Abstract
Background and Aims
The OVATE gene encodes a nuclear-localized regulatory protein belonging to a distinct family of plant-specific proteins known as the OVATE family proteins (OFPs). OVATE was first identified as a key regulator of fruit shape in tomato, with nonsense mutants displaying pear-shaped fruits. However, the role of OFPs in plant development has been poorly characterized.
Methods
Public databases were searched and a total of 265 putative OVATE protein sequences were identified from 13 sequenced plant genomes that represent the major evolutionary lineages of land plants. A phylogenetic analysis was conducted based on the alignment of the conserved OVATE domain from these 13 selected plant genomes. The expression patterns of tomato SlOFP genes were analysed via quantitative real-time PCR. The pattern of OVATE gene duplication resulting in the expansion of the gene family was determined in arabidopsis, rice and tomato.
Key Results
Genes for OFPs were found to be present in all the sampled land plant genomes, including the early-diverged lineages, mosses and lycophytes. Phylogenetic analysis based on the amino acid sequences of the conserved OVATE domain defined 11 sub-groups of OFPs in angiosperms. Different evolutionary mechanisms are proposed for OVATE family evolution, namely conserved evolution and divergent expansion. Characterization of the AtOFP family in arabidopsis, the OsOFP family in rice and the SlOFP family in tomato provided further details regarding the evolutionary framework and revealed a major contribution of tandem and segmental duplications towards expansion of the OVATE gene family.
Conclusions
This first genome-wide survey on OFPs provides new insights into the evolution of the OVATE protein family and establishes a solid base for future functional genomics studies on this important but poorly characterized regulatory protein family in plants.
Keywords: OVATE family proteins, OFP, land plants, angiosperm, phylogenetic analyses, Arabidopsis thaliana, Oryza sativa, Solanum lycopersicum, segmental duplication, tandem duplication
INTRODUCTION
The OVATE gene was first identified as an important regulator of fruit shape in tomato, in which a naturally occurring premature stop codon in OVATE results in pear-shaped fruit with longitudinal elongation and neck constriction (Liu et al., 2002). This revealed a previously uncharacterized class of regulatory genes in plant development, which encode proteins with a conserved 70 amino acid C-terminal domain. This domain was designated as the OVATE domain, also known as DUF623 (Domain of Unknown Function 623), and proteins containing this domain were designated OVATE family proteins (OFPs), which are found exclusively in plants (Hackbusch et al., 2005; Wang et al., 2007, 2011).
To date, OFPs have been primarily characterized in arabidopsis (AtOFPs) and demonstrated to regulate plant growth and development (Hackbusch et al., 2005; Pagnussat et al., 2007; Wang et al., 2007, 2010; Li et al., 2011). AtOFPs were shown to have close functional interactions with three amino acid loop extension (TALE) homeodomain proteins, and AtOFP1 and AtOFP5 regulate the sub-cellular localization of TALE homeoproteins (Hackbusch et al., 2005). The plant TALE proteins are a conserved superclass of homeodomain proteins characterized by an extension of three amino acids between helices 1 and 2 of the homeodomain (Bertolino et al., 1995), and comprise two sub-classes called the KNOTTED-like homeobox (KNOX) and BEL1-like homeodomain (BELL) proteins (Hay and Tsiantis, 2009, 2010; Hamant and Pautot, 2010). Furthermore, it has been well documented that interactions between KNOX and BELL proteins result in heterodimers regulating plant development in a connected and complex network (Bellaoui et al., 2001; Smith et al., 2002; Smith and Hake, 2003; Chen et al., 2004; Hackbusch et al., 2005; Cole et al., 2006). AtOFP1 has been reported to function as an active transcriptional repressor of AtGA20ox1 in the gibberellin (GA) biosynthesis pathway, suppressing cell elongation (Wang et al., 2007). A recent study also indicated that AtOFP1 interacts with AtKu70, a protein involved in DNA repair through the non-homologous end-joining pathway (Wang et al., 2010). Similar to AtOFP1, AtOFP4 acts as a transcriptional repressor and has been proposed to form a functional complex with KNAT7, one of four class II arabidopsis KNOTTED1-like Arabidopsis thaliana (KNAT) members (Hake et al., 2004; Hamant and Pautot, 2010), in regulating secondary cell wall formation (Li et al., 2011). AtOFP5, reported to interact with both BLH1 and KNAT3, which belong to BELL and KNOX sub-classes of TALE homeodomain proteins, respectively (Hackbusch et al., 2005), can act as regulator of the BELL–KNOX TALE complex involved in normal embryo sac development in arabidopsis (Pagnussat et al., 2007). Recently, a genome-wide analysis of AtOFPs revealed conserved functions as transcriptional repressors, with overexpression leading to a number of abnormal phenotypes, implying novel roles in regulating plant growth and development (Wang et al., 2011).
Gene homologues containing the conserved OVATE domain have been found in tomato, arabidopsis and rice (Liu et al., 2002). Furthermore, OVATE-like genes appear to be conserved in many plant species, with conserved genomic microsynteny discovered not only between Solanaceae relatives (Wang et al., 2008), but also in distantly related species (arabidopsis, snapdragon, papaya, poplar, grape and coffee tree) (Causier et al., 2010; Guyot et al., 2012), suggesting ancestral synteny of these regions in plants. In pepper (Capsicum annuum), a relative of tomato in the Solanaceae family, an OVATE family member (CaOvate) was also shown to be involved in determining fruit shape by negatively affecting the expression of CaGA20ox1 (Tsaballa et al., 2011).
Despite the aforementioned analyses of OFPs, our knowledge concerning this protein family remains relatively poor. In this study, we focused on the evolution of OFPs in land plants by performing, for the first time, a genome-wide comparative analysis of sequences encoding putative OFPs in a wide variety of plant genomes. Furthermore, we provided a detailed understanding of the evolutionary framework of the OVATE family in the model species arabidopsis, rice and tomato, including investigation of the expression patterns of SlOFP genes.
MATERIALS AND METHODS
Retrieval of putative OVATE proteins in selected plant genomes
A total of 13 sequenced plant genomes that represent the major evolutionary lineages of land plants and are available from public databases (Supplementary Data Table S1) were selected for the phylogenetic analysis of OVATE proteins. These were: Solanum lycopersicum, S. tuberosum and Mimulus guttatus from the asterid clade of core eudicots; Arabidopsis thaliana, Vitis vinifera, Populus trichocarpa, Prunus persica and Carica papaya from the rosid clade; Aquilegia coerulea representing the basal eudicots; Oryza sativa and Zea mays representing the monocots; and finally Selaginella moellendorffii and Physcomi-trella patens representing lycophytes (seedless vascular plants) and bryophytes (mosses; non-vascular plants), respectively. Two strategies, a BLAST search and a keyword search, were adopted to identify the putative OVATE proteins. The amino acid sequences of well-studied OVATE proteins, namely AtOFPs in arabidopsis (Wang et al., 2011) and the wild-type OVATE protein in round-fruited tomato (AAN17752; designated SlOFP1) were used as queries for TBLASTN searches of the selected plant genomes. A BLAST search against the maize genome was also carried out using putative rice OVATE protein sequences as queries. All homologues with E < 10 were retrieved for subsequent analyses. A key word search in the phytozome (v8.0) database (http://www.phytozome.net/) for putative OVATE proteins was conducted by searching ontologies with the term ‘PF04844’.
From all searches, two previously unpublished AtOFPs were identified: At2g36026 (designated AtOFP19) was identified from a TBLASTN search against the TAIR (The Arabidopsis Information Resource 10.0) database (http://www.arabidopsis.org/index.jsp) using SlOFP1 as the query; while At1g06923 (designated AtOFP20) was detected in a segmental duplication block by the PGDD (Plant Genome Duplication Database, http://chibba.agtec.uga.edu/duplication/) as a putative paralogue of At2g30395 (AtOFP17; see below for details). All search results were collated and manually curated to produce a non-redundant data set that was then subjected to a CDD (Conserved Domain Database) search (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml; Marchler-Bauer et al., 2011) for the verification of the OVATE domain (PF04844). All sequences were thus verified, with the exception of SlOFP16, as well as AtOFP17, AtOFP20 and their orthologues. SlOFP16 no longer seems to possess the OVATE domain, whereas the AtOFP17/20-like proteins have a divergent but still recognizable OVATE domain in the multiple alignment with other OVATE proteins (Supplementary Data File S2). Excluding SlOFP16, a total of 265 putative OVATE protein sequences were obtained from the 13 selected plant genomes. The nomenclature of all the putative OVATE proteins is listed in Supplementary Data Table S2.
Phylogenetic reconstruction and motif analysis
Multiple sequence alignment of all 265 full-length putative OVATE protein sequences was performed using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/; Edgar, 2004). PtOFP5 and ZmOFP42 were re-annotated because of an insertion of 59 amino acids in the conserved OVATE domain (see Supplementary Data File S1). Given the high degree of diversity among the full-length OVATE protein sequences, phylogenetic analysis was conducted on the alignment of the conserved OVATE domain. Alignments were manually revised in BioEdit (Hall, 1999), and the resulting alignment of 86 amino acid sites was used for maximum likelihood (ML) analysis using RAxML 7.0.4 (Stamatakis, 2006), with the BLOSUM62 amino acid substitution matrix and CAT approximation, and 200 bootstrap replicates. The phylogenetic relationships of OVATE proteins in arabidopsis, rice and tomato were also estimated separately using MEGA 4.0 (Kumar et al., 2008) with the Neighbor–Joining (NJ) method based on the p-distance model, pairwise deletion and 1000 bootstraps. Trees were visually inspected and edited in Dendroscope (Huson et al., 2007). Since there was no prior information on placement of the root, trees were rooted with midpoint rooting. The MEME (version 4.9.0) motif search tool (http://meme.nbcr.net/meme/intro.html; Bailey et al., 2009) was used to detect conserved motifs within related OFPs with the following parameters: maximum number of motifs: 10; 4 ≤ motif width ≤ 70; 2 ≤ number of sites ≤ 300. The sequence logos and E-values for each motif are indicated in Supplementary Data Fig. S1.
Molecular evolutionary analysis
In order to estimate the evolutionary rate for each sub-group, first whole cDNA sequences from each sub-group were retrieved and aligned with the corresponding alignment of protein sequences as reference using PAL2NAL (Suyama et al., 2006). Then, aligned codon sequences encoding the OVATE domain were selected for estimating the ratio of dN (the number of non-synonymous substitutions per non-synonymous site) to dS (the number of synonymous substitutions per synonymous site), i.e. the ω ratio, by the CODEML implemented in PAML 4.6 (Yang, 2007). The average ω ratio of each group was estimated with the one-ratio site model M0 (NSsites = 0, model = 0), which assumes that all sites across the phylogeny evolve with the same ω ratio. To determine whether some branches evolve under positive selection (ω > 1), the ω ratio of each branch was estimated under the assumption of a free ratios model (NSsites = 0 and model = 1).
Inference of duplication and loss of OVATE family genes
The ML tree based on the conserved OVATE domain was reconciled with the known species tree in Notung 2.6 (Durand et al., 2006; Vernot et al., 2008) to infer possible gene duplication and loss events. Rearrangement producing alternative event histories with a minimum duplication/loss score was performed to avoid overestimation of gene turnover along all lineages.
Chromosomal localization of OVATE genes in arabidopsis, rice and tomato
The chromosomal localization of AtOFP genes in arabidopsis was visualized using the Chromosome Map Tool available at TAIR (http://www.arabidopsis.org/jsp/ChromosomeMap/tool.jsp). The chromosomal locations of OsOFP genes in rice and SlOFP genes in tomato were generated by the rice genome browser at the MSU RGAP (Rice Genome Annotation Project) database (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/), and the tomato genome browser at the SGN (Sol Genome Network) database (http://solgenomics.net/gbrowse/bin/gbrowse/ITAG2_genomic/), respectively.
Determination of gene duplication pattern
The pattern of OVATE gene duplication resulting in the expansion of the OVATE gene family was determined in arabidopsis, rice and tomato. If paralogues were either adjacent or separated by ≤ 5 genes along a chromosome they were assigned as duplicates by tandem duplication (Zhao et al., 2010; Xia et al., 2011). If paralogues were within known genomic duplication blocks, they were considered to be duplicated through segmental duplication. The Locus Search Tool at the PGGD (Plant Genome Duplication Database, http://chibba.agtec.uga.edu/duplication/) was used to determine if the AtOFP genes, OsOFP genes and SlOFP genes existed within genomic duplication blocks. The presence of OsOFP genes in segmental duplication blocks was also investigated using the rice segmental duplication block database at MSU RGAP (http://rice.plantbiology.msu.edu/segmental_dup/index.shtml). Given that synonymous substitution rates (Ks) within duplicated genes are postulated to be similar, Ks values can be used as the proxy for time (Shiu et al., 2004). The Ks values of paralogues in segmental duplication blocks were retrieved from the PGDD or, in the case of OsOFP23 and OsOFP30, from the rice segmental duplication block database at MSU RGAP computed by the KaKs_calculator (Zhang et al., 2006). The timing of duplication events can be estimated using the Ks value and a given clock-like rate, λ, through the formula T = Ks/2λ, where for arabidopsis and tomato, λ = 1·5 × 10–8 substitutions per synonymous site per year, while for rice, λ = 6·5 × 10–9 substitutions per synonymous site per year (Blanc and Wolfe, 2004; Liu et al., 2010).
Quantitative real-time PCR of SlOFP genes
Quantitative real-time PCR was performed on a round-fruited tomato variety ‘Stupicke’. Total RNA was isolated from root, leaf, sepal, petal, stamen, pistil, green fruit and red fruit tissue (Trizol, Invitrogen). First-strand cDNA was synthesized using the PrimeScript® RT reagent Kit with gDNA Eraser (TaKaRa). Quantitative real-time PCRs were carried out in 20 μL with 2 μL of first-strand cDNA (20 ng μL–1), 10 μL of 2 × SYBR Premix® ExTaq™ (TaKaRa), 0·5 μL of 50 × ROX reference dye II (TaKaRa) and 0·8 μL of both forward and reverse primers (10 μm) in an ABI PRISM 7500 system (Applied Biosystems). Cycling conditions were 95 °C for 10 s followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. Melting curve analysis was used to confirm the specificity of amplification. Tomato Actin (Chen et al., 2007) was used as the endogenous control. The primers used in the assay are listed in Supplementary Data Table S3. Three technical replicates were carried out for each PCR. Relative expression was calculated using the 2−ΔΔCT method as previously described (Livak and Schmittgen, 2001). All expression levels were shown as relative to the expression of tomato SlOFP9 in sepals, which exhibited an average expression level. A heatmap was visualized using Genesis software (Sturn et al., 2002) representing the log2 value of the relative expression level. Bar charts for the relative expression level of each SlOFP gene are shown in Supplementary Data Fig. S2. There is a possibility that the differences in relative expression between the different genes are due to unequal efficiencies in the amplification of the target sequences.
RESULTS AND DISCUSSION
Distribution of OVATE proteins in land plants
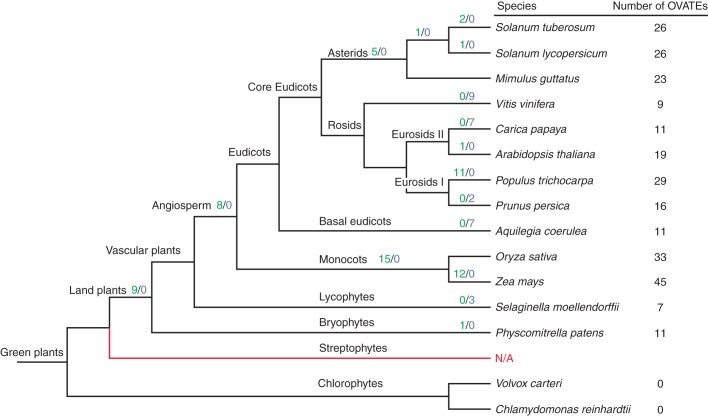
OVATE-like proteins were identified in all 13 plant genomes selected to represent the major evolutionary lineages of land plants including the early-diverged land plants Physcomitrella patens (moss) and Selaginella moellendorffii (spikemoss) (Fig. 1). In general, monocots had more OFPs than eudicots, with Zea mays (maize) containing the largest number (i.e. 45) of OVATE proteins. With respect to the core eudicots, thegenomes of Mimulus guttatus (monkey flower), Solanum lycopersicum (tomato) and Solanum tuberosum (potato) from the asterid clade contained similar numbers of OVATE proteins. In the rosid clade, the OVATE protein numbers in different species vary from nine in Vitis vinifera (grape vine) to 29 in Populus trichocarpa (poplar) (Fig. 1). It is worth noting that a previously unnamed AtOFP, At2g36026 (designated AtOFP19), and an uncharacterized AtOFP member, At1g06923 (designated AtOFP20), were included in this study. AtOFP19 possesses the conserved OVATE domain and is annotated as an OVATE family protein in TAIR (http://www.arabidopsis.org/). AtOFP20 is annotated as being most closely related to AtOFP17 (At2g30395), consistent with both genes being putative paralogues occurring within segmental duplication blocks detected using the PGDD. Although AtOFP17 is annotated as an OVATE family protein, the presence of a conserved OVATE domain was not detected in a CDD search. The absence of the OVATE domain is prevalent in both its paralogue AtOFP20 and their putative orthologues in other species, which cluster with AtOFP17 into a distinct group (sub-group 8; Fig. 2). Here, AtOFP17-like members are included as OVATE proteins on the basis that the OVATE domain in the AtOFP17 sub-group seems to be quite obvious from the alignment that was used for the phylogenetic analysis (Supplementary Data File S2), as well as the fact that AtOFP17 was annotated as an OVATE-containing protein despite relatively low sequence similarity with other AtOFPs determined by Wang et al. (2011).
Fig. 1.
A simplified cladogram of the species selected for screening putative OVATE proteins in this study. Figures in the right column indicate the total numbers of OVATE proteins found in each species. Figures above branches represent the estimated numbers of gene duplication (green before the forward slash) and loss (blue after the forward slash) events in different species and species split nodes using Notung 2.6, with no estimation of gene losses/duplications for the lineages of Mimulus guttatus and Oryza sativa. ‘N/A’ in red indicates the current lack of genome sequence data for streptophytes.
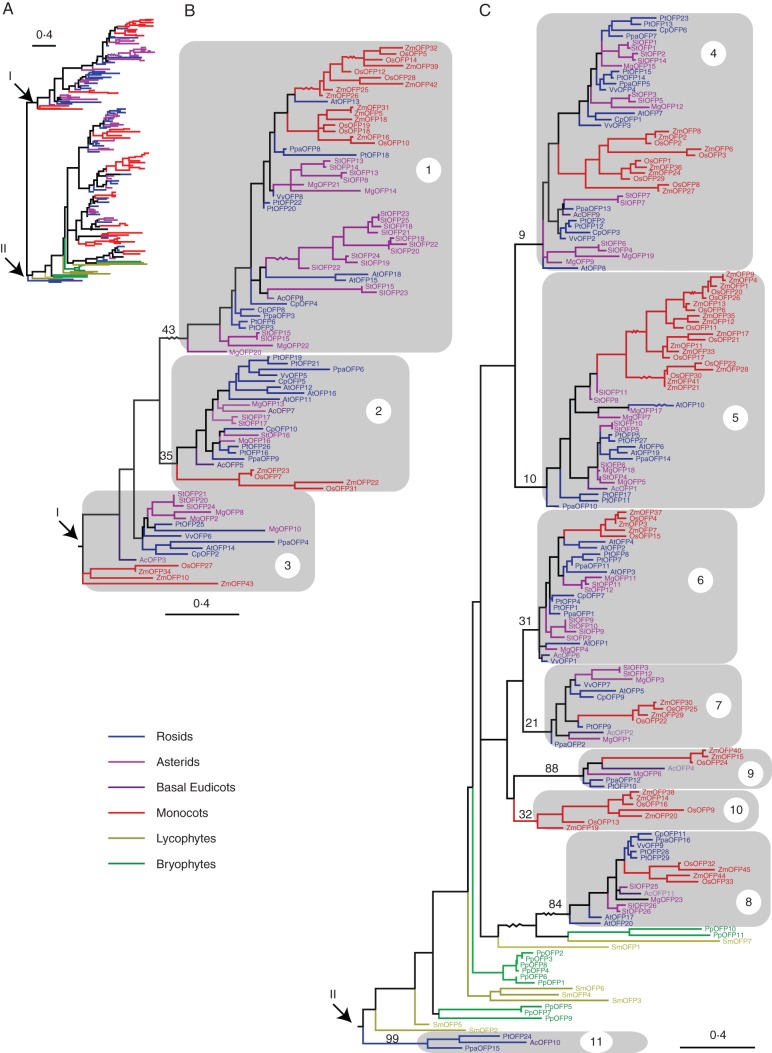
Fig. 2.
Maximum likelihood analysis of 265 plant OVATE proteins from 13 plant genomes. (A) An overall maximum likelihood phylogram of 265 OVATE protein sequences, with midpoint rooting. (B) Full view of part I from (A) showing sub-groups 1–3. (C) Full view of part II from (A) showing sub-groups 4–11. The grey rectangles delineate 11 sub-groups of OVATE proteins in angiosperms. Bootstrap values above the nodes that define the sub-groups are indicated. The zig-zag lines indicate branches that are not drawn to scale. Coloured branches and labels indicate the clade to which the species belong [blue, Carica papaya, Prunus persica, Vitis vinifera, Populus trichocarpa, Arabidopsis thaliana (rosids); violet, Mimulus guttatus, Solanum lycopersicum, Solanum tuberosum (asterids); purple, Aquilegia coerulea (basal eudicots); red, Oryza sativa, Zea mays (monocots); olive, Selaginella moellendorffii (lycophytes); green, Physcomitrella patens (bryophytes)].
AtOFP9 has been re-annotated to include an additional open reading frame (ORF) encoding a new protein of 129 amino acids (NP_191312) that is distinct from the 411 amino acid protein in the study of Wang et al. (2011). Moreover, this newly annotated AtOFP9 protein has lost the conserved OVATE domain that characterizes the OFPs. Given this, the newly annotated AtOFP9 should no longer be considered as an OVATE protein in arabidopsis, and we propose that there are 19 arabidopsis OVATE proteins instead of the 18 originally described (Wang et al., 2007, 2011).
To elucidate further the evolutionary history and origin of the OVATE protein family in green plants (Viridiplantae), we also screened the genomes of the chlorophytes, Volvox carteri and Chlamydomonas reinhardtii, for OVATE proteins. No proteins with similarity to OFPs were found in the two chlorophyte genomes, which represent the ancestral algal relatives of the Viridiplantae.
Phylogenetic relationships of OVATE family proteins in land plants
To obtain an overview of the phylogenetic relationships between plant OFPs, phylogenetic trees were reconstructed based on the alignment of the conserved OVATE regions (Fig. 2). OVATE proteins from different flowering plant (angiosperms) species cluster together, while bryophyte and lycophyte OVATE proteins form distinct, predominantly lineage-specific clades (Fig. 2). Eleven sub-groups of angiosperm OVATE proteins were defined according to the topology of the phylogenetic trees (Fig. 2). These sub-groups were also supported by the presence of conserved motifs that are outside the OVATE domain and unique to each sub-group detected by the MEME program (Fig. 3; Supplementary Data Fig. S1). Although some of the 11 sub-groups have low bootstrap support due to the fact that only 86 amino acid sites in the OVATE domain were used for inferring the phylogeny, the presence of conserved signature motifs unique to each sub-group corroborates our phylogenetic analysis (Fig. 3; Supplementary Data Fig. S1). The OVATE proteins from mosses and lycophytes were not classified further, as they do not form well-defined sub-groups with the angiosperm OVATE proteins.
Fig. 3.
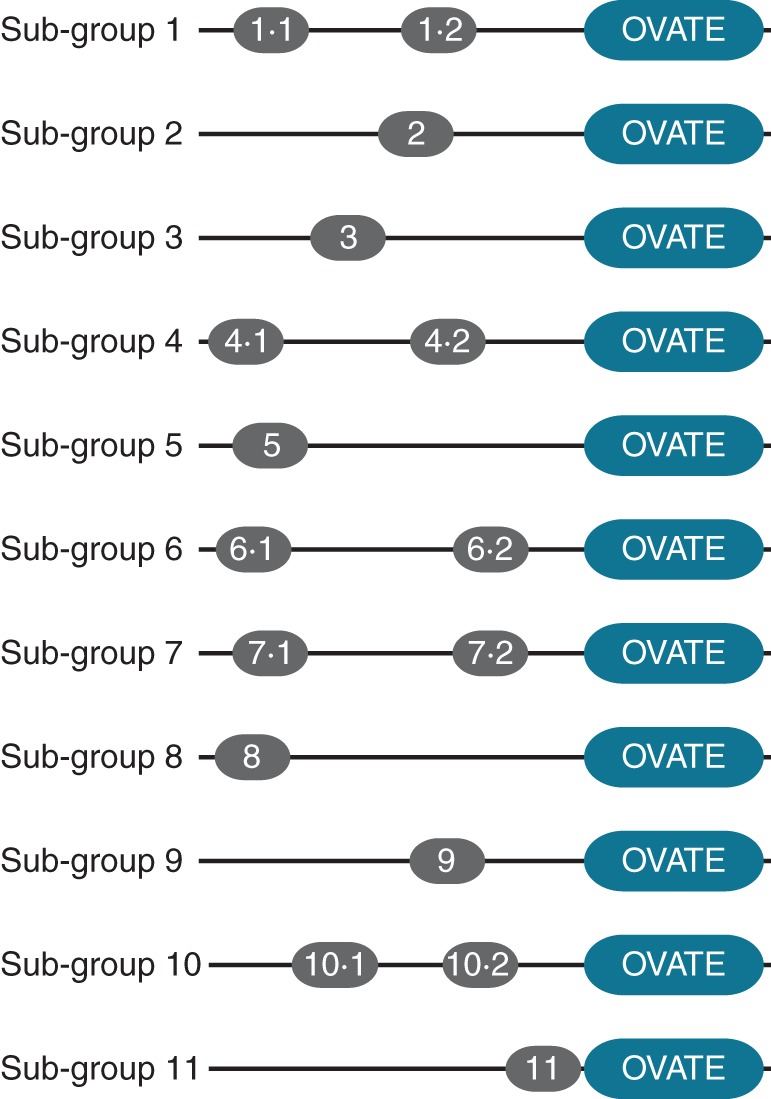
A schematic diagram of the organization of OVATE proteins outside the conserved OVATE domain and signature motifs defining each OVATE sub-group. The diagram is not drawn to scale. The sequence logos and E-values for each motif are given in Supplementary Data Fig, S1. The sequence information of motifs is as follows: motif 1·1, SC[TGK][HNQ]P[RK]TLSFRAE[DN][ND]; motif 1·2, [RS]S[EDK]R[LF]FFEPGETSS[IS]; motif 2, PD[FL][AS]T[AI][FI]AS[QR]RFFFSSPGRSNSI[IV][DE]S[PS]PS; motif 3, [GD]E[GD][DH]AATLSD[VI]DRFLFENF[RK]SLYIKDDD[EN]; motif 4·1, SF[GQ]SCRS[KR][DN]PSD[LV][PI][EQ]; motif 4·2, [ED]D[ED]xD[ED][ED][ED]TET[LF][FV]SS[RS][SR][FLS]SSD; motif 5, [CS][GNS]C[RG][RK][PA]KL[SV]S[VI]Fx[PS][KS]; motif 6·1, MG[NR][YH][KR]F[RK]LSDM[MI]PNAWFYKL[KR]DMSK[SPT]R[NGK]H; motif 6·2, R[RS]S[VS]SS[SA]R[GR][VL][KR][LT]R[TA]N[ST]PR[IL]AS; motif 7·1, P[LV]SW[LF][SA]K[FL]; motif 7·2, [ST][KA][VK][EM][IE]C[KR]I[KR]A[LI]ED[ML]; motif 8, [MR]K[VLK][KRS][STV][LFI][IG][ARS][FL]K[SC]KL[FLS][KN][PS]C[KNR][KR][FIL][LV][QRS][LI]F[RK]F[RK]; motif 9, MVQ[EA]RL[DQ][QS]MI[RD][EA][RA][AQ]E[AR]; motif 10·1, [PAC][CAP][PACS][CPRVY][SCF]PNR[AE]SYY[FVL][APN]S[ARQ][DE]R[AC][RILV][PQ]; motif 10·2, [AFT][PQ][ED]L[KQ]LRPI[LRV]TR; motif 11, SASLP[DE]DV[CQ]G[AIV][FY][AS][GD].
Angiosperm OVATE proteins from different species grouped into compact clades. This possibly resulted from rapid expansion of OVATE proteins in early seed plants or angiosperms following their divergence from the early-diverged land plant species. Many gene families within angiosperms have experienced rapid expansion during evolution and adaption to various environments (Corrêa et al., 2008). Both monocot and eudicot OVATE proteins expanded rapidly within sub-groups 1, 4 and 5 to form a lineage-specific clade (e.g. a Solanum-specific clade containing exclusively OVATE proteins from S. lycopersicum and S. tuberosum; Fig. 2). Of the 11 angiosperm OVATE sub-groups, sub-group 10 was specific to monocots, while sub-group 11 was an ‘orphan’ group specific to eudicots and included only three members, AcOFP10, PpaOFP15 and PtOFP24 (Fig. 2).
Differential evolutionary pattern of the OVATE family
In order to clarify the evolution of the OVATE family further, we examined the distribution of OFPs from different plant species within each sub-group (Table 1). We suggest two different mechanisms to account for the observed evolutionary pattern, namely conserved evolution and divergent expansion, similar to what was proposed for the evolution of F-box genes in plants (Xu et al., 2009). Within sub-group 8, sequences are conserved in all angiosperm species, with only one or two members from each species. Sub-groups 3, 7 and 9 also resemble this, comprising primarily one or two OVATE proteins from each plant species, with phylogenetic relationships partially mirroring the plant phylogeny (Fig. 2; Table 1). As such, the OVATE members of these sub-groups appear to have remained relatively conserved without large-scale gene expansions during plant divergence. This suggests that they may have crucial functions that are vital to plant development. For example, AtOFP5, a previously characterized protein interacting with the two TALE homeodomain proteins BLH1 and KNAT3 (Hackbusch et al., 2005), was reported to act as a regulator of the BELL–KNOX TALE complex required for normal embryo sac development in arabidopsis (Pagnussat et al., 2007). In this study, phylogenetic analysis placed AtOFP5 in the highly conserved sub-group 7, consistent with a conserved role in the essential process of female gametophyte development.
Table 1.
Classification and distribution of OVATE proteins in different sub-families of each plant species
| Zm | Os | Ac | Ppa | Pt | At | Cp | Vv | Mg | St | Sl | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 45 | 33 | 11 | 16 | 29 | 19 | 11 | 9 | 23 | 26 | 25 | |
| Sub-group1 | 9 | 7 | 1 | 2 | 5 | 3 | 2 | 1 | 4 | 9 | 9 |
| Sub-group2 | 2 | 2 | 2 | 2 | 4 | 3 | 2 | 1 | 2 | 2 | 1 |
| Sub-group3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 2 | 1 |
| Sub-group4 | 6 | 5 | 1 | 3 | 6 | 2 | 3 | 3 | 4 | 5 | 5 |
| Sub-group5 | 12 | 8 | 1 | 2 | 4 | 2 | – | – | 4 | 3 | 3 |
| Sub-group6 | 3 | 2 | 1 | 2 | 4 | 4 | 1 | 1 | 2 | 3 | 3 |
| Sub-group7 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 |
| Sub-group8 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 2 |
| Sub-group9 | 2 | 1 | 1 | 1 | 1 | – | – | – | 1 | – | – |
| Sub-group10 | 4 | 3 | – | – | – | – | – | – | – | – | – |
| Sub-group11 | – | – | 1 | 1 | 1 | – | – | – | – | – | – |
Pp, Physcomitrella patens; Sm, Selaginella moellendorffii; Zm, Zea mays; Os, Oryza sativa; Ac, Aquilegia coerulea; Ppa, Prunus persica; Pt, Populus trichocarpa; At, Arabidopsis thaliana; Cp, Carica papaya; Vv, Vitis vinifera; Mg, Mimulus guttatus; St, Solanum tuberosum; Sl, Solanum lycopersicum.
Divergent expansion of protein sub-families occurs when members are not under such tight functional constraints and thus may diverge more rapidly between taxa, often adopting species-specific functions (Hamilton et al., 2006). Such sub-families appear to have evolved for more OVATE proteins, with each containing 3–12 members from some plant species examined and in some cases forming lineage-specific sub-clades (Fig. 2; Table 1). AtOFP1 and AtOFP4, the two best-characterized OVATE proteins (Wang et al., 2007, 2010; Li et al., 2011), were assigned to sub-group 6 (Fig. 2). This sub-group has four AtOFPs, namely AtOFP1, AtOFP2, AtOFP3 and AtOFP4. AtOFP1 is suggested to function as an active transcriptional repressor of AtGA20ox1 expression in the GA biosynthesis pathway, suppressing cell elongation (Wang et al., 2007). AtOFP4 is also a transcriptional repressor and has been proposed to form a functional complex with KNAT7, an arabidopsis KNOX homeodomain protein, to regulate secondary cell wall formation (Li et al., 2011). AtOFP1, AtOFP2 and AtOFP4 were shown to have similar expression patterns, with all expressed in the roots, inflorescent stem, flower buds and young siliques (Wang et al., 2011). Promoter–GUS (β-glucuronidase) fusions for AtOFP1 and AtOFP4 revealed strong GUS activity in the root vascular cylinder and inflorescent stem (Li et al., 2011). Furthermore, arabidopsis plants overexpressing AtOFP4 had similar pleiotropic phenotypes to those overexpressing AtOFP1, including dwarfism, ovate-shaped organs and reduced fertility (Li et al., 2011). Nevertheless, it was suggested that AtOFP4 may play a more specific role in the differentiation of xylary fibres and interfascicular fibres based on the irx phenotype (Brown et al., 2005) and the thicker interfascicular fibre cell walls seen in the ofp4–2 loss-of-function mutant but not in the ofp1–1 loss-of-function mutant (Li et al., 2011). The irx phenotype, described as irregular xylem (irx), is characterized by a collapse of xylem vessels causing defects in the secondary wall and might be indicative of any secondary cell wall mutation (Brown et al., 2005). Thus, functional divergence of AtOFP1 and AtOFP4 from a common ancestral gene may have occurred during arabidopsis genome evolution.
Sub-group 1 contained the largest and most diverse group of OVATE proteins among the 11 sub-groups. Within this sub-group, the monocot OVATE proteins expanded rapidly, while the eudicots expanded into a Solanum species-specific clade, containing OVATE proteins from only S. lycopersicum and S. tuberosum (Fig. 2). Sub-group 4 contained the first identified OVATE gene (SlOFP1), controlling fruit shape in tomato (Liu et al., 2002). AtOFP7 was the most closely related AtOFP protein to SlOFPs, although little is known about the function of this arabidopsis protein. Within sub-group 4, the SlOFP1/AtOFP7 clade does not include A. coerulea OVATE proteins, compared with the existence of AcOFP9 in the SlOFP7 group (Fig. 2). This suggests that a duplication event occurred within eudicots following their divergence from monocots, followed by a loss of the SlOFP1/AtOFP7 orthologue in the basal eudicot A. coerulea.
ω (dN/dS) ratios were analysed to investigate selection acting upon OVATE genes within each sub-group. Average ω ratios of each group showed no significant difference under the assumption of the one-ratio model, with the highest and the lowest value being 0·23 (sub-group 1) and 0·07 (sub-group 9), respectively (Supplementary Data Table S4). However, ω ratios estimated under the assumption of free ratio models suggested that some sub-groups were conservative while others were divergent. As expected, most genes from sub-groups 3, 7, 8 and 9, in which only one or two copies are retained for most species, evolved under strong purifying selection (ω < 0·3; Supplementary Data Figs S5, S9–S11), which is suggestive of their conservative functions. In contrast, ω values of each branch from the sub-groups which experienced expansion (sub-groups 1, 4, 5 and 6) diversified significantly; usually one copy of duplicated genes evolves under strong purifying selection, while the other one evolves under relaxed purifying selection (0·3 < ω < 1) or even positive selection (ω > 1; Supplementary Data Figs S3, S6–S8), which gives the indication of sub-functionalization or neofunctionalization. Further functional analyses are needed to verify the functional diversification among these duplicated genes. Only three genes were found in sub-group 11; therefore, we did not estimate the dN/dS of this lineage.
Estimation of gene duplication and loss events in the OVATE gene family
Reconciliation of the gene tree (Fig. 2) with the species tree (Fig. 1) resulted in an estimation of 66 duplications and 28 losses (D/L score = 127) during plant evolution in the rearrange mode in Notung analysis. The result showed that in early land plant evolution, the OVATE genes already experienced nine duplications in the common ancestor of land plants. Eight duplication events occurred after the divergence of lycophytes from early vascular plants and before the split of monocots and eudicots (Fig. 1). That might possibly be related to the two ancestral whole-genome duplications (WGDs), post-dating diversification of S. moellendorffii and shortly before the diversification of extant seed plants (ζ) and extant angiosperms (ε), respectively (Jiao et al., 2011). After the split of monocots and eudicots, 15 duplications were inferred in monocots, before the diversification of rice and maize (Fig. 1). Similarly, two generally accepted polyploidy events (named σ and ρ) in the monocot lineage have pre-dated the diversification of major grasses (shared by Poaceae) (Paterson et al., 2004; Wang et al., 2005; Salse et al., 2008; Tang et al., 2010). Twelve duplications were estimated following the split of rice and maize (Fig. 1). In the diversification of asterids, there were five estimated duplications after the asterid–rosid split, preceding the split of Lamiales (M. guttatus) and Solanales (S. lycopersicum and S. tuberosum), followed by one duplication shared by S. lycopersicum and S. tuberosum, which underwent one and two duplications, respectively, after they split from each other (Fig. 1). In rosids, duplication events of OVATE genes were not inferred to be ancestral duplications. Up to 11 duplications were revealed to be lineage specific in the diversification of P. trichocarpa, after the split from the last common ancestor of P. trichocarpa and P. persica (Fig. 1). This duplication timing fits the recent WGD referred as the ‘salicoid’ duplication event (Tuskan et al., 2006).
Extensive gene loss events in the OVATE family were not inferred in land plant evolution. Gene loss events were estimated in lineage-specific clades only in five species: S. moellendorffii, A. coerulea, P. persica, C. papaya and V. vinifera.
Characterization of the OVATE family in rice, arabidopsis and tomato
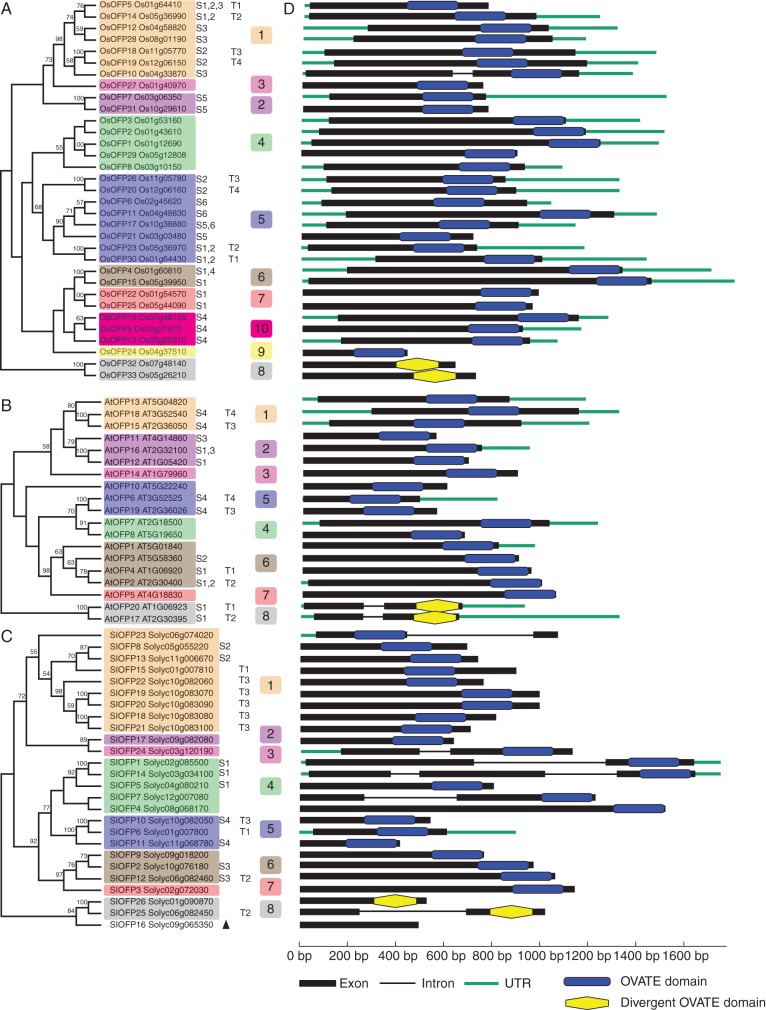
The rice genome has 33 OVATE-encoding genes designated OsOFP genes. These OsOFP genes are distributed unevenly across the 12 chromosomes of rice, with the highest density on chromosome 1 (eight genes), followed by chromosome 5 with seven OsOFP genes, while chromosomes 6 and 9 have no OFP genes (Fig. 4A). The phylogenetic relationships of OsOFPs within rice conformed to the analysis based on the conserved OVATE region in all 13 plant genomes (Fig. 5A). The OsOFP family could be divided into ten sub-groups, of which sub-group 10 was a monocot-specific clade, and sub-group 5 contained eight members accounting for a quarter of the OsOFP family in rice (Fig. 5A). The genic structures of all OsOFP-coding genes were also examined. Nearly all OsOFP genes are intronless, except OsOFP10, which contains a small intron (Fig. 5D).
Fig. 4.
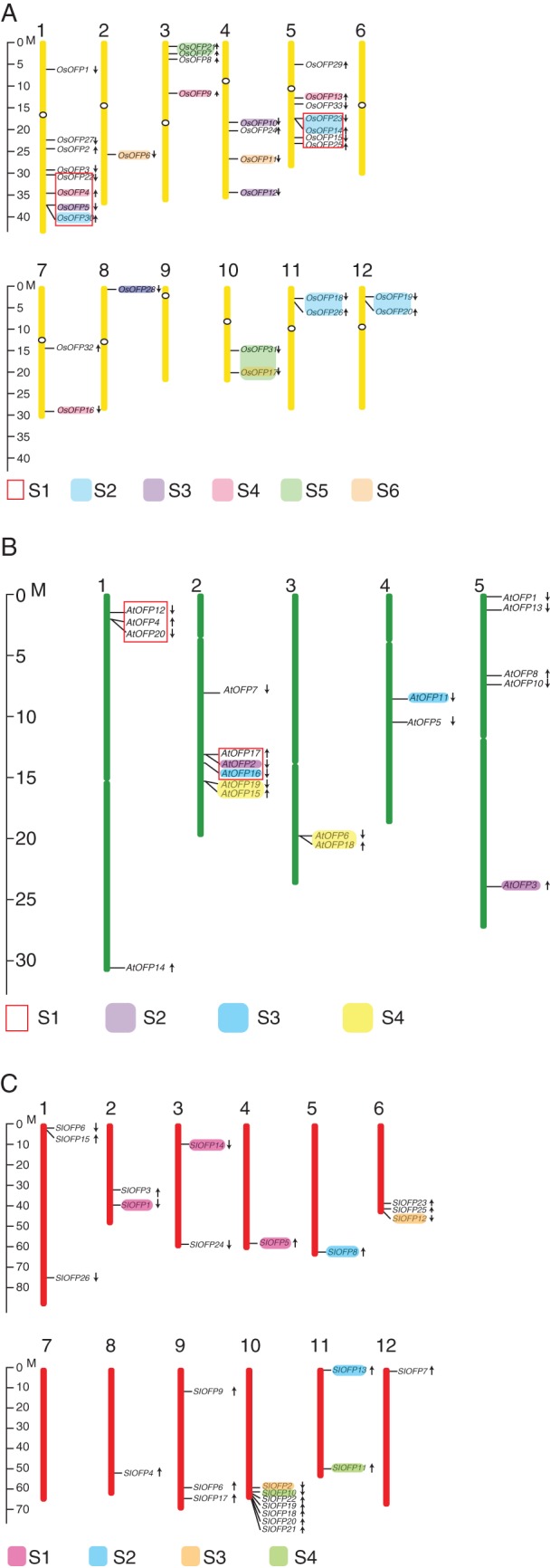
Chromosomal locations of OsOFP genes in rice (A), AtOFP genes in arabidopsis (B) and SlOFP genes in tomato (C). The orientation of each gene is shown by the arrows. Colours and red boxes represent the segmental duplication blocks of the OVATE genes in rice (A), arabidopsis (B) and tomato (C) genomes.
Fig. 5.
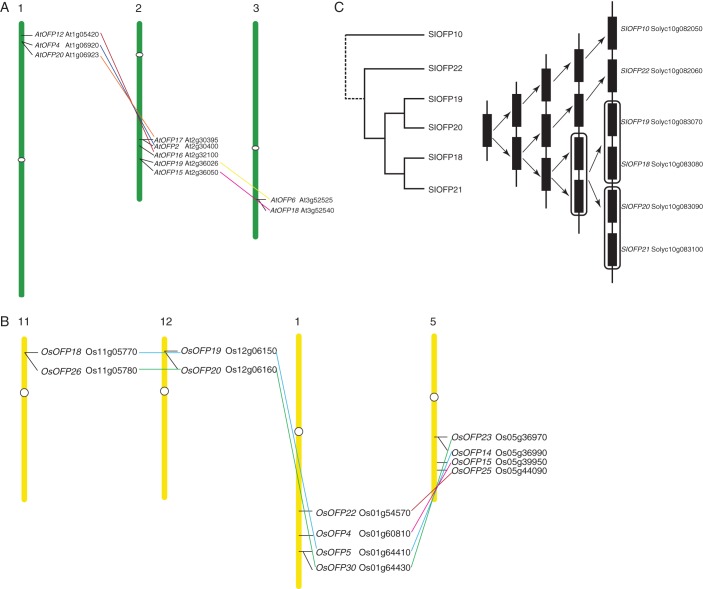
Phylogenetic analyses of OsOFPs in rice (A), AtOFPs in arabidopsis (B) and SlOFPs in tomato (C). The genomic organization of the corresponding OVATE protein-coding genes is shown in (D). Colours represent the sub-groups to which the OVATE proteins belong in the analysis of the 13 plant genomes. ‘S1’ to ‘S6’ indicate segmentally duplicated genes in segmental duplication blocks corresponding to Fig. 4; ‘T1’ to ‘T4’ indicate tandemly duplicated gene arrays in the chromosomes. Bootstrap values > 50 % are indicated above the branch. The black triangle in (C) marks SlOFP16, which was excluded from the phylogenetic analysis of all OVATE proteins from the 13 plant genomes. Blue rectangles mark the conserved OVATE domains, while yellow hexagons mark the divergent OVATE domains.
In the arabidopsis genome, 19 AtOFP genes are dispersed among all five chromosomes in clusters of two or more, with the majority clustered on chromosome 2 (Fig. 4B). Phylogenetic analysis of only the AtOFPs revealed three major clades containing eight out of the 11 sub-groups of plant OVATE proteins in the multispecies analysis (Figs 2 and 5B). Clade C1 comprised sub-groups 1, 2 and 3 (AtOFP11, 12, 13, 14, 15, 16 and 18), whereas sub-groups 4, 5, 6 and 7 belonged to clade C2 (AtOFP1, 2, 3, 4, 5, 6, 7, 8, 10 and 19). Clade C3 contained just the paralogues AtOFP17 and AtOFP20, which represent sub-group 8 and are more distantly related to other AtOFPs, being the only AtOFP genes that contain an intron (Fig. 5D).
It is of interest that overexpression of AtOFP genes with close phylogenetic relationships was previously shown to produce similar phenotypes. Plants overexpressing AtOFP1, AtOFP2, AtOFP4, AtOFP5 and AtOFP7 resulted in kidney-shaped cotyledons, and round and curled leaves (class I); overexpression of AtOFP6 and AtOFP8 caused flat, thick and cyan leaves (class II); and overexpression of AtOFP13, AtOFP15, AtOFP16 and AtOFP18 led to blunt-end siliques (class III) (Wang et al., 2011). Class I and II AtOFPs just fell into clade C2 while class III corresponded to clade C1 according to our study. Thus it appears that the evolution of the AtOFP family represented here is consistent with their probable patterns of functional divergence.
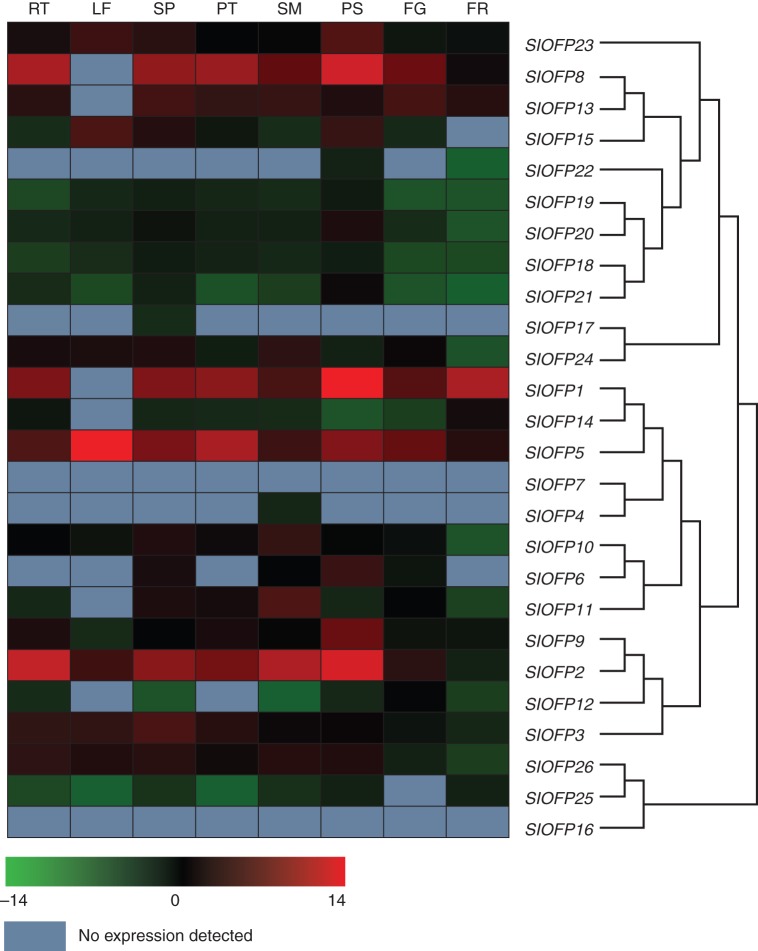
In the tomato genome, over a quarter of the SlOFP family genes (7/26) are located in a single gene cluster on chromosome 10, with the remaining genes dispersed across all 11 remaining chromosomes except chromosome 7 (Fig. 4C). Nine members of the SlOFP family are located in sub-group 1, including five of those linked in the cluster at the end of chromosome 10 (Figs 4C and 5C). To investigate further the diversity of SlOFP genes, their expression patterns in a round-fruit tomato variety were examined by quantitative real-time PCR. Of the 26 SlOFP genes, 12 (SlOFP2, SlOFP3, SlOFP5, SlOFP9, SlOFP10, SlOFP18, SlOFP19, SlOFP20, SlOFP21, SlOFP23, SlOFP24 and SlOFP26) were found to be expressed in all tissues examined, while expression of SlOFP7 and SlOFP16 ws undetectable (Fig. 6). One possibility is that their transcripts are present at a level below the limit of detection, or are only induced in response to certain conditions or treatments or at specific developmental stages. Alternatively, SlOFP16 may be a pseudogene, which is consistent with the fact that the OVATE region of SlOFP16 does not align with that of the other OVATE protein sequences. Tissue-specific expression profiles were discovered for three genes, with SlOFP4, SlOFP17 and SlOFP22 specific to stamens, sepals and pistils within the floral organs, respectively. Meanwhile, SlOFP5 was most highly transcribed in the leaf, while relatively high expression of SlOFP1, SlOFP2 and SlOFP8 was detected in pistils and a high abundance of SlOFP2 and SlOFP8 transcripts was also detected in roots. Preferential or tissue-specific transcription of SlOFP genes may be indicative of tissue-specific functions in plant growth and development.
Fig. 6.
Expression profile of SlOFP family genes by quantitative real-time PCR assays in a round-fruited tomato variety ‘Stupicke’. The colour key represents the log2 value of the relative expression level. Grey indicates that no expression was detected. RT, root; LF, leaf; SP, sepal; PT, petal; SM, stamens; PS, pistil; FG, green fruit; FR, red mature fruit.
From the phylogenetic analysis of SlOFPs, nine pairs of paralogues could be identified, eight of which were well supported by bootstrap analysis (Fig. 5C). Their phylogenetic relationships and expression patterns revealed two fates of the duplicated paralogues, i.e. retention–retention (RR) indicating that two paralogues retain the same original expression pattern or function, and retention–divergence (RD) indicating that one paralogue retains but the other diverges in expression pattern or function. Four pairs of paralogues (SlOFP8 and SlOFP13, SlOFP19 and SlOFP20, SlOFP18 and SlOFP21, and SlOFP2 and SlOFP9) were consistent with the RR mode, as they exhibited the same expression patterns.The other four pairs of paralogues (SlOFP17 and SlOFP 24, SlOFP1 and SlOFP 14, SlOFP6 and SlOFP10, and SlOFP25 and SlOFP26) were considered to be RD mode owing to their diverged expression patterns (Fig. 6). For example, SlOFP24 was found to be ubiquitously expressed in all tissues, whereas the transcript of its most closely related paralogue SlOFP17 was restricted to the sepal (Fig. 6). Both sub-functionalization and neofunctionalization might follow the divergence.
Duplication and OVATE gene family expansion in arabidopsis, rice and tomato
Plant genomes contain a higher proportion of duplicated genes than most other eukaryotic genomes and this has been argued to be a robust evolutionary force (Lockton and Gaut, 2005; Shan et al., 2007). Gene duplication is speculated to provide the raw genetic materials for the phenotypic or functional novelty necessary for adaptive evolution (Flagel and Wendel, 2009; Jiao et al., 2011). Segmental duplication, tandem duplication and retrotransposition or other transposition events are three major mechanisms for gene duplication (Kong et al., 2007). Analysis of 50 large gene families in arabidopsis combining information about genomic segmental duplications, gene family phylogeny and gene positions revealed that some gene families expanded mainly through tandem duplication, while some were oriented towards segmental duplication (Cannon et al., 2004). Based on the chromosomal location and phylogenetic relationships of OVATE family genes, we analysed the relative contributions of tandem duplication and segmental duplication to OVATE family expansion in arabidopsis, rice and tomato. Of the 19 AtOFP genes, eight (42·1 %) and 12 (63·2 %) appear to have arisen by tandem duplication and segmental duplication (Figs 4B and 5B), respectively. Within the rice OsOFP family, eight (24·2 %) have resulted from tandem duplication, whereas the majority (24/33; 72·7 %) were present within segmental duplication blocks (Figs 4A and 5A). In terms of the SlOFP genes, ten (38·5 %) were tandemly duplicated and nine (34·6 %) were located within segmental duplication blocks (Figs 4C and 5C). Therefore, tandem duplication and segmental duplication appear to play a principle role in SlOFP gene family expansion in all of the three sampled plant genomes, with segmental duplication particularly important for expansion of the arabidopsis AtOFP and rice OsOFP gene family (Fig. 7A, B). This is similar to the observation in other large gene families such as the bHLH (basic helix–loop–helix) gene family in rice (Li et al., 2006). Also, the largest tandem-arrayed SlOFP gene cluster, exhibiting a single-lineage expansion, on tomato chromosome 10 arose from a tandem duplication event (Fig. 7C). It is reported that stress-responsive gene sets are enriched for tandemly duplicated genes, thus tandemly duplicated genes involved in stress response are suggested to be preferably retained (Rizzon et al., 2006; Hanada et al., 2008).
Fig. 7.
Typical duplication events underlying expansion of the OVATE family in arabidopsis, rice and tomato. (A) Major expansions of AtOFP genes by segmental duplications in arabidopsis; (B) major expansions of OsOFP genes by segmental duplications in rice; (C) expansion of the largest tandem-arrayed SlOFP gene cluster by tandem duplication in tomato.
To better understand the evolutionary history of the OVATE family in arabidopsis, rice and tomato, we estimated the timing of segmental duplication events using values for Ks as the proxy for time (Tables 2–4). Ks values of AtOFP parologues centre around two ranges, from 0·70 to 0·84 and from 1·51 to 1·82. This suggests that two major duplication events occurred in arabidopsis, one around 23·33–28·00 million years ago (Mya) and a second around 50·33–60·67 Mya. These dates are approximately in line with the two WGD events (α and β) reported for arabidopsis, subsequent to the divergence of arabidopsis and C. papaya around 70 Mya (Bowers et al., 2003; Lockton and Gaut, 2005; Ming et al., 2008; Tang et al., 2008; Abrouk et al., 2010; Jiao et al., 2011). These results hint that the duplications in AtOFP sequences may be a consequence of the two WGDs, or of isolated, local segmental duplication events that occurred within those two periods. For the rice OsOFP genes, most of the segmental duplication events were estimated to occur between 50 and 70 Mya, consistent with the ρ WGD event pre-dating the diversification of major cereal clades (Paterson et al., 2004; Wang et al., 2005; Salse et al., 2008). The duplication time of paralogous SlOFP genes via segmental duplication was estimated to occur between 29 and 78·33 Mya, which may have involved the recent triplication event (52–91 Mya) shared by both tomato and potato in the Solanaceae, preceding their divergence about 7·3 Mya (Tomato Genome Consortium, 2012).
Table 2.
Duplicated AtOFP paralogues in segmental duplication blocks in arabidopsis
| Duplicated AtOFP gene in block 1 |
Duplicated AtOFP gene in block 2 |
Ks | Data (Mya) | ||
|---|---|---|---|---|---|
| Gene name | Gene locus | Gene name | Gene locus | ||
| AtOFP2 | At2g30400 | AtOFP3 | At5g58360 | 1·76 | 58·67 |
| AtOFP2 | At2g30400 | AtOFP4 | At1g06920 | 0·79 | 26·33 |
| AtOFP4 | At1g06920 | AtOFP3 | At5g58360 | 1·51 | 50·33 |
| AtOFP6 | At3g52525 | AtOFP19 | At2g36026 | 0·78 | 26·00 |
| AtOFP11 | At4g14860 | AtOFP16 | At2g32100 | 1·82 | 60·67 |
| AtOFP12 | At1g05420 | AtOFP16 | At2g32100 | 0·77 | 25·67 |
| AtOFP15 | At2g36050 | AtOFP18 | At3g52540 | 0·84 | 28·00 |
| AtOFP17 | At2g30395 | AtOFP20 | At1g06923 | 0·70 | 23·33 |
Table 3.
Duplicated OsOFP paralogues in segmental duplication blocks in rice
| Duplicated OsOFP gene in block 1 |
Duplicated OsOFP gene in block 2 |
Ks | Date (Mya) | ||
|---|---|---|---|---|---|
| Gene name | Gene locus | Gene name | Gene locus | ||
| OsOFP4 | LOC_Os01g60810 | OsOFP9 | LOC_Os03g21870 | 0·84 | 64·62 |
| OsOFP4 | LOC_Os01g60810 | OsOFP13 | LOC_Os05g25910 | 0·97 | 74·62 |
| OsOFP4 | LOC_Os01g60810 | OsOFP15 | LOC_Os05g39950 | 0·51 | 39·23 |
| OsOFP5 | LOC_Os01g64410 | OsOFP10 | LOC_Os04g33870 | 0·87 | 66·92 |
| OsOFP5 | LOC_Os01g64410 | OsOFP12 | LOC_Os04g58820 | 0·97 | 74·62 |
| OsOFP5 | LOC_Os01g64410 | OsOFP14 | LOC_Os05g36990 | 0·63 | 48·46 |
| OsOFP5 | LOC_Os01g64410 | OsOFP18 | LOC_Os11g05770 | 0·67 | 51·54 |
| OsOFP5 | LOC_Os01g64410 | OsOFP19 | LOC_Os12g06150 | 0·72 | 55·38 |
| OsOFP6 | LOC_Os02g45620 | OsOFP11 | LOC_Os04g48830 | 0·54 | 41·54 |
| OsOFP6 | LOC_Os02g45620 | OsOFP17 | LOC_Os10g38880 | 0·67 | 51·54 |
| OsOFP7 | LOC_Os03g06350 | OsOFP31 | LOC_Os10g29610 | 0·60 | 46·15 |
| OsOFP9 | LOC_Os03g21870 | OsOFP15 | LOC_Os05g39950 | 0·93 | 71·54 |
| OsOFP9 | LOC_Os03g21870 | OsOFP16 | LOC_Os07g48150 | 0·62 | 47·69 |
| OsOFP10 | LOC_Os04g33870 | OsOFP14 | LOC_Os05g36990 | 0·85 | 65·38 |
| OsOFP10 | LOC_Os04g33870 | OsOFP18 | LOC_Os11g05770 | 0·86 | 66·15 |
| OsOFP10 | LOC_Os04g33870 | OsOFP19 | LOC_Os12g06150 | 0·90 | 69·23 |
| OsOFP11 | LOC_Os04g48830 | OsOFP17 | LOC_Os10g38880 | 0·60 | 46·15 |
| OsOFP11 | LOC_Os04g48830 | OsOFP21 | LOC_Os03g03480 | 0·67 | 51·54 |
| OsOFP12 | LOC_Os04g58820 | OsOFP14 | LOC_Os05g36990 | 0·68 | 52·31 |
| OsOFP12 | LOC_Os04g58820 | OsOFP18 | LOC_Os11g05770 | 0·75 | 57·69 |
| OsOFP12 | LOC_Os04g58820 | OsOFP28 | LOC_Os08g01190 | 0·63 | 48·46 |
| OsOFP13 | LOC_Os05g25910 | OsOFP15 | LOC_Os05g39950 | 1·19 | 91·54 |
| OsOFP14 | LOC_Os05g36990 | OsOFP19 | LOC_Os12g06150 | 0·66 | 50·77 |
| OsOFP15 | LOC_Os05g39950 | OsOFP16 | LOC_Os07g48150 | 1·02 | 78·46 |
| OsOFP17 | LOC_Os10g38880 | OsOFP21 | LOC_Os03g03480 | 0·51 | 39·24 |
| OsOFP18 | LOC_Os11g05770 | OsOFP19 | LOC_Os12g06150 | 0·53 | 40·77 |
| OsOFP20 | LOC_Os12g06160 | OsOFP26 | LOC_Os11g05780 | 0·51 | 39·23 |
| OsOFP22 | LOC_Os01g54570 | OsOFP25 | LOC_Os05g44090 | 0·79 | 60·77 |
| OsOFP23 | LOC_Os05g36970 | OsOFP30 | LOC_Os01g64430 | 0·78* | 60·00 |
*The Ks value was calculated by the KaKs_ calculator.
Table 4.
Duplicated SlOFP paralogues in segmental duplication blocks in tomato
| Duplicated SlOFP gene in block 1 |
Duplicated SlOFP gene in block 2 |
Ks | Date (Mya) | ||
|---|---|---|---|---|---|
| Gene name | Gene locus | Gene name | Gene locus | ||
| SlOFP1 | Solyc02g085500·2 | SlOFP14 | Solyc03g034100·2 | 1·3 | 43·33 |
| SlOFP2 | Solyc10g076180·1 | SlOFP12 | Solyc06g082460·1 | 1·37 | 45·67 |
| SlOFP5 | Solyc04g080210·1 | SlOFP14 | Solyc03g034100·2 | 2·35 | 78·33 |
| SlOFP8 | Solyc05g055220·1 | SlOFP13 | Solyc11g006670·1 | 0·87 | 29·00 |
| SlOFP10 | Solyc10g082050·1 | SlOFP11 | Solyc11g068780·1 | 1·81 | 60·33 |
Conclusions
In this study, we performed the first comparative genomic analysis of the OVATE protein family, a recently discovered plant-specific gene regulatory family in land plants. OVATE family proteins are present in all major lineages of land plants including the early-diverged species. Our phylogenetic analysis of 13 available plant genomes spanning major evolutionary lineages defined 11 sub-groups of OVATE family proteins in angiosperms. Two different mechanisms, namely conserved evolution and divergent expansion, are proposed to be involved in OVATE family evolution in plants. Detailed characterization of the AtOFP family in arabidopsis, the OsOFP family in rice and the SlOFP family in tomato provided a deeper understanding of the evolutionary framework and revealed a principle role for tandem duplication and segmental duplication in expansion of the OVATE gene family. This study has established a solid base for subsequent functional genomics studies on this important gene family in plants, which has been poorly characterized to date.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Dr Shucai Wang for communications on the OVATE gene family in arabidopsis, and Drs Hongwen Huang and Ming Kang for experimental assistance. This work was supported by a grant from the Major State Basic Research Development Program of China (973 Program) (no. 2010CB126603), the National Natural Science Foundation of China (31270340 and 30570172) and the Knowledge Innovation Project of The Chinese Academy of Sciences (KSCX2-EW-J-20).
LITERATURE CITED
- Abrouk M, Murat F, Pont C, et al. Palaeogenomics of plants: synteny-based modelling of extinct ancestors. Trends in Plant Science. 2010;15:479–487. doi: 10.1016/j.tplants.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaoui M, Pidkowich MS, Samach A, et al. The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. The Plant Cell. 2001;13:2455–2470. doi: 10.1105/tpc.010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. Journal of Biological Chemistry. 1995;270:31178–31188. doi: 10.1074/jbc.270.52.31178. [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. The Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. The Plant Cell. 2005;17:2281–2295. doi: 10.1105/tpc.105.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon S, Mitra A, Baumgarten A, Young N, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biology. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Castillo R, Xue Y, Schwarz-Sommer Z, Davies B. Tracing the evolution of the floral homeotic B- and C-function genes through genome synteny. Molecular Biology and Evolution. 2010;27:2651–2664. doi: 10.1093/molbev/msq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Hu J, Sun S, Xu G. Conservation and divergence of both phosphate- and mycorrhiza-regulated physiological responses and expression patterns of phosphate transporters in solanaceous species. New Phytologist. 2007;173:817–831. doi: 10.1111/j.1469-8137.2006.01962.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Banerjee AK, Hannapel DJ. The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. The Plant Journal. 2004;38:276–284. doi: 10.1111/j.1365-313X.2004.02048.x. [DOI] [PubMed] [Google Scholar]
- Cole M, Nolte C, Werr W. Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Research. 2006;34:1281–1292. doi: 10.1093/nar/gkl016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa LGG, Riaño-Pachón DM, Schrago CG, Vicentini dos Santos R, Mueller-Roeber B, Vincentz M. The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS One. 2008;3:e2944. doi: 10.1371/journal.pone.0002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand D, Halldórsson BV, Vernot B. A hybrid micro-macroevolutionary approach to gene tree reconstruction. Journal of Computational Biology. 2006;13:320–335. doi: 10.1089/cmb.2006.13.320. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytologist. 2009;183:557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- Guyot R, Lefebvre-Pautigny F, Tranchant-Dubreuil C, et al. Ancestral synteny shared between distantly-related plant species from the asterid (Coffea canephora and Solanum Sp.) and rosid (Vitis vinifera) clades. BMC Genomics. 2012;13:103. doi: 10.1186/1471-2164-13-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbusch J, Richter K, Muller J, Salamini F, Uhrig JF. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proceedings of the National Academy of Sciences; USA. 2005. pp. 4908–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Smith HMS, Holtan H, Magnani E, Mele G, Ramirez J. The role of KNOX genes in plant development. Annual Review of Cell and Developmental Biology. 2004;20:125–151. doi: 10.1146/annurev.cellbio.20.031803.093824. [DOI] [PubMed] [Google Scholar]
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT,. 41 (1999) Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hamant O, Pautot V. Plant development: a TALE story. Comptes Rendus Biologies. 2010;333:371–381. doi: 10.1016/j.crvi.2010.01.015. [DOI] [PubMed] [Google Scholar]
- Hamilton AT, Huntley S, Tran-Gyamfi M, Baggott DM, Gordon L, Stubbs L. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Research. 2006;16:584–594. doi: 10.1101/gr.4843906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu S-H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiology. 2008;148:993–1003. doi: 10.1104/pp.108.122457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. A KNOX family TALE. Current Opinion in Plant Biology. 2009;12:593–598. doi: 10.1016/j.pbi.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. KNOX genes: versatile regulators of plant development and diversity. Development. 2010;137:3153–3165. doi: 10.1242/dev.030049. [DOI] [PubMed] [Google Scholar]
- Huson D, Richter D, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Kong H, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, DePamphilis CW. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. The Plant Journal. 2007;50:873–885. doi: 10.1111/j.1365-313X.2007.03097.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Wang S, Liu Y, Chen JG, Douglas CJ. OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. The Plant Journal. 2011;67:328–341. doi: 10.1111/j.1365-313X.2011.04595.x. [DOI] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, et al. Genome-wide analysis of basic/helix–loop–helix transcription factor family in rice and Arabidopsis. Plant Physiology. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Van Eck J, Cong B, Tanksley SD. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proceedings of the National Academy of Sciences; USA. 2002. pp. 13302–13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang C, Yang Y, Hu X. Genome-wide and molecular evolution analyses of the phospholipase D gene family in poplar and grape. BMC Plant Biology. 2010;10:117. doi: 10.1186/1471-2229-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lockton S, Gaut BS. Plant conserved non-coding sequences and paralogue evolution. Trends in Genetics. 2005;21:60–65. doi: 10.1016/j.tig.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Research. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Hou S, Feng Y, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu H-J, Sundaresan V. Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-Like homeodomain gene BLH1. The Plant Cell. 2007;19:3578–3592. doi: 10.1105/tpc.107.054890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proceedings of the National Academy of Sciences; USA. 2004. pp. 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Computational Biology. 2006;2:e115. doi: 10.1371/journal.pcbi.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, et al. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. The Plant Cell. 2008;20:11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H, Zhang N, Liu C, et al. Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Molecular Phylogenetics and Evolution. 2007;44:26–41. doi: 10.1016/j.ympev.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Shiu S-H, Karlowski WM, Pan R, Tzeng Y-H, Mayer KFX, Li W-H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. The Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HMS, Hake S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. The Plant Cell. 2003;15:1717–1727. doi: 10.1105/tpc.012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HMS, Boschke I, Hake S. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proceedings of the National Academy of Sciences; USA. 2002. pp. 9579–9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Research. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Paterson AH. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proceedings of the National Academy of Sciences; USA. 2010. pp. 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaballa A, Pasentsis K, Darzentas N, Tsaftaris AS. Multiple evidence for the role of an Ovate-like gene in determining fruit shape in pepper. BMC Plant Biology. 2011;11:46. doi: 10.1186/1471-2229-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Vernot B, Stolzer M, Goldman A, Durand D. Reconciliation with non-binary species trees. Journal of Computational Biology. 2008;15:981–1006. doi: 10.1089/cmb.2008.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chang Y, Guo J, Chen JG. Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. The Plant Journal. 2007;50:858–72. doi: 10.1111/j.1365-313X.2007.03096.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Chang Y, Guo J, Zeng Q, Ellis BE, Chen JG. Arabidopsis ovate family proteins, a novel transcriptional repressor family, control multiple aspects of plant growth and development. PLoS One. 2011;6:e23896. doi: 10.1371/journal.pone.0023896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Shi X, Hao B, Ge S, Luo J. Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytologist. 2005;165:937–946. doi: 10.1111/j.1469-8137.2004.01293.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Diehl A, Wu F, et al. Sequencing and comparative analysis of a conserved syntenic segment in the Solanaceae. Genetics. 2008;180:391–408. doi: 10.1534/genetics.108.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-K, Chang W-C, Liu P-F, et al. Ovate family protein 1 as a plant Ku70 interacting protein involving in DNA double-strand break repair. Plant Molecular Biology. 2010;74:453–466. doi: 10.1007/s11103-010-9685-5. [DOI] [PubMed] [Google Scholar]
- Xia K, Liu T, Ouyang J, Wang R, Fan T, Zhang M. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.) DNA Research. 2011;18:363–377. doi: 10.1093/dnares/dsr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Ma H, Nei M, Kong H. Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proceedings of the National Academy of Sciences; USA. 2009. pp. 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao X-Q, Wang J, Wong GK-S, Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics, Proteomics and Bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Huang J, Yu H, Wang L, Xie W. Genomic survey, characterization and expression profile analysis of the peptide transporter family in rice (Oryza sativa L.) BMC Plant Biology. 2010;10:92. doi: 10.1186/1471-2229-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.