Abstract
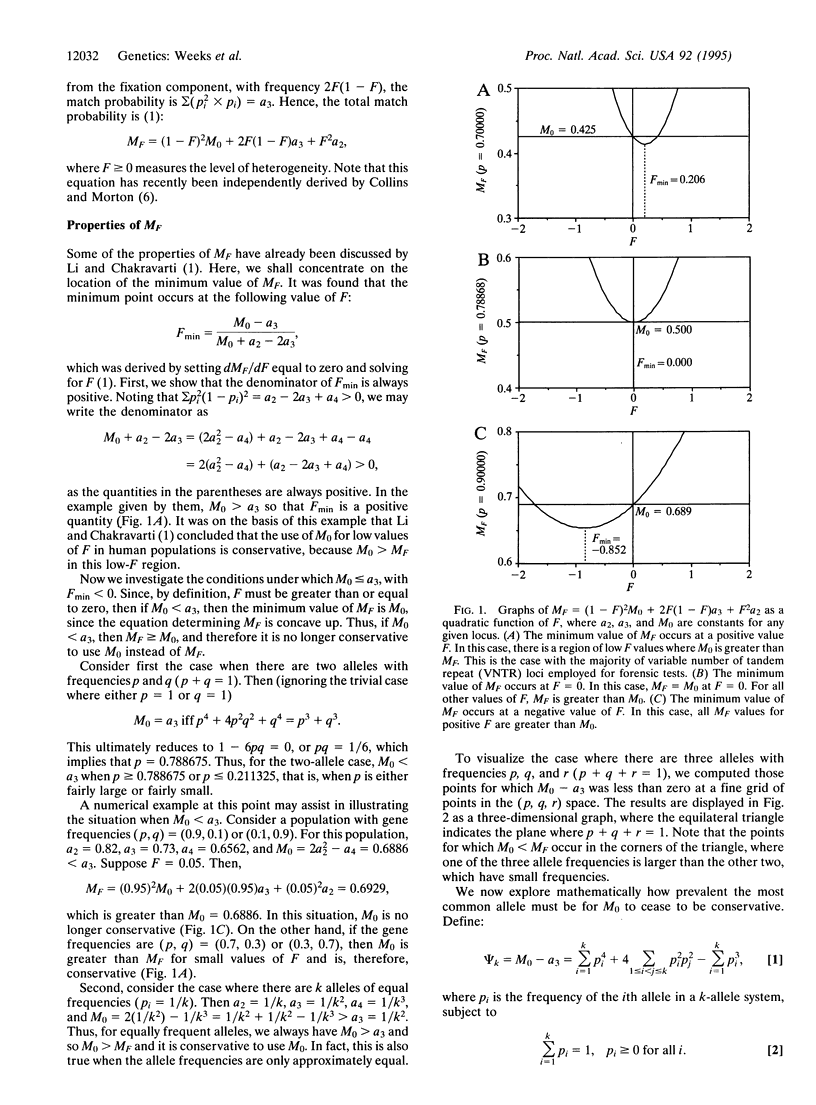
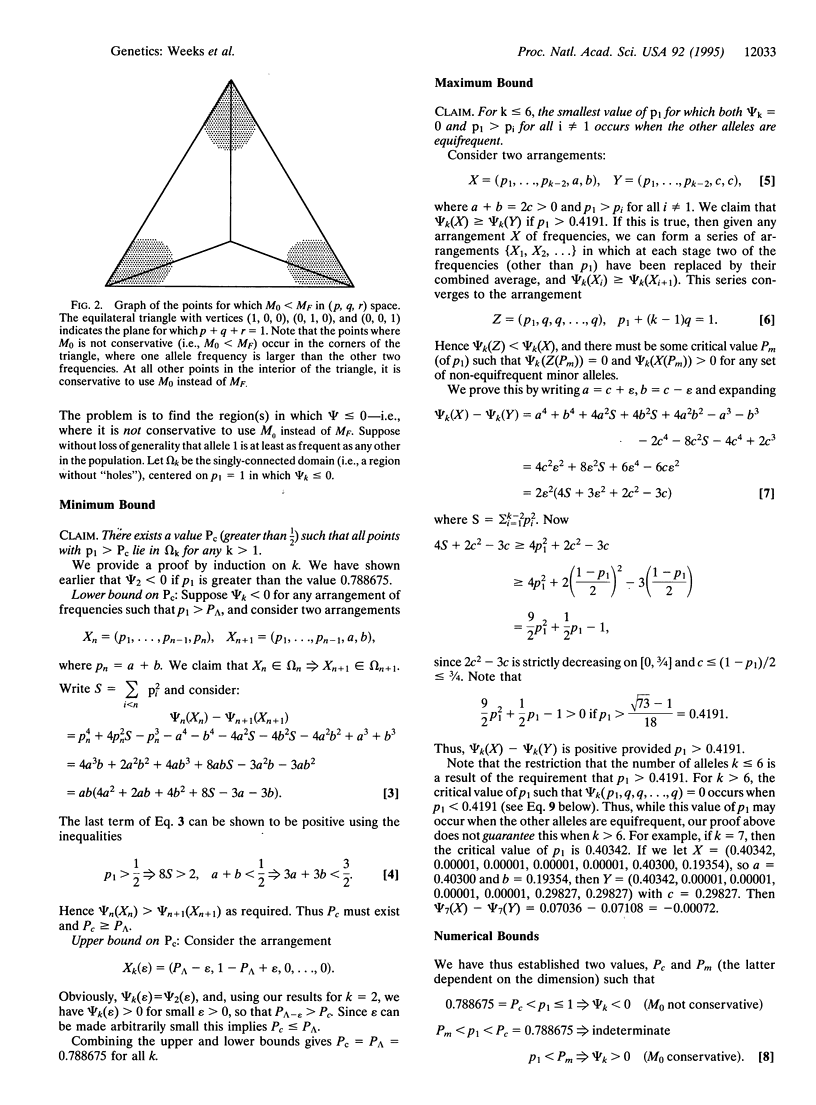
Li and Chakravarti [Li, C.C. & Chakravarti, A. (1994) Hum. Hered. 44, 100-109] compared the probability (MO) of a random match between the two DNA profiles of a pair of individuals drawn from a random-mating population to the probability (MF) of the match between a pair of random individuals drawn from a subdivided population. The level of heterogeneity in this subdivided population is measured by the parameter F, where there is no subdivision when F = 0 and increasing values of F indicate increasing subdivisions. Li and Chakravarti concluded that it is conservative to use the match probability MO, which is derived under the assumption that the two individuals are drawn from a homogeneous random-mating population without subdivision. However, MO may not be always greater than MF, even for biologically reasonable values of F. We explore here those mathematical conditions under which MO is less than MF, and we find that MO is not conservative mainly when there is an allele with a much higher frequency than all the other alleles. When empirical data for both variable number of tandem repeat (VNTR) and short tandem repeat (STR) systems are evaluated, we find that in the majority of cases MO represents a conservative probability of a match, and so the subdivision of human populations may usually be ignored for a random match, although not, of course, for relatives. Loci for which MO is not conservative should be avoided for forensic inference.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Collins A., Morton N. E. Likelihood ratios for DNA identification. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):6007–6011. doi: 10.1073/pnas.91.13.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Risch N. Ethnic differentiation at VNTR loci, with special reference to forensic applications. Am J Hum Genet. 1992 Sep;51(3):534–548. [PMC free article] [PubMed] [Google Scholar]
- Hammond H. A., Jin L., Zhong Y., Caskey C. T., Chakraborty R. Evaluation of 13 short tandem repeat loci for use in personal identification applications. Am J Hum Genet. 1994 Jul;55(1):175–189. [PMC free article] [PubMed] [Google Scholar]
- Lange K. The affected sib-pair method using identity by state relations. Am J Hum Genet. 1986 Jul;39(1):148–150. [PMC free article] [PubMed] [Google Scholar]
- Li C. C., Chakravarti A. DNA profile similarity in a subdivided population. Hum Hered. 1994 Mar-Apr;44(2):100–109. doi: 10.1159/000154199. [DOI] [PubMed] [Google Scholar]
- Li C. C., Weeks D. E., Chakravarti A. Similarity of DNA fingerprints due to chance and relatedness. Hum Hered. 1993 Jan-Feb;43(1):45–52. doi: 10.1159/000154113. [DOI] [PubMed] [Google Scholar]
- Morton N. E. Genetic structure of forensic populations. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2556–2560. doi: 10.1073/pnas.89.7.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]


