Abstract
Bacteria have a great capacity for adjusting their metabolism in response to environmental changes by linking extracellular stimuli to the regulation of genes by transcription factors. By working in a co-operative manner, transcription factors provide a rapid response to external threats, allowing the bacteria to survive. This review will focus on transcription factors MarA, SoxS and Rob in Escherichia coli, three members of the AraC family of proteins. These homologous proteins exemplify the ability to respond to multiple threats such as oxidative stress, drugs and toxic compounds, acidic pH, and host antimicrobial peptides. MarA, SoxS and Rob recognize similar DNA sequences in the promoter region of more than 40 regulatory target genes. As their regulons overlap, a finely tuned adaptive response allows E. coli to survive in the presence of different assaults in a co-ordinated manner. These regulators are well conserved amongst Enterobacteriaceae and due to their broad involvement in bacterial adaptation in the host, have recently been explored as targets to develop new anti-virulence agents. The regulators are also being examined for their roles in novel technologies such as biofuel production.
Keywords: antibiotic resistance, AraC family regulators, Escherichia coli, Enterobacteriaceae, global regulators, drug development
1. Introduction
Bacteria are found in every possible habitat on the planet and adaptation to changing external factors is essential for their survival and growth. This ability to adapt resides in multiple regulatory networks that control gene expression in a co-ordinated manner in response to environmental stimuli. Signals are transmitted to transcriptional regulators that interact with target DNA in the promoter region of a specific open reading frame (ORF), activating or repressing the expression of the target gene [1]. In certain cases, transcription regulators control genes and operons that belong to different metabolic pathways. These transcription factors are known as global regulators. By working in a co-operative manner on several promoters, global regulators provide flexible and finely tuned responses to external signals [1].
This review will focus on MarA, SoxS and Rob of Escherichia coli. These homologous regulators are excellent examples of global regulators that are part of multiple regulatory mechanisms necessary for the adaptive response. MarA, SoxS and Rob, which are all members of the AraC family of proteins, respond to many stimuli, including changing pH, the presence of antibiotics, oxidative stressors and organic solvents, all of which threaten survival. Several studies have shown that these three transcriptional regulators (TRs) are closely related and bind to a degenerate consensus sequence creating considerable overlap in the genes they control [2–7]. Because the genes of the regulons are often under the control of all three TRs to various degrees, a network of pathways is formed that allows small adjustments in response to different extracellular threats [6, 7]. One of their principal roles is to mediate multidrug resistance by up-regulating expression of the AcrAB-TolC multidrug efflux pump [8] and of MicF [9], a small inhibitory RNA that down-regulates the outer membrane porin OmpF. In addition to their contribution to drug resistance, MarA, SoxS and Rob have roles at the host-pathogen interface [10]. Furthermore, a recent study has identified a number of attractive candidates for MarA/SoxS/Rob-controlled loci found to play a role in persistence of E. coli in a mouse model of kidney infection [11].
With this great ability to adapt to environmental threats, it is not surprising that bacteria become resistant to all classes of antimicrobial drugs. Antimicrobial drug development has focused on targeting essential proteins that prevent growth of the micro-organism or which are bacteriocidal [12]. However, exposure of bacteria to antibiotics, especially at levels below the minimal inhibitory concentration (MIC), allows selection of adaptive responses and mutations so that micro-organisms can grow in the presence of the drug. Recent studies have brought forward the concept of targeting virulence factors instead of essential growth determinants [13, 14]. By targeting virulence, the fitness of a bacterium within the host is impaired, allowing, in theory, the host immune system to combat the infection. If new compounds could target non-essential processes, selective pressure would be reduced and the development of bacteria with antibiotic resistance phenotypes decreased. Due to their broad involvement in bacterial adaptation within the host, the three TRs and homologous regulators in different bacterial genera have been used as targets to develop new anti-virulence agents. Furthermore, in recent years, a novel application of the TRs has been their use in increasing tolerance to organic solvents during the production of biofuels.
2.MarA and the marRAB operon
The mar locus was first identified by a Tn5 transposon insertion that reversed a multiple drug resistance (MDR) phenotype of E. coli isolates selected by growth on subinhibitory levels of tetracycline or chloramphenicol [15]. Later, genetic analysis showed that the mar locus consisted of two divergently transcribed units: marC and the marRAB operon [16]. However, MarC, a putative integral inner membrane protein, does not contribute to the MDR phenotype [17]. MarA, encoded by the second gene in the marRAB operon, is a member of the AraC family of transcription regulators [16, 18, 19]. The direct function of this 127 residue protein in antibiotic resistance was first identified through genetic screening [20]. marB, located just downstream of marA in the operon, is of unknown function, although it has been shown to somehow increase the level of MarA messenger [21, 22]. MarB has a predicted periplasmic signal sequence, suggesting that it acts post-transcriptionally [22]. Interestingly, it has recently been reported to be involved in the transcriptional regulation of inaA, a MarA regulon gene (see section 5.4.2).
In the absence of a signal, the transcription of the whole operon is repressed by MarR, encoded by the first gene of the marRAB operon [16, 23]. The MarR regulator forms dimers and the DNA-binding domain of the protein consists of a winged helix-turn-helix motif [24], which promotes binding to two specific palindromic sites within the promoter region marO, named site 1 and site 2 (Figure 1A) [25]. The MarR-marO interaction is highly specific with an apparent Kd of ~10−9M [25]. Binding of MarR to the marO promoter region is relieved when a ligand, such as the phenolic sodium salicylate, the naphthoquinones menadione and plumbagin, 2,4-dinitrophenol, or the metabolite 2,3-dihydroxybenzoate, interacts with MarR, inactivating it and allowing transcription of the marRAB operon [26–29]. Once de-repression occurs, MarA activates or represses the transcription of the genes in its regulon [6, 7]. MarA also activates its own transcription by binding as a monomer to the 20-bp marbox located upstream of the −35 hexamer in the promoter region of the marRAB operon [30]. Transcription of marRAB leads to the production of two transcripts of 1.1 and 0.9 kb length [16, 31]. The source of the 0.9kb transcript could be an internal promoter located within marR or a result of processing the 1.1 kb transcript. In the absence of a molecular signal, MarR binds again to the operator and repression of transcription resumes. Additional regulation is provided by the Lon protease that rapidly degrades MarA [32]; the half-life of MarA is extremely short (~3 min), which ensures that the response cascade is rapidly removed once the stress signal disappears. Thus, in the absence of signal, MarA levels are extremely low [32]. Activation of marRAB has been shown to increase in the presence of Fis (Factor for inversion stimulation), a small DNA binding and bending protein [33]. Fis binds to the marO promoter region at a DNA sequence located upstream of the marbox. Binding of Fis stimulates the transcription of the marRAB operon only in the presence of MarA, SoxS or Rob, acting as an auxiliary protein [33]. MarA, which lacks the effector domain characteristic of other AraC-like proteins, was co-crystallized with the marbox found in marO [18]. The tridimensional structure showed that MarA contains two helix-turn-helix (HTH) motifs, of which recognition helices 3 and 6 bind two successive major grooves in the DNA, consequently bending the DNA [18]. Alanine-scanning mutagenesis has identified the amino acid residues within the recognition helices that are involved in DNA binding [34]. The structural requirements for marbox function in transcriptional regulation are discussed below (Section 5.1).
Figure 1. Schematic representation of marRAB (A), soxRS (B) and rob (C) loci.
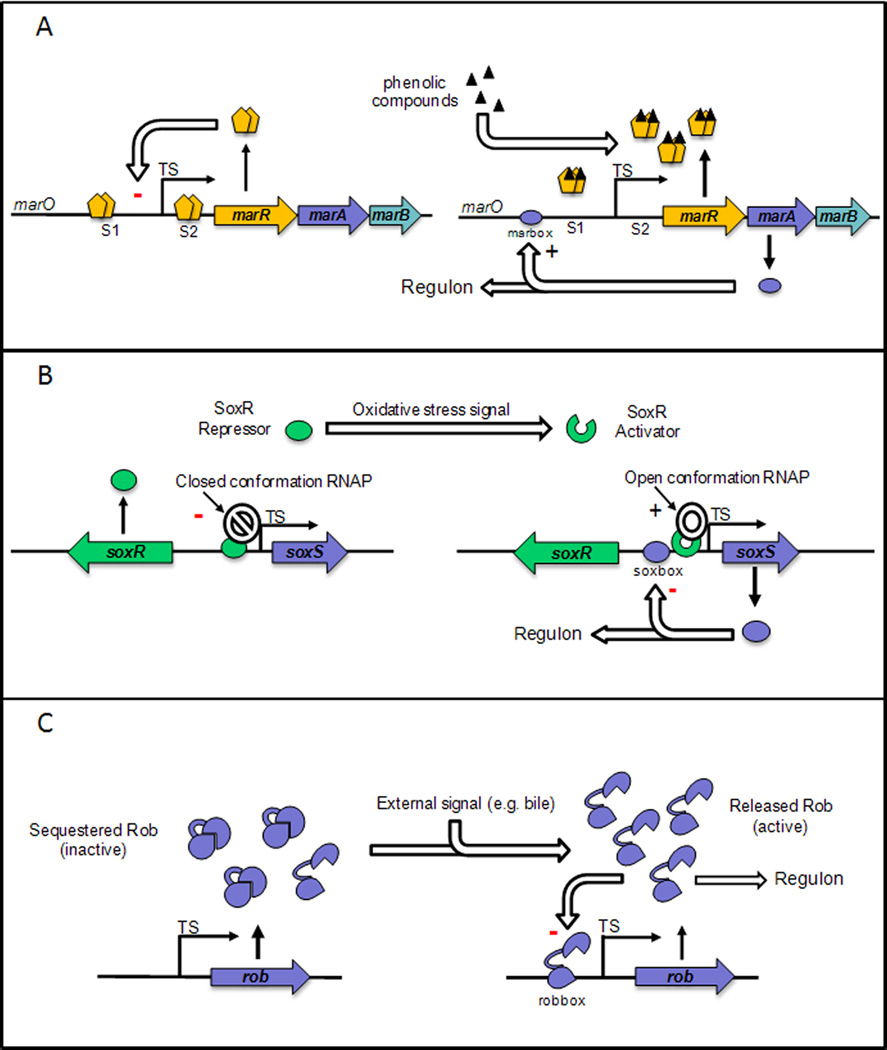
Respective mechanism of transcription activation are described in the text. TS, transcription start; S1, site 1; S2, site 2; marO, promoter region of the marRAB operon.
Several transcription factors, other than MarR and MarA, have been shown to regulate the transcription of marRAB. Overexpression of homologous regulator, SoxS, activates transcription of the operon [35]. Martin and colleagues also demonstrated that purified SoxS was able to bind the marbox located within marO [30]. Basal levels of Rob were also shown to be responsible for basal levels of marRAB transcription [2, 36]. Later, in vitro studies demonstrated that Rob also binds to the marbox as a monomer [37]. However, cross talk between the mar and sox systems is limited under physiological conditions. For example, SoxS, when expressed, is not expected to activate marRAB transcription in the absence of salicylate, when MarR still represses the promoter [38]. In the case of the rob and mar systems, the situation is different as salicylate is able to activate marRAB through Rob [38]. The mechanism by which Rob is activated by salicylate is still unknown. Another regulator, EmrR, has been reported to repress the marRAB operon upon overexpression from a multicopy plasmid [39, 40]. EmrR, a MarR homolog also known as MprA, is the transcriptional repressor of the EmrAB operon that codes for an efflux pump [41, 42]. Whether or not there are environmental conditions where levels of EmrR are high enough to repress marRAB has not been demonstrated. Additionally, two regulators of the carbon metabolism, CRP and Cra, have been shown to control marRAB transcription. The activity of these TRs varies with glucose concentration: CRP (cyclic AMP [cAMP] receptor protein) is activated after binding to cAMP, which is synthesized in the absence of glucose [43], while Cra (catabolite repressor activator) is inactivated upon interaction with inducers such as D-fructose-1-phosphate or D-fructose-1,6-biphosphate, both generated in the presence of glucose [44]. These regulators have opposing effects on marRAB transcription: CRP positively regulates marRAB transcription [45], whereas Cra represses marRAB [46].
3. SoxS and SoxR
Early on in the characterization of the marRAB operon, soxR and soxS loci were identified as an oxidative response system [47–49]. soxR, which encodes a regulator of the MerR family, is divergently transcribed from soxS (Figure 1B). The SoxR promoter is embedded within the soxS structural gene [47, 48]. In the absence of an oxidative stress signal, the SoxR homodimer binds to the promoter region of soxS and prevents enhanced transcription [49, 50]. SoxR repression, combined with the action of the Lon protease and to a lesser extent, the FtsH protease, leads to very low levels of SoxS protein in the absence of an induction signal [32]. When the SoxR [2Fe-2S] cluster is oxidized, SoxR becomes an activator of soxS transcription [51–55]. This activation has long been considered to be the result of oxidization by the superoxide anion generated by compounds such as the napthoquinones, xenobiotics and paraquat [7, 56, 57] or by nitric oxide generated for example by macrophages [58]. However, it has recently been shown that superoxide is not the oxidizer of SoxR; rather the redox cycling drugs themselves oxidize SoxR as they in turn are reduced [59]. Once synthesized, SoxS is able to repress its own transcription [60]. MarA and Rob have also been shown to repress soxS expression when ectopically overexpressed [38].
The SoxS protein is closely related to MarA (41 % identity, 67 % similarity), although slightly shorter in length (107 amino acids). Like MarA, SoxS lacks the dimerization domain of AraC-like proteins [19, 61] and binds to a 20-bp sequence (soxbox) as a monomer [3, 4, 62]. The SoxS regulon was found to have a remarkable degree of overlap with the MarA regulon [6, 7]. However, because of the difference in affinities of MarA and SoxS for different promoters and also because of their different methods of activation, SoxS is the first responder to oxidative stress and MarA is the first to react to antibiotic assault. The regulons of both MarA and SoxS are discussed below (Section 5).
4. Rob
Rob was first identified by its ability to bind to the right border of oriC DNA [63] but it appears to have no function in replication and the physiological meaning of its binding to the oriC sequence is not known [2]. Rob’s function in resistance to antibiotics, organic solvents and superoxide-generating agents was first shown by overexpression of Rob through a plasmid [2, 36]. Later, Rob was found to activate transcription of multiple antibiotic and superoxide resistance genes by interacting as a monomer with the highly degenerate sequence recognized by MarA and SoxS [37, 64]. Unlike MarA and SoxS that are synthesized de novo in the presence of the activating signal, Rob is constitutively expressed (Figure 1C). However, while abundant in the cell, Rob remains in an inactive state [37, 63] due to its sequestration in intracellular foci [65]. A recent study suggests that a fraction of Rob is in an active form even in the absence of inducer [38]. Transcription of Rob has been shown to be repressed by MarA, SoxS [66–68] and by Rob itself [38].
Rob is a 289 amino acid protein of the AraC family of regulators. Its N-terminal DNA-binding domain has 51 % identity and 71 % similarity with MarA [19, 69]. Rob differs from MarA and SoxS by the presence of a C-terminal domain [70]. The crystal structure of Rob in complex with the robbox of the micF promoter revealed two HTH motifs within the N-terminal domain, which, not surprisingly, is similar to that of MarA [70]. However, unlike MarA, only one HTH motif interacts with the major groove. The other HTH motif interacts with the backbone of the DNA instead of lying in the adjacent major groove [70]. Consequently, the DNA appears unbent [70]. For a long period of time, the function of Rob’s C-terminal domain was unknown, until several studies showed its role in sequestration and release from sequestration, and consequent activation upon interaction with bile salts or dipyridyl [71–73] (Figure 1C). The C-terminus also acts to protect the protein from degradation by proteases Lon and, to a lesser extent, ClpYQ [71].
5. The regulons and phenotypes associated in E. coli
The DNA-binding domain of MarA, SoxS and Rob is exceptionally similar in terms of primary sequence homology, DNA sequence recognition and transcriptional activation properties. However, the expression (MarA and SoxS) or activity (Rob) of the three TRs is stimulated by specific signals, suggesting that the three TRs diverge to generate an adequate response. Although each system can function independently, they are closely interlinked and their physiological responses are comparable. In this section, we will discuss in detail the characteristics of a functional MarA/SoxS/Rob DNA-binding site and the genes that belong to their regulons.
5.1. Structural requirements for marbox function and discrimination between the regulon promoters
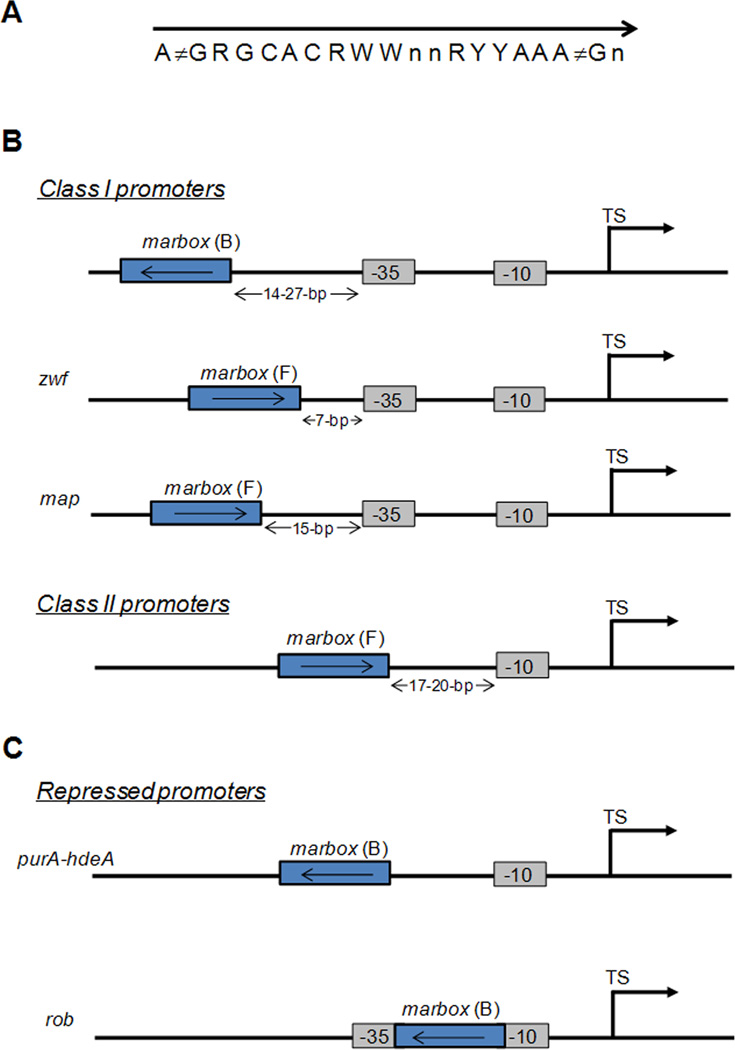
The DNA binding site recognized by MarA, SoxS and Rob consists of an asymmetric and degenerate sequence that we will call the marbox for simplicity. The consensus sequence is illustrated in Figure 2A. Martin and colleagues defined two classes of activated promoters, class I and II, that are distinct by the orientation and position of the marbox relative to the −35 and −10 recognition sequences of the RNA polymerase (RNAP) [5, 74]. In most class I promoters, the regulator binds upstream of the −35 hexamer to a marbox oriented backward (B). Among the class I promoters, zwf and map promoters are exceptions for which the marbox orientation is forward (F) [5, 74]. In class II promoters, the regulator binding site is oriented in the forward direction (F) and overlaps with the −35 hexamer [5, 75]. The different classes of promoters are summarized in Figure 2B.
Figure 2. Sequence and structural characteristics of the regulon promoters.
A. Consensus sequence for the MarA, SoxS and Rob DNA binding sites. R, A/G; Y, C/T; W, A/T; n, any base. B. Schematic representation of class I and II promoters activated by MarA, SoxS and Rob proteins. For class II promoters, the marbox overlaps −35 hexamer sequence recognized by the RNA polymerase, which is not indicated. C. Schematic representation of promoters repressed by MarA, SoxS and Rob proteins. For purA and hdeA promoters, the marbox overlaps −35 hexamer sequence recognized by the RNA polymerase, which is not indicated. The marbox is represented by a blue box. The −10 and −35 sequences are represented by grey boxes. TS, transcription start site.
The different orientations and positions of the marbox relative to the −35 and −10 signals of the RNAP imply two configurations of the ternary complex Regulator-RNAP-DNA. Transcriptional activation at class I promoters was shown to require interaction of the regulator with the C-terminal domain of the RNAP α-subunit (RNAP α-CTD), while activation at class II promoters has been shown to involve interaction with RNAP α-CTD and with region 4 of the sigma factor 70 [4, 5, 37, 76–79]. NMR studies have identified the specific residues of RNAP α-CTD that interact with MarA, and it is considered likely that MarA, SoxS and Rob bind similar residues [80]. Two pathways to the formation of the ternary complex Regulator-RNAP-DNA have been put forward. In the RNA capture pathway, the regulator binds the cognate DNA first and then recruits RNAP [81]. In the DNA-scanning pathway, the regulator binds first to RNAP and this binary complex scans the DNA for binding sites [75]. The latter is favored for the three TRs because MarA, SoxS and Rob interact with RNAP in solution [4, 37, 75, 76] and the ternary complex (Regulator-RNAP-DNA) is more stable than the binary complex (Regulator-RNAP) [75]. It has been also suggested that binding of the regulator to RNAP increases the kinetics of the polymerase, resulting in the need for a lower number of binary complexes to produce the same amount of transcripts [82].
Although different environmental signals induce the expression of MarA, SoxS and Rob, these three homologous regulators activate a common group of promoters, which results in MDR, oxidative stress response and organic solvent tolerance. Nevertheless, these regulators do not activate each of the regulon promoters to the same extent [2, 83]. For instance, compared to SoxS, MarA displayed lower binding affinity and lower transcription activity at promoters of genes involved in superoxide defense [83]. Small differences in the marbox sequences are the main factors responsible for discrimination [83]. An example given by Martin and Rosner shows that in MarA, steric hindrance occurs between the side chain of glutamic acid at position 89 (Glu-89) and the phosphate located between position 12 and 13 of the marboxes of class I promoters [84]. The steric hindrance lowers the affinity of the regulator for this DNA sequence. A thymine at position 12 in the marbox leads to higher affinity, suggesting that it accommodates the side chain of Glu-89. Affinity was also increased by changing Glu-89 to an alanine [84]. This phenomenon implies that, with different affinities for DNA, the binding of each regulator to a specific promoter is also dependent on protein concentration [85]. For example, high concentrations of MarA are necessary to bind promoters such as the acrAB promoter. This is potentially because lower concentrations are necessary to activate other responses involved in antibiotic resistance such as micF transcription that turns down translation of the outer membrane porin ompF (see below). Moreover, high expression of AcrAB-TolC pump is costly in terms of the cell’s energy resources [85].
The three TRs also act as transcription repressors. MarA was shown to directly repress purA, hdeA and rob transcription [67, 86]. The marboxes lie in backward orientation within all three repressed promoters. The marboxes of hdeA and purA promoters overlap the −35 recognition sequence of the RNAP [86], while in the rob promoter, it lies between the −10 and −35 sites [67]. It has been first reported that MarA represses transcription of rob via a RNAP-DNA-MarA ternary complex [67]. However, the same research group later demonstrated that non-specific binding of RNAP to DNA was responsible for the ternary complexes, and that repression appears to occur by steric hindrance of RNAP [68].
5.2. Multidrug resistance and organic solvent tolerance
One of the principal phenotypes of expression of MarA, SoxS and of activation of Rob is MDR. During the initial discovery of the marA locus, spontaneous drug resistant mutants were found to be less susceptible to several other antibiotics; this susceptibility could be further decreased by stepwise growth on increasing antibiotic concentrations [15, 31, 87]. Transposon insertion in marR led to a 3–8-fold increase in the minimum inhibitory concentration (MIC) of several antibiotics [16]. Additionally, expression of MarA from a multicopy plasmid led to increases in MIC of 4–5 fold [20]. Deletion or mutation of marA, however, resulted only in small decreases in MIC of just under 2 fold or less [26]. Therefore, increasing expression of MarA leads to a greater effect than deleting it. Reduced antibiotic susceptibility by SoxRS was first described in the early 1990’s [48, 88] after treatment of E. coli with oxidizing agents. Control was found to occur through SoxR activating SoxS transcription (Figure 1B). Decreased antibiotic susceptibility due to higher Rob expression was identified in 1995 when its N-terminal sequence similarity with the MarA and SoxS sequences was noted [2]. The mechanism by which the three TRs cause decreased antibiotic susceptibility is through activation of similar genes.
5.2.1. The AcrAB-TolC efflux pump
All three TRs regulate transcription of the efflux pump AcrAB-TolC, a member of the resistance-nodulation-division (RND) family of proteins. acrA and acrB are part of the same operon and are regulated through the acrA promoter. AcrB provides a channel between the cytoplasm and the periplasm and interacts with TolC via the adaptor protein AcrA. TolC provides a continuation of the AcrB channel from the periplasm to the extracellular space. Compounds may also enter the pump from the periplasm via channels in AcrB. The pump is one of the most important in antibiotic resistance and, as the interior of the channel is hydrophobic, in efflux of a diverse array of lipophilic compounds. An excellent review is given by Nikaido and Takatsuka [89].
Several studies [8, 64, 90, 91] have shown that MDR and organic solvent tolerance (see below) phenotypes of the three TRs are in large part due to this pump. For the marRAB operon, elevated transcription of acrAB occurs in marR mutants and deletion of acrAB results in loss of MDR in such mutants, leading to a 2–3 fold decrease in MICs compared with the isogenic wild type strain [8, 92]. The MICs of the marR mutants were further elevated in the absence of acrR, the main repressor of acrAB, suggesting that constitutively expressed marA raises the MICs again, through a derepressed acrAB operon [93]. More directly, microarray data of MarA induced by salicylate or constitutively expressed in a marR mutant showed 2–3 and 3–4 fold increases in acrA and tolC transcript levels respectively [6]. Furthermore, MarA has been shown to bind the tolC promoter in vitro [94] and substrate accumulation in strains with mutations in tolC led to the upregulation of marRAB, suggesting a regulatory feedback loop [95]. Interestingly, 10-fold variations in the levels of TolC in either direction from wild type levels do not appear to affect MICs, suggesting that, of the TolC protein produced, only a small fraction is operational [96].
Microarray studies showed that SoxS increases the transcription of acrA by 2 fold upon exposure of cells to paraquat [7] and directly binds to the promoter sequence of acrA and tolC [74, 85, 97]. Rob also depends largely on the pump for decreasing antibiotic susceptibility [64] and has been shown to activate the transcription of an acrAp-lacZ promoter fusion in the presence of bile salts, independently of marA and soxS, and to bind to the acrA promoter [73].
There are several layers of control for the pump as well as the acrA promoter. TolC has 4 promoters that are bound by different regulators. MarA, SoxS and Rob bind the third and fourth promoters, while other transcriptional regulators bind the first and second [97]. Recently it has been shown that a small 49 residue membrane protein, AcrZ, also regulates the pump as acrZ deletion changes the substrate specificity of the pump. Furthermore, AcrZ co-purifies with the pump and is also regulated by all three TRs [98].
While AcrAB is the only pump known to be regulated by these TRs, it is has been shown that pumps from different families can work synergistically and have a greater than additive effect against certain antibiotics [99]. AcrAB upregulation by the TRs and upregulation of other pumps by alternative pathways would then decrease antibiotic susceptibility more substantially.
AcrAB-TolC efflux of other substrates than antibiotics
The AcrAB-TolC efflux pump is also important for the efflux of other components such as triclosan and pine oil [100]. Overexpression of the mar operon resulted in reduced triclosan susceptibility, presumably by upregulation of the acrAB/tolC genes. In a few cases, MDR strains with triclosan resistance have been identified with overexpression of marA or soxS [101]. With respect to pine oil, deletion of acrAB made strains hypersusceptible suggesting active pine oil efflux; increased susceptibility was dependent on MarA as deletion of SoxS or Rob only caused increased susceptibility if the marA locus was removed. Strains overexpressing MarA and showing decreased pine oil susceptibility also exhibited MDR [100].
Upregulation of the AcrAB-TolC pump by MarA (SoxS and Rob) may also have other consequences as AcrAB-TolC has been shown to efflux mammalian steroidal hormones, which bacteria are exposed to in the urinary, vaginal and gastrointestinal tract [102]. AcrAB-TolC has also been demonstrated to partner with the YojI protein (previously thought to be an ATP binding cassette type pump) to efflux microcin J25. Without yojI or tolC, E. coli become very susceptible to microcin J25 [103]. Microcins are small bacteriocins (peptides) of 10kDa or less, produced by bacteria as antimicrobials to outcompete other bacteria. Cationic antimicrobial peptides (CAMPs) on the other hand are larger amphipathic peptides that are part of the host defense against invading bacteria. CAMPs can upregulate MarA with the assistance of Rob, thus decreasing susceptibility to CAMPs via AcrAB [104]. Efflux of steroidal hormones, CAMPs, peptides such as microcin J25 and enterobactin could all affect how bacteria survive in human hosts either as commensals or as pathogens.
AcrAB-TolC in organic solvent tolerance
Organic solvents are present in different environments where E. coli can be found, such as soil and waste water, and present a challenge to bacterial growth. Organic solvents disrupt the membrane and inactivate bacterial proteins leading to cell lysis and death [105].
Loss of marA or marORAB causes E. coli to grow less well in n-hexane, while over-expression of marORAB results in better growth in the presence of n-hexane and growth in cyclo-hexane where previously no growth occurred [90, 106]. The organic solvent susceptibility in the marORAB deletion can be complemented by expression of the marCORAB operon, soxS or rob but none of the TRs can complement organic solvent sensitivity due to AcrB deletion. That the different regulators may depend on each other with respect to AcrAB-TolC expression was separately demonstrated by Nakajima and colleagues [36], who showed that the over-expression of Rob that leads to organic solvent tolerance is partially dependent on the presence of SoxS.
It is not known how organic solvents activate the transcription of marA or soxS or how they activate Rob. As for antibiotics, increases in organic solvent tolerance mediated by the three TRs could allow adaptation to higher levels of solvents by mutation. Since the same regulon genes are involved, it seems likely that increasing organic solvent tolerance would also decrease antibiotic susceptibility and might also increase the number of drug resistant bacteria in the environment.
Recently, genetic manipulation of TRs has been applied to biofuel production where organic solvent tolerance is a necessity. Oh and co-workers [107] showed that marR deletion in E. coli, together with deletion of fadR, which changes the composition of the inner membrane to make it less permeable to organic solvents, caused an increase in growth and cell viability with organic solvents, and also decreased organic solvent accumulation within the bacteria. In a similar vein, Watanabe and Doukyu [108] identified mutations in AcrR and MarR that maximize the tolerance of the bacteria to organic solvents. However, in a different biofuel study [109], overexpression of the TRs (MarA, SoxS and Rob) in strains that overproduce free fatty acids (FFAs) that are potential precursors for high density biofuels did not lead to an increased tolerance for FFAs.
In the general theme of pollution, Rob and SoxS have also been found to mediate resistance to some heavy metals in E. coli [36], though whether or not this is dependent on AcrA-TolC is unknown as currently there is no known efflux of metals by this pump in E. coli. Resistance to some heavy metals could occur through the superoxide dismutases, one of which, sodA, is a member of all three TR regulons (Section 5.3.2).
5.2.2. Outer membrane permeability
The outer membrane of Gram negative bacteria is a physical barrier to hydrophobic and hydrophilic compounds and many toxic molecules [110]. The major outer membrane proteins F and C of E. coli (OmpF/C) form pores in the outer membrane and facilitate diffusion of nutrients and excretion of toxic products [110]. Since OmpF and OmpC are central to the accumulation of antibiotics such as penicillin, cephalosporin and tetracycline, decreased OmpF/C levels results in reduced sensitivity to antibiotics [111–113]. The environment-dependent regulation of these porins is complex and involves a network of transcription regulators and small non-coding RNAs [114–118].
micF, a small inhibitory RNA of 93 nucleotides [119], regulates OmpF expression at the post-transcriptional level by binding the 5’ untranslated region of the mRNA, such that it shields the promoter region, the Shine-Dalgarno sequence and the first AUG codon [120]. Transcription of micF is controlled by several global regulators [121–123], and by the EnvZ/OmpR two component system [124, 125]. MicF levels are known to increase with osmolarity via the OmpR regulator, and at higher temperatures, although the mechanism by which this occurs is not clear [126]. Oxidative and toxic compounds also activate micF, leading to an adaptive response that protects bacteria from toxic compounds by decreasing membrane permeability. Cohen and co-workers first found that strains with a MDR phenotype due to mutations in the mar locus had decreased levels of OmpF in their membranes and that this phenomenon required intact micF [9, 127]. MarA was later shown to bind to sequences adjacent to and slightly overlapping the −35 position of the micF promoter [34]. SoxS is also able to promote micF transcription in vivo [7] and purified SoxS has been shown to activate in vitro transcription of micF, indicating direct binding to the promoter [3, 4]. Moreover, Rob up-regulates micF [2, 37, 70, 128] and it is likely that this would also be part of the mechanism of bile tolerance by E. coli [128]. During salicylate induction, both MarA and Rob are responsible for the MicF-dependent decrease in OmpF levels [38]. Also, these two regulators have been shown to regulate OmpF translation through an unknown MicF-independent pathway [38].
YedS porin has also been shown to be regulated by MarA in a carbapenem resistant clinical isolate of E. coli [129]. The study revealed that a mutation found in marR led to increased expression of MarA, which resulted in activation of micF transcription. While OmpF and OmpC were absent in this isolate due to mutation in the corresponding genes, YedS, which shows high homology with OmpF, appeared to be functional and translation of its messenger was repressed by MicF.
5.3. Oxidative stress
Oxidative stress is generated by oxidizing chemicals (e.g. paraquat, plumbagin, menadione), antibacterial compounds including certain antibiotics, host defense mechanisms and normal metabolic processes resulting in the formation of the reactive oxygen species (ROS), superoxide, hydrogen peroxide and the hydroxyl radical. These molecules are able to damage RNA, DNA, proteins and lipids. Superoxide dismutases convert superoxides to hydrogen peroxide and water. Catalases/peroxidases convert hydrogen peroxide to water and oxygen. There is no enzyme available to convert hydroxyl radicals to a safe compound and this ROS is therefore highly toxic.
Of the three TRs, SoxS is the first responder to oxidative stress. There is some controversy as to the mechanism of SoxR activation and as mentioned in section 3, it is in fact redox-cycling drugs themselves that oxidize SoxR rather than superoxide. The reduced redox-cycling drugs then donate the electron acquired from SoxR to a respiratory molecule such as a quinone, becoming reoxidised and able to continue to oxidize further SoxR proteins [59]. It is has also been shown that high oxygen concentrations activate SoxR in the absence redox-cycling drugs [130] potentially through the decrease of NADPH. This molecule has been suggested as a source of electrons for SoxR reducing enzymes (rsxABCDGE operon and rseC) [130–133].
There is also evidence that under anaerobic conditions, SoxR can be activated by redox cycling drugs if nitrate is present [59]. In this case, nitrate acts as an electron acceptor to reoxidise reduced redox-cycling drugs. Additionally, SoxR is activated by nitric oxide (NO) [134]. NO nitrosylates the iron sulfur cluster forming dinitrosyl–iron clusters and this occurs more efficiently in the absence of oxygen. SoxR is not a response factor of NO under aerobic conditions [135]. We speculate that SoxR could be a defense mechanism against NO produced by macrophages during an infection [136] under anaerobic conditions such as in the gut.
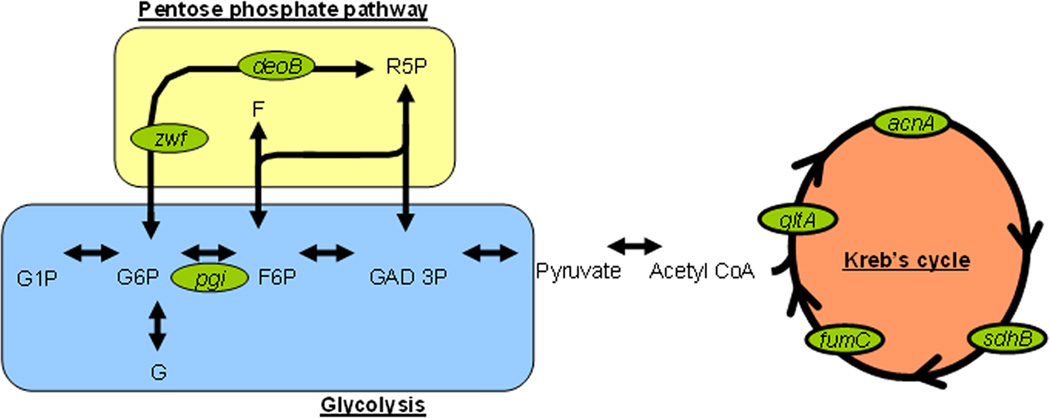
The genes regulated by SoxS, MarA and Rob that are involved in the respiratory pathway and in oxidative stress response are shown in Table 1. Several of the genes are involved in maintaining the pool of NADH and NADPH that is depleted during oxidative stress. These genes are part of the respiratory pathways: glycolysis, the Krebs cycle (also called the citric acid cycle or tricarboxylic acid (TCA) cycle) and the pentose phosphate pathway (Table 1, Figure 3). Others contribute to lowering levels of superoxide and are oxidative stress response genes. Other proteins of the regulons, such as AcrAB and OmpF, act to keep out redox-cycling drugs (discussed in section 5). Here, we present an overview of these genes that are regulated by MarA, SoxS and Rob.
Table 1.
Respiratory and oxidative stress genes regulated by MarA/SoxS/Roba
| Gene | Protein encoded | Pathwayb | Reaction | Regulated by | |
|---|---|---|---|---|---|
| From | To | ||||
| acnA | Aconitase A | Krebs | Citrate | Isocitrate | MarA, SoxS, Rob |
| deoB | Deoxytibouratase, phosphopentomutase | PPP | 2 deoxy-D-ribose 6-P D-ribose 5-P |
2 deoxy-D-ribose 1-P D-ribose 1-P |
MarA, SoxS |
| fumC | Fumarase C | Krebs | Fumarate | S-Malate | MarA, SoxS, Rob |
| fpr | Ferredoxin NADP+ reductase | 2 reduced ferredoxin + NADP+ + H+ | 2 oxidized ferredoxin + NADPH | MarA, SoxS, Rob | |
| gltA | Citrate synthase | Krebs | Oxaloacetate | Citrate | MarA, SoxS |
| pgi | Glucose-6-phosphate isomerase | Glycolysis | β-D-glucose 6-P | D-fructose 6-P | MarA, SoxS |
| sdhB | Subunit of Succinate dehydrogenase iron-sulfur protein | Krebs | Succinate | Fumarate | SoxS |
| sodA | Mn Superoxide Dismutase | Superoxide | Oxygen + hydrogen peroxide | MarA, SoxS, Rob | |
| zwf | Glucose-6-phoshate dehydrogenase | PPP | Glucose-6-P | 6-P-D-gluconolactone | MarA, SoxS, Rob |
References in text.
PPP, pentose phosphate pathway; Krebs, Krebs Citric Acid Cycle.
Figure 3. MarA, SoxS and Rob regulated enzymes in simplified respiratory pathway.
Enzymes shown are regulated by one or more of MarA, SoxS and Rob (see Table 1). acnA – aconitase A, sucD - succinyl CoA synthase, sdhB – succinyl dehydrogenase B, fumC – fumarase C, gltA – citrate synthase, pgi – glucose-6-phosphate isomerase, zwf – glucose-6-phosphate dehydrognease, deoB – phosphopentomutase. G – glucose, F – fructose, P – phosphate, R – ribulose, GAD – glyceraldehyde.
5.3.1. Respiratory enzymes
Figure 3 shows a simplified representation of the respiratory pathways: glycolysis, the Krebs cycle and the pentose phosphate pathway (PPP). Highlighted are the enzymes regulated by one or more of the three TRs. For some, the evidence is based only on microarray studies and for others, more experiments have been performed. Because of their connection within the respiratory pathway, all will be mentioned, if only briefly.
zwf and fpr
zwf is an important enzyme in the PPP that codes for glucose-6-phosphate dehydrognease (G6PDH) (Figure 3). It converts glucose-6-phosphate to 6-phosphogluconolactone and concurrently converts NADP+ to NADPH; it is the major source for NADPH in E. coli [137]. During oxidative stress, levels of G6PDH rapidly increase as a result of SoxS induction increasing NADPH that can then be used to reduce oxidized molecules and proteins [137]. However, increasing levels of G6PDH lead to a decrease in SoxS. Giro and colleagues [137] attribute this to the increased levels of NADPH used by SoxR reducing enzymes (rsxABCDGE operon and rseC) [131, 132] leading to SoxR reduction and SoxS repression. In effect, this provides a negative feedback loop between zwf/G6PDH and SoxS.
Ferredoxin NADP+ reductase (Fpr) on the other hand promotes the conversion of NADPH to NADP+ by reducing ferredoxin and flavodoxin; the latter is also a member of the SoxS regulon [7, 138]. Ferredoxin and flavodoxin act to reduce the iron sulfur clusters of various enzymes (e.g. ribonucleotide reductase, methionine synthase and pyruvate/formate lyase) oxidized during oxidative stress conditions, restoring their function [137, 139]. Giro and colleagues [137] found that following oxidative stress induction, a 30 minute lag occurred before Fpr levels increased. At this time, oxidative damage would already have occurred and therefore Fpr may be used to repair enzymes damaged by oxidation after the fact rather than as an immediate response to limit damage. A balance between SoxR reduction/oxidation, G6DPH and Fpr expression must be in place as the levels of each varies with the redox state of the bacteria.
Further complexity occurs in this balance as fpr and zwf are also part of the MarA and Rob regulons, though their influence is weaker [85, 139] and there are other global transcriptional regulators such as OxyR and Fur that respond to oxidative stress; the latter also being a SoxS regulon member and involved in iron regulation [56, 140].
Pgi and DeoB
In the same theme as zwf regulation, glucose-6-phosphate isomerase (Pgi) is at the start of the glycolysis pathway and deoxyribouratase (DeoB – a phosphopentomutase) is part of the PPP (Figure 3). Both have been shown by microarray to be regulated by MarA and SoxS [6, 7, 138]. As noted above, NADPH is produced by the PPP and glucose-6-phosphate is the starting point for this pathway and the second step of the glycolysis pathway. If zwf is triggered to initiate the PPP, then we speculate that deoB is also upregulated to ensure completion of the pathway and the replenishment of NADPH levels depleted under oxidizing condition. How SoxS and MarA balance this is not known but it is likely that it is influenced by the oxidizing state of the cytosol and the resulting effect on SoxR and other regulators that control these enzymes.
FumC, GltA, AcnA and SdhB
Fumarase C (FumC), Citrate synthase (GltA), aconitase A (AcnA) and succinate dehydrogenase B (SdhB) are all enzymes or subunits of enzymes of the Krebs cycle (Figure 3, Table 1). With the exception of FumC, only microarray evidence exists demonstrating regulation by MarA or SoxS [2, 6, 7, 138]. FumC and AcnA are additionally regulated by Rob [37,74].
FumC is one of three fumarases in E. coli. In comparison with FumA and FumB, FumC is thermostable and stable under oxidative attack, perhaps because it does not contain a Fe-S cluster. Fumarase interconverts fumarate to L-malate in the Krebs cycle. It was first identified as a SoxS regulon member by Liochev and Fridovich in 1992 [141]. It is intimately connected with zwf as strains where zwf gene is deleted show increased expression of fumC.
5.3.2. Oxidative stress response genes
SodA and YggX
SodA and YggX are two of the most important components of the oxidative stress response. Superoxide dismutase A (SodA) converts superoxide ions to hydrogen peroxide [142]. It is induced by all three TRs and by heat shock, DNA binding drugs, ethanol, high salt, and heavy metals [143] (references therein), and by high oxygen conditions [130] via SoxS. Since the hydroxyl radical is highly lethal and not convertible, SodA (and SodB and SodC) acts in concert with catalase/peroxidases KatG, KatE and AhpC to convert superoxide molecules to hydrogen peroxide and the latter to water. In particular, SodA captures superoxide molecules being produced by the electron transport chain. From microarray data, katE and ahpC are also part of the SoxS regulon but no further studies have been done [7].
SoxS binds directly to the yggX promoter to produce the transcript for an 11kDa protein. MarA and Rob have no effect on yggX transcription [144]. Deletion of yggX almost abolished the ability of E. coli to protect against the effects of paraquat and it is suggested that YggX is involved in repair of Fe-S clusters [59].
NfsA (MdaA) and NfsB (NfnB)
NfsA and NfsB are oxygen insensitive flavodoxin monocleotide (FMN) binding nitroreductases with broad specificity for electron acceptors and use NAD(P)H as the electron donor [145, 146]. They are involved in resistance to nitrofurans such as nitrofurantoin [147]. NfsA is the major component while NfsB plays a more minor role [146]. Studies have shown that that nfsA and nfsB were upregulated by the constitutive or induced expression of MarA and that MarA, SoxS and Rob bind directly to the nfsA and nfsB promoters but with different affinities [74, 148]. Another study also showed that paraquat was able to upregulate the expression of nfsA in a soxR (and therefore presumably in a soxS) dependent manner [149]. Mutations in nfsA and nfsB have been found in clinical strains highly resistant to nitrofurantoin [147]. Nitrofurans are a second line of treatment for infections such as UTIs, where resistance to commonly used antibiotics is an increasing problem [147]. These strains were not analyzed for expression of the TRs, so it is not known if they were responsible for the resistance.
An interesting aspect of these two proteins is that together with NemA, they are also part of 2,4,6-trinitrotoluene (TNT) degradation. Though no-longer produced in the United States, TNT is a major soil contaminant at some industrials sites where explosives were manufactured. NemA, NfsA and NfsB enable E. coli to extract nitrogen from TNT [150]. If organic solvents that might typically be expected to be part of soil contamination at such industrial sites are also present, MarA might contribute to TNT breakdown through upregulation of nfsA and nfsB. It is not known if TNT increases expression of marA.
5.4 Acid tolerance
5.4.1. HdeAB
HdeA and B are chaperone proteins that prevent denaturation of proteins at low pH [151]. Their expression levels also vary according to the bacterial growth phase and the operon is regulated by a host of different factors. MarA has been shown to act as a repressor of hdeA in microarray studies [6, 7]. It directly binds to the hdeA promoter to repress transcription in vitro [86]. In vivo, MarA enhances the repressor activity of H-NS on hdeAB during exponential and stationary phases at neutral and acidic pH and inhibits the activity of the hdeAB activator GadE during stationary phase, particularly at an acidic pH [152]. MarA also affects other hdeAB regulators such as RpoS, Lrp, GadX and GadW, though the effect was not as large as detected for H-NS and GadE. Many of the regulators of hdeAB themselves affect the transcription of other proteins and together with the regulation by MarA, results in a highly complex regulatory network, able to respond to the many different conditions that the cell might encounter.
5.4.2. InaA
Weak acids induce inaA expression [153] in a Rob, MarA and SoxS-dependent manner [7, 154, 155]. Although little is known about the protein product of inaA, it has recently been used as a marker for compounds toxic to micro-organisms present in lignocelluolose hydrolysate. Lignocellulose is a potential feedstock for biofuel production. Lee and coworkers [156] showed through qPCR and microarray studies of E. coli that marR, marA, marB and inaA were upregulated and occasionally downregulated in response to chemicals present as byproducts of lignocellulose breakdown. Using this information, Lee and Mitchell [157] fused the inaA promoter to the luxCDABE reporter system to detect toxic compounds present in lignocellulose hydrolysate; deletion of marA prevented this response which is in agreement with MarA acting as an inaA activator. Interestingly the authors found that deletion of marB, about which very little is known, increased the reporter signal. Use of such a reporter system to detect the buildup of toxic compounds during the conversion of lignocellulose to biofuel would be useful in adapting strains to be resistant to the toxins and to identify their need for removal, to enhance the efficiency of the process.
5.5. Metabolism
5.5.1. PurA
PurA is an adenylsuccinate synthase and is necessary for de novo purine synthesis. Deletion of purA in E. coli has also been shown to decrease the ability of the bacteria to invade human brain microvascular endothelial cells that make up the blood-brain barrier, which E. coli is able to cross to cause new born meningitis [158]. Microarray studies show a 2-fold reduction of purA levels with constitutive MarA expression [6] and MarA is able to directly bind and repress the purA promoter, resulting in a 56% decrease in transcription in vitro compared with the wild type strain [86]. In the study of urinary tract infections, mutation of purA caused a decrease to 1/5 of the WT growth rate and was strongly outcompeted by the WT strain in human urine. The effect on UTI was not tested for this mutant [159]. However, it was shown that deletion of purA in Salmonella typhimurium decreased bacterial persistence in mice infected orally [160, 161].
5.5.2. Map
Methionine aminopeptidase (map) is an essential metallooligopeptidase that catalyzes the removal of the N-terminal methione [162]. It is considered as an antibacterial target candidate since its inhibition or loss causes decrease in viability or death respectively [163]. Activation through binding to the map promoter has been shown through lacZ fusions during treatment by paraquat and dipyridyl implicating SoxS and Rob in transcriptional activation [74]. Sodium salicylate, and therefore by implication MarA, had a smaller and perhaps insignificant effect. Activation of map by SoxS has also been identified in two different microarray studies [7, 138]. The consequences of this regulation are unknown.
5.6. Virulence and biofilm
With their extensive regulons the three TRs might be expected to have a role in virulence during infections and we have touched on this point in some of the preceding sections through the functions of the regulon members. An example of their involvement during infections comes from a triple deletion mutant (marA-soxS-rob) failing to persist in an E. coli mouse model of ascending pylenophritis [10]. Warner et al. have further used microarrays to demonstrate how the zinc transport system znuABC is part of the SoxS operon and necessary for colonization of the mouse kidneys in this mouse model [11]. It is likely that the decreased susceptibility to CAMPs mediated by MarA and Rob also plays a role in virulence [104].
Biofilms are an aspect of virulence and their formation is a great problem during infections, as they are more resistant to antibiotics than planktonically growing bacteria. Upregulation of MarA by salicylate or via mutation in marR in a UPEC strain was shown to down-regulate the expression of fimB and therefore results in decreased biofilm formation. This could have implications for treatment of UPEC infections, where biofilms are problematic [164]. However, caution should be used as an earlier study showed that overexpression of the mar operon only protected non-pathogenic E. coli against very low levels of ciprofloxacin [165]. Additionally, Duo et al. [166] showed that overexpression of Rob caused increased resistance of biofilms against the aminoglycoside, tobramycin[166]. No involvement of SoxS in biofilms has been demonstrated.
5.7. Overlap with other global regulators
MarA also regulates proteins that have been shown to be involved in other stress adaptation mechanisms. For example, many of the genes that are regulated as a function of entry into stationary phase, hypertonic tension, acid shock and cold shock also appear in the regulon of MarA [167–169]. The genes in the regulon of RpoS are also involved in response to these stress factors [169] and many of the genes found in the regulon of RpoS are also found in the regulon of SoxS and MarA. This has been directly confirmed in the acid response where MarA and RpoS work antagonistically in the regulation of HdeA and B expression [152]. Paraquat induces not only the expression of soxS but also oxyR, fur and several other regulatory genes. Mapping out the different conditions when each transcriptional regulator is active is beyond the scope of this review but it is apparent that the network of response genes is able to minutely adjust the bacterial responses to altering conditions.
6. Homologs in Enterobacteriaceae
Cohen and colleagues [170] identified 9 different genera including 3 Enterobacter spp., Klebsiella, Shigella, Citrobacter, Salmonella spp., and E. coli, in which they experimentally identified marA related sequences. Analysis of the marRAB operon with BLAST and FastA also identifies marA in these as well as in Cronobacter. The percentage amino acid identities of the encoded proteins with the E. coli counterparts are shown in Table 2. Many of these Gram negative micro-organisms are pathogenic and, over recent years in the case of Salmonella, MarA, SoxS/R and Rob-like proteins have been shown to be involved in pathogenesis (see below). Dietrich confirmed that the regulators are confined to the Enterobacteriaceae [171].
Table 2.
MarA, SoxS and Rob homologs in Enterobacteriacea.a
| Species | Gene | % Identity with E. coli geneb | |
|---|---|---|---|
| Salmonella enterica subsp. Enterica serovar Typhimurium str. LT2 | MarA | 95(120/126) | |
| MarR | 92 (133/144) | ||
| MarB | 46 (31/71) | ||
| SoxS | 95 (102/107) | ||
| SoxR | 96 (146/152) | ||
| Rob | 93(268/289) | ||
| RamA | 45 (45/101 MarA) 45 (45/100 SoxS) |
||
| Shigella flexneri 2a str. 2457T | MarA | 100 (127/127) | |
| MarR | 99 (142/144) | ||
| MarB | 97 (70/72) | ||
| SoxS | 99 (106/107) | ||
| SoxR | 100 (154/154) | ||
| Rob | 100 (289/289) | ||
| RamA | not present | ||
| Klebsiella pneumoniae 342 | MarA | 93(115/124) | |
| MarR | 83 (120/144) | ||
| MarB | 44 (70/72) | ||
| SoxS | 88 (95/107) | ||
| SoxR | 89 (136/152) | ||
| Rob | 81 (236/290) | ||
| RamA | 47 (47/104 MarA) 49 (49/100 SoxS) |
||
| Citrobacter koseri ATCC BAA-895 | MarA | 96 (122/127) | |
| MarR | 93 (134/144) | ||
| MarB | 51 (33/65) | ||
| SoxS | 95 (102/107) | ||
| SoxR | 95 (145/152) | ||
| Rob | 90 (261/289) | ||
| RamA (hypothetical)a | 45 (47/104 MarA) 47 (47/100 SoxS) |
||
| Enterobacter cloacae subsp. cloacae ATCC 13047 | MarA | 94 (116/124) | |
| MarR | 86 (124/144) | ||
| MarB | 48 (28/58) | ||
| SoxS | 90 (96/107) | ||
| SoxR | 92 (142/152) | ||
| Rob | 83 (240/289) | ||
| RamA | 49 (51/104) MarA) 46 (46/100 SoxS) |
||
| Cronobacter sakazakii ATCC BAA894 | MarA | 90 (111/124) | |
| MarR | 88 (126/144) | ||
| MarB | - | ||
| SoxS | 88 (91/104) | ||
| SoxR | 87 (132/152) | ||
| Rob | 78 (224/287) | ||
| RamA | not present | ||
| Yersinia pestis KIM | % MarA identity | % Rob Identity | |
| MarA47 | 47 (43/91) | 32 (84/259) | |
| MarA48 | 48 (49/101) | 67 (195/289) | |
| LcrF | 24 (23/96) | 28 (26/92) | |
MarA, MarR, MarB, SoxS, SoxR and Rob sequences used were those of E. coli K12, accession number NP_416047
RamA sequences identified using BLAST search with K. pneumonia 342 RamA protein.
6.1. MarA, SoxS and Rob in Salmonella, Shigella, Klebsiella, Citrobacter, Enterobacter and Yersinia
During food poisoning, Salmonella enterica causes intestinal inflammation with diarrhea. Salmonella is similar to E. coli in that it has a complete marRAB operon, soxRS loci and a rob gene. Table 2 shows the high identity that these proteins have with their E. coli counterparts. There have been several studies on MarA, SoxS and Rob in Salmonella and their involvement with regulating expression of the AcrAB efflux pump [172–175]. Most of the studies show that marRAB and soxS are induced by sodium salicylate and paraquat respectively and that this leads to increased MICs via the Salmonella AcrAB pump [173–176].
The situation changes with respect to bile induction. In Salmonella, bile can induce marRAB but does not induce or activate Rob, as it does in E. coli. In Salmonella, bile induction of marRAB leads to a decrease in susceptibility to antibiotics but this is not thought to be a major pathway for decreased susceptibility. Some of AcrAB upregulation through bile occurs independently of MarR and MarA, and it is likely that AcrAB still forms part of the major mechanism of antibiotic and bile resistance [172].
MarA, SoxS and Rob behave in a very similar way to their counterparts in E. coli in response to the oxidative stress compound sodium hypochlorite (NaOCl). Expression of MarA and SoxS are increased and deletion mutants cause a 20-fold increase in sensitivity to the compound [177]. All phagocytic cells, with the exception of macrophages, produce NaOCl via the myeloperoxidase enzyme. This could be one mechanism by which MarA and SoxS favor the survival of Salmonella during infections. Studies have shown that MarA was important for the colonization of chickens by Salmonella and played a role in antibiotic resistance development to tetracycline [178]. In contrast, deletion of marA did not affect the oral inoculation LD50 values of Salmonella in mice [176]. Additionally, in WT strains, MarA and SoxS co-operatively induce the expression of ompW upon exposure of the bacteria to menadione [179]. OmpW is a minor immunogenic membrane porin that has been implicated in paraquat resistance, osmoprotection and hydrogen peroxide, and hypochlorous acid influx [179 and references therein]. OmpW has not been shown to be regulated by the TRs in E. coli, but other porins are under their control, suggesting a similar theme for both species (see section 5.2.2).
Outside of the hospital, Klebsiella pneumoniae causes some form of respiratory disease such as pneumonia, bronchitis or bronchopneumonia. In a hospital setting, it often colonizes the lungs of patients causing inflammation and hemorrhage, particularly in immunocompromised patients. K. pneumoniae can also colonize the urinary tract, lower biliary tract and surgical wound sites. Infections are difficult to treat and MDR is increasingly seen. Klebsiella, like Salmonella, has all three TRs and they bear high identity to their E. coli counterparts (Table 2). Together with AcrR, MarA, SoxS and Rob, are thought to be able to regulate AcrAB in Klebsiella [180–182]. Bratu et al. showed correlation between MarA/SoxS expression and antibiotic susceptibilities (levofloxacin and tigecycline) in clinical isolates [183].
Enterobacter cloacae is not a primary pathogen, but has been known to cause UTI and respiratory tract infections. It has marRAB, soxS and rob with high identity to the E. coli homologs (Table 2). E. cloacae Rob was shown to cause 2–32 fold increases in antibiotic MICs when plasmid borne. It also downregulated OmpF expression, similarly to in E. coli [184]. Additionally, Perez et al. [185] found that Rob and SoxS were able to increase expression of acrAB to different extents in the presence of sodium decanoate (a bile salt) and paraquat in a situation similar to that in E. coli.
Yersinia pestis and pseudotuberculosis are also Enterobacteriaceae and are the causative agents of plague and stomach infections respectively [186]. They have two proteins with 47 and 48 % identity to MarA (named MarA47 and MarA48 respectively). MarA48 has 67% overall identity and 54% identity with the C-terminal domain (regulatory domain, residues 122–289) of E. coli Rob (Table 2). Studies showed that they were involved in colonization in a mouse pneumonic plague model [187]. Mar47 also increased transcriptional levels of acrAB, and was shown to be involved in antibiotic susceptibility [188]. No MarR exists in Yersiniae spp. so it is uncertain how MarA48 and MarA47 themselves are regulated in response to external stimuli [187, 188].
Little literature is available for Citrobacter and Shigella flexneri, other than sequence data identifying the presence of marA, soxS and rob (Table 2). In Shigella flexneri, there is little evidence of their role in the antibiotic resistance of this pathogen [189, 190].
6.2. RamA in Salmonella, Klebsiella, Citrobacter and Enterobacter
Salmonella has a second MarA/SoxS-like protein called RamA. This protein was first identified in Klebsiella pneumoniae as being responsible for an MDR phenotype [191] and having 47% and 49% identity with E. coli MarA and E. coli SoxS (Table 2). While not present in E. coli, RamA is present in a wide range of Enterobacteriaceae. ramA is regulated by transcriptional repressor RamR, located just upstream of ramA. Mutation or inactivation of RamR results in upregulation of ramA and an MDR phenotype [192–194]. RamA mediates its effects through AcrAB-TolC. Paraquat, bile and indole (an intercellular signaling molecule) upregulate expression of RamA and therefore AcrAB, but bile does not alter the transcription of RamA indicating post transcriptional or post translational regulation [195]. Recently, an alternative induction mechanism has been proposed; many antibiotics that are substrates for AcrAB do not cause increased expression of RamA. However, if AcrAB is inhibited or disrupted and the strain subjected to these antibiotics, RamA expression increases. It is suggested that internal metabolites or stress signals resulting from the absence of the pump or a decrease in efflux, rather than the antibiotics themselves, causes the increased ramA transcription [196]. This effect is similar to the mechanism proposed for MarA, SoxS and Rob activation in E. coli in a tolC strain (see section 5.2.1).
In Salmonella, RamA also plays a role in nitric oxide metabolism via the flavohemoglobin, Hmp. NO produced at nanomolar concentrations can act as a signaling molecule whereas higher concentrations damage non-target enzymes [197]. Inactivation of RamA decreases expression of hmp and increases susceptibility to NO [198]. In E. coli, the gene hmpA is also upregulated by SoxS and is important in NO metabolism [56, 135]. In terms of virulence, while RamA appears to be involved in multiple signaling pathways, there is so far little evidence that it is important for the in vivo virulence properties of Salmonella [199].
RamA is also found in Klebsiella and is 63% identical to the Salmonella RamA. Additionally, it is also regulated by repressor RamR and mutations in this gene have been found in clinical strains with decreased susceptibility to tigecycline [182, 200]. Interestingly, a tigecycline resistant strain has also been identified with increased RamA levels, but with no mutation in RamR suggesting an alternative method of RamA regulation [182]. Unlike Salmonella, Klebsiella also has a gene named RomA, which lies in between RamA and RamR but the regulation of RomA by RamR is uncertain. It appears that RomA and RamA are independently regulated as their expression levels do not correlate [182]. Other studies show that RamR mutations, like SoxR and MarR mutations, also lead to decreased susceptibility to various antibiotics including ciprofloxacin, chloramphenicol, cefuroxime and trimethoprim/sulfmethexazole as well as tigecycline and this may also be mediated by cumulative effects of a decrease in porin OmpK35 and an increase in AcrAB [181, 201, 202].
Enterobacter cloacae has also been found to have a ramA gene regulated by an upstream ramR that can be activated to induce AcrAB expression in the presence of sodium salicylate [185]. Interestingly, the authors found that MarA was not affected and it was RamA that induced the increase in MIC through AcrAB. A subsequent study found that decreased susceptibility to tigecycline in clinical isolates was almost always due to RamR mutations causing an increase in RamA and therefore AcrAB expression [203].
In Citrobacter, only sequence data identifying the presence of RamA, RomA and RamR are published [182]. A homolog of RamA is not present in Shigella or in Cronobacter
6.3. LcrF in Yersiniae spp
A second transcription factor, LcrF, is present in Yersinia spp (Y. pestis, Y. pseudotuberculosi and Y. enterolitica). Although LcrF has a much lower identity with MarA, SoxS and Rob (Table 2), it falls into the AraC family of transcription factors. LcrF regulates the transcription of the type three secretion system that protrudes, like a needle, from the surface of the bacteria to make contact with immune system cells such as neutrophils and macrophages; the effector proteins known as Yersinia Outer Proteins (YOPs) can then be injected into the eukaryotic cell [204, 205]. The YOPs interfere with cellular processes in these cells, preventing their normal function such as the secretion of immune signaling molecules, the cytokines. This allows the bacteria to evade the immune system and continues the infection. Deletion of lcrF results in an absence of disease [206, 207]. LcrF has therefore been pursued as a drug target (see below).
7. Development of novel anti-virulence agents
Since antibiotics target bacterial processes essential for the growth of the micro-organism, these molecules exert a strong selective pressure on resistance development, based on the ability of the bacteria to survive in the presence of the drug. With a few exceptions, most antibiotics developed in the past decades are improved derivatives of existing chemical classes [208, 209]. It is reasonable to expect that these drugs will eventually be subject to a pre-existing resistance mechanism. Undoubtedly, fighting bacterial resistance will require a more innovative research approach.
As an alternative to traditional antimicrobials, research targeting pathogen virulence rather than growth has been reported in the literature [13, 14]. Paratek Pharmaceutical Inc. used this approach to develop inhibitors of MarA, SoxS and Rob that can be used as anti-virulence compounds since studies have shown that these regulators controlled the virulence of E. coli in an animal model of ascending pyelonephritis infection [10]. Using the structures of MarA-DNA and Rob-DNA complexes, this group constructed a model of the conserved DNA-binding domain. Docking methodology followed by in vitro screening of commercially available molecules was then used to identify compounds with inhibitory activity against these regulators [210]. N-hydroxybenzimidazole and derivative compounds were shown to prevent the binding to DNA by SoxS, MarA, and Rob. Moreover, one analog was shown to decrease the bacterial load of E. coli in an animal model of infection [210].
More recent studies have also demonstrated that N-hydroxybenzamidazole derivatives were active against other regulators of virulence. Inhibitors of the SoxS-DNA interaction were modified to develop compounds targeting LcrF [211], a regulator of the AraC family protein which controls virulence in Y. pestis and Y. pseudoturbeculosis (see above) [204, 205, 211, 212]. These compounds were also shown to reduce the virulence of Yersinia in a whole cell assay. Later, Grier and colleagues used the same strategy to identify new N-hydroxybenzimidazole derivatives active against ExsA [213], a regulator for which the DNA-binding domain displays 85% of identity with Yersinia LcrF. ExsA is necessary for full virulence in Pseudomonas aeruginosa, an opportunistic Gram negative pathogen [214–216]. The study also demonstrated that these compounds significantly reduced cytotoxicity of P. aeruginosa to infected macrophages [213]. Furthermore, encouraging results have shown that some of these compounds displayed good metabolic stability with in vitro human liver microsomes [213].
8. Conclusion
MarA, SoxS and Rob are three global TRs that through their hierarchical response to external stimuli such as antibiotics, oxidative stress, pH and host signals such as bile and CAMPs and through the complex network of interactions with other transcriptional regulators, can steer E. coli through changing environments, to produce a finely tuned but robust response mechanism [217]. The importance of their roles can perhaps be understood because of their conserved nature in Gram negative Enterobacteriacae [171] and by the finding that in other Gram negative and a few Gram positive bacteria, where direct homologs are not found, genes that code for similar proteins exist. For example, in Pseudomonas spp., a SoxR protein is involved in metabolism of endogenous molecules that act as an intercellular signaling molecule, though a SoxS protein is absent. In another example, Shin and colleagues [59] report that the SoxR homolog in S. coelicolor has five identified regulon members, and requires the presence of secreted antibiotic acthinorhodin to cause regulon member transcription. Other Gram positive bacteria have proteins with fairly high identity to MarA, SoxS or Rob. A FASTA search with MarA reveals a Bacillus spp. protein with 41% identity but little work has been done with these proteins. In this period of increasing antibiotic resistance, these three TRs and their relatives also show potential for innovative therapeutics to combat resistant infections. Like MarA, SoxS and Rob in E. coli, a subset of the AraC family of proteins regulate the expression of virulence genes in many clinically important bacterial species. Given that these regulators are not required for bacterial survival outside of a host, inhibitors of these proteins are less likely to apply the selection pressure for resistance development.
Acknowledgments
We thank Dr Stuart B. Levy for helpful comments in the preparation of this manuscript. VD is supported by a grant AI56021 to S.B. Levy from the National Institutes of Health.
References
- 1.Ishihama A. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol Rev. 2010;34:628–645. doi: 10.1111/j.1574-6976.2010.00227.x. http://dx.doi.org/10.1111/j.1574-6976.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- 2.Ariza RR, Li Z, Ringstad N, Demple B. Activation of multiple antibiotic resistance and binding of stress- inducible promoters by Escherichia coli Rob protein. J Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Z, Demple B. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J Biol Chem. 1994;269:18371–18377. [PubMed] [Google Scholar]
- 4.Jair KW, Fawcett WP, Fujita N, Ishihama A, Wolf RE. Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxide-inducible genes. Mol Microbiol. 1996;19:307–317. doi: 10.1046/j.1365-2958.1996.368893.x. http://dx.doi.org/10.1046/j.1365-2958.1996.368893.x. [DOI] [PubMed] [Google Scholar]
- 5.Martin RG, Gillette WK, Rhee S, Rosner JL. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol Microbiol. 1999;34:431–441. doi: 10.1046/j.1365-2958.1999.01599.x. http://dx.doi.org/10.1046/j.1365-2958.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- 6.Barbosa TM, Levy SB. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol. 2000;182:3467–3474. doi: 10.1128/jb.182.12.3467-3474.2000. http://dx.doi.org/10.1128/JB.182.12.3467-3474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli r esponses to superoxide stress and sodium salicylate. J Bacteriol. 2001;183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. http://dx.doi.org/10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen SP, McMurry LM, Levy SB. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casaz P, Garrity-Ryan L, McKenney D, et al. MarA, SoxS, and Rob function as virulence factors in an Escherichia coli murine model of ascending pyelonephritis. Microbiology. 2006;152:3643–3650. doi: 10.1099/mic.0.2006/000604-0. http://dx.doi.org/10.1099/mic.0.2006/000604-0. [DOI] [PubMed] [Google Scholar]
- 11.Warner DM, Levy SB. SoxS increases the expression of the zinc uptake system ZnuACB in an Escherichia coli murine pyelonephritis model. J Bacteriol. 2012;194:1177–1185. doi: 10.1128/JB.05451-11. http://dx.doi.org/10.1128/JB.05451-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alekshun MN. New advances in antibiotic development and discovery. Expert Opin Investig Drugs. 2005;14:117–134. doi: 10.1517/13543784.14.2.117. http://dx.doi.org/10.1517/13543784.14.2.117. [DOI] [PubMed] [Google Scholar]
- 13.Alksne LE, Projan SJ. Bacterial virulence as a target for antimicrobial chemotherapy. Curr Opin Biotechnol. 2000;11:625–636. doi: 10.1016/s0958-1669(00)00155-5. http://dx.doi.org/10.1016/S0958-1669(00)00155-5. [DOI] [PubMed] [Google Scholar]
- 14.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. http://dx.doi.org/10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 15.George AM, Levy SB. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J Bacteriol. 1983;155:541–548. doi: 10.1128/jb.155.2.541-548.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen SP, Hachler H, Levy SB. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDermott PF, McMurry LM, Podglajen I, et al. The marC gene of Escherichia coli is not involved in multiple antibiotic resistance. Antimicrob Agents Chemother. 2008;52:382–383. doi: 10.1128/AAC.00930-07. http://dx.doi.org/10.1128/AAC.00930-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee S, Martin RG, Rosner JL, Davies DR. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. http://dx.doi.org/10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin RG, Rosner JL. The AraC transcriptional activators. Curr Opin Microbiol. 2001;4:132–137. doi: 10.1016/s1369-5274(00)00178-8. http://dx.doi.org/10.1016/S1369-5274(00)00178-8. [DOI] [PubMed] [Google Scholar]
- 20.Gambino L, Gracheck SJ, Miller PF. Overexpression of the MarA positive regulator is sufficient to confer multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1993;175:2888–2894. doi: 10.1128/jb.175.10.2888-2894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nichols RJ, Sen S, Choo YJ, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. http://dx.doi.org/10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinue L, McMurry LM, Levy SB. The 216 bp marB gene of the marRAB operon in Escherichia coli encodes a periplasmic protein which reduces the transcription rate of marA. FEMS Microbiol Lett. 2013;345:49–55. doi: 10.1111/1574-6968.12182. http://dx.doi.org/10.1111/1574-6968.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ariza RR, Cohen SP, Bachhawat N, Levy SB, Demple B. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1994;176:143–148. doi: 10.1128/jb.176.1.143-148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat Struct Biol. 2001;8:710–714. doi: 10.1038/90429. http://dx.doi.org/10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 25.Martin RG, Rosner JL. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci U S A. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. http://dx.doi.org/10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen SP, Levy SB, Foulds J, Rosner JL. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alekshun MN, Levy SB. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J Bacteriol. 1999;181:4669–4672. doi: 10.1128/jb.181.15.4669-4672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chubiz LM, Rao CV. Aromatic acid metabolites of Escherichia coli K-12 can induce the marRAB operon. J Bacteriol. 2010;192:4786–4789. doi: 10.1128/JB.00371-10. http://dx.doi.org/10.1128/JB.00371-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duval V, McMurry LM, Foster K, Head JF, Levy SB. A mutational analysis of the multiple antibiotic resistance regulator MarR reveals a ligand binding pocket at the interface between the dimerization and DNA-binding domains. J Bacteriol. 2013;195:3341–3351. doi: 10.1128/JB.02224-12. http://dx.doi.org/10.1128/JB.02224-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin RG, Jair KW, Wolf RE, Rosner JL. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J Bacteriol. 1996;178:2216–2223. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hachler H, Cohen SP, Levy SB. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1991;173:5532–5538. doi: 10.1128/jb.173.17.5532-5538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffith KL, Shah IM, Wolf RE. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol Microbiol. 2004;51:1801–1816. doi: 10.1046/j.1365-2958.2003.03952.x. http://dx.doi.org/10.1046/j.1365-2958.2003.03952.x. [DOI] [PubMed] [Google Scholar]
- 33.Martin RG, Rosner JL. Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or Rob. J Bacteriol. 1997;179:7410–7419. doi: 10.1128/jb.179.23.7410-7419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillette WK, Martin RG, Rosner JL. Probing the Escherichia coli transcriptional activator MarA using alanine-scanning mutagenesis: residues important for DNA binding and activation. J Mol Biol. 2000;299:1245–1255. doi: 10.1006/jmbi.2000.3827. http://dx.doi.org/10.1006/jmbi.2000.3827. [DOI] [PubMed] [Google Scholar]
- 35.Miller PF, Gambino LF, Sulavik MC, Gracheck SJ. Genetic relationship between soxRS and mar loci in promoting multiple antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother. 1994;38:1773–1779. doi: 10.1128/aac.38.8.1773. http://dx.doi.org/10.1128/AAC.38.8.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakajima H, Kobayashi K, Kobayashi M, Asako H, Aono R. Overexpression of the robA gene increases organic solvent tolerance and multiple antibiotic and heavy metal ion resistance in Escherichia coli. Appl Environ Microbiol. 1995;61:2302–2307. doi: 10.1128/aem.61.6.2302-2307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jair KW, Yu X, Skarstad K, et al. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J Bacteriol. 1996;178:2507–2513. doi: 10.1128/jb.178.9.2507-2513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chubiz LM, Glekas GD, Rao CV. Transcriptional cross talk within the mar-sox-rob regulon in Escherichia coli is limited to the rob and marRAB operons. J Bacteriol. 2012;194:4867–4875. doi: 10.1128/JB.00680-12. http://dx.doi.org/10.1128/JB.00680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sulavik MC, Gambino LF, Miller PF. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med. 1995;1:436–446. [PMC free article] [PubMed] [Google Scholar]
- 40.Brooun A, Tomashek JJ, Lewis K. Purification and ligand binding of EmrR, a regulator of a multidrug transporter. J Bacteriol. 1999;181:5131–5133. doi: 10.1128/jb.181.16.5131-5133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lomovskaya O, Lewis K, Matin A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol. 1995;177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong A, Gottman A, Park C, et al. The EmrR protein represses the Escherichia coli emrRAB multidrug resistance operon by directly binding to its promoter region. Antimicrob Agents Chemother. 2000;44:2905–2907. doi: 10.1128/aac.44.10.2905-2907.2000. http://dx.doi.org/10.1128/AAC.44.10.2905-2907.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fic E, Bonarek P, Gorecki A, et al. cAMP receptor protein from Escherichia coli as a model of signal transduction in proteins-a review. J Mol Microbiol Biotechnol. 2009;17:1–11. doi: 10.1159/000178014. http://dx.doi.org/10.1159/000178014. [DOI] [PubMed] [Google Scholar]
- 44.Saier MH., Jr Cyclic AMP-independent catabolite repression in bacteria. FEMS Microbiol Lett. 1996;138:97–103. doi: 10.1111/j.1574-6968.1996.tb08141.x. http://dx.doi.org/10.1111/j.1574-6968.1996.tb08141.x. [DOI] [PubMed] [Google Scholar]
- 45.Zheng D, Constantinidou C, Hobman JL, Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. http://dx.doi.org/10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimada T, Yamamoto K, Ishihama A. Novel members of the Cra regulon involved in carbon metabolism in Escherichia coli. J Bacteriol. 2011;193:649–659. doi: 10.1128/JB.01214-10. http://dx.doi.org/10.1128/JB.01214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu J, Weiss B. Two divergently transcribed genes soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amabile-Cuevas CF, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. http://dx.doi.org/10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu J, Weiss B. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J Bacteriol. 1992;174:3915–3920. doi: 10.1128/jb.174.12.3915-3920.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hidalgo E, Leautaud V, Demple B. The redox-regulated SoxR protein acts from a single DNA site as a repressor and an allosteric activator. EMBO J. 1998;17:2629–2636. doi: 10.1093/emboj/17.9.2629. http://dx.doi.org/10.1093/emboj/17.9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nunoshiba T, Hidalgo E, Amabile Cuevas CF, Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J Bacteriol. 1992;174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hidalgo E, Demple B. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 1994;13:138–146. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu J, Dunham WR, Weiss B. Overproduction and physical characterization of SoxR, a [2Fe-2S] protein that governs an oxidative response regulon in Escherichia coli. J Biol Chem. 1995;270:10323–10327. doi: 10.1074/jbc.270.17.10323. http://dx.doi.org/10.1074/jbc.270.17.10323. [DOI] [PubMed] [Google Scholar]
- 54.Ding H, Hidalgo E, Demple B. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J Biol Chem. 1996;271:33173–33175. doi: 10.1074/jbc.271.52.33173. http://dx.doi.org/10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- 55.Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci U S A. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pomposiello PJ, Demple B. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 2001;19:109–114. doi: 10.1016/s0167-7799(00)01542-0. http://dx.doi.org/10.1016/S0167-7799(00)01542-0. [DOI] [PubMed] [Google Scholar]
- 57.Demple B. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon - a review. Gene. 1996;179:53–57. doi: 10.1016/s0378-1119(96)00329-0. http://dx.doi.org/10.1016/S0378-1119(96)00329-0. [DOI] [PubMed] [Google Scholar]
- 58.Ding H, Demple B. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc Natl Acad Sci USA. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu M, Imlay JA. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol Microbiol. 2011;79:1136–1150. doi: 10.1111/j.1365-2958.2010.07520.x. http://dx.doi.org/10.1111/j.1365-2958.2010.07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nunoshiba T, Hidalgo E, Li Z, Demple B. Negative autoregulation by the Escherichia coli SoxS protein: a dampening mechanism for the soxRS redox stress response. J Bacteriol. 1993;175:7492–7494. doi: 10.1128/jb.175.22.7492-7494.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fawcett WP, Wolf RE. Purification of a MalE-SoxS fusion protein and identification of the control sites of Escherichia coli superoxide-inducible genes. Mol Microbiol. 1994;14:669–679. doi: 10.1111/j.1365-2958.1994.tb01305.x. http://dx.doi.org/10.1111/j.1365-2958.1994.tb01305.x. [DOI] [PubMed] [Google Scholar]
- 63.Skarstad K, Thony B, Hwang DS, Kornberg A. A novel binding protein of the origin of the Escherichia coli chromosome. J Biol Chem. 1993;268:5365–5370. [PubMed] [Google Scholar]
- 64.Tanaka T, Horii T, Shibayama K, et al. RobA-induced multiple antibiotic resistance largely depends on the activation of the AcrAB efflux. Microbiol Immunol. 1997;41:697–702. doi: 10.1111/j.1348-0421.1997.tb01913.x. [DOI] [PubMed] [Google Scholar]
- 65.Azam TA, Hiraga S, Ishihama A. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells. 2000;5:613–626. doi: 10.1046/j.1365-2443.2000.00350.x. http://dx.doi.org/10.1046/j.1365-2443.2000.00350.x. [DOI] [PubMed] [Google Scholar]
- 66.Michan C, Manchado M, Pueyo C. SoxRS down-regulation of rob transcription. J Bacteriol. 2002;184:4733–4738. doi: 10.1128/JB.184.17.4733-4738.2002. http://dx.doi.org/10.1128/JB.184.17.4733-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schneiders T, Levy SB. MarA-mediated transcriptional repression of the rob promoter. J Biol Chem. 2006;281:10049–10055. doi: 10.1074/jbc.M512097200. http://dx.doi.org/10.1074/jbc.M512097200. [DOI] [PubMed] [Google Scholar]
- 68.McMurry LM, Levy SB. Evidence that regulatory protein MarA of Escherichia coli represses rob by steric hindrance. J Bacteriol. 2010;192:3977–3982. doi: 10.1128/JB.00103-10. http://dx.doi.org/10.1128/JB.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alekshun MN, Levy SB. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kwon HJ, Bennik MH, Demple B, Ellenberger T. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat Struct Biol. 2000;7:424–430. doi: 10.1038/75213. http://dx.doi.org/10.1038/75213. [DOI] [PubMed] [Google Scholar]
- 71.Griffith KL, Fitzpatrick MM, Keen EF, Wolf RE. Two functions of the C-terminal domain of Escherichia coli Rob: mediating "sequestration-dispersal" as a novel off-on switch for regulating Rob's activity as a transcription activator and preventing degradation of Rob by Lon protease. J Mol Biol. 2009;388:415–430. doi: 10.1016/j.jmb.2009.03.023. http://dx.doi.org/10.1016/j.jmb.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosner JL, Dangi B, Gronenborn AM, Martin RG. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J Bacteriol. 2002;184:1407–1416. doi: 10.1128/JB.184.5.1407-1416.2002. http://dx.doi.org/10.1128/JB.184.5.1407-1416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol. 2003;48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. http://dx.doi.org/10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 74.Martin RG, Rosner JL. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol Microbiol. 2002;44:1611–1624. doi: 10.1046/j.1365-2958.2002.02985.x. http://dx.doi.org/10.1046/j.1365-2958.2002.02985.x. [DOI] [PubMed] [Google Scholar]
- 75.Martin RG, Gillette WK, Martin NI, Rosner JL. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in Escherichia coli. Mol Microbiol. 2002;43:355–370. doi: 10.1046/j.1365-2958.2002.02748.x. http://dx.doi.org/10.1046/j.1365-2958.2002.02748.x. [DOI] [PubMed] [Google Scholar]
- 76.Jair KW, Martin RG, Rosner JL, et al. Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J Bacteriol. 1995;177:7100–7104. doi: 10.1128/jb.177.24.7100-7104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zafar MA, Shah IM, Wolf RE. Protein-protein interactions between sigma(70) region 4 of RNA polymerase and Escherichia coli SoxS, a transcription activator that functions by the prerecruitment mechanism: evidence for "off-DNA" and "on-DNA" interactions. J Mol Biol. 2010;401:13–32. doi: 10.1016/j.jmb.2010.05.052. http://dx.doi.org/10.1016/j.jmb.2010.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shah IM, Wolf RE. Novel protein-protein interaction between Escherichia coli SoxS and the DNA binding determinant of the RNA polymerase alpha subunit: SoxS functions as a co-sigma factor and redeploys RNA polymerase from UP-element-containing promoters to SoxS-dependent promoters during oxidative stress. J Mol Biol. 2004;343:513–532. doi: 10.1016/j.jmb.2004.08.057. http://dx.doi.org/10.1016/j.jmb.2004.08.057. [DOI] [PubMed] [Google Scholar]
- 79.Zafar MA, Sanchez-Alberola N, Wolf RE. Genetic evidence for a novel interaction between transcriptional activator SoxS and region 4 of the sigma(70) subunit of RNA polymerase at class II SoxS-dependent promoters in Escherichia coli. J Mol Biol. 2011;407:333–353. doi: 10.1016/j.jmb.2010.12.037. http://dx.doi.org/10.1016/j.jmb.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dangi B, Gronenborn AM, Rosner JL, Martin RG. Versatility of the carboxy-terminal domain of the alpha subunit of RNA polymerase in transcriptional activation: use of the DNA contact site as a protein contact site for MarA. Mol Microbiol. 2004;54:45–59. doi: 10.1111/j.1365-2958.2004.04250.x. http://dx.doi.org/10.1111/j.1365-2958.2004.04250.x. [DOI] [PubMed] [Google Scholar]
- 81.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. http://dx.doi.org/10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 82.Wall ME, Markowitz DA, Rosner JL, Martin RG. Model of transcriptional activation by MarA in Escherichia coli. PLoS Comput Biol. 2009;5:e1000614. doi: 10.1371/journal.pcbi.1000614. http://dx.doi.org/10.1371/journal.pcbi.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martin RG, Gillette WK, Rosner JL. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol Microbiol. 2000;35:623–634. doi: 10.1046/j.1365-2958.2000.01732.x. http://dx.doi.org/10.1046/j.1365-2958.2000.01732.x. [DOI] [PubMed] [Google Scholar]
- 84.Martin RG, Rosner JL. Promoter discrimination at class I MarA regulon promoters mediated by glutamic acid 89 of the MarA transcriptional activator of Escherichia coli. J Bacteriol. 2011;193:506–515. doi: 10.1128/JB.00360-10. http://dx.doi.org/10.1128/JB.00360-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martin RG, Bartlett ES, Rosner JL, Wall ME. Activation of the Escherichia coli marA/soxS/rob regulon in response to transcriptional activator concentration. J Mol Biol. 2008;380:278–284. doi: 10.1016/j.jmb.2008.05.015. http://dx.doi.org/10.1016/j.jmb.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schneiders T, Barbosa TM, McMurry LM, Levy SB. The Escherichia coli transcriptional regulator MarA directly represses transcription of purA and hdeA. J Biol Chem. 2004;279:9037–9042. doi: 10.1074/jbc.M313602200. http://dx.doi.org/10.1074/jbc.M313602200. [DOI] [PubMed] [Google Scholar]
- 87.George AM, Levy SB. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983;155:531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. http://dx.doi.org/10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta. 2009;1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. http://dx.doi.org/10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.White DG, Goldman JD, Demple B, Levy SB. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fralick JA. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ma D, Cook DN, Alberti M, et al. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. http://dx.doi.org/10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 93.Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. http://dx.doi.org/10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 94.Aixia Z, Judah LR, Robert GM. Transcriptional activation by MarA, SoxS and Rob of two tolC promoters using one binding site: a complex promoter configuration for tolC in Escherichia coli. Mol Microbiol. 2008;69:1450–1455. doi: 10.1111/j.1365-2958.2008.06371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rosner JL, Martin RG. An excretory function for the Escherichia coli outer membrane pore TolC: Upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J Bacteriol. 2009;191:5283–5292. doi: 10.1128/JB.00507-09. http://dx.doi.org/10.1128/jb.00507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krishnamoorthy G, Tikhonova EB, Dhamdhere G, Zgurskaya HI. On the role of TolC in multidrug efflux: the function and assembly of AcrAB-TolC tolerate significant depletion of intracellular TolC protein. Mol Microbiol. 2013;87:982–997. doi: 10.1111/mmi.12143. http://dx.doi.org/10.1111/mmi.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang A, Rosner JL, Martin RG. Transcriptional activation by MarA, SoxS and Rob of two tolC promoters using one binding site: a complex promoter configuration for tolC in Escherichia coli. Mol Microbiol. 2008;69:1450–1455. doi: 10.1111/j.1365-2958.2008.06371.x. http://dx.doi.org/10.1111/j.1365-2958.2008.06371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc Natl Acad Sci U S A. 2012;109:16696–16701. doi: 10.1073/pnas.1210093109. http://dx.doi.org/10.1073/pnas.1210093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee A, Mao W, Warren MS, et al. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol. 2000;182:3142–3150. doi: 10.1128/jb.182.11.3142-3150.2000. http://dx.doi.org/10.1128/jb.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moken MC, McMurry LM, Levy SB. Selection of multiple-antibiotic-resistant (mar) mutants of Escherichia coli by using the disinfectant pine oil: roles of the mar and acrAB loci. Antimicrob Agents Chemother. 1997;41:2770–2772. doi: 10.1128/aac.41.12.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McMurry LM, Oethinger M, Levy SB. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol Lett. 1998;166:305–309. doi: 10.1111/j.1574-6968.1998.tb13905.x. http://dx.doi.org/10.1111/j.1574-6968.1998.tb13905.x. [DOI] [PubMed] [Google Scholar]
- 102.Elkins CA, Mullis LB. Mammalian steroid hormones are substrates for the major RND- and MFS-type tripartite multidrug efflux pumps of Escherichia coli. J Bacteriol. 2006;188:1191–1195. doi: 10.1128/JB.188.3.1191-1195.2006. http://dx.doi.org/10.1128/jb.188.3.1191-1195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Delgado MA, Vincent PA, Farias RN, Salomon RA. YojI of Escherichia coli functions as a microcin J25 efflux pump. J Bacteriol. 2005;187:3465–3470. doi: 10.1128/JB.187.10.3465-3470.2005. http://dx.doi.org/10.1128/jb.187.10.3465-3470.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Warner DM, Levy SB. Different effects of transcriptional regulators MarA, SoxS and Rob on susceptibility of Escherichia coli to cationic antimicrobial peptides (CAMPs): Rob-dependent CAMP induction of the marRAB operon. Microbiology. 2010;156:570–578. doi: 10.1099/mic.0.033415-0. http://dx.doi.org/10.1099/mic.0.033415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Isken S, de Bont JA. Bacteria tolerant to organic solvents. Extremophiles. 1998;2:229–238. doi: 10.1007/s007920050065. [DOI] [PubMed] [Google Scholar]
- 106.Aono R, Tsukagoshi N, Yamamoto M. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J Bacteriol. 1998;180:938–944. doi: 10.1128/jb.180.4.938-944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oh HY, Lee JO, Kim OB. Increase of organic solvent tolerance of Escherichia coli by the deletion of two regulator genes, fadR and marR. Appl Microbiol Biotechnol. 2012;96:1619–1627. doi: 10.1007/s00253-012-4463-8. http://dx.doi.org/10.1007/s00253-012-4463-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Watanabe R, Doukyu N. Contributions of mutations in acrR and marR genes to organic solvent tolerance in Escherichia coli. AMB Express. 2012;2:58. doi: 10.1186/2191-0855-2-58. http://dx.doi.org/10.1186/2191-0855-2-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lennen RM, Kruziki MA, Kumar K, et al. Membrane stresses induced by overproduction of free fatty acids in Escherichia coli. Appl Environ Microbiol. 2011;77:8114–8128. doi: 10.1128/AEM.05421-11. http://dx.doi.org/10.1128/AEM.05421-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Masi M, Pages JM. Structure, function and regulation of outer membrane proteins involved in drug transport in Enterobactericeae: the OmpF/C - TolC Case. Open Microbiol J. 2013;7:22–33. doi: 10.2174/1874285801307010022. http://dx.doi.org/10.2174/1874285801307010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mortimer PG, Piddock LJ. The accumulation of five antibacterial agents in porin-deficient mutants of Escherichia coli. J Antimicrob Chemother. 1993;32:195–213. doi: 10.1093/jac/32.2.195. http://dx.doi.org/10.1093/jac/32.2.195. [DOI] [PubMed] [Google Scholar]
- 112.Vidal S, Bredin J, Pages JM, Barbe J. Beta-lactam screening by specific residues of the OmpF eyelet. J Med Chem. 2005;48:1395–1400. doi: 10.1021/jm049652e. http://dx.doi.org/10.1021/jm049652e. [DOI] [PubMed] [Google Scholar]
- 113.Duval V, Nicoloff H, Levy SB. Combined inactivation of lon and ycgE decreases multidrug susceptibility by reducing the amount of OmpF porin in Escherichia coli. Antimicrob Agents Chemother. 2009;53:4944–4948. doi: 10.1128/AAC.00787-09. http://dx.doi.org/10.1128/AAC.00787-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. http://dx.doi.org/10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- 115.Dupont M, James CE, Chevalier J, Pages JM. An early response to environmental stress involves regulation of OmpX and OmpF, two enterobacterial outer membrane pore-forming proteins. Antimicrob Agents Chemother. 2007;51:3190–3198. doi: 10.1128/AAC.01481-06. http://dx.doi.org/10.1128/AAC.01481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yoshida T, Qin L, Egger LA, Inouye M. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J Biol Chem. 2006;281:17114–17123. doi: 10.1074/jbc.M602112200. http://dx.doi.org/10.1074/jbc.M602112200. [DOI] [PubMed] [Google Scholar]
- 117.Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol. 2006;9:605–611. doi: 10.1016/j.mib.2006.10.006. http://dx.doi.org/10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 118.De la Cruz MA, Calva E. The complexities of porin genetic regulation. J Mol Microbiol Biotechnol. 2010;18:24–36. doi: 10.1159/000274309. http://dx.doi.org/10.1159/000274309. [DOI] [PubMed] [Google Scholar]
- 119.Andersen J, Delihas N, Ikenaka K, et al. The isolation and characterization of RNA coded by the micF gene in Escherichia coli. Nucleic Acids Res. 1987;15:2089–2101. doi: 10.1093/nar/15.5.2089. http://dx.doi.org/10.1093/nar/15.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Delihas N, Forst S. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J Mol Biol. 2001;313:1–12. doi: 10.1006/jmbi.2001.5029. http://dx.doi.org/10.1006/jmbi.2001.5029. [DOI] [PubMed] [Google Scholar]
- 121.Deighan P, Free A, Dorman CJ. A role for the Escherichia coli H-NS-like protein StpA in OmpF porin expression through modulation of micF RNA stability. Mol Microbiol. 2000;38:126–139. doi: 10.1046/j.1365-2958.2000.02120.x. http://dx.doi.org/10.1046/j.1365-2958.2000.02120.x. [DOI] [PubMed] [Google Scholar]
- 122.Huang L, Tsui P, Freundlich M. Integration host factor is a negative effector of in vivo and in vitro expression of ompC in Escherichia coli. J Bacteriol. 1990;172:5293–5298. doi: 10.1128/jb.172.9.5293-5298.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ferrario M, Ernsting BR, Borst DW, et al. The leucine-responsive regulatory protein of Escherichia coli negatively regulates transcription of ompC and micF and positively regulates translation of ompF. J Bacteriol. 1995;177:103–113. doi: 10.1128/jb.177.1.103-113.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Aiba H, Matsuyama S, Mizuno T, Mizushima S. Function of micF as an antisense RNA in osmoregulatory expression of the ompF gene in Escherichia coli. J Bacteriol. 1987;169:3007–3012. doi: 10.1128/jb.169.7.3007-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Takayanagi K, Maeda S, Mizuno T. Expression of micF involved in porin synthesis in Escherichia coli: two distinct cis-acting elements respectively regulate micF expression positively and negatively. FEMS Microbiol Lett. 1991;67:39–44. doi: 10.1016/0378-1097(91)90440-l. http://dx.doi.org/10.1111/j.1574-6968.1991.tb04385.x. [DOI] [PubMed] [Google Scholar]
- 126.Andersen J, Forst SA, Zhao K, Inouye M, Delihas N. The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J Biol Chem. 1989;264:17961–17970. [PubMed] [Google Scholar]
- 127.Cohen SP, McMurry LM, Hooper DC, Wolfson JS, Levy SB. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989;33:1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oh JT, Cajal Y, Skowronska EM, et al. Cationic peptide antimicrobials induce selective transcription of micF and osmY in Escherichia coli. Biochim Biophys Acta. 2000;1463:43–54. doi: 10.1016/s0005-2736(99)00177-7. http://dx.doi.org/10.1016/S0005-2736(99)00177-7. [DOI] [PubMed] [Google Scholar]
- 129.Warner DM, Yang Q, Duval V, et al. Involvement of MarR and YedS in carbapenem resistance in a clinical isolate of Escherichia coli from China. Antimicrob Agents Chemother. 2013;57:1935–1937. doi: 10.1128/AAC.02445-12. http://dx.doi.org/10.1128/AAC.02445-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Baez A, Shiloach J. Escherichia coli avoids high dissolved oxygen stress by activation of SoxRS and manganese-superoxide dismutase. Microb Cell Fact. 2013;12:23. doi: 10.1186/1475-2859-12-23. http://dx.doi.org/10.1186/1475-2859-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Koo MS, Lee JH, Rah SY, et al. A reducing system of the superoxide sensor SoxR in Escherichia coli. Embo J. 2003;22:2614–2622. doi: 10.1093/emboj/cdg252. http://dx.doi.org/10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kobayashi K, Tagawa S. Isolation of reductase for SoxR that governs an oxidative response regulon from Escherichia coli. FEBS Lett. 1999;451:227–230. doi: 10.1016/s0014-5793(99)00565-7. http://dx.doi.org/10.1016/S0014-5793(99)00565-7. [DOI] [PubMed] [Google Scholar]
- 133.Poole K. Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria. Trends Microbiol. 2012;20:227–234. doi: 10.1016/j.tim.2012.02.004. http://dx.doi.org/10.1016/j.tim.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 134.Ding H, Demple B. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc Natl Acad Sci U S A. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. http://dx.doi.org/10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Forrester MT, Foster MW. Protection from nitrosative stress: a central role for microbial flavohemoglobin. Free Radic Biol Med. 2012;52:1620–1633. doi: 10.1016/j.freeradbiomed.2012.01.028. http://dx.doi.org/10.1016/j.freeradbiomed.2012.01.028. [DOI] [PubMed] [Google Scholar]
- 136.Vasil'eva SV, Stupakova MV, Lobysheva II, Mikoyan VD, Vanin AF. Activation of the Escherichia coli SoxRS-regulon by nitric oxide and its physiological donors. Biochemistry (Mosc) 2001;66:984–988. doi: 10.1023/a:1012317508971. http://dx.doi.org/10.1023/A:1012317508971. [DOI] [PubMed] [Google Scholar]
- 137.Giro M, Carrillo N, Krapp AR. Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology. 2006;152:1119–1128. doi: 10.1099/mic.0.28612-0. http://dx.doi.org/10.1099/mic.0.28612-0. [DOI] [PubMed] [Google Scholar]
- 138.Blanchard JL, Wholey WY, Conlon EM, Pomposiello PJ. Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional networks. PLoS One. 2007;2:e1186. doi: 10.1371/journal.pone.0001186. http://dx.doi.org/10.1371/journal.pone.0001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Niazi JH, Kim BC, Gu MB. Characterization of superoxide-stress sensing recombinant Escherichia coli constructed using promoters for genes zwf and fpr fused to lux operon. Appl Microbiol Biotechnol. 2007;74:1276–1283. doi: 10.1007/s00253-006-0758-y. http://dx.doi.org/10.1007/s00253-006-0758-y. [DOI] [PubMed] [Google Scholar]
- 140.Zheng M, Doan B, Schneider TD, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liochev SI, Fridovich I. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci U S A. 1992;89:5892–5896. doi: 10.1073/pnas.89.13.5892. http://dx.doi.org/10.1073/pnas.89.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. http://dx.doi.org/10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 143.Geslin C, Llanos J, Prieur D, Jeanthon C. The manganese and iron superoxide dismutases protect Escherichia coli from heavy metal toxicity. Res Microbiol. 2001;152:901–905. doi: 10.1016/s0923-2508(01)01273-6. http://dx.doi.org/10.1016/S0923-2508(01)01273-6. [DOI] [PubMed] [Google Scholar]
- 144.Pomposiello PJ, Koutsolioutsou A, Carrasco D, Demple B. SoxRS-regulated expression and genetic analysis of the yggX gene of Escherichia coli. J Bacteriol. 2003;185:6624–6632. doi: 10.1128/JB.185.22.6624-6632.2003. http://dx.doi.org/10.1128/JB.185.22.6624-6632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zenno S, Koike H, Tanokura M, Saigo K. Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri. J Biochem. 1996;120:736–744. doi: 10.1093/oxfordjournals.jbchem.a021473. [DOI] [PubMed] [Google Scholar]
- 146.Zenno S, Koike H, Kumar AN, et al. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J Bacteriol. 1996;178:4508–4514. doi: 10.1128/jb.178.15.4508-4514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sandegren L, Lindqvist A, Kahlmeter G, Andersson DI. Nitrofurantoin resistance mechanism and fitness cost in Escherichia coli. J Antimicrob Chemother. 2008;62:495–503. doi: 10.1093/jac/dkn222. http://dx.doi.org/10.1093/jac/dkn222. [DOI] [PubMed] [Google Scholar]
- 148.Barbosa TM, Levy SB. Activation of the Escherichia coli nfnB gene by MarA through a highly divergent marbox in a class II promoter. Mol Microbiol. 2002;45:191–202. doi: 10.1046/j.1365-2958.2002.03006.x. http://dx.doi.org/10.1046/j.1365-2958.2002.03006.x. [DOI] [PubMed] [Google Scholar]
- 149.Liochev SI, Hausladen A, Fridovich I. Nitroreductase A is regulated as a member of the soxRS regulon of Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:3537–3539. doi: 10.1073/pnas.96.7.3537. http://dx.doi.org/10.1073/pnas.96.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ju KS, Parales RE. Nitroaromatic compounds, from synthesis to biodegradation. Microbiol Mol Biol Rev. 2010;74:250–272. doi: 10.1128/MMBR.00006-10. http://dx.doi.org/10.1128/MMBR.00006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hong W, Wu YE, Fu X, Chang Z. Chaperone-dependent mechanisms for acid resistance in enteric bacteria. Trends Microbiol. 2012;20:328–335. doi: 10.1016/j.tim.2012.03.001. http://dx.doi.org/10.1016/j.tim.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 152.Ruiz C, McMurry LM, Levy SB. Role of the multidrug resistance regulator MarA in global regulation of the hdeAB acid resistance operon in Escherichia coli. J Bacteriol. 2008;190:1290–1297. doi: 10.1128/JB.01729-07. http://dx.doi.org/10.1128/JB.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.White S, Tuttle FE, Blankenhorn D, Dosch DC, Slonczewski JL. pH dependence and gene structure of inaA in Escherichia coli. J Bacteriol. 1992;174:1537–1543. doi: 10.1128/jb.174.5.1537-1543.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bennik MH, Pomposiello PJ, Thorne DF, Demple B. Defining a rob regulon in Escherichia coli by using transposon mutagenesis. J Bacteriol. 2000;182:3794–3801. doi: 10.1128/jb.182.13.3794-3801.2000. http://dx.doi.org/10.1128/JB.182.13.3794-3801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rosner JL, Slonczewski JL. Dual regulation of inaA by the multiple antibiotic resistance (mar) and superoxide (soxRS) stress response systems of Escherichia coli. J Bacteriol. 1994;176:6262–6269. doi: 10.1128/jb.176.20.6262-6269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee S, Nam D, Jung JY, et al. Identification of Escherichia coli biomarkers responsive to various lignin-hydrolysate compounds. Bioresour Technol. 2012;114:450–456. doi: 10.1016/j.biortech.2012.02.085. http://dx.doi.org/10.1016/j.biortech.2012.02.085. [DOI] [PubMed] [Google Scholar]
- 157.Lee S, Mitchell RJ. Detection of toxic lignin hydrolysate-related compounds using an inaA::luxCDABE fusion strain. J Biotechnol. 2012;157:598–604. doi: 10.1016/j.jbiotec.2011.06.018. http://dx.doi.org/10.1016/j.jbiotec.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 158.Hoffman JA, Badger JL, Zhang Y, Sik Kim K. Escherichia coli K1 pur A and sor C are preferentially expressed upon association with human brain microvascular endothelial cells. Microbial Pathogenesis. 2001;31:69–79. doi: 10.1006/mpat.2001.0451. [DOI] [PubMed] [Google Scholar]
- 159.Vejborg RM, de Evgrafov MR, Phan MD, et al. Identification of genes important for growth of asymptomatic bacteriuria Escherichia coli in urine. Infect Immun. 2012;80:3179–3188. doi: 10.1128/IAI.00473-12. http://dx.doi.org/10.1128/IAI.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dunstan SJ, Simmons CP, Strugnell RA. Comparison of the abilities of different attenuated Salmonella typhimurium strains to elicit humoral immune responses against a heterologous antigen. Infect Immun. 1998;66:732–740. doi: 10.1128/iai.66.2.732-740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.O'Callaghan D, Maskell D, Liew FY, Easmon CS, Dougan G. Characterization of aromatic-and purine-dependent Salmonella typhimurium: attention, persistence, and ability to induce protective immunity in BALB/c mice. Infect Immun. 1988;56:419–423. doi: 10.1128/iai.56.2.419-423.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ben-Bassat A, Bauer K, Chang SY, et al. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chai SC, Wang WL, Ding DR, Ye QZ. Growth inhibition of Escherichia coli and methicillin-resistant Staphylococcus aureus by targeting cellular methionine aminopeptidase. Eur J Med Chem. 2011;46:3537–3540. doi: 10.1016/j.ejmech.2011.04.056. http://dx.doi.org/10.1016/j.ejmech.2011.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vila J, Soto SM. Salicylate increases the expression of marA and reduces in vitro biofilm formation in uropathogenic Escherichia coli by decreasing type 1 fimbriae expression. Virulence. 2012;3:280–285. doi: 10.4161/viru.19205. http://dx.doi.org/10.4161/viru.19205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Maira-Litran T, Allison DG, Gilbert P. An evaluation of the potential of the multiple antibiotic resistance operon (mar) and the multidrug efflux pump acrAB to moderate resistance towards ciprofloxacin in Escherichia coli biofilms. J Antimicrob Chemother. 2000;45:789–795. doi: 10.1093/jac/45.6.789. http://dx.doi.org/10.1093/jac/45.6.789. [DOI] [PubMed] [Google Scholar]
- 166.Duo M, Hou S, Ren D. Identifying Escherichia coli genes involved in intrinsic multidrug resistance. Appl Microbiol Biotechnol. 2008;81:731–741. doi: 10.1007/s00253-008-1709-6. http://dx.doi.org/10.1007/s00253-008-709-6. [DOI] [PubMed] [Google Scholar]
- 167.Stancik LM, Stancik DM, Schmidt B, et al. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J Bacteriol. 2002;184:4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002. http://dx.doi.org/10.1128/JB.184.15.4246-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. http://dx.doi.org/10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.White-Ziegler CA, Um S, Perez NM, et al. Low temperature (23 degrees C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology. 2008;154:148–166. doi: 10.1099/mic.0.2007/012021-0. http://dx.doi.org/10.1099/mic.0.2007/012021-0. [DOI] [PubMed] [Google Scholar]
- 170.Cohen SP, Yan W, Levy SB. A multidrug resistance regulatory chromosomal locus is widespread among enteric bacteria. J Infect Dis. 1993;168:484–488. doi: 10.1093/infdis/168.2.484. [DOI] [PubMed] [Google Scholar]
- 171.Dietrich LE, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321:1203–1206. doi: 10.1126/science.1160619. http://dx.doi.org/10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Prouty AM, Brodsky IE, Falkow S, Gunn JS. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology. 2004;150:775–783. doi: 10.1099/mic.0.26769-0. http://dx.doi.org/10.1099/mic.0.26769-0. [DOI] [PubMed] [Google Scholar]
- 173.Hartog E, Ben-Shalom L, Shachar D, Matthews KR, Yaron S. Regulation of marA, soxS, rob, acrAB and micF in Salmonella enterica serovar Typhimurium. Microbiol Immunol. 2008;52:565–574. doi: 10.1111/j.1348-0421.2008.00075.x. [DOI] [PubMed] [Google Scholar]
- 174.Hartog E, Menashe O, Kler E, Yaron S. Salicylate reduces the antimicrobial activity of ciprofloxacin against extracellular Salmonella enterica serovar Typhimurium, but not against Salmonella in macrophages. J Antimicrob Chemother. 2010;65:888–896. doi: 10.1093/jac/dkq077. http://dx.doi.org/10.1093/jac/dkq077. [DOI] [PubMed] [Google Scholar]
- 175.Nikaido E, Shirosaka I, Yamaguchi A, Nishino K. Regulation of the AcrAB multidrug efflux pump in Salmonella enterica serovar Typhimurium in response to indole and paraquat. Microbiology. 2011;157:648–655. doi: 10.1099/mic.0.045757-0. http://dx.doi.org/10.1099/mic.0.045757-0. [DOI] [PubMed] [Google Scholar]
- 176.Sulavik MC, Dazer M, Miller PF. The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J Bacteriol. 1997;179:1857–1866. doi: 10.1128/jb.179.6.1857-1866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Collao B, Morales EH, Gil F, et al. Differential expression of the transcription factors MarA, Rob, and SoxS of Salmonella Typhimurium in response to sodium hypochlorite: down-regulation of rob by MarA and SoxS. Arch Microbiol. 2012;194:933–942. doi: 10.1007/s00203-012-0828-8. http://dx.doi.org/10.1007/s00203-012-0828-8. [DOI] [PubMed] [Google Scholar]
- 178.Randall LP, Woodward MJ. Role of the mar locus in virulence of Salmonella enterica serovar Typhimurium DT104 in chickens. J Med Microbiol. 2001;50:770–779. doi: 10.1099/0022-1317-50-9-770. [DOI] [PubMed] [Google Scholar]
- 179.Collao B, Morales EH, Gil F, Calderon IL, Saavedra CP. ompW is cooperatively upregulated by MarA and SoxS in response to menadione. Microbiology. 2013;159:715–725. doi: 10.1099/mic.0.066050-0. http://dx.doi.org/10.1099/mic.0.066050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ruzin A, Visalli MA, Keeney D, Bradford PA. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2005;49:1017–1022. doi: 10.1128/AAC.49.3.1017-1022.2005. http://dx.doi.org/10.1128/AAC.49.3.1017-1022.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schneiders T, Amyes SG, Levy SB. Role of AcrR and ramA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob Agents Chemother. 2003;47:2831–2837. doi: 10.1128/AAC.47.9.2831-2837.2003. http://dx.doi.org/10.1128/AAC.47.9.2831-2837.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rosenblum R, Khan E, Gonzalez G, Hasan R, Schneiders T. Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. Int J Antimicrob Agents. 2011;38:39–45. doi: 10.1016/j.ijantimicag.2011.02.012. http://dx.doi.org/10.1016/j.ijantimicag.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bratu S, Landman D, George A, Salvani J, Quale J. Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J Antimicrob Chemother. 2009;64:278–283. doi: 10.1093/jac/dkp186. http://dx.doi.org/10.1093/jac/dkp186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lee EH, Collatz E, Podglajen I, Gutmann L. A rob-like gene of Enterobacter cloacae affecting porin synthesis and susceptibility to multiple antibiotics. Antimicrob Agents Chemother. 1996;40:2029–2033. doi: 10.1128/aac.40.9.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Perez A, Poza M, Aranda J, et al. Effect of transcriptional activators SoxS, RobA, and RamA on expression of multidrug efflux pump AcrAB-TolC in Enterobacter cloacae. Antimicrob Agents Chemother. 2012;56:6256–6266. doi: 10.1128/AAC.01085-12. http://dx.doi.org/10.1128/AAC.01085-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Achtman M, Zurth K, Morelli G, et al. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lister IM, Mecsas J, Levy SB. Effect of MarA-like proteins on antibiotic resistance and virulence in Yersinia pestis. Infect Immun. 2010;78:364–371. doi: 10.1128/IAI.00904-09. http://dx.doi.org/10.1128/IAI.00904-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Udani RA, Levy SB. MarA-like regulator of multidrug resistance in Yersinia pestis. Antimicrob Agents Chemother. 2006;50:2971–2975. doi: 10.1128/AAC.00015-06. http://dx.doi.org/10.1128/AAC.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mehata S, Duan GC. Molecular mechanism of multi-drug resistance in Shigella isolates from rural China. Nepal Med Coll J. 2011;13:27–29. [PubMed] [Google Scholar]
- 190.Pu XY, Zhang Q, Pan JC, Shen Z, Zhang W. Spontaneous mutation frequency and molecular mechanisms of Shigella flexneri fluoroquinolone resistance under antibiotic selective stress. World J Microbiol Biotechnol. 2013;29:365–371. doi: 10.1007/s11274-012-1190-3. http://dx.doi.org/10.1007/s11274-012-1190-3. [DOI] [PubMed] [Google Scholar]
- 191.George AM, Hall RM, Stokes HW. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology. 1995;141:1909–1920. doi: 10.1099/13500872-141-8-1909. http://dx.doi.org/10.1099/13500872-141-8-1909. [DOI] [PubMed] [Google Scholar]
- 192.Abouzeed YM, Baucheron S, Cloeckaert A. ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. 2008;52:2428–2434. doi: 10.1128/AAC.00084-08. http://dx.doi.org/10.1128/AAC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ricci V, Busby SJ, Piddock LJ. Regulation of RamA by RamR in Salmonella enterica serovar Typhimurium: isolation of a RamR superrepressor. Antimicrob Agents Chemother. 2012;56:6037–6040. doi: 10.1128/AAC.01320-12. http://dx.doi.org/10.1128/AAC.01320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ricci V, Piddock LJ. Only for substrate antibiotics are a functional AcrAB-TolC efflux pump and RamA required to select multidrug-resistant Salmonella typhimurium. J Antimicrob Chemother. 2009;64:654–657. doi: 10.1093/jac/dkp234. http://dx.doi.org/10.1093/jac/dkp234. [DOI] [PubMed] [Google Scholar]
- 195.Nikaido E, Yamaguchi A, Nishino K. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J Biol Chem. 2008;283:24245–24253. doi: 10.1074/jbc.M804544200. http://dx.doi.org/10.1074/jbc.M804544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lawler AJ, Ricci V, Busby SJ, Piddock LJ. Genetic inactivation of acrAB or inhibition of efflux induces expression of ramA. J Antimicrob Chemother. 2013;68:1551–1557. doi: 10.1093/jac/dkt069. http://dx.doi.org/10.1093/jac/dkt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Vallance P, Charles I. Nitric oxide as an antimicrobial agent: does NO always mean NO? Gut. 1998;42:313–314. doi: 10.1136/gut.42.3.313. [comment] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hernandez-Urzua E, Zamorano-Sanchez DS, Ponce-Coria J, et al. Multiple regulators of the Flavohaemoglobin (hmp) gene of Salmonella enterica serovar Typhimurium include RamA, a transcriptional regulator conferring the multidrug resistance phenotype. Arch Microbiol. 2007;187:67–77. doi: 10.1007/s00203-006-0175-8. http://dx.doi.org/10.1007/s00203-006-0175-8. [DOI] [PubMed] [Google Scholar]
- 199.van der Straaten T, Zulianello L, van Diepen A, et al. Salmonella enterica serovar Typhimurium RamA, intracellular oxidative stress response, and bacterial virulence. Infect Immun. 2004;72:996–1003. doi: 10.1128/IAI.72.2.996-1003.2004. http://dx.doi.org/10.1128/iai.72.2.996-1003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hentschke M, Wolters M, Sobottka I, Rohde H, Aepfelbacher M. ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrob Agents Chemother. 2010;54:2720–2723. doi: 10.1128/AAC.00085-10. http://dx.doi.org/10.1128/AAC.00085-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kallman O, Motakefi A, Wretlind B, et al. Cefuroxime non-susceptibility in multidrug-resistant Klebsiella pneumoniae overexpressing ramA and acrA and expressing ompK35 at reduced levels. J Antimicrob Chemother. 2008;62:986–990. doi: 10.1093/jac/dkn296. http://dx.doi.org/10.1093/jac/dkn296. [DOI] [PubMed] [Google Scholar]
- 202.Bialek-Davenet S, Marcon E, Leflon-Guibout V, et al. In vitro selection of ramR and soxR mutants overexpressing efflux systems by fluoroquinolones as well as cefoxitin in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2011;55:2795–2802. doi: 10.1128/AAC.00156-11. http://dx.doi.org/10.1128/AAC.00156-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Veleba M, De Majumdar S, Hornsey M, Woodford N, Schneiders T. Genetic characterization of tigecycline resistance in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Antimicrob Chemother. 2013;68:1011–1018. doi: 10.1093/jac/dks530. http://dx.doi.org/10.1093/jac/dks530. [DOI] [PubMed] [Google Scholar]
- 204.Skurnik M, Toivanen P. LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J Bacteriol. 1992;174:2047–2051. doi: 10.1128/jb.174.6.2047-2051.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cornelis GR, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. http://dx.doi.org/10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 206.Jackson MW, Silva-Herzog E, Plano GV. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol Microbiol. 2004;54:1364–1378. doi: 10.1111/j.1365-2958.2004.04353.x. http://dx.doi.org/10.1111/j.1365-2958.2004.04353.x. [DOI] [PubMed] [Google Scholar]
- 207.Garrity-Ryan LK, Kim OK, Balada-Llasat JM, et al. Small molecule inhibitors of LcrF, a Yersinia pseudotuberculosis transcription factor, attenuate virulence and limit infection in a murine pneumonia model. Infect Immun. 2010;78:4683–4690. doi: 10.1128/IAI.01305-09. http://dx.doi.org/10.1128/IAI.01305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Talbot GH, Bradley J, Edwards JE, Jr, et al. Bad bugs need drugs: an update on the development pipeline from the antimicrobial availability task force of the Infectious Diseases Society of America. Clin Infect Dis. 2006;42:657–668. doi: 10.1086/499819. http://dx.doi.org/10.1086/499819. [DOI] [PubMed] [Google Scholar]
- 209.Projan SJ, Bradford PA. Late stage antibacterial drugs in the clinical pipeline. Curr Opin Microbiol. 2007;10:441–446. doi: 10.1016/j.mib.2007.08.007. http://dx.doi.org/10.1016/j.mib.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 210.Bowser TE, Bartlett VJ, Grier MC, et al. Novel anti-infection agents: small-molecule inhibitors of bacterial transcription factors. Bioorg Med Chem Lett. 2007;17:5652–5655. doi: 10.1016/j.bmcl.2007.07.072. http://dx.doi.org/10.1016/j.bmcl.2007.07.072. [DOI] [PubMed] [Google Scholar]
- 211.Kim OK, Garrity-Ryan LK, Bartlett VJ, et al. N-hydroxybenzimidazole inhibitors of the transcription factor LcrF in Yersinia: novel antivirulence agents. J Med Chem. 2009;52:5626–5634. doi: 10.1021/jm9006577. http://dx.doi.org/10.1021/jm9006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Thoerner P, Bin Kingombe CI, Bogli-Stuber K, et al. PCR detection of virulence genes in Yersinia enterocolitica and Yersinia pseudotuberculosis and investigation of virulence gene distribution. Appl Environ Microbiol. 2003;69:1810–1816. doi: 10.1128/AEM.69.3.1810-1816.2003. http://dx.doi.org/10.1128/AEM.69.3.1810-1816.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Grier MC, Garrity-Ryan LK, Bartlett VJ, et al. N-Hydroxybenzimidazole inhibitors of ExsA MAR transcription factor in Pseudomonas aeruginosa: in vitro anti-virulence activity and metabolic stability. Bioorg Med Chem Lett. 2010;20:3380–3383. doi: 10.1016/j.bmcl.2010.04.014. http://dx.doi.org/10.1016/j.bmcl.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 214.Finck-Barbancon V, Goranson J, Zhu L, et al. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. http://dx.doi.org/10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 215.Laskowski MA, Osborn E, Kazmierczak BI. A novel sensor kinase-response regulator hybrid regulates type III secretion and is required for virulence in Pseudomonas aeruginosa. Mol Microbiol. 2004;54:1090–1103. doi: 10.1111/j.1365-2958.2004.04331.x. http://dx.doi.org/10.1111/j.1365-2958.2004.04331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Yahr TL, Wolfgang MC. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol. 2006;62:631–640. doi: 10.1111/j.1365-2958.2006.05412.x. http://dx.doi.org/10.1111/j.1365-2958.2006.05412.x. [DOI] [PubMed] [Google Scholar]
- 217.Ruiz C, Levy SB. Many chromosomal genes modulate MarA-mediated multidrug resistance in Escherichia coli. Antimicrob Agents Chemother. 2010;54:2125–2134. doi: 10.1128/AAC.01420-09. http://dx.doi.org/10.1128/AAC.01420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]