected with PBS at 3 hpi (panel 1.1). Scale bars, 100 μm (A and C) and 50 μm (B) (insets, 20 μm [A, panels 1.1 and 1.2] and 10 μm [A, panels 2.1 and 2.2]).
It has been shown that zebrafish embryonic neutrophils prefer a solid surface for efficient phagocytosis of E. coli (54), and we could substantiate this finding using fluorescent E. coli. Tg(mpx:EGFP)i114 zebrafish embryos were then injected with F. noatunensis subsp. orientalis-mCh into the tail muscle at 3 days postfertilization (dpf) to investigate whether Francisella spp. can be phagocytized by neutrophils more efficiently than after intravenous injection. Tail muscle infection was followed by several hours of in vivo high-resolution confocal microscopy imaging. We observed a strong attraction of neutrophils to the injection site, consistent with an inflammatory response (Fig. 2B), and indeed the activated neutrophils had a higher bacterial burden than was the case when the fish were infected via the circulatory system (Fig. 2B). The ability of F. noatunensis subsp. orientalis-mCh to attract neutrophils was even more evident when F. noatunensis subsp. orientalis-mCh was injected into the otic vesicle (ear), which prior to the infection was devoid of neutrophils (Fig. 2C). A strong neutrophil attraction to the vicinity of the otic vesicle was observed at 3 hpi (Fig. 2C).
In summary, F. noatunensis subsp. orientalis-mCh was capable of entering both lineages of zebrafish phagocytes. The macrophages were clearly the main phagocytes for taking up the bacteria. Nevertheless neutrophils were also rapidly attracted to the bacteria but were able to phagocytize them efficiently only when F. noatunensis subsp. orientalis-mCh were present on a surface (muscle tissue).
Model 2, infection of zebrafish with F. noatunensis subsp. noatunensis at 22°C: F. noatunensis subsp. noatunensis infection of zebrafish embryos causes chronic disease.
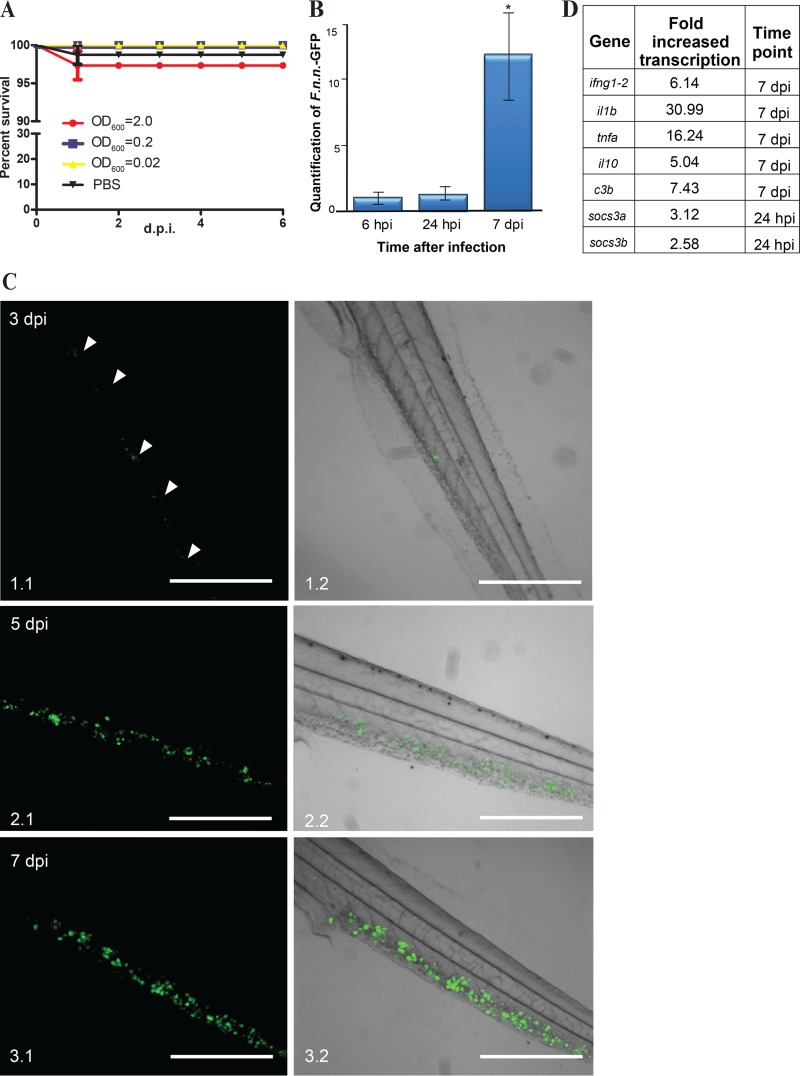
F. noatunensis subsp. noatunensis has an optimal growth temperature of 20 to 22°C. Therefore, the infected embryos were kept at 22°C after infection and at 28°C prior to it. In striking contrast to infections with F. noatunensis subsp. orientalis-mCh, the F. noatunensis subsp. noatunensis-infected zebrafish embryos showed no significant mortality regardless of the bacterial load used for infection (OD600 of 0.2 or 2.0) (Fig. 3A). F. noatunensis subsp. noatunensis-GFP was detected in infected embryos by RT-qPCR at 6 hpi (Fig. 3B), and although no significant bacterial growth was observed by 24 hpi, there was an approximately 13-fold increase in the amount of bacteria in the fish by 7 dpi, yielding an estimated doubling time of 43 h. The lower growth rate in vivo of F. noatunensis subsp. noatunensis compared to F. noatunensis subsp. orientalis likely reflects the lower in vitro growth rate of this strain. This absence of mortality is similar to the situation in the natural host, the Atlantic cod, when experimentally challenged with F. noatunensis subsp. noatunensis. In this host, even when injecting high doses of bacteria, there is often a lack of mortality although formation of granulomas is visible by the naked eye (5, 29, 56).
FIG 3.
F. noatunensis subsp. noatunensis-GFP infection of zebrafish embryos. (A) Kaplan-Meier representation of cumulative survival of zebrafish embryos infected with approximately 9 × 103 (OD600 = 2), 9 × 102 (OD600 = 0.2), and 9 × 10 (OD600 = 0.02) CFU of F. noatunensis subsp. noatunensis-GFP at 22°C. (B) Time-dependent growth of F. noatunensis subsp. noatunensis-GFP in zebrafish embryos infected with 9 × 103 CFU. The asterisk indicates a statistically significant difference (P < 0.05). (C) Development of F. noatunensis subsp. noatunensis-GFP granuloma-like structures in zebrafish embryos over time: 3 dpi (panels 1.1 and 1.2), 5 dpi (panels 2.1 and 2.2), and 7 dpi (panels 3.1 and 3.2). (D) The most upregulated genes of zebrafish embryos infected with F. noatunensis subsp. noatunensis-GFP. Scale bars, 200 μm.
(i) Imaging of GLS development in F. noatunensis subsp. noatunensis-GFP-infected zebrafish embryos.
It was possible to detect GLS formation within 3 days postinfection of embryos with F. noatunensis subsp. noatunensis-GFP via the intravenous route. The GLS were distributed throughout the fish (Fig. 3C). The size, number, and fluorescent intensity of GLS increased with time, indicating growth of the bacteria in infected embryos. The development of the GLS (Fig. 3C) resembled that seen with F. noatunensis subsp. orientalis-mCh infection; however, consistent with the lower temperature used for F. noatunensis subsp. noatunensis, the formation of GLS was observed 1 to 2 days later than in the case with F. noatunensis subsp. orientalis-mCh infection.
When embryos were kept at their optimal growth temperature of 28°C, it was not possible to detect significant amounts of F. noatunensis subsp. noatunensis-GFP by fluorescent imaging after intravascular injections, suggesting that this temperature is not compatible with bacterial proliferation. This is in agreement with the in vitro growth characteristics of F. noatunensis subsp. noatunensis (57).
(ii) Immune response of zebrafish embryos infected with F. noatunensis subsp. noatunensis-GFP.
The immune response of zebrafish embryos to F. noatunensis subsp. noatunensis-GFP infection was dominated by the same pattern as the immune response to F. noatunensis subsp. orientalis-mCh, although it was somewhat delayed and reduced (Fig. 3D; see Fig. S3 in the supplemental material). The transcription levels of tnfa increased by 15-fold, those of il10 increased by 4-fold, those of complement factor increased by 3- to 6-fold, those of ifng1-2 increased by 5.1-fold, and those of il1b reached a 30-fold increase on 7 dpi and were slightly elevated at 24 hpi (approximately a 2-fold increase). With regard to the socs genes, no change was detected for socs1; however, statistically significant differences were detected for socs3a at 24 hpi and socs3b at 7 dpi (2.1- and 0.85-fold increases, respectively). The slight elevation of socs3 was markedly reduced compared to the increase observed in zebrafish embryos infected with F. noatunensis subsp. orientalis-mCh, probably reflecting a lower bacterial burden and/or a temperature dependency in the host response.
(iii) Interaction of F. noatunensis subsp. noatunensis-mCh with zebrafish embryo immune cells.
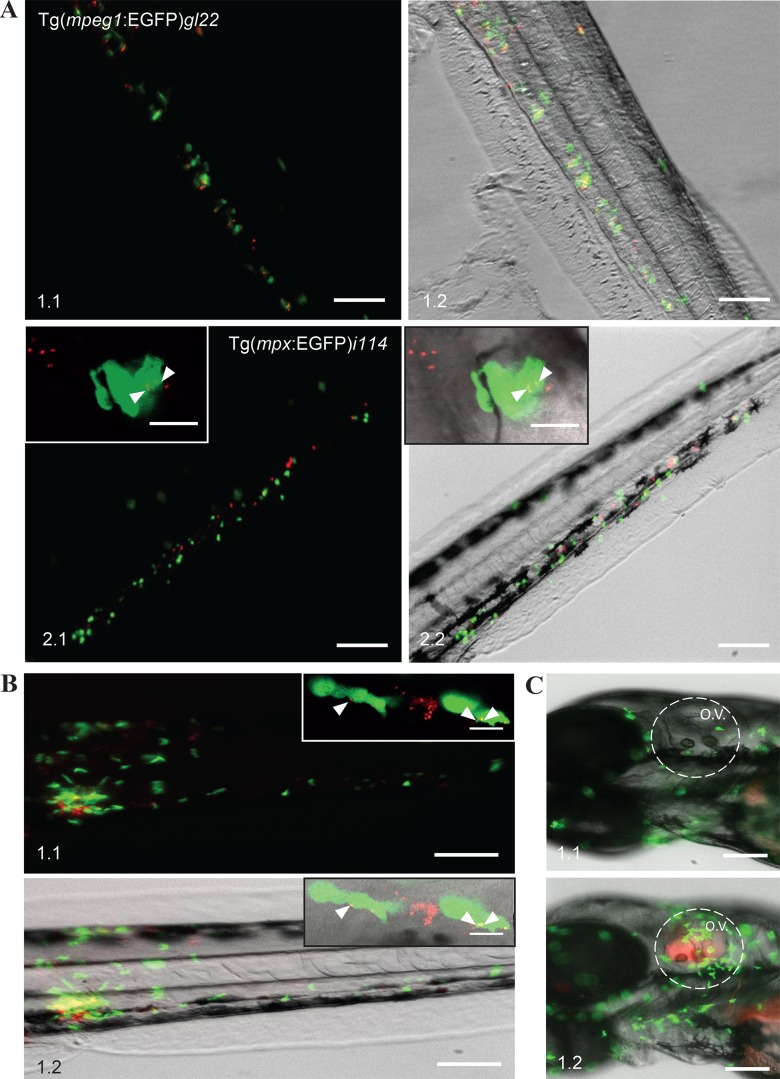
To allow for colocalization studies of F. noatunensis subsp. noatunensis with transgenic zebrafish strains expressing EGFP-labeled immune cells, wild-type F. noatunensis subsp. noatunensis was transformed with the pKK289Km plasmid containing the mCherry gene in exchange for gfp. The zebrafish embryos infected with F. noatunensis subsp. noatunensis-mCh showed the same survival curves as the wild-type fish (data not shown). Colocalization of a majority of the bacteria and macrophages was observed as early as 3 hpi (Fig. 4A) and lasted for at least another 5 days. Clusters of bacteria not colocalized with EGFP-expressing macrophages were also visible at 4 dpi (data not shown). This observation suggests that these bacteria were able to kill the macrophages and reside free in the tissue or, more likely, represent intracellular bacteria in macrophages which have lost their fluorescence as a result of downregulation of EGFP expression at later time points of infection, as discussed above. Similar to the case for F. noatunensis subsp. orientalis-mCh infection, the amount of bacteria colocalizing with neutrophils was much less than that for macrophages (Fig. 4A), although small amounts of F. noatunensis subsp. noatunensis-mCh could be detected inside neutrophils after intravascular injections when observed by confocal microscopy (Fig. 4A, panels 2.1 and 2.2, insets). Therefore, macrophages are the main cells responsible for phagocytosis of intravascularly injected F. noatunensis subsp. noatunensis-mCh as well. Infection of Tg(mpx:EGFP)i114 zebrafish embryos with F. noatunensis subsp. noatunensis-mCh was also performed by injections into the tail muscle (Fig. 4B) and otic vesicle (Fig. 4C). Imaging of the fish at 3 hpi revealed that neutrophils were strongly attracted to the sites of injections. Confocal microscopy identified small numbers of intracellular bacteria in neutrophils when F. noatunensis subsp. noatunensis-mCh was injected into tissue, verifying that F. noatunensis subsp. noatunensis also is able to infect this phagocytic cell (Fig. 4B, panels 1.1. and 1.2, insets).
FIG 4.
Interaction of F. noatunensis subsp. noatunensis-mCh with innate immune cells. (A) Colocalization of F. noatunensis subsp. noatunensis-mCh and macrophages in Tg(mpeg1:EGFP)gl22 zebrafish embryos (panels 1.1 and 1.2) and no or little colocalization between F. noatunensis subsp. noatunensis-mCh and neutrophils in the Tg(mpx:EGFP)i114 zebrafish line at 3 hpi (panels 2.1 and 2.2; insets show neutrophils with few phagocytized bacteria). (B) Injection of Tg(mpx:EGFP)i114 zebrafish embryos with F. noatunensis subsp. noatunensis-mCh suspension into the tail muscle demonstrates attraction of neutrophils to the bacteria followed by phagocytosis at 3 hpi (panels 1.1 and 1.2). Insets show slightly loaded neutrophils and a macrophage with a high red fluorescent bacterial burden. (C) The otic vesicle of Tg(mpx:EGFP)i114 zebrafish embryos injected with PBS (panel 1.1) excludes neutrophils, while they are attracted in the case of F. noatunensis subsp. noatunensis-mCh infection (panel 1.2). Scale bars, 100 μm (insets, 10 μm).
Model 3, infection of zebrafish embryos with F. tularensis subsp. novicida at 32°C: F. tularensis subsp. novicida causes 60% mortality in zebrafish embryos.
When injected into the duct of Cuvier, F. tularensis subsp. novicida-GFP was capable of causing mortality of embryos kept at both the optimal temperature for the embryos (28°C) and an elevated temperature more adapted to bacterial growth (32°C). F. tularensis subsp. novicida-GFP infection developed at a lower rate than F. noatunensis subsp. orientalis-mCh infection. Cumulative mortality after 6 days for the embryos infected with F. tularensis subsp. novicida-GFP at an OD600 of 2.0 reached 60% and 50% at 28°C and 32°C, respectively (Fig. 5A). The onset of mortality occurred earlier at 32°C even though the cumulative mortality was almost equal at 32°C and 28°C. Zebrafish embryos tolerated the elevated temperature well, showing no abnormalities in behavior or development. Taking these data into consideration, the remaining experiments were carried out at 32°C to approach the normal growth conditions of F. tularensis subsp. novicida. F. tularensis subsp. novicida grows faster than the Francisella fish pathogens in vitro, which was also reflected in the bacterial growth rate in infected fish at 32°C as determined by RT-qPCR. F. tularensis subsp. novicida-GFP increased by 16-fold from 6 hpi to 24 hpi and by more than 55-fold by 48 hpi (Fig. 5B), yielding an estimated doubling time of 7 h.
FIG 5.
F. tularensis subsp. novicida-GFP infection of zebrafish embryos. (A) Kaplan-Meier representation of cumulative survival of zebrafish embryos infected with approximately 3 × 104 (OD600 = 2.0) and 3 × 103 (OD600 = 0.2) CFU of F. tularensis subsp. novicida-GFP and kept at 28°C (upper graph) or 32°C (lower graph). (B) Time-dependent growth of F. tularensis subsp. novicida-GFP in zebrafish embryos infected with 3 × 104 CFU. The asterisk indicates a statistically significant difference (P < 0.05). (C). Granuloma-like structures in zebrafish infected with F. tularensis subsp. novicida-mCh at 4 dpi. (D) The most upregulated genes of zebrafish embryos infected with F. tularensis subsp. novicida-GFP. Scale bar, 100 μm.
(i) Imaging of zebrafish embryos infected with F. tularensis subsp. novicida-GFP.
A peculiarity of F. tularensis subsp. novicida-GFP infection led to moderate GLS formation compared to that with F. noatunensis infections. A common observation was several small bacterial aggregations rather than the larger aggregates seen with the two other strains of Francisella used in this study (Fig. 5C). Even though zebrafish embryos heavily loaded with granulomas were observed in the infected groups, those were not numerous (data not shown). The factors responsible for the observed differences in disease development remain to be investigated even though this can arguably be explained by the fact that the infection studies were performed at a temperature 5°C lower than optimal for growth of F. tularensis subsp. novicida.
(ii) Immune response of zebrafish embryos infected with F. tularensis subsp. novicida-GFP.
The zebrafish embryo host immune response to F. tularensis subsp. novicida-GFP infection, as evaluated by RT-qPCR, revealed increased transcription of key immune molecules, dominated by il1b and tnfa, starting as early as 6 hpi (Fig. 5D; see Fig. S4 in the supplemental material). Other immune-related genes showed less response, though c3b, mxa, and il12a also showed statistically significantly increased transcription. Additionally, nos2a was increased at 48 hpi, although only by a factor of 2.5. Interestingly, the socs3 genes exhibited a fast and strong response, especially socs3b (189-fold increase at 6 hpi and 94-fold increase at 24 hpi). The socs3a gene responded slower, with a 4-fold increase (not statistically significant) at 6 hpi and a 96-fold increase at 24 hpi. No transcriptional differences were observed for socs1, except for a slight elevation (0.75-fold) at 48 hpi.
(iii) Interaction of F. tularensis subsp. novicida with host immune cells.
Infection of the previously mentioned transgenic zebrafish embryos with F. tularensis subsp. novicida-mCh resulted in an outcome similar to those for F. noatunensis subsp. orientalis-mCh and F. noatunensis subsp. noatunensis-mCh. Intravascularly injected F. tularensis subsp. novicida-mCh preferably entered macrophages (Fig. 6A). Neutrophils were strongly attracted to the sites of bacterial location (Fig. 6B and C), though they were even less efficient in phagocytizing F. tularensis subsp. novicida-mCh (Fig. 6A; see Video S2 in the supplemental material) than in the case of F. noatunensis subsp. orientalis-mCh and F. noatunensis subsp. noatunensis-mCh when the bacteria were injected into circulation. F. tularensis subsp. novicida-mCh was taken up by neutrophils in the tail muscle infection (Fig. 6B; see Video S3 in the supplemental material), as seen for the two F. noatunensis subspecies. Interestingly, F. tularensis subsp. novicida-mCh was detected in neutrophils also at later stages of the infection (4 dpi), when GLS are formed (see Video S4 in the supplemental material), though the majority of the bacteria remained colocalized with macrophages (Fig. 6B, panels 1.1 and 1.2, insets). This suggests that F. tularensis subsp. novicida-mCh is able to survive in neutrophils after phagocytosis.
FIG 6.
Interaction of F. tularensis subsp. novicida-mCh with innate immune cells. (A) EGFP-expressing macrophages in Tg(mpeg1:EGFP)gl22 zebrafish embryos show a high degree of colocalization with F. tularensis subsp. novicida-mCh (panels 1.1 and 1.2), but EGFP-expressing neutrophils in Tg(mpx:EGFP)i114 zebrafish embryos and F. tularensis subsp. novicida-mCh are not often found to be colocalized (panels 2.1 and 2.2). Insets show single neutrophils at 3 hpi. (B) Tg(mpx:EGFP)i114 zebrafish embryos injected with F. tularensis subsp. novicida-mCh suspension into tail muscle. Neutrophils are attracted to the site of injection and are able to phagocytize the bacteria at 3 hpi. (C) Neutrophils are attracted to F. tularensis subsp. novicida-mCh-infected otic vesicle of Tg(mpx:EGFP)i114 zebrafish embryos (panel 1.2) but not in the case of PBS injection (panel 1.1). Scale bars, 100 μm (insets, 10 μm).
DISCUSSION
Due to the highly transparent embryonic stage, the zebrafish is increasingly used as a robust model system for the analysis of various bacterial infections (31, 54, 58). Our present study increases the value of this model by showing that zebrafish embryos can be adapted to significantly different temperatures to monitor infections with three different strains of Francisella. Two of these strains are important pathogens of aquaculture fish. F. noatunensis subsp. noatunensis is a pathogen of “cold-water” fish such as Atlantic cod and Atlantic salmon, and it could infect but not kill the embryos at 22°C. F. noatunensis subsp. orientalis, which infects “warm-water” commercial fish such as tilapia and has the same optimal growth temperature as zebrafish (28°C), caused rapidly developing disease and lethality in zebrafish embryos. For the third strain, F. tularensis subsp. novicida, a mouse pathogen that is widely used as a model for the human pathogen F. tularensis subsp. tularensis, the embryos were kept at 32°C in a compromise between optimal growth conditions for the bacteria and maintaining good health of the zebrafish embryos. Such a compromise could be the reason for a relatively mild infection with intermediate mortality, in spite of strong bacterial growth.
Although the preferred growth temperature of the zebrafish is 28°C, the embryos adapted well to both increased and decreased temperatures, in agreement with earlier studies focusing on viral infections of embryos at 24°C and for short periods of time at 15°C (49, 59). No abnormalities were evident in the appearance, behavior, or mortality profiles of noninfected embryos at the different temperatures. At all temperatures tested, the three bacterial strains could successfully infect the fish regardless the route of infection (circulatory system, tail muscle, or otic vesicle). The qPCR analysis showed that all three bacterial strains were able to multiply in zebrafish embryos infected via the intravascular route. The generation intervals for the different Francisella strains reflected their in vitro growth characteristics. The fastest-growing F. tularensis subsp. novicida has a generation interval of about 5 h in broth (60), which is only slightly less than the estimated 7 h observed in vivo in this study. The shorter doubling time in zebrafish of F. noatunensis subsp. orientalis (9 h) than of F. noatunensis subsp. noatunensis (43 h) is in agreement with the in vitro growth rates of these strains (37) also estimated by CFU counts (unpublished data).
The responses of the embryos to the different Francisella species were similar, based on a number of criteria. First, there was significant replication of all three bacterial strains as determined by RT-qPCR, despite a pronounced response by the fish immune system. Second, mCherry-labeled bacteria of all three species interacted similarly with zebrafish immune cells. These data were revealed by taking advantage of the availability and ease of imaging of different marker cell types in transgenic zebrafish lines. The EGFP-fluorescing macrophage transgenic fish line Tg(mpeg1:EGFP)gl22 has been previously used in other studies for determining the association of pathogens with these cells (see, e.g., reference 61). Here, the use of this transgenic fish (38) clearly demonstrated that the macrophage is the main Francisella-phagocytizing cell type, which is in agreement with earlier studies with mouse and human macrophages (62, 63). Imaging of the formed GLS showed that macrophages were the major site of Francisella residence, and even though the bacteria persisted in neutrophils, the amount was still limited at 4 dpi. Since the half-life of zebrafish neutrophils is 120 h (64), we observed the same cells over a few days. Therefore, it can be argued that Francisella does not multiply in neutrophils and that the main site of replication is probably within macrophages.
Using the EGFP-expressing neutrophil line Tg(mpx:EGFP)i114 (39) allowed observation of strong neutrophil attraction to the sites of bacterial location, though in comparison to macrophages, these cells were less efficient at phagocytosis of Francisella spp., particularly from the circulatory system. Previously, Colucci-Guyon et al. (54) reported that zebrafish neutrophils required a surface for efficient phagocytosis of nonpathogenic E. coli. We also observed efficient neutrophil uptake of E. coli pmCherry injected into tissue, and we extended the observation by showing that this striking behavior is also valid for pathogenic Francisella strains (Fig. 2A and B, 4A and B, and 6A and B). In the case of intravenous infections, neutrophils showed only modest phagocytic activity toward F. noatunensis subsp. orientalis and F. noatunensis subsp. noatunensis and were not found to colocalize at all with F. tularensis subsp. novicida Even though neutrophils appeared to be more successful in taking up Francisella subsp. from the tail muscle substrate, they were still less efficient in overall phagocytosis than macrophages. Our findings are consistent with existing reports of bacterial interaction with immune cells in their natural hosts. For example, previous studies of F. noatunensis subsp. noatunensis in Atlantic cod showed bacteria in the close proximity of neutrophils but never or rarely inside them (22, 56). F. tularensis subsp. novicida has been demonstrated to be efficiently taken up by neutrophils in a mouse pulmonary infection model (63), similar to the results for F. tularensis subsp. novicida injected into muscle tissue observed in this study. Regardless of the potential efficiency of phagocytosis, the results show that all three strains of Francisella investigated in this study were taken up by both lineages of zebrafish phagocytes. The efficiency of zebrafish neutrophils in phagocytizing bacteria in blood is likely to be dependent on the specific bacterial species, as intravascularly injected Pseudomonas aeruginosa is efficiently taken up and killed by zebrafish neutrophils (65).
The third criterion that was similar for the three bacterial infections was the fact that the infected macrophages could assemble into what we operationally define as granuloma-like structures (GLS). Differences were noted in the strength of the granulomatous response, with the largest GLS seen with F. noatunensis subsp. orientalis at 28°C and the least developed ones seen with F. tularensis subsp. novicida at 32°C. We hypothesize that the studied Francisella species have different potentials for causing GLS formation in zebrafish, which might also be influenced by the differences in temperature. The fourth criterion of similarity came from the analysis of the innate immune response of the embryos to the three bacterial strains, as similar patterns of gene transcription were seen. Increased mRNAs levels for il1b, tnfa, and socs3b in all three strains were observed, with less but still significantly increased transcription of c3b, il10, il12a, and ifng1-2 for some strains. The fifth criterion indicative of a successful infection by two of the three pathogens was their ability to kill all of F. noatunensis subsp. orientalis-infected, and a significant fraction of F. tularensis subsp. novicida-infected, embryos. An exception was seen with F. noatunensis subsp. noatunensis, which failed to kill any embryos in the time frame of the experiment at 22°C. This bacterium induced a chronic rather than a lethal infection. Interestingly, F. noatunensis subsp. noatunensis causes similar chronic infections in its natural host, the Atlantic cod (4, 29, 55), highlighting the strong potential of the zebrafish model. Another significant difference in the immune response of F. noatunensis subsp. noatunensis that was not seen with the other two bacteria was an induction of the anti-inflammatory cytokine IL-10; this might contribute to the absence of lethality with F. noatunensis subsp. noatunensis, as well as the lower growth rate of F. noatunensis subsp. noatunensis compared to the two other strains in infected fish. A lack of IL-10 production in systemic infections can result in higher levels of proinflammatory cytokines, more tissue damage, and earlier death, while excessive production of IL-10 can cause decreased ability to clear pathogens, resulting in chronic or lethal infections (66).
An earlier study by Vojtech et al. (36) showed that adult zebrafish at 28°C could be infected with a strain of F. noatunensis subsp. orientalis that induced impressive early GLS formation, as seen by histological examination. While the adult fish has both the innate and adaptive immune systems operating, the embryos have only innate immunity. Thus, it is notable that two of the immune gene products that were strongly upregulated in the analysis of adult fish, namely, il1b and tnfa, were also prominently elevated in our infected zebrafish embryos. One clear difference was that whereas the transcription of ifng1-2 was increased in adult fish, we saw no significant elevation in the embryos in response to infection with F. noatunensis subsp. orientalis (although it was upregulated with F. noatunensis subsp. noatunensis and F. tularensis subsp. novicida). The increased transcription of il1b, il10, and ifng1-2 at 7 dpi for F. noatunensis subsp. noatunensis also resembles the gene transcription results seen in Atlantic cod (29) and primary macrophages isolated from Atlantic cod (21). This argues that immune responses initiated upon recognition of Francisella spp. are conserved between different fish species and Francisella strains. Obviously, the additional activity of the adaptive immune system is crucial for full defense against Francisella, as Vojtech and colleagues found that 106 CFU of F. noatunensis subsp. orientalis was needed to kill 100% of adult fish (36), whereas only 9 × 102 CFU led to the same result for zebrafish embryos.
The question of how Francisella spp. are able to persist in infected macrophages, despite their potential to kill and degrade phagocytized pathogens, is crucial for understanding the pathogenesis of Francisella infections. It has previously been shown for Francisella that TNF-α and IFN-γ are responsible for inducing inhibition of bacterial growth and activation of bacterial killing by macrophages and that this effect is directly related to increased production of nitric oxide (NO) (67). NO is generated in macrophages by the inducible NO synthase (iNOS), the transcription of which is induced through the JAK-STAT signaling pathway by IFN-γ and through NF-κB activation by TNF-α and IFN-γ (68). In spite of our observations of increased transcription of these genes, neither we nor Vojtech et al. (36) could observe strong increases in other expected downstream effector systems, such as iNOS. This bacterium-induced inhibition is thought to be mediated by active inhibition by a bacterial heat-stable factor that has been shown to work in mammals by inhibiting STAT-dependent signaling by increased transcription of socs3 (53). The SOCS proteins have been identified in many fish species (69), including zebrafish, and increased transcription of socs3 was observed in response to infection of zebrafish embryos with all three Francisella strains. This argues that the active suppression of the host immune response in infection by Francisella spp. is an ancient mechanism that is conserved between different Francisella species and between mammals and fish. Mycobacterium marinum and S. Typhimurium have previously been reported to induce socs3 in zebrafish (55, 70), opening up the possibility that this is a common pathway used by facultative intracellular bacteria to inhibit activation of zebrafish macrophages.
In conclusion, we have expanded the strength of zebrafish embryos as a powerful and robust model for studying bacterial pathogenesis, adding three Francisella subspecies to the list of microorganisms to which the embryos are susceptible. This opens up new possibilities for studying Francisella pathogenesis due to the capability for in vivo imaging of early events that happen right after infection. Zebrafish embryos appeared to be a reliable model at three different temperatures without apparent adverse effects on their health. We clearly showed the major role of macrophages in the phagocytosis of invading Francisella cells. Neutrophils were capable of quickly responding to the infection by migrating to the bacterial locations, though they had a variable degree of success in phagocytizing bacteria, depending on the route of infection. Our data also demonstrate that the host response to Francisella infections is similar for different species. Conclusively, the data obtained in this study lay a solid background for further investigation of the infections caused by different Francisella species.
Supplementary Material
ACKNOWLEDGMENTS
We greatly appreciate the support of Jan-Roger Torp Sørby and Peter Aleström in setting up the zebrafish facility in the Institute of Biological Sciences and the help of Jan-Roger Torp Sørby and Ana Carolina Sulen Tavara in maintaining our fish stocks. We thank Stephen Renshaw and Graham Leischke for permission to use the Tg(mpx1:EGFP)i114 and Tg(mpeg1:EGFP)gl22 transgenic strains, respectively, and Annemarie Meijer for sending us these strains. The pKK289Km/gfp plasmid was a kind gift from Emelie N. Salomonsson, and the F. tularensis subsp. novicida wt strain was a kind gift from Tina Guina and Åke Forsberg. The F. noatunensis subsp. orientalis wt strain was a kind gift from Duncan J. Colquhoun. We also thank Tor Gjøen for lending us the primers to carry out some of the zebrafish immune response studies.
This work was supported by funds from the Norwegian School of Veterinary Sciences (E.B.), the University of Oslo (L.S.U. and E.O.L), and the Research Council of Norway.
E.B. planned and performed some of the experiments and was involved in writing the paper. L.S.U. planned and performed some of the experiments and was involved in writing the paper. E.O.L. planned and performed some of the experiments. A.-L.R. performed some of the experiments. G.G.'s group set up the zebrafish facility and contributed to planning the experiments and writing the paper. H.C.W-L. performed some of the experiments and contributed to the design of the experiments and writing the paper.
Footnotes
Published ahead of print 10 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00077-14.
REFERENCES
- 1. Ellis J, Oyston PC, Green M, Titball RW. 2002. Tularemia. Clin. Microbiol. Rev. 15:631–646. 10.1128/CMR.15.4.631-646.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Keim P, Johansson A, Wagner DM. 2007. Molecular epidemiology, evolution, and ecology of Francisella. Ann. N. Y. Acad. Sci. 1105:30–66. 10.1196/annals.1409.011 [DOI] [PubMed] [Google Scholar]
- 3. Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Tonat K. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285:2763–2773. 10.1001/jama.285.21.2763 [DOI] [PubMed] [Google Scholar]
- 4. Mikalsen J, Olsen AB, Rudra H, Moldal T, Lund H, Djønne B, Bergh O, Colquhoun DJ. 2009. Virulence and pathogenicity of Francisella philomiragia subsp. noatunensis for Atlantic cod, Gadus morhua L., and laboratory mice. J. Fish Dis. 32:377–381. 10.1111/j.1365-2761.2008.00987.x [DOI] [PubMed] [Google Scholar]
- 5. Colquhoun DJ, Duodu S. 2011. Francisella infections in farmed and wild aquatic organisms. Vet. Res. 42:47. 10.1186/1297-9716-42-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamaishi T, Fukuda Y, Nishiyama M, Kawakami M, Matsuyama T, Yoshinaga T, Oseko N. 2005. Identification and pathogenicity of intracellular Francisella bacterium in three-line grunt Parapristipoma trilineatum. Fish Pathol. 40:67–71. 10.3147/jsfp.40.67 [DOI] [Google Scholar]
- 7. Mauel MJ, Miller DL, Styer E, Pouder DB, Yanong RP, Goodwin AE, Schwedler TE. 2005. Occurrence of piscirickettsiosis-like syndrome in tilapia in the continental United States. J. Vet. Diagn. Invest. 17:601–605. 10.1177/104063870501700616 [DOI] [PubMed] [Google Scholar]
- 8. Mauel MJ, Soto E, Moralis JA, Hawke J. 2007. A piscirickettsiosis-like syndrome in cultured Nile tilapia in Latin America with Francisella spp. as the pathogenic agent. J. Aquat. Anim. Health 19:27–34. 10.1577/H06-025.1 [DOI] [PubMed] [Google Scholar]
- 9. Jeffery KR, Stone D, Feist SW, Verner-Jeffreys DW. 2010. An outbreak of disease caused by Francisella sp. in Nile tilapia Oreochromis niloticus at a recirculation fish farm in the UK. Dis. Aquat. Organ. 91:161–165. 10.3354/dao02260 [DOI] [PubMed] [Google Scholar]
- 10. Birkbeck TH, Bordevik M, Frøystad MK, Baklien A. 2007. Identification of Francisella sp. from Atlantic salmon, Salmo salar L., in Chile. J. Fish Dis. 30:505–507. 10.1111/j.1365-2761.2007.00837.x [DOI] [PubMed] [Google Scholar]
- 11. Olsen AB, Mikalsen J, Rode M, Alfjorden A, Hoel E, Straum-Lie K, Haldorsen R, Colquhoun DJ. 2006. A novel systemic granulomatous inflammatory disease in farmed Atlantic cod, Gadus morhua L., associated with a bacterium belonging to the genus Francisella. J. Fish Dis. 29:307–311. 10.1111/j.1365-2761.2006.00714.x [DOI] [PubMed] [Google Scholar]
- 12. Nylund A, Ottem KF, Watanabe K, Karlsbakk E, Krossøy B. 2006. Francisella sp. (Family Francisellaceae) causing mortality in Norwegian cod (Gadus morhua) farming. Arch. Microbiol. 185:383–392. 10.1007/s00203-006-0109-5 [DOI] [PubMed] [Google Scholar]
- 13. Jones RM, Nicas M, Hubbard A, Sylvester MD, Reingold A. 2005. The infectious dose of Francisella tularensis (tularemia). Appl. Biosaf. 10:227–239 [Google Scholar]
- 14. Soto E, Fernandez D, Hawke JP. 2009. Attenuation of the fish pathogen Francisella sp. by mutation of the iglC* gene. J. Aquat. Anim. Health 21:140–149. 10.1577/H08-056.1 [DOI] [PubMed] [Google Scholar]
- 15. Kamaishi T, Miwa S, Goto E, Matsuyama T, Oseko N. 2010. Mass mortality of giant abalone Haliotis gigantea caused by a Francisella sp. bacterium. Dis. Aquat. Organ. 89:145–154. 10.3354/dao02188 [DOI] [PubMed] [Google Scholar]
- 16. Soto E, Hawke JP, Fernandez D, Morales JA. 2009. Francisella sp., an emerging pathogen of tilapia, Oreochromis niloticus (L.), in Costa Rica. J. Fish Dis. 32:713–722. 10.1111/j.1365-2761.2009.01070.x [DOI] [PubMed] [Google Scholar]
- 17. Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjöstedt A. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 71:5940–5950. 10.1128/IAI.71.10.5940-5950.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chong A, Celli J. 2010. The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front. Microbiol. 1:138 doi:. 10.3389/fmicb.2010.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. 2006. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. U. S. A. 103:14578–14583. 10.1073/pnas.0601838103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soto E, Fernandez D, Thune R, Hawke JP. 2010. Interaction of Francisella asiatica with tilapia (Oreochromis niloticus) innate immunity. Infect. Immun. 78:2070–2078. 10.1128/IAI.01308-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bakkemo KR, Mikkelsen H, Bordevik M, Torgersen J, Winther-Larsen HC, Vanberg C, Olsen R, Johansen LH, Seppola M. 2011. Intracellular localisation and innate immune responses following Francisella noatunensis infection of Atlantic cod (Gadus morhua) macrophages. Fish Shellfish Immunol. 31:993–1004. 10.1016/j.fsi.2011.08.020 [DOI] [PubMed] [Google Scholar]
- 22. Furevik A, Pettersen EF, Colquhoun D, Wergeland HI. 2011. The intracellular lifestyle of Francisella noatunensis in Atlantic cod (Gadus morhua L.) leucocytes. Fish Shellfish Immunol. 30:488–494. 10.1016/j.fsi.2010.11.019 [DOI] [PubMed] [Google Scholar]
- 23. Lyons CR, Wu TH. 2007. Animal models of Francisella tularensis infection. Ann. N. Y. Acad. Sci. 1105:238–265. 10.1196/annals.1409.003 [DOI] [PubMed] [Google Scholar]
- 24. Cross AS, Calia FM, Edelman R. 2007. From rabbits to humans: the contributions of Dr. Theodore E. Woodward to tularemia research. Clin. Infect. Dis. 45:S61–S67. 10.1086/518150 [DOI] [PubMed] [Google Scholar]
- 25. Pechous RD, McCarthy TR, Zahrt TC. 2009. Working toward the future: insights into Francisella tularensis pathogenesis and vaccine development. Microbiol. Mol. Biol. Rev. 73:684–711. 10.1128/MMBR.00028-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hepburn MJ, Purcell BK, Lawler JV, Coyne SR, Petitt PL, Sellers KD, Norwood DA, Ulrich MP. 2006. Live vaccine strain Francisella tularensis is detectable at the inoculation site but not in blood after vaccination against tularemia. Clin. Infect. Dis. 43:711–716. 10.1086/506348 [DOI] [PubMed] [Google Scholar]
- 27. Fuller CL, Brittingham KC, Hepburn MJ, Martin JW, Petitt PL, Pittman PR, Bavari S. 2006. Dominance of human innate immune responses in primary Francisella tularensis live vaccine strain vaccination. J. Allergy Clin. Immunol. 117:1186–1188 (Letter.) 10.1016/j.jaci.2006.01.044 [DOI] [PubMed] [Google Scholar]
- 28. Soto E, Wiles J, Elzer P, Macaluso K, Hawke JP. 2011. Attenuated Francisella asiatica iglC mutant induces protective immunity to francisellosis in tilapia. Vaccine 29:593–598. 10.1016/j.vaccine.2010.06.040 [DOI] [PubMed] [Google Scholar]
- 29. Ellingsen T, Inami M, Gjessing MC, Van Nieuwenhove K, Larsen R, Seppola M, Lund V, Schrøder MB. 2011. Francisella noatunensis in Atlantic cod (Gadus morhua L.); waterborne transmission and immune responses. Fish Shellfish Immunol. 31:326–333. 10.1016/j.fsi.2011.05.021 [DOI] [PubMed] [Google Scholar]
- 30. Phelps HA, Neely MN. 2005. Evolution of the zebrafish model: from development to immunity and infectious disease. Zebrafish 2:87–103. 10.1089/zeb.2005.2.87 [DOI] [PubMed] [Google Scholar]
- 31. Meijer AH, Spaink HP. 2011. Host-pathogen interactions made transparent with the zebrafish model. Curr. Drug Targets 12:1000–1017. 10.2174/138945011795677809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Allen JP, Neely MN. 2010. Trolling for the ideal model host: zebrafish take the bait. Future Microbiol. 5:563–569. 10.2217/fmb.10.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lam SH, Chua HL, Gong Z, Lam TJ, Sin YM. 2004. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev. Comp. Immunol. 28:9–28. 10.1016/S0145-305X(03)00103-4 [DOI] [PubMed] [Google Scholar]
- 34. Herbomel P, Thisse B, Thisse C. 1999. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126:3735–3745 [DOI] [PubMed] [Google Scholar]
- 35. van der Sar AM, Musters RJ, van Eeden FJ, Appelmelk BJ, Vandenbroucke-Grauls CM, Bitter W. 2003. Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell. Microbiol. 5:601–611. 10.1046/j.1462-5822.2003.00303.x [DOI] [PubMed] [Google Scholar]
- 36. Vojtech LN, Sanders GE, Conway C, Ostland V, Hansen JD. 2009. Host immune response and acute disease in a zebrafish model of Francisella pathogenesis. Infect. Immun. 77:914–925. 10.1128/IAI.01201-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brudal E, Winther-Larsen HC, Colquhoun DJ, Duodu S. 2013. Evaluation of reference genes for reverse transcription quantitative PCR analyses of fish-pathogenic Francisella strains exposed to different growth conditions. BMC Res. Notes 6:76. 10.1186/1756-0500-6-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. 2011. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117:e49–e56. 10.1182/blood-2010-10-314120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. 2006. A transgenic zebrafish model of neutrophilic inflammation. Blood 108:3976–3978. 10.1182/blood-2006-05-024075 [DOI] [PubMed] [Google Scholar]
- 40. Westerfield M. 2000. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio), 4th ed. University of Oregon Press, Eugene, OR [Google Scholar]
- 41. Matthews M, Varga ZM. 2012. Anesthesia and euthanasia in zebrafish. ILAR J. 53:192–204. 10.1093/ilar.53.2.192 [DOI] [PubMed] [Google Scholar]
- 42. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van der Vaart M, Spaink HP, Meijer AH. 2012. Pathogen recognition and activation of the innate immune response in zebrafish. Adv. Hematol. 2012:159807. 10.1155/2012/159807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang DC, Shao YQ, Huang YQ, Jiang SG. 2005. Cloning, characterization and expression analysis of interleukin-10 from the zebrafish (Danio rerio). J. Biochem. Mol. Biol. 38:571–576. 10.5483/BMBRep.2005.38.5.571 [DOI] [PubMed] [Google Scholar]
- 45. Wang Z, Zhang S, Wang G. 2008. Response of complement expression to challenge with lipopolysaccharide in embryos/larvae of zebrafish Danio rerio: acquisition of immunocompetent complement. Fish Shellfish Immunol. 25:264–270. 10.1016/j.fsi.2008.05.010 [DOI] [PubMed] [Google Scholar]
- 46. Igawa D, Sakai M, Savan R. 2006. An unexpected discovery of two interferon gamma-like genes along with interleukin (IL)-22 and -26 from teleost: IL-22 and -26 genes have been described for the first time outside mammals. Mol. Immunol. 43:999–1009. 10.1016/j.molimm.2005.05.009 [DOI] [PubMed] [Google Scholar]
- 47. Altmann SM, Mellon MT, Johnson MC, Paw BH, Trede NS, Zon LI, Kim CH. 2004. Cloning and characterization of an Mx gene and its corresponding promoter from the zebrafish, Danio rerio. Dev. Comp. Immunol. 28:295–306. 10.1016/j.dci.2003.09.001 [DOI] [PubMed] [Google Scholar]
- 48. Yoshimura A, Naka T, Kubo M. 2007. SOCS proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 7:454–465. 10.1038/nri2093 [DOI] [PubMed] [Google Scholar]
- 49. Dios S, Romero A, Chamorro R, Figueras A, Novoa B. 2010. Effect of the temperature during antiviral immune response ontogeny in teleosts. Fish Shellfish Immunol. 29:1019–1027. 10.1016/j.fsi.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 50. Tang R, Dodd A, Lai D, McNabb WC, Love DR. 2007. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim. Biophys. Sin. (Shanghai) 39:384–390. 10.1111/j.1745-7270.2007.00283.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McCurley AT, Callard GV. 2008. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 9:102. 10.1186/1471-2199-9-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ottem KF, Nylund A, Isaksen TE, Karlsbakk E, Bergh Ø. 2008. Occurrence of Francisella piscicida in farmed and wild Atlantic cod, Gadus morhua L., in Norway. J. Fish Dis. 31:525–534. 10.1111/j.1365-2761.2008.00930.x [DOI] [PubMed] [Google Scholar]
- 53. Parsa KV, Butchar JP, Rajaram MV, Cremer TJ, Gunn JS, Schlesinger LS, Tridandapani S. 2008. Francisella gains a survival advantage within mononuclear phagocytes by suppressing the host IFNgamma response. Mol. Immunol. 45:3428–3437. 10.1016/j.molimm.2008.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Colucci-Guyon E, Tinevez JY, Renshaw SA, Herbomel P. 2011. Strategies of professional phagocytes in vivo: unlike macrophages, neutrophils engulf only surface-associated microbes. J. Cell Sci. 124:3053–3059. 10.1242/jcs.082792 [DOI] [PubMed] [Google Scholar]
- 55. Kanwal Z, Zakrzewska A, den Hertog J, Spaink HP, Schaaf MJM, Meijer AM. 2013. Deficiency in hematopoietic phosphatase ptpn6/Shp1 hyperactivates the innate immune system and impairs control of bacterial infections in zebrafish embryos. J. Immunol. 190:1631–1645. 10.4049/jimmunol.1200551 [DOI] [PubMed] [Google Scholar]
- 56. Gjessing MC, Inami M, Weli SC, Ellingsen T, Falk K, Koppang EO, Kvellestad A. 2011. Presence and interaction of inflammatory cells in the spleen of Atlantic cod, Gadus morhua L., infected with Francisella noatunensis. J. Fish Dis. 34:687–699. 10.1111/j.1365-2761.2011.01284.x [DOI] [PubMed] [Google Scholar]
- 57. Mikalsen J, Olsen AB, Tengs T, Colquhoun DJ. 2007. Francisella philomiragia subsp. noatunensis subsp. nov., isolated from farmed Atlantic cod (Gadus morhua L.). Int. J. Syst. Evol. Microbiol. 57:1960–1965. 10.1099/ijs.0.64765-0 [DOI] [PubMed] [Google Scholar]
- 58. Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. 2002. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 17:693–702. 10.1016/S1074-7613(02)00475-2 [DOI] [PubMed] [Google Scholar]
- 59. Ludwig M, Palha N, Torhy C, Briolat V, Colucci-Guyon E, Bremont M, Herbomel P, Boudinot P, Levraud JP. 2011. Whole-body analysis of a viral infection: vascular endothelium is a primary target of infectious hematopoietic necrosis virus in zebrafish larvae. PLoS Pathog. 7:e1001269. 10.1371/journal.ppat.1001269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cowley SC, Gray CJ, Nano FE. 2000. Isolation and characterization of Francisella novicida mutants defective in lipopolysaccharide biosynthesis. FEMS Micriobiol. Lett. 182:63–67. 10.1111/j.1574-6968.2000.tb08874.x [DOI] [PubMed] [Google Scholar]
- 61. Benard EL, van der Sar AM, Ellett F, Lieschke GJ, Spaink HP, Meijer AH. 2012. Infection of zebrafish embryos with intracellular bacterial pathogens. J. Vis. Exp. 2012:e3781. 10.3791/3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Clemens DL, Lee BY, Horwitz MA. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 72:3204–3217. 10.1128/IAI.72.6.3204-3217.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hall JD, Woolard MD, Gunn BM, Craven RR, Taft-Benz S, Frelinger JA, Kawula TH. 2008. Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect. Immun. 76:5843–5852. 10.1128/IAI.01176-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dixon G, Elks PM, Loynes CA, Whyte MK, Renshaw SA. 2012. A method for the in vivo measurement of zebrafish tissue neutrophil lifespan. ISRN Hematol. 2012:915868. 10.5402/2012/915868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brannon MK, Davis JM, Mathias JR, Hall CJ, Emerson JC, Crosier PS, Huttenlocher A, Ramakrishnan L, Moskowitz SM. 2009. Pseudomonas aeruginosa type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cell. Microbiol. 11:755–768. 10.1111/j.1462-5822.2009.01288.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Couper KN, Blount DG, Riley EM. 2008. IL-10: the master regulator of immunity to infection. J. Immunol. 180:5771–5777 [DOI] [PubMed] [Google Scholar]
- 67. Fortier AH, Polsinelli T, Green SJ, Nacy CA. 1992. Activation of macrophages for destruction of Francisella tularensis: identification of cytokines, effector cells, and effector molecules. Infect. Immun. 60:817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aktan F. 2004. iNOS-mediated nitric oxide production and its regulation. Life Sci. 75:639–653. 10.1016/j.lfs.2003.10.042 [DOI] [PubMed] [Google Scholar]
- 69. Wang T, Gorgoglione B, Maehr T, Holland JW, Vecino JL, Wadsworth S, Secombes CJ. 2011. Fish suppressors of cytokine signaling (SOCS): gene discovery, modulation of expression and function. J. Signal Transduct. 2011:905813. 10.1155/2011/905813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Veneman WJ, Stockhammer OW, de Boer L, Zaat SA, Meijer AH, Spaink HP. 2013. A zebrafish high throughput screening system used for Staphylococcus epidermidis infection marker discovery. BMC Genomics 14:255. 10.1186/1471-2164-14-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.